Abstract
microRNA (miRNA) are small non-coding RNA targeting mRNAs leading to their instability and diminished translation. Altered expression of miRNA is associated with cancer. Inflammation and nitric oxide modulates the development of lymphomas in p53 knockout mice and there exists a negative feedback loop between p53 and NOS2. Using a genetic strategy, we tested the hypothesis that inflammation-induced oxidative and nitrosative stress modulates miRNA expression in mouse model deficient in either p53 or NOS2. Mice treated with C. parvum, to induce inflammation, clearly separated from controls by their miRNA profiles in wild-type, p53- and NOS2-knockout genetic backgrounds. C. parvum-induced inflammation significantly (p<0.005) increased miR-21, miR-29b and miR-34a/b/c and decreased (p<0.005) mir-29c and mir-181a/c expression in the spleen of C57BL mice. However, p53-knockout C57BL mice did not show a significant increase in the mir-34b/c or a decrease in mir-29c expression following C.parvum-induced inflammation. Expression of mir-21, mir-29b and mir-181a was independent of p53-status. NOS2-knockout C57BL mice showed a significant increase in miR-21 and miR-34a/b/c and decrease in miR-181a similar to the wild-type (WT) mice following C.parvum-induced inflammation. However, in contrast to the WT mice, miR-29b/c expression was not affected following C. parvum-induced inflammation in NOS2 knockout mice. N-acetyl cysteine, an anti-oxidant, reduced the expression of miR-21 and miR-29b in C.parvum-treated WT mice (p<0.005) as compared with control C.parvum-treated mice. These data are consistent with the hypothesis that inflammation modulates miRNA expression in vivo and the alteration in specific miRNA under an inflammatory microenvironment, can be influenced by p53 (miR-34b/c) and NO• (29b/c).
Keywords: Inflammation, microRNA, nitric oxide, nitric oxide synthase-2 (NOS2), p53 and cytokine
Introduction
miRNAs are small (18–24 bp ) non-coding RNAs, functioning as regulator of mRNA expression at post transcription level and subsequent translation of a wide variety of target genes that are involved in numerous biological processes 1. The fact that a single miRNA can target a large number of different gene transcripts, any change in miRNA expression may cause alteration in a range of crucial biological processes including but not limited to apoptosis, DNA repair, cellular proliferation and immune response leading to the disruption of cellular homeostasis. miRNAs expression is modulated by a number of mechanisms, which involves transcriptional activation or repression, genomic alterations, and epigenetic modifications of genome. Furthermore, single nucleotide polymorphisms in miRNA genes, their processing complex and target binding site can alter their function and affect cancer risk, treatment efficacy and patient outcome 2. Most interestingly, recent studies have provided evidence indicating the regulation of miRNAs by inflammatory stimuli and existence of feedback loops between a number of different microRNAs and inflammatory components in particular pro-inflammatory cytokines [reviewed in 3, 4] 5. Furthermore, altered miRNAs expression has been reported in a number of different chronic inflammatory diseases including psoriasis, rheumatoid arthritis, primary billiary cirrhosis, ulcerative colitis and pancreatitis 6, 7. An altered expression of miRNAs is implicated in the development of human cancer 1. The aberrant expression of specific miRNAs can both, induce or suppress tumor development. Whereas an overexpression of miR-155 or miR-21 could induce tumors, the overexpression of let-7a reduced lung cancer in mouse models 8–10.
Approximately 1/4th of all cancer is associated with infection and chronic inflammation and the recent studies have further strengthened the link between inflammation and cancer by deciphering the underlying molecular mechanism [reviewed in 11, 12]. The emergence of the concept that miRNA alteration is an important molecular link between inflammation and cancer is most interesting and requires further investigation. We have earlier reported that TP53 is a molecular node in coordinating the inflammatory stress response pathways and regulates specific set of genes following stimulation with NO•, hydrogen peroxide, hydroxyurea (DNA replication stress) or hypoxia, inducing 4 different stress conditions commonly found during chronic inflammation 13. An increased level of nitric oxide, largely due to the induction of NOS2 during inflammation, is associated with tumorigenesis12 and there is a negative feedback regulation between p53 and NOS214. Our previous studies have shown that inflammation and NO• modulates the development of lymphomas in p53 knockout mice15. In the present study, we investigated the influence of inflammation on miRNA expression in the wild type, p53- or NOS2-knockout C57BL mice and tested the hypothesis that inflammation induced oxidative and nitrosative stress modulates miRNA expression.
