Abstract
Alzheimer’s disease (AD) is the most common type of senile dementia in aging populations. Amyloid β (Aβ)-mediated dysregulation of ionic homeostasis is the prevailing underlying mechanism leading to synaptic degeneration and neuronal death. Aβ-dependent ionic dysregulation most likely occurs either directly via unregulated ionic transport through the membrane or indirectly via Aβ binding to cell membrane receptors and subsequent opening of existing ion channels or transporters. Receptor binding is expected to involve a high degree of stereospecificity. Here, we investigated whether an Aβ peptide enantiomer, whose entire sequence consists of d-amino acids, can form ion-conducting channels; these channels can directly mediate Aβ effects even in the absence of receptor–peptide interactions. Using complementary approaches of planar lipid bilayer (PLB) electrophysiological recordings and molecular dynamics (MD) simulations, we show that the d-Aβ isomer exhibits ion conductance behavior in the bilayer indistinguishable from that described earlier for the l-Aβ isomer. The d isomer forms channel-like pores with heterogeneous ionic conductance similar to the l-Aβ isomer channels, and the d-isomer channel conductance is blocked by Zn2+, a known blocker of l-Aβ isomer channels. MD simulations further verify formation of β-barrel-like Aβ channels with d- and l-isomers, illustrating that both d- and l-Aβ barrels can conduct cations. The calculated values of the single-channel conductance are approximately in the range of the experimental values. These findings are in agreement with amyloids forming Ca2+ leaking, unregulated channels in AD, and suggest that Aβ toxicity is mediated through a receptor-independent, nonstereoselective mechanism.
Introduction
Alzheimer’s disease (AD) is the most common form of dementia and accounts for 60–80% of the cases. In the United States, AD is the sixth leading cause of death and predicted to grow with the increasing life expectancy.1 Prevailing evidence suggests that amyloid β (Aβ) peptides are cytotoxic to cells and play a role in the pathogenesis of AD.2 Aβ-induced neuronal degeneration is most likely preceded by dysregulation of cellular ionic homeostasis, especially [Ca2+].3 Loss of cellular ionic homeostasis could occur via several mechanisms, including Aβ-specific membrane receptors and amyloid-specific membrane channels.4,5 Arispe et al.6 reported the Aβ-induced nongated ion channels in model membranes that showed cation selectivity and could be inhibited by Tris (tromethamine) and zinc.7 These results prompted the amyloid channel hypothesis where Aβ directly forms channels toxic to neurons when sufficient peptide is available.8,9
Aβ channels are characterized by heterogeneous conductances, suggesting a dynamic conformation of the pore structures. Recently, similar conclusions were obtained for Aβ1–42 by the single-channel Ca2+ imaging technique.10 Figure 1 shows an example where various size steps, bursts, and spikes are apparent.11 Channel conductances ranging from 10 pS to 2 nS have been reported.9 Channel-like structures embedded in membranes with mostly tetramer to hexamer arrangements have been observed by atomic force microscopy (AFM).12,13 Molecular dynamics (MD) simulations based on solid state nuclear magnetic resonance (ssNMR) based structures also indicated channel structures,14−16 fitting well the dimensions and subunit arrangements observed in AFM12,13 and electron microscopy (EM)17 experiments. Furthermore, simulations suggested that subunits (defined as oligomers with 2–5 monomers embedded in the bilayer) are mobile and form heterogeneous structures. This dynamic behavior could explain the typical heterogeneous conductances of amyloid channels.9,18
Figure 1.
Current vs time trace showing Aβ1–42 has pore-forming activity in artificial bilayers. Aβ1–42 activity is characterized by heterogeneous conductances and exhibits channel-like steps, spikes, and bursts. Final Aβ1–42 concentration was 9 μM. The 20 min trace was held to an applied potential of −50 mV. Predominant steps on the trace have conductances of 140, 81, 91, and 127 pS. Bilayer was prepared by the painted technique from a 1:1 (w/w) lipid mixture of DOPS/POPE. Both sides of the bilayer chamber contained as electrolyte 150 mM KCl, 1 mM MgCl2, and 10 mM Hepes pH 7.4. l-Aβ was added to the cis side. The trans side of the chamber was virtual ground. This trace was subjected to low-pass Gaussian filtering set at 50 Hz.
In order to elucidate the direct vs indirect mechanism of Aβ-mediated toxicity, it is important to distinguish a receptor-mediated from a nonreceptor-mediated (ion channel) pathway. We reasoned that using Aβ peptides whose sequence consists entirely of true mirror image d-amino acids (d-Aβ) we may discern the relative contributions to toxicity by ligand-specific receptors vs channel formation; putative cellular receptors will not bind the d-Aβ because of a lack of conformational fitting; however, d-Aβ should be able to form pores and bind to nonstereo-specific targets (e.g., electrostatically to negative lipid headgroups and hydrophobic interactions with lipid tails).
Here we show that the d-Aβ peptide forms channels that retain the properties known for l-amino acids Aβ (l-Aβ) channels, including heterogeneous conductances and blockage by Zn2+. Furthermore, to understand the chiral molecular details we modeled the d- and l-Aβ barrels in the anionic lipid bilayer using atomistic MD simulations. The models support and expand the experimental observations. Both isomeric Aβs form β-barrel-like ion-permeable channels in an anionic lipid bilayer.19 The dynamic channel assembly in the fluid lipid bilayer suggests an explanation for the heterogeneous conductances observed for both d- and l-Aβ channel-like structures. MD simulations show that for both d- and l-Aβ barrels the calculated values of the conductance for cations in the solvated pores are approximately in the range of the experimental values.
Materials and Methods
Materials
We purchased all l-amino acids Aβ1–42 and their true mirror images, all d-amino acids Aβ1–42, from Bachem and the following lipids from Avanti Polar Lipids: 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE), 1,2-dioleoyl-sn-glycero-3-phosphoserine (DOPS). All other chemicals were purchased from Sigma–Aldrich. For all experiments both d- and l-Aβ1–42 peptides were dissolved in water to 0.5 or 1 mM stock concentration, aliquoted, and kept frozen at −80 °C. Samples were thawed only once.