Materials and Methods
Wild-type, p53-and NOS2-knockout mice, and treatment of heat-inactivated Corynebacterium- parvum (C.parvum) and N-acetyl cysteine (NAC)
p53-knockout and NOS2-knockout mice were back-crossed more than 8 times with C57BL6 wild-type mice to obtain >99% C57BL6 strain15. Animals were bred and used under approved ACUC, NCI-Frederick, protocol. 7–8 week old mice were treated either with a single i.p. dose of 100 mg/kg body weight of heat-inactivated C. parvum (Van Kampen Gr., Inc, USA) or saline control as described earlier 15. A group of wild-type mice were also treated with an antioxidant, NAC (Sigma, USA) at a dose of 1gm/Kg body weight, in drinking water, starting a day prior to C.parvum treatment and continued through the end of experiment. Mice were maintained in a climate control facility with food and water ad libitum at NCI-Frederick animal facilty. On the 10th day following C. parvum treatment, mice were sacrificed and the spleen snap-frozen in liquid nitrogen for analyses.
RNA isolation and miRNA Analysis
Total RNA was extracted from spleen using TRIZOL (Invitrogen, cat. no. 15596-026), according to the manufacturer’s procedures. miRNA expression levels were measured using Ohio State University miRNA microarray chips version 4, that include 474 human and 373 mouse miRNA, (based on the December 2006 Sanger miRNA database). Total RNA (5 ug) was converted to biotin-labeled first strand cDNA, hybridized onto the chips, and processed by direct detection of the biotin-containing transcripts by streptavidin-Alexa 647 conjugate. Slides were subsequently scanned with the Axon 4000B Scanner (Molecular Device, Inc) and spot intensities were quantified with Genepix (version Pro 6.0.1.00). In compliance with the MIAME guidelines, microarray data is submitted to the Gene Expression Omnibus (GEO number= GSE30218).
Expression of miRNAs with statistically significant altered levels was validated using qRT-PCR with Taqman miRNA reverse transcription assays (Applied Biosystems, cat. no. 4366596) and appropriate primers, following the manufacturer’s instructions. For each miRNA, reactions were performed in triplicate using the Applied Biosystems 7500 RT-PCR system. The endogenous control sno202 (Applied Biosystems) was used as a normalization control. miRNA expression values are presented as mean ± SD. Differential expression was assessed using two-sided unpaired Student’s t-tests. Statistical significance was achieved when P < 0.005 (taking into account a Bonferroni correction for multiple comparisons) and borderline statistical significance was achieved when P fell between 0.05 and 0.005.
Results and Discussion
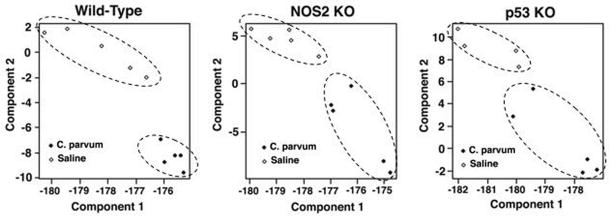
To investigate the influence of inflammation on the modulation of miRNA expression in vivo, we induced systemic inflammation in wild type, p53- or NOS2-knockout C57BL mice by treating them with a single dose of heat-inactivated bacteria, C. parvum. We tested the hypothesis that inflammation induced oxidative and nitrosative stress modulates miRNA expression by performing global miRNA microarray expression profiling of the spleen from these mice. Principal component analysis revealed that mice treated with C. parvum and controls can be clearly separated by their miRNA profiles in all three genetic backgrounds (Figure 1). The miRNA microarray results were then validated using quantitative PCR. Administration of a single dose of heat-inactivated C. parvum (100 mg/kg body weight) causes the stimulation of reticuloendothelial system and an increase in the level of cytokines, NOS2 expression and NO• production, which peaked on day 10 following treatment, in mice 14, 15. Spleen was found to be one of the principal organs for an increased NOS2 expression in p53 knockout mice following C. parvum-induced inflammation and splenectomy in these mice caused a major decrease in overall NO• production 14.
Figure 1.
Principal components analysis using miRNA microarray expression, revealing distinct groupings of C. parvum treated and control C57BL WT mice (a), NOS2 KO mice (b), and p53 KO mice (c). Mice treated with C. parvum and controls can be clearly separated by their miRNA profiles in all three genetic backgrounds.
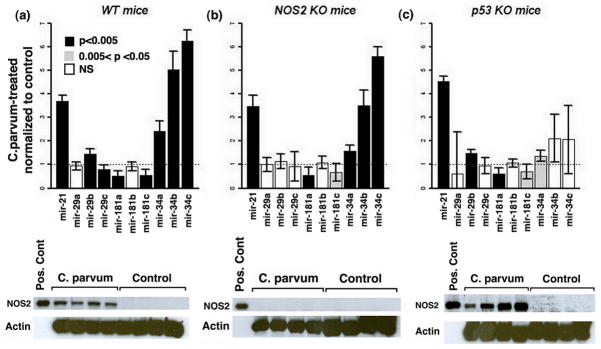
A significant increase in miR-21, miR-29b and miR-34a/b/c and decrease in miR-29c and miR-181a/c expression was observed in the spleen following C. parvum-induced inflammation in C57BL mice as compared to the controls (p<0.005) (Figure 2 and Table 1; supplemental Tables 1–3). Altered expression of these miRNAs is reported to be associated with cancer and some of them are associated with inflammation and immune response. miR-21 is one of the most commonly altered miRNA in almost all cancer types examined so far and its increase is considered to be protumorigenic 16. miR-21 expression is associated with expression of several cytokines including IL-6, IL-8, IL-10 and IL-12a in colon cancer 17. In addition, miR-21 is induced by IL-6, a pro-inflammatory cytokine, in a STAT3-dependent manner and is present at a higher level in chronic inflammatory diseases 18. Furthermore, epidermal growth factor receptor (EGFR) signaling pathways positively regulate miR-21 expression in lung cancer 19. miR-34a/b/c are partially regulated by TP53 tumor suppressor and they play a role in mediating p53-induced apoptosis, cell cycle arrest and senescence [reviewed in 20]. Anti-sense inhibition of miR-34a increases population doubling of normal human fibroblasts and thus delays replicative senescence 21. However, p53-independent upregulation of miR-34a is also reported during oncogene-induced senescence 22.
Figure 2.
(a) Mice with wild-type p53 and NOS2 show increased miR-21, miR-29b, and miR-34a/b/c expression and decreased miR-29c, miR-181a/c expression in response to inflammation (p-val < 0.005). (b) NOS2 KO mice show increased expression of miR-21, miR-34a/b/c and decreased expression of miR-181a in mice treated with C. parvum compared to control mice (pval < 0.005). Reduced expression of miR-181c in response to inflammation is borderline statistically significant (0.05 < pval < 0.005), (c) Increased expression of mir-21, mir-29b and reduced expression of miR-181a is observed in response to inflammation in p53 KO mice (pval < 0.005). Reduced expression of miR-181c and increased expression of mir-34a in response to inflammation is borderline statistically significant (0.05 < pval < 0.005) in this group of mice. Western blot analysis (bottom panel) showed enhanced expression of NOS2 following C. parvum-induced inflammation in wild type and p53-knockout mice. As expected, NOS2 knockout mice did not show any NOS2 expression.
Table 1.
qRT-PCR validation results of miRNAs altered in C. parvum-treated as compared with control wild-type, NOS2 KO and p53 KO mice
| NOS2 and p53 WT (C. Parv: N=5 No treatment: N = 4) | NOS2 KO (C. Parv: N=5 No treatment: N = 5) | p53 KO (C. Parv: N=5 No treatment: N = 5) | |
|---|---|---|---|
| mir-21 | UP (3.69 +/− 0.24) | UP (3.49 +/− 0.46) | UP (4.55 +/− 0.2) |
| mir-29a | no change | no change | no change |
| mir-29b | UP (1.44 +/− 0.23) | no change | UP (1.48 +/− 0.15) |
| mir-29c | DOWN (0.78 +/− 0.19) | no change | no change |
| mir-181a | DOWN (0.51 +/− 0.24) | DOWN (0.55 +/− 0.35) | DOWN (0.6 +/− 0.24) |
| mir-181b | no change | no change | no change |
| mir-181c | DOWN (0.55 +/− 0.26) | DOWN * (0.67 +/− 0.38) | DOWN * (0.7 +/− 0.3) |
| mir-34a | UP (2.43 +/− 0.41) | UP (1.58 +/− 0.26) | UP * (1.37 +/− 0.24) |
| mir-34b | UP (5.02 +/− 0.79) | UP (3.5 +/− 0.67) | no change |
| mir-34c | UP (6.25 +/− 0.48) | UP (5.6 +/− 0.39) | no change |
Two-sided unpaired t-test applied to determine differential expression.