Planar Lipid Bilayers
We prepared planar lipid bilayers (PLB) either by the so-called “folding technique”,20,21 by apposition of lipid monolayers over a pore (diameter ≈ 120 μm) in a Teflon film, or by the so-called “painting technique”,22,23 which applied lipids dissolved in heptane over a pore with a diameter of ∼250 μm in a Delrin septum. For the folded bilayers we pretreated the Teflon film with 5% (v/v) hexadecane dissolved in pentane and then formed bilayers using 5 μL from a 20 mg/mL 1:1 (w/w) mixture of DOPS and DOPE solution dissolved in pentane. For the painted bilayers, we used a bilayer cup (Warner Instruments, Delrin perfusion cup, volume 1 mL) and a 1:1 (w/w) mixture of lipids DOPS/POPE dissolved in heptane or hexane. The lipid concentrations were 20 mg/mL. The electrolyte contained either 50 or 150 mM KCl and 1 mM MgCl2 buffered with 10 mM HEPES pH 7.4 or MES pH 6.4. In order to promote fusion of Aβ proteoliposomes into painted bilayers, we used a KCl gradient formed by adding to the cis side (the side of proteoliposome addition) KCl to 350 mM and leaving the trans side with 150 mM KCl. For these experiments, we prepared proteoliposomes using DOPS or POPS containing Aβ as described previously.6
We performed all recordings using custom-made software with either a BC-535 or a EPC-7 amplifier and Ag/AgCl electrodes directly into the electrolyte or (for reversal potential experiments) with 1% agarose (ultra-high purity) agar salt bridges containing 1 M KCl. We used amplifiers with an inbuilt filter cutoff frequency of 2 (BC-535) or 3 kHz (EPC-7) and a sampling frequency of 15 kHz for all bilayer recordings. For representation in figures, we filtered the current versus time traces with a digital Gaussian low-pass filter with a cutoff frequency of 50 or 100 Hz. A positive potential indicates a higher potential in the cis side of the PLB chamber; hence, a positive current moves cations from the cis to the trans side. The trans side of the PLB chamber was grounded.
Before adding Aβ, we verified that both painted and folded bilayers were stable for several minutes and that capacitance was above 120 or 70 pF, respectively. When both criteria were fulfilled, we added Aβ1–42 to the cis side either directly or via Aβ-liposome fusion and stirred for 3–5 min every 15 min. The final Aβ1–42 concentration in the bilayer chamber was between 0.5 and 18 μM, with generally lower concentrations needed when using Aβ-liposomes. Bilayer stability was monitored by periodical capacitance measurements during the course of experiments. Experiments with painted bilayers were terminated 90–120 min after peptide addition, regardless of whether or not channel activity was present. This was done because the stability of bilayer-only experiments showed that DOPS/POPE-painted membranes are generally stable for ∼90 min while folded DOPS/DOPE experiments exceeded 4 h.
Reversal potential (Vrev) experiments were carried out in painted bilayers made with DOPS/POPE phospholipids. Bilayers were formed using as electrolyte a solution containing 50 mM KCl, 1 mM MgCl2, and 10 mM Hepes pH 7.4 on both sides of the PLB chamber (symmetric solutions). After verifying the bilayers stability, 4.5 μM d- or l-Aβ1–42 dissolved in water was added to the cis side (hot wire) and an equal volume of water to maintain electrolyte symmetry was added to the trans side, which is the virtual ground. Once Aβ activity was established a current (I) vs voltage (V) plot (IV plot) is taken. We next made asymmetric the concentration of KCl across the two PLB compartments. Specifically, a 1:10 KCl gradient is established by adding a small volume of a concentrated KCl solution to achieve 500 mM in the cis (hot wire) side. An equal volume of 50 mM KCl is added to the trans side. Complete mixing to establish the KCl gradient is essential. Cation selectivity is expected to produce an upward drift in current at zero voltage. To determine the reversal potential (or Vrev, defined as the electrical potential giving zero current) a negative voltage is required. The PLB setup was also verified using the potassium-selective valinomycin.
Atomistic Molecular Dynamics Simulations
d- and l-Aβ1–42 barrels embedded in an anionic lipid bilayer containing DOPS and POPE were simulated using explicit all-atom lipid simulations. The CHARMM program24 using the revised CHARMM27 (C27r) force field for lipids25 and the modified TIP3P water model26 were used to construct the set of starting points and to relax the systems to a production-ready stage. For production runs, the NAMD code27 on a Biowulf cluster (http://biowulf.nih.gov) at the NIH was used for the starting point with the same CHARMM27 force field.
The standard CHARMM force field is primarily designed for l-amino acids. To simulate d-amino acids, we need a protein force field for asymmetric isomers. d-Amino acid is a mirror image of the l-amino acid; thus, except for their backbone chirality they are identical. Because the l- and d-isomers have the same backbone bonding and angles, in the simulations we adapted the standard l-amino acid parameters to d-amino acids. However, the parameters include the dihedral angle cross-term map (CMAP) correction,28 which was created for only l-amino acids, and cannot be directly applied to d-amino acids. Thus, in our simulation we used a mirror-image CMAP term for d-amino acids by reflecting the phi–psi CMAP matrix.19
Two U-shaped monomer conformations of Aβ, Aβ1–42 as defined in the fibril structure based on hydrogen/deuterium-exchange NMR data, side-chain packing constraints from pairwise mutagenesis, ssNMR and EM (PDB code 2BEG),29 and Aβ1–40 based on the ssNMR model of small protofibrils30 were used to construct Aβ barrels. However, the N-terminal coordinates of both conformers are missing due to disorder. We used the Aβ1–16 coordinates in the absence of Zn2+ (PDB code 1ZE7)31 for the missing portions of the peptides. For each combination of the N-terminal structure with the U-shaped motifs two Aβ1–42 conformers were generated (Supporting Information, Figure S1A). Conformer 1 has a turn at Ser26-Ile31 and conformer 2 at Asp23-Gly29. In the latter conformer two C-terminal residues, Ile41 and Ala42, were added to create Aβ1–42. The coordinates of d-Aβ1–42 are mirror images of l-Aβ1–42 and can be obtained by reflecting the coordinates with respect to the reference plane. The Ramachandran plot for d-Aβ1–42 clearly indicates asymmetric dihedral distributions compared to l-Aβ1–42 (Supporting Information, Figure S1B). The d-Aβ1–42 conformers still retain the U-shaped β-strand-turn-β-strand motif as the l-Aβ1–42 conformers, regardless of their isomeric forms. To construct the β-barrel structure, these Aβ conformers were inclined ∼37° relative to the pore axis32 and then rotated 18 times with respect to the pore axis creating Aβ barrels (Supporting Information, Figure S1C and S1D).19 We modeled 18-mer Aβ barrels, that is, 18 β-strands enclosing the solvated pore. This number is within the range of the 8–22 β-strands observed for biological β-barrels.33,34 Further, previously we simulated Aβ channels with different sizes (12–36 β-strands) and compared the obtained morphologies and outer channel and pore dimensions with those measured by AFM.16,32,35 We observed that the preferred size range of Aβ channels is 16–24 β-strands. We further observed that this range holds for other toxic β-sheet channels; the K3 fragment of β2-microglobulin forms channels with 24 β-strands,36 and PG-1 channels with 16–20 β-strands.23,37 In both of these K3 and PG-1 cases we also compared our simulations with our AFM measurements; in both it is expected that given the heterogeneous landscape of amyloids, variable channel sizes within this range can form.