In parenthesis, the fold changes +/− pooled standard deviation are reported.
Borderline statistical significance (0.005 < pval < 0.05)
C. parvum-induced inflammation activates the p53 pathway in mice and there exist a negative feed back loop between p53 and NOS214. NO• is an important signaling molecule and plays a crucial role during inflammatory response. However, a sustained and high level of NO•, as seen during chronic inflammation, can be deleterious causing DNA alterations including point mutations in cancer related genes and post-translational modifications in important cellular proteins 12. C. parvum-induced inflammation in p53 knockout mice accelerates the development of lymphomas as compared with saline-treated control mice, however, p53 and NOS2 double knockout mice did not show any difference in tumor latency following C. parvum treatment as compared with saline-treated controls, indicating the role of NO• in rapid lymphomagenesis in p53 knockout mice following inflammation 15.
Based on the evidence of a cross-talk between p53 and NO• in inflammation and cancer, we further investigated the role of p53 and nitrosative stress in the regulation of miRNA. We analyzed miRNA expression in p53-knockout as well as NOS2-knockout mice following C. parvum-induced inflammation. TP53-knockout mice did not show a significant increase in miR-34b/c expression or a decrease in miR-29c expression associated with C. parvum-induced inflammation (Figure 2 and Table 1; Supplemental Tables 1–3). However, we did observe a minimal increase in miR-34a expression with borderline statistical significance. These data are consistent with earlier in vitro report of p53-mediated regulation of miR-3423. Increase in miR-21 and miR-29b and decrease in miR-181a expression was independent of p53 status and could have been associated with oxidative and/or nitrosative stress induced during inflammation. Therefore, we treated the wild-type mice with N-acetyl cysteine (NAC), an anti-oxidant, and determined the miRNA expression following C. parvum-induced inflammation. Interestingly, expression of both miR-21 and miR-29b was significantly inhibited as compared with control mice (p<0.005) (Figure 3). However, no change in the expression of mirR-181a was noted in NAC treated mice. These observations indicated that oxidative stress due to the generation of reactive oxygen species might have contributed to the increase in the expression of miR-21 and miR-29b. Hydrogen peroxide (H2O2) has been earlier shown to upregulate miR-21 in vascular smooth muscle cells24. However, the exact mechanism is not clearly known. Whereas, IL-6 can induce the expression of miR-21, we did not find any significant increase in IL-6 expression or any change in phospho-Stat3 in the spleen of wild-type mice following C. parvum treatment (data not shown). An increased level of IL-6 is earlier reported by us in the spleen, thymus and serum of p53-knockout and p53/NOS2 double knockout mice following C.parvum-induced imflammation15. However, in the present study, we don’t know the contribution of IL-6 in miR-21 expression under inflammatory condition in p53 knockout mice.
Figure 3.
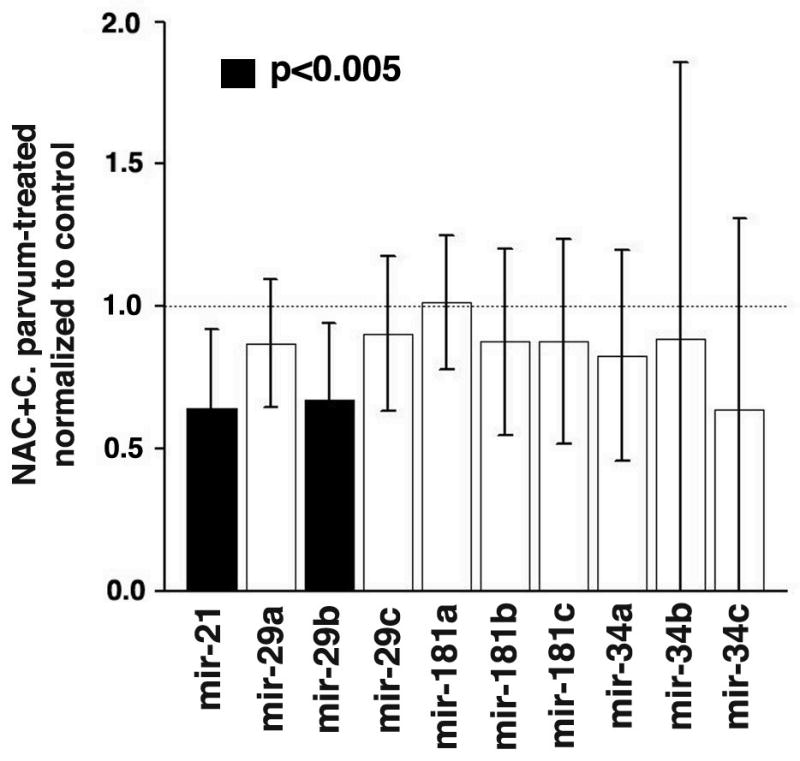
Effect of NAC on miRNA expression in C. parvum treated p53 and NOS2 wild-type mice. MiR-21 and miR-29b expression is reduced (pval < 0.005) in mice treated with both NAC and C. parvum, compared to mice treated with only C. parvum. miRNA expression values are presented as mean ± SD.
Because C. parvum-induced inflammation increases nitrosative stress in inflammatory microenvironment, we extended our investigation in NOS2-knockout mice to investigate the role of NO• in regulation of miRNA expression. NOS2- knockout mice showed a significant increase in the expression of miR-21 and miR-34a/b/c and a decrease in miR-181a following C. parvum-induced inflammation as compared with the control mice (Figure 2 and Table 1; Supplemental Table 1–3). These results are similar to the wild-type mice indicating that NO• may not contribute to the regulation of miR-21, miR-34a/b/c, miR-181a under this inflammatory condition in mice. In NOS2 knockout mice the alteration in miR-21 could be due to the oxidative stress similar to the wild-type mice, however, these mice were not treated with NAC. The increase in miR-34a/b/c in NOS2-knockout mice is consistent with p53 mediated regulation of these microRNA as in the wild-type mice. However, in contrast to the wild-type mice, there was no significant change in the expression of miR-29b/c in NOS2-knockout mice following C. parvum-induced inflammation as compared to controls. Further studies are needed to investigate the association between NO• and regulation of miR-29a/b/c. Both tumor suppressive and tumorigenic role of miR-29 have been reported which seems to be tissue and cellular context dependent. miR-29s target DNA methyltransferase-3A and -3B and have been found to be down-regulated in lung cancer suggesting its role as tumor suppressor 25. In contrast, however, overexpression of miR-29 in mouse myeloid cells caused acute myeloid leukemia26.
In the present study, an increased level of miR-21, the most commonly altered microRNA in many cancer types 16, in the WT, p53- and NOS2-knockout mice following inflammation is interesting in relation to its oncogenic function. Whereas, p53 knockout mice are cancer-prone, NOS2 knockout or wild-type C57BL mice do not exhibit cancer-prone phenotype. One of the possible explanations is the presence of an increased expression level of miR-34a/b/c in wild type and NOS2 knockout mice as compared to p53 knockout mice, where we observed only a minimal increase in miR-34a. However, it is not currently understood how and if an imbalance in the miRNA expression with oncogenic and tumor suppressive function will influence tumor development. Future studies using anti-miRNA and miRNA-knockout mice are warranted to answer this question.
This is the first in vivo study to our knowledge that investigates the role of inflammation, p53 and nitric oxide in the regulation of miRNA expression. The data are consistent with the hypothesis that inflammation regulates miRNA expression in vivo and can be modulated by p53 and NO•. C. parvum-induced inflammation accelerates the development of lymphomas in p53 knockout mice, however, there is no effect of inflammation on tumor latency in p53 and NOS2 double knockout mice 15. Further mechanistic studies are needed to determine the possible connection between inflammation-associated alteration in miRNA expression and development of cancer.
Supplementary Material
Acknowledgments
We would like to thank Dr. Curtis Harris for his support and constructive criticism. We would also like to thank Dr. Victor Laubach for providing us the NOS2 knockout mice. We acknowledge the help of Ms. Terry Sweeney in managing the animal colony and C. parvum treatment. Dr. Draginja Djurickovic’s technical advice is greatly appreciated. This work was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institute of Health.