A unit cell containing two layers of lipids was constructed. In the middle of the unit cell lipid molecules were randomly selected from the library of the pre-equilibrated state and replaced by pseudo-vdW spheres at the positions of the lipid headgroups, constituting the lipid bilayer topology.38,39 For DOPS, the cross-section areas per lipid and the headgroup distance are 65.3 Å2 and 38.4 Å at 303 K, respectively.40 For POPE, they are 56.0 Å2 and 41.3 Å at 303 K, respectively.41 For the bilayer construction, we closely follow previous β-sheet channel simulations.16,35,42 For a given number of lipid molecules the optimal value of lateral cell dimensions can be determined. An anionic lipid bilayer composed of DOPS/POPE (mole ratio 1:2) containing a total of 420 lipids constitutes the unit cell with TIP3P waters added at both sides. The system contains Mg2+, K+, Ca2+, and Zn2+ at the same concentration of 25 mM to satisfy a total cation concentration near 100 mM. In our simulations, the Lennard–Jones parameters for these ions were taken from theoretical studies.43−45 The bilayer system containing an Aβ barrel, lipids, salts, and waters has almost 190 000 atoms.
We generated many different initial configurations for the relaxation process in order to obtain the best initial configuration toward a starting point. A series of minimizations was performed for the initial configurations to remove overlaps of the alkane chains in the lipids and to gradually relax the solvents around the Aβ barrel, which was held rigid. The initial configurations were gradually relaxed through dynamic cycles with electrostatic cutoffs (12 Å). In the subsequent pre-equilibrium stages, a series of dynamic cycles was performed with the harmonically restrained peptides in the channels, and then the harmonic restraints were gradually diminished until gone with the full Ewald electrostatics calculation. The entire pre-equilibration cycle took 5 ns to yield the starting point. A Nosé–Hoover thermostat/barostat was used to maintain a constant temperature of 303 K. Simulations for the pre-equilibrations and production runs were performed on the NPAT (constant number of atoms, pressure, surface area, and temperature) ensemble. Production runs of 120 ns for the starting points with the NAMD code27 were performed on a Biowulf cluster at the NIH. Averages were taken after 20 ns, discarding initial transients. Analysis was performed with the CHARMM programming package.24
Results
Both d- and l-Aβ1–42 Isomers Form Channel-Like Pores in the Bilayer
The l-Aβ channel activity was previously studied using planar lipid bilayers (PLBs).6,11,21,46 We investigate whether the d-Aβ forms a conducting channel similar to its l mirror image. Figure 2 demonstrates that both d- and l-Aβ1–42 show ion channel-like activity (Figure 2A and 2B). At concentrations in the range of 0.5–18 μM and constant voltage both isomers exhibit step-like current jumps typical of Aβ channels. In both cases, the current jump steps are heterogeneous, as previously reported for l-Aβ.46 We confirm this behavior for l-Aβ and present it for d-Aβ (Figures 2 and 3 and Supporting Information, Figure S2). The Aβ ‘spiky’ behavior is similar to that described for β-sheet-rich antimicrobial peptides (AMPs) such as protegrin-1 (PG-1).23,47,48 Both d- and l-Aβ peptides were sensitive to Zn2+ addition (Figure 2C and 2D), showing that this inhibition is achiral. Both d- and l-Aβ showed linear current (I) vs voltage (V) plots (IV plots) in the ±80 mV range tested (Supporting Information, Figure S3). Linear IV plots results were also observed using 50 and 150 mM KCl or electrolyte at pH 6.4 (data not shown). To investigate ion selectivity, both d- and l-Aβ isomers were tested for their reversal potential in asymmetric KCl solutions where a 10-fold difference in KCl concentration was established across the two PLB compartments (experimental details in the Materials and Methods section). The results are summarized in Figure S4 and Table S1 (Supporting Information). They show a range of reversal potentials, indicating a variety of conducting structural entities. The PK+/PCl– ratios indicate a modest to moderate cation selectivity preference with ratios ranging from a minimum recorded of a PK+/PCl– ratio of 1.44 to a maximum 6.35. These findings for both d- and l-Aβ isomers are in agreement with the reported literature for l-Aβ.6,11,21,46,49 Additionally, we tested whether the activity of d-Aβ might be affected by pH and found that at pH 6.4 both isomers retain the channel activity characteristics described for pH 7.4 (Supporting Information, Figure S2).
Figure 2.
Both d- and l-Aβ1–42 isomers form channel-like pores in painted bilayer membranes and are inhibited by addition of Zn2+. (A) Representative current vs time trace of d-Aβ1–42 isomer activity. (B) Representative current vs time trace of l-Aβ1–42 isomer activity. (C) Inhibition of d-Aβ1–42 activity by addition of Zn2+. (D) Inhibition of l-Aβ1–42 activity by Zn2+ addition. Time of Zn2+ addition (2 mM) is marked by tilted arrows on panels C and D. In panels C and D, the increased noise between the two vertical arrows shows when stirring begins and ends. The C letters on top of vertical lines show monitoring of bilayer capacitance during these recordings. Changes in the applied voltage are indicated by the voltage–time plot under the current traces. Final Aβ1–42 peptide concentrations were 9 (A), 8.5 (B), 5 (C), and 5 μM (D). Lipids and electrolyte solution are the same as in Figure 1; either Aβ1–42 peptide was added to the cis (hot wire) side. All traces were subjected to low-pass Gaussian filtering set at 50 Hz.
Figure 3.
Both d- and l-Aβ1–42 isomers are membrane active in ‘solvent-free’ folded DOPS/DOPE bilayers. (A) Representative activity of l-Aβ1–42 in ‘folded’ membrane bilayers. (B) Representative activity of d-Aβ1–42 in equivalent bilayer membrane. Note that in this type of bilayer both d- and l-Aβ1–42 isomers show predominantly short-lived spikes and bursts of activity with less frequent stepwise current jumps when compared to the painted bilayers shown in Figure 2. Peptide concentrations in the bilayer chamber were 4.5 μM. As electrolyte we used 150 mM KCl, 1 mM MgCl2, 10 mM Hepes, pH 7.4. Both Aβ1–42 were added to the cis side. Trans side was the virtual ground. Both 30 s current traces are shown with the same scaling of y and x axes after low-pass Gaussian filtering with a cutoff frequency of 50 Hz. Both traces shown are at 70 mV.
We also investigated the channel behavior of both Aβ isomers using ‘solvent-free’ folded membranes composed of DOPS/DOPE (1:1 w/w).20,21 Figure 3 shows representative current versus time traces for both d- and l-Aβ isomers. In folded bilayers, channel-like activity was observed in 41% (7/17) of the experiments with l-Aβ and in 36.3% (4/11) of the experiments with d-Aβ. The channel activity was generally short lived and appears mostly as spikes or spikes–bursts. Stepwise current jumps are present but appear with lesser frequency when compared to painted bilayers. In folded bilayers, however, the channel activity usually had lower current amplitudes for both d- and l-Aβ1–42 isomers. In painted bilayers using 150 mM KCl channel activity was observed in 82% (9/11) of the experiments for l-Aβ and 87% (7/8) of the experiments for d-Aβ. These results demonstrate nearly identical Aβ1–42 channel behavior for both d- and l-Aβ isomers in both types of bilayers.
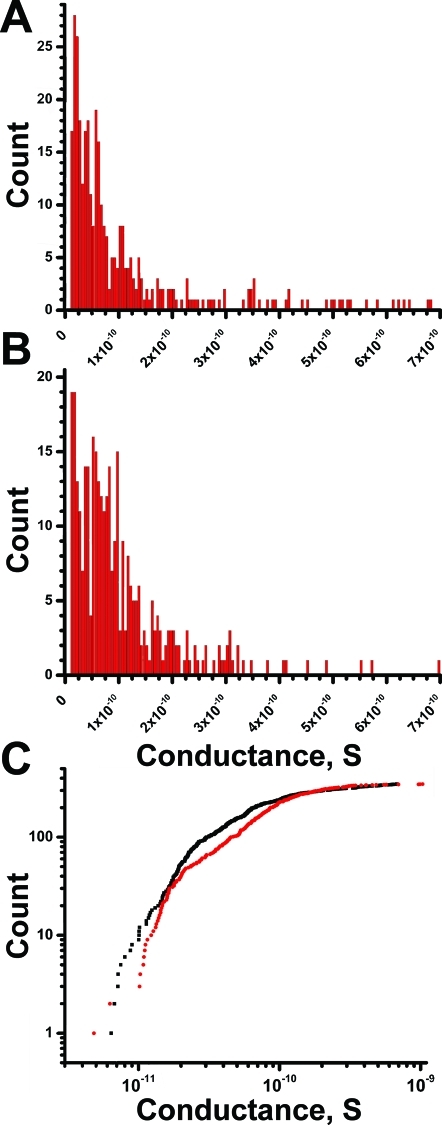
We next examined whether a difference existed among d- and l-Aβ channel conductances in DOPS/POPE bilayers. Figure 4 shows the histogram distribution of d- and l-Aβ1–42 single-channel conductances. While for both peptides the histogram distribution illustrates a wide range of conductances, 65% of them are below 100 pS and 85% are between 5 and 200 pS (Figure 4A and 4B). Overall, no significant difference was observed in conductance values among the d- and l-Aβ1–42 (Figure 4C). Additionally, the results in the figure show that Aβ binding to phosphatydyl-serine is electrostatic, since no difference was observed regardless of the chiral nature of l-Ser in the PS headgroup. The combined results show that the permeabilization activity of d-Aβ in anionic bilayers is indistinguishable from its natural l mirror image. We further tested whether d-Aβ behaves similarly to l-Aβ at atomistic resolution by simulating the d- and l-Aβ isomers.
Figure 4.
Histogram analysis of (A) d-Aβ1–42 and (B) l-Aβ1–42 conductances. Both histograms are binned at 5 pS. (C) Plot of both sets of conductances in log scale shows a similar trend with slight overrepresentation for l-Aβ1–42 around ∼50 pS conductances. Both sets of Aβ1–42 conductances can be sorted into three groups; the most frequent occurrence in the interval up to 100 pS has an average conductance of ∼52 ± 27 pS for l-Aβ1–42 and 42 ± 23 pS for d-Aβ1–42 and contains 65% of all samples. Second group of conductances in the range from ∼100 to 200 pS shows an average of 142 ± 29 pS for l-Aβ1–42 and 134 ± 28 pS for d-Aβ1–42. Third group shows sparse conductances above the 300 pS range. There is a continuum of conductances between 5 and 200 pS, with an 85% representation of the entire sample in this interval. Note that the wide distribution in pore conductance and their reduced frequency above a certain threshold are expected for the Aβ1–42 channel structures proposed in this study. Data sample size was 347 for d-Aβ1–42 and 345 for l-Aβ1–42. Data were collected in the ±80 mV range, where IV plots are linear. Experiments were performed using both peptides in the concentration range from 0.5 to 18 μM. Electrolyte used was 150 mM KCl, 1 mM MgCl2, and 10 mM Hepes pH 7.4. Bilayers were made with DOPS/POPE lipids dissolved in heptane.
MD Simulations of d- and l-Aβ1–42 Barrels Support Ion Channel Features
We modeled Aβ barrels with the β-sheet structure by mimicking naturally occurring β-barrels observed in transmembrane proteins that are found frequently in the outer membranes of bacteria, mitochondria, and chloroplasts. The β-barrel motif is a large β-sheet composed of an even number of β-strands. Some known structures of β-barrel membrane proteins have β-strands ranging from 8 to 2233,34 and induce cytotoxicity.50 While functional gated channels contain mostly α-helices, physiological toxic amyloid channels contain mostly β-sheets. A substantial body of evidence has already indicated that amyloid channels consist of the β-sheet structure.46,51 More recently, it has been demonstrated that the presence of lipid bilayer membranes can also catalyze β-sheet formation.52−55 Recently, we reviewed the evidence which supports that amyloid channels consist of the β-sheet structure.52
We performed 120 ns explicit MD simulations on d- and l-Aβ1–42 barrels embedded in an anionic lipid bilayer composed of DOPS/POPE (mole ratio 1:2). Both isomer barrels contain two Aβ conformers: with a turn at Ser26-Ile31 (conformer 1) and Asp23-Gly29 (conformer 2). Our conceptual design was inspired by an initial shape of the Aβ barrel with a perfect annular shape.32 The initial annular conformation is gradually lost via relaxation of the lipid bilayer, and environmentally relaxed peptides can be observed after 30 ns (Supporting Information, Figure S5). The membrane-embedded portions of the Aβ barrels reach equilibration after the initial transient state, while the extramembranous N-termini of the peptides are disordered. Snapshots representing the averaged barrel conformations clearly indicate the relaxed Aβ barrel conformations (Supporting Information, Figure S6). In the relaxed Aβ barrels, the amino acids still retain their original chirality. Contour maps representing the high probability of backbone dihedral angles of phi (φ) and psi (ψ) clearly indicate the β-sheet secondary structure and asymmetric dihedral angle distributions between d- and l-amino acid chiralities (Supporting Information, Figure S7). Secondary structure analysis indicates that our Aβ barrels preserve the β-sheet structure, especially in the pore-lining strands (Supporting Information, Figure S8). These results are in agreement with our previous analyses of Aβ channel simulations that we carried out for Aβ1–42, Aβ17–42 (p3), and Aβ9–42 (N9) across a range of channel sizes.16,19,32,35,42
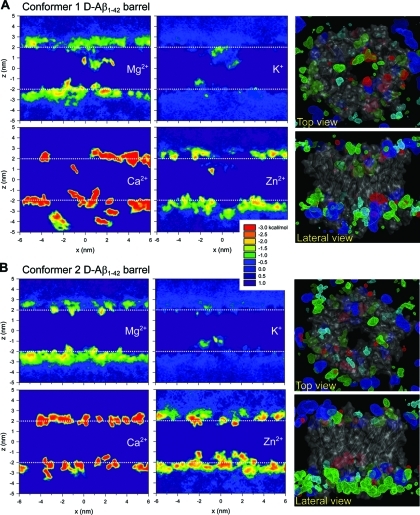
Regardless of the d- and l-amino acid chirality, both Aβ conformers form barrels with the membrane-embedded β-strands (central domain of the peptides) lining the solvated pore and the C-terminal β stands interact with lipid tails, whereas the N-terminal portions are disordered and extramembranous. Recent structural modeling of Aβ18–41 dimer with the N-terminal Aβ1–16 binding sites in a membrane environment suggests that residues ∼21–29 and the N-terminal sites are exposed to solution while the C-terminal hydrophobic residues are involved in dimer–dimer interactions and buried in the lipid hydrophobic core.56 In our model, residues ∼21–29 are involved in the central domain or turn and also exposed to solution. For convenience, we define the upper bilayer leaflet where the turn residues in the barrel are and the lower bilayer leaflet where both termini are located. The hydrophobic center of the lipid bilayer is located in the center of the pore axis z. In the conformer 1 Aβ barrel, the negatively charged Glu22 side chains (z = ∼4.3 Å) are located just above the bilayer center pointing toward the water pore, while the positively charged Lys16 side chains (z = ∼−13.0 Å) are located near the channel mouth in the lower bilayer leaflet. For the conformer 2 barrels, while the negatively charged Glu22 side chains (z = ∼12.4 Å) are located near the channel mouth in the upper bilayer leaflet, the positively charged Lys16 side chains (z = ∼−3.9 Å) are located just below the bilayer center. These different charge distributions in the pore reflect different turn motifs between the Aβ conformers. The charged side chains in the pore can serve as ionic binding sites, facilitating ionic permeation through the solvated pore. Figure 5 shows the two-dimensional (2D) potential of mean force (PMF) of ions for the d-Aβ1–42 barrels. The contour maps clearly indicate the populated cationic binding sites. In the calculation of the 2D PMF map, the barrels are translated into the bilayer center for each simulation time frame and the occupancy probabilities of salts at each grid point were calculated. Thus, any contour map plotted in the ranges −2 < z < 2 nm and −1.5 < x < 1.5 nm can be regarded as cations interacting with pore residues. The details of the PMF calculation are described in the Supporting Information. In the conformer 1 d-Aβ barrel, cations can bind to the interaction sites, including the negatively charged side chains in the pore and negative polar residues in the extramembranous N-termini and even including the anionic DOPS headgroups. Compared to other cations, K+ is very mobile while Ca2+ binds to the interaction sites rather strongly, producing a relatively low free energy profile at these sites. Zn2+ provides a similar map as Mg2+ but binds to the interaction sites relatively more strongly than Mg2+. In the conformer 2 d-Aβ barrel, the cations behavior is similar except that, as indicated by the contour maps, the Glu22 binding sites are located closer to the upper bilayer leaflet. For l-Aβ barrels, 2D PMFs also provide similar binding sites for cations as seen for their d counterparts (Supporting Information, Figure S9).
Figure 5.
Two-dimensional (2D) potential of mean force (PMF) representing the relative free energy profile for Mg2+, K+, Ca2+, and Zn2+ as a function of the position on the x–z plane for d-Aβ1–42 barrels (A) with conformer 1 (turn at Ser26-Ile31) and (B) conformer 2 (turn at Asp23-Gly29). In the 2D PMF map, dotted lines at z = ∼2 nm indicate the upper bilayer leaflet while at z = ∼−2 nm denote the lower bilayer leaflet. Right column shows the three-dimensional density map of Mg2+ (green mesh), K+ (red mesh), Ca2+ (blue mesh), and Zn2+ (cyan mesh) for the d-Aβ1–42 barrels in the top and lateral views. Averaged channel structure is shown as the ribbon and transparent surface representations in gray. Density map indicates populated interaction sites for the cations, each with the same probability of 0.01.
The Aβ barrel pore is wide enough for multiple ions to enter and exit at the same time. The calculated pore diameters by the HOLE program57 are ∼1.9 and ∼2.1 nm for conformer 1 and 2 d-Aβ barrels and ∼2.2 and ∼2.0 nm for conformer 1 and 2 l-Aβ barrels, respectively. We observed that few ions cross through the water pore, but most ions spend time at the binding sites and are frequently near the channel mouth during the simulation. To observe ion fluctuation across the pore, we calculated the change in total charge in the pore as a function of the simulation time (Supporting Information, Figure S10). Four different pore lengths with different cutoffs along the pore axis were used in the calculation. For d-Aβ barrels, the fluctuations of the total charge change in the pore increase as the pore length cutoff increases. The degree of charge fluctuations can be defined as the standard deviations of the averaged changes in the total charge in the pore. For the conformer 1 d-Aβ barrel, the standard deviations of the changes in total charge are ±7.96, ±11.35, ±19.18, and ±25.55 C/ns with pore length cutoffs along the pore axis, |z| < 1.0, 1.5, 1.8, and 2.0 nm, respectively. For the conformer 2 d-Aβ barrel, these are ±5.43, ±11.43, ±19.92, and ±27.29 C/ns. We also observed similar fluctuations in the total charges in the l-Aβ barrel pores. Note that with the larger pore length cutoff |z| < 2 nm the charge fluctuations may involve contributions of ions interacting with lipid headgroups, since the thickness of the anionic bilayer is ∼40.3 Å.
For the equilibrium all-atom MD simulations in the absence of membrane potentials the maximum conductance, gmax,58 representing ion transport can be described as
| 1 |
where e is the elementary charge, kB denotes the Boltzmann’s constant, T is the simulation temperature, and L represents the pore length. In brackets, D(z) and GPMF(z) denote the one-dimensional diffusion coefficient and the one-dimensional potential of mean force for ions, respectively. The bracket averages over the pore length L (38 Å). Using eq 1, for Mg2+, K+, Ca2+, and Zn2+ the maximum conductances are 350, 230, 87, and 170 pS and 82, 200, 53, and 100 pS in the pores of conformer 1 and 2 d-Aβ barrels, respectively. In the l-Aβ barrel pores they are 280, 290, 130, and 210 pS and 150, 130, 76, and 100 pS for Mg2+, K+, Ca2+, and Zn2+, respectively. The theoretical value of the maximal conductance only provides a rough estimate of the conductance, because it neglects the effect of multiple ion occupancy at high concentration.58 Nevertheless, our estimated values of the single-channel conductance are approximately in the range of the experimental values.
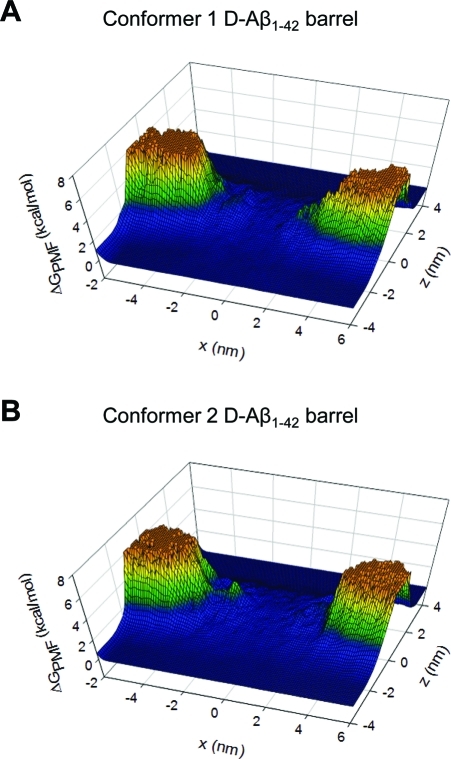
Water is an important carrier for ion permeation through the pore. For d-Aβ barrels, Figure 6 shows the 2D PMF of water on the three-dimensional mesh graph. It is clear from the figure that the hydrophobic core of the lipid bilayer provides high barriers for water on the PMF surface. In the pore, however, the Aβ barrels provide a low free energy profile for water, indicating that water can move freely through the center of the pore. The behavior of water in the pore is similar to bulk water, suggesting that water is a good carrier to deliver ions across the pore. No difference is found in the l-Aβ barrels (Supporting Information, Figure S11), illustrating that both Aβ isomers form channel-like structures and exhibit channel-like activity.
Figure 6.
Two-dimensional potential of mean force (PMF) representing the relative free energy profile for water plotted as a three-dimensional mesh graph as a function of the position on the x–z plane for (A) conformer 1 and (B) conformer 2 d-Aβ1–42 barrels.
Discussion
Lipid–peptide interactions are complex and key to amyloid structures.59−62 Here we show that the d enantiomer of Aβ1–42 composed entirely from mirror image d-amino acids, spontaneously forms channels in model bilayers, with characteristics indistinguishable from those of l-amino acids Aβ1–42 (Figure 2A and 2B). Both peptides form channel-like structures over a range of concentrations, show linear current vs voltage relationships (Supporting Information, Figure S3), and have a broad distribution of channel conductances (Figure 4). Both Aβ isomers show similar lipid preferences and are inhibited by Zn2+ (Figure 2C and 2D). For both l- and d-peptides, we observed that ∼85% of the recorded conductances fall below 200 pS with the remaining higher conductances appearing less frequently (Figure 4). Both appear to bind and insert equally in bilayers made with 50% DOPS, indicating a purely electrostatic interaction with the l-Ser in the PS headgroup, in agreement with l-Aβ activity in bilayers containing PG, another anionic lipid.46 Channel activity for both d- and l-Aβ isomers is short lived when using folded membranes (Figure 3), which may suggest a role for membrane fluidity in peptide and oligomer membrane insertion and stability of the channel ensembles in these thinner63 and more fluid bilayers.64 Gafni and co-workers showed an Aβ role for bilayer permeation and fluidity.65 Reversal potential experiments showed cation selectivity with variable PK+/PCl– ratios (Supporting Information, Figure S4). Unlike more stable and well-defined β barrel structures like α-hemolysin,66,67 the Aβ results further suggest structural pore models where Aβ can assemble into various conducting and dynamic structures and where cation selectivity could depend on the arrangement of Aβ monomers in the channel and on the number of subunits. Lipid headgroups are also likely to be involved in the evolution of the channel pore.
MD simulations also show that d-Aβ1–42 forms ion-conducting channels in an anionic lipid bilayer composed of DOPS/POPE. In the simulations, the d-Aβ barrels modeled with two different conformers (conformer 1 with turn at Ser26-Ile31 and conformer 2 with turn at Asp23-Gly29) preserve the solvated pore, wide enough for ions and water to cross through.19 The pores enclose the cationic binding sites, providing low free energy profiles for cations, hence supporting ion-permeable Aβ channels. The behavior of d-Aβ barrels is almost identical to the l-Aβ barrels, even though constituted by peptides with different backbone chiralities. In our previous simulations of the truncated Aβ channels (p3 (Aβ17–42) and N9 (Aβ9–42) channels),14−16,32,35,42 the channels similarly presented strong attraction for cations in the solvated pore. This suggests that the U-shaped C-terminal domain of Aβ1–42 is indeed the membrane-embedded portion which is responsible for channel formation and conductance. Missing the N-terminal portion of Aβ1–42, the truncated Aβ channels are still conducting, strongly suggesting that the polar/charged N-termini could be extramembranous.
The d-enantiomer of Aβ1–40 and Aβ1–42 peptides has been previously studied and shown to have biophysical properties such as fibril formation, CD spectra, and aggregation similar to their l mirror images;68−71 however, differences in the rate of aggregation when using Thioflavin-T fluorescence68 and Aβ1–42 fiber chirality70 were reported. Functional and structural similarities between Aβ and AMPs have been noticed earlier. Recently, it was proposed that Aβ is an AMP72 and that β-rich AMP can be amyloidogenic.73 AFM imaging, MD simulations, and functional assays such as PLB recordings and fluorescence imaging10 suggest that Aβ is capable of forming channel-like structures in cells. However, they cannot quantify this effect or exclude the possibility that Aβ might act via other mechanisms. Studies on the mechanism of AMPs faced similar difficulties, which were addressed using AMPs composed of d-amino acids. For several AMPs, there are no chiral receptors and the cell membrane is the sole target.47,74,75 The results presented here suggest that cellular studies might be able to evaluate the relative contribution of Aβ channel-mediated versus receptor-mediated toxicities. Differences, if present, can be attributed to stereospecific mechanisms. However, studies using d-Aβ1–42 to test cellular toxicity reported conflicting results.70,71 The earlier study by Cribbs et al. showed similar toxicity for both Aβ1–42 isomers,71 while a more recent study by Cioccotosto et al. showed lack of cell toxicity for the d-Aβ isomer.70 Aβ cellular toxicity could be cell-type dependent as shown for some AMPs;47,76−78 alternatively, differences could also relate to sample preparation and experimental approaches.
To summarize, using planar lipid bilayers and MD simulations we show that the d-Aβ1–42 forms channel-like structures with behavior indistinguishable from the naturally occurring l-Aβ1–42. The combined results support the view that excessive Aβ in the brain can be neurotoxic via direct membrane permeation with a mechanism consistent with channel formation, as compared to stereospecific receptor binding.
Acknowledgments
This research was supported by the National Institutes of Health (National Institute on Aging AG028709 to R.L.). This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract no. HHSN261200800001E. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. All simulations were performed using the high-performance computational facilities of the Biowulf PC/Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov).
Supporting Information Available
Details of PMF calculations; monomer conformations and initial Aβ barrel structures of MD simulations; additional d- and l-Aβ1–42 current traces at pH 6.4; IV plots showing linear relationship in current vs voltage reversal potential experiments; peptide rmsd and averaged peptide interaction energy with DOPS and POPE lipids as a function of time; snapshots representing averaged barrel conformations; backbone dihedral angle distributions; secondary structure for Aβ barrels and 2D PMF for ions for l-Aβ1–42 barrels; change in total charge in the pore as a function of the simulation time; 2D PMF for water for l-Aβ1–42 barrels; reversal potential permeability ratios and conductance values. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
This paper was published ASAP on February 23, 2012. A change has been made in the seventh paragraph of the Results section. The correct version was published on February 27, 2012.
Author Contributions
⊥ Both authors contributed equally to this work.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Ballard C.; Gauthier S.; Corbett A.; Brayne C.; Aarsland D.; Jones E. Lancet 2011, 377, 1019–1031. [DOI] [PubMed] [Google Scholar]
- Khachaturian Z. S. Ann. N.Y. Acad. Sci. 1994, 747, 1–11. [DOI] [PubMed] [Google Scholar]
- Mattson M. P.; Cheng B.; Davis D.; Bryant K.; Lieberburg I.; Rydel R. E. J. Neurosci. 1992, 12, 376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons M. A.; Schneider C. R. Neurosci. Lett. 1993, 150, 133–136. [DOI] [PubMed] [Google Scholar]
- Yankner B. A.; Duffy L. K.; Kirschner D. A. Science 1990, 250, 279–282. [DOI] [PubMed] [Google Scholar]
- Arispe N.; Rojas E.; Pollard H. B. Proc. Natl. Acad. Sci. U.S.A. 1993, 90, 567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arispe N.; Pollard H. B.; Rojas E. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, 1710–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard H. B.; Rojas E.; Arispe N. Ann. N.Y. Acad. Sci. 1993, 695, 165–168. [DOI] [PubMed] [Google Scholar]
- Kagan B. L.; Azimov R.; Azimova R. J. Membr. Biol. 2004, 202, 1–10. [DOI] [PubMed] [Google Scholar]
- Demuro A.; Smith M.; Parker I. J. Cell Biol. 2011, 195, 515–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourie J.; Henry C.; Farrelly P. Cell Mol. Neurobiol. 2001, 21, 255–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H. A. I.; Bhatia R.; Lal R. FASEB J. 2001, 15, 2433–2444. [DOI] [PubMed] [Google Scholar]
- Quist A.; Doudevski I.; Lin H.; Azimova R.; Ng D.; Frangione B.; Kagan B.; Ghiso J.; Lal R. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 10427–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.; Zheng J.; Nussinov R. Biophys. J. 2007, 93, 1938–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.; Zheng J.; Lal R.; Nussinov R. Trends Biochem. Sci. 2008, 33, 91–100. [DOI] [PubMed] [Google Scholar]
- Jang H.; Arce F. T.; Capone R.; Ramachandran S.; Lal R.; Nussinov R. Biophys. J. 2009, 97, 3029–3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafrir Y.; Durell S.; Arispe N.; Guy H. R. Proteins 2010, 78, 3473–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourie J.; Culverson A.; Farrelly P.; Henry C.; Laohachai K. Cell Biochem. Biophys. 2002, 36, 191–207. [DOI] [PubMed] [Google Scholar]
- Connelly L.; Jang H.; Teran Arce F.; Capone R.; Kotler S. A.; Ramachandran S.; Kagan B. L.; Nussinov R.; Lal R. J. Phys. Chem. B 2012, 116, 1728–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal M.; Mueller P. Proc. Natl. Acad. Sci. U.S.A. 1972, 69, 3561–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone R.; Quiroz F. G.; Prangkio P.; Saluja I.; Sauer A. M.; Bautista M. R.; Turner R. S.; Yang J.; Mayer M. Neurotox. Res. 2009, 16, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P.; Rudin D. O.; Tien H. T.; Wescott W. C. Nature 1962, 194, 979–980. [DOI] [PubMed] [Google Scholar]
- Capone R.; Mustata M.; Jang H.; Arce F. T.; Nussinov R.; Lal R. Biophys. J. 2010, 98, 2644–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks B. R.; Bruccoleri R. E.; Olafson B. D.; States D. J.; Swaminathan S.; Karplus M. J. Comput. Chem. 1983, 4, 187–217. [Google Scholar]
- Klauda J. B.; Brooks B. R.; MacKerell A. D. Jr.; Venable R. M.; Pastor R. W. J. Phys. Chem. B 2005, 109, 5300–5311. [DOI] [PubMed] [Google Scholar]
- Durell S. R.; Brooks B. R.; Bennaim A. J. Phys. Chem. 1994, 98, 2198–2202. [Google Scholar]
- Phillips J. C.; Braun R.; Wang W.; Gumbart J.; Tajkhorshid E.; Villa E.; Chipot C.; Skeel R. D.; Kale L.; Schulten K. J. Comput. Chem. 2005, 26, 1781–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackerell A. D.; Feig M.; Brooks C. L. J. Comput. Chem. 2004, 25, 1400–1415. [DOI] [PubMed] [Google Scholar]
- Luhrs T.; Ritter C.; Adrian M.; Riek-Loher D.; Bohrmann B.; Doeli H.; Schubert D.; Riek R. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 17342–17347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova A. T.; Yau W. M.; Tycko R. Biochemistry 2006, 45, 498–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zirah S.; Kozin S. A.; Mazur A. K.; Blond A.; Cheminant M.; Segalas-Milazzo I.; Debey P.; Rebuffat S. J. Biol. Chem. 2006, 281, 2151–2161. [DOI] [PubMed] [Google Scholar]
- Jang H.; Arce F. T.; Ramachandran S.; Capone R.; Lal R.; Nussinov R. J. Mol. Biol. 2010, 404, 917–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz G. E. Biochim. Biophys. Acta 2002, 1565, 308–317. [DOI] [PubMed] [Google Scholar]
- Marsh D.; Pali T. Biophys. J. 2001, 80, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.; Arce F. T.; Ramachandran S.; Capone R.; Lal R.; Nussinov R. J. Phys. Chem. B 2010, 114, 9445–9451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustata M.; Capone R.; Jang H.; Arce F. T.; Ramachandran S.; Lal R.; Nussinov R. J. Am. Chem. Soc. 2009, 131, 14938–14945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.; Ma B.; Lal R.; Nussinov R. Biophys. J. 2008, 95, 4631–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T. B.; Roux B. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 11631–11635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolf T. B.; Roux B. Proteins 1996, 24, 92–114. [DOI] [PubMed] [Google Scholar]
- Petrache H. I.; Tristram-Nagle S.; Gawrisch K.; Harries D.; Parsegian V. A.; Nagle J. F. Biophys. J. 2004, 86, 1574–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rand R. P.; Parsegian V. A. Biochim. Biophys. Acta 1989, 988, 351–376. [Google Scholar]
- Jang H.; Arce F. T.; Ramachandran S.; Capone R.; Azimova R.; Kagan B. L.; Nussinov R.; Lal R. Proc. Natl. Acad. Sci. U.S.A. 2010, 107, 6538–6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beglov D.; Roux B. J. Chem. Phys. 1994, 100, 9050–9063. [Google Scholar]
- Stote R. H.; Karplus M. Proteins 1995, 23, 12–31. [DOI] [PubMed] [Google Scholar]
- Marchand S.; Roux B. Proteins 1998, 33, 265–284. [PubMed] [Google Scholar]
- Hirakura Y.; Lin M. C.; Kagan B. L. J. Neurosci. Res. 1999, 57, 458–466. [DOI] [PubMed] [Google Scholar]
- Yeaman M. R.; Yount N. Y. Pharmacol. Rev. 2003, 55, 27–55. [DOI] [PubMed] [Google Scholar]
- Sokolov Y.; Mirzabekov T.; Martin D. W.; Lehrer R. I.; Kagan B. L. Biochim. Biophys. Acta 1999, 1420, 23–29. [DOI] [PubMed] [Google Scholar]
- Kagan B. L. Science 2005, 307, 42–43 author reply pp 42–43. [DOI] [PubMed] [Google Scholar]
- Shulga N.; Wilson-Smith R.; Pastorino J. G. Cell Cycle 2009, 8, 3355–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzabekov T.; Lin M. C.; Yuan W. L.; Marshall P. J.; Carman M.; Tomaselli K.; Lieberburg I.; Kagan B. L. Biochem. Biophys. Res. Commun. 1994, 202, 1142–1148. [DOI] [PubMed] [Google Scholar]
- Kagan B. L.; Jang H.; Capone R.; Teran Arce F.; Ramachandran S.; Lal R.; Nussinov R. Mol. Pharm. 2012, 10.1021/mp200419b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan B. L.; Thundimadathil J. Adv. Exp. Med. Biol. 2010, 677, 150–167. [DOI] [PubMed] [Google Scholar]
- de Planque M. R.; Raussens V.; Contera S. A.; Rijkers D. T.; Liskamp R. M.; Ruysschaert J. M.; Ryan J. F.; Separovic F.; Watts A. J. Mol. Biol. 2007, 368, 982–997. [DOI] [PubMed] [Google Scholar]
- Lau T. L.; Ambroggio E. E.; Tew D. J.; Cappai R.; Masters C. L.; Fidelio G. D.; Barnham K. J.; Separovic F. J. Mol. Biol. 2006, 356, 759–770. [DOI] [PubMed] [Google Scholar]
- Streltsov V. A.; Varghese J. N.; Masters C. L.; Nuttall S. D. J. Neurosci. 2011, 31, 1419–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smart O. S.; Goodfellow J. M.; Wallace B. A. Biophys. J. 1993, 65, 2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T. W.; Andersen O. S.; Roux B. Proc. Natl. Acad. Sci. U.S.A. 2004, 101, 117–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahse K.; Garvey M.; Kovermann M.; Vogel A.; Balbach J.; Fändrich M.; Fahr A. J. Mol. Biol. 2010, 403, 643–659. [DOI] [PubMed] [Google Scholar]
- Davis C. H.; Berkowitz M. L. Proteins 2010, 78, 2533–2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sani M.-A.; Gehman J. D.; Separovic F. FEBS Lett. 2011, 585, 749–754. [DOI] [PubMed] [Google Scholar]
- Ravault S.; Soubias O.; Saurel O.; Thomas A.; Brasseur R.; Milon A. Protein Sci. 2005, 14, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuks B.; Homblé F. Biophys. J. 1994, 66, 1404–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemela P. S.; Ollila S.; Hyvonen M. T.; Karttunen M.; Vattulainen I. PLoS Comput. Biol. 2007, 3, 304–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P. T.; Schauerte J. A.; Wisser