Footnotes
Conflict of Interest Statement: Authors have no conflict of interest to declare.
References
- 1.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–14. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain SP, Harris CC. Inflammation and cancer: an ancient link with novel potentials. Int J Cancer. 2007;121:2373–80. doi: 10.1002/ijc.23173. [DOI] [PubMed] [Google Scholar]
- 4.Schetter AJ, Heegaard NH, Harris CC. Inflammation and cancer: interweaving microRNA, free radical, cytokine and p53 pathways. Carcinogenesis. 2010;31:37–49. doi: 10.1093/carcin/bgp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F, Zikusoka M, Trindade A, Dassopoulos T, Harris ML, Bayless TM, Brant SR, Chakravarti S, Kwon JH. MicroRNAs are differentially expressed in ulcerative colitis and alter expression of macrophage inflammatory peptide-2 alpha. Gastroenterology. 2008;135:1624–35. e24. doi: 10.1053/j.gastro.2008.07.068. [DOI] [PubMed] [Google Scholar]
- 7.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 8.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, Jacks T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105:3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–9. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medina PP, Nolde M, Slack FJ. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010 doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 11.Tlsty TD, Coussens LM. Tumor stroma and regulation of cancer development. Annu Rev Pathol. 2006;1:119–50. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 12.Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–85. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- 13.Staib F, Robles AI, Varticovski L, Wang XW, Zeeberg BR, Sirotin M, Zhurkin VB, Hofseth LJ, Hussain SP, Weinstein JN, Galle PR, Harris CC. The p53 tumor suppressor network is a key responder to microenvironmental components of chronic inflammatory stress. Cancer Res. 2005;65:10255–64. doi: 10.1158/0008-5472.CAN-05-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ambs S, Ogunfusika MO, Merriam WG, Bennett WP, Billiar TR, Harris CC. Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice. Proc Natl Acad Sci U S A. 1998;95:8823–8. doi: 10.1073/pnas.95.15.8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussain SP, He P, Subleski J, Hofseth LJ, Trivers GE, Mechanic L, Hofseth AB, Bernard M, Schwank J, Nguyen G, Mathe E, Djurickovic D, et al. Nitric oxide is a key component in inflammation-accelerated tumorigenesis. Cancer Res. 2008;68:7130–6. doi: 10.1158/0008-5472.CAN-08-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schetter AJ, Nguyen GH, Bowman ED, Mathe EA, Yuen ST, Hawkes JE, Croce CM, Leung SY, Harris CC. Association of inflammation-related and microRNA gene expression with cancer-specific mortality of colon adenocarcinoma. Clin Cancer Res. 2009;15:5878–87. doi: 10.1158/1078-0432.CCR-09-0627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, Burger R, Gramatzki M, Blumert C, Bauer K, Cvijic H, Ullmann AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–3. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- 19.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, Gemma A, Kudoh S, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci U S A. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermeking H. p53 enters the microRNA world. Cancer Cell. 2007;12:414–8. doi: 10.1016/j.ccr.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Fujita K, Mondal AM, Horikawa I, Nguyen GH, Kumamoto K, Sohn JJ, Bowman ED, Mathe EA, Schetter AJ, Pine SR, Ji H, Vojtesek B, et al. p53 isoforms Delta133p53 and p53beta are endogenous regulators of replicative cellular senescence. Nat Cell Biol. 2009;11:1135–42. doi: 10.1038/ncb1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Christoffersen NR, Shalgi R, Frankel LB, Leucci E, Lees M, Klausen M, Pilpel Y, Nielsen FC, Oren M, Lund AH. p53-independent upregulation of miR-34a during oncogene-induced senescence represses MYC. Cell Death Differ. 2010;17:236–45. doi: 10.1038/cdd.2009.109. [DOI] [PubMed] [Google Scholar]
- 23.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin Y, Liu X, Cheng Y, Yang J, Huo Y, Zhang C. Involvement of MicroRNAs in hydrogen peroxide-mediated gene regulation and cellular injury response in vascular smooth muscle cells. J Biol Chem. 2009;284:7903–13. doi: 10.1074/jbc.M806920200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–10. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–89. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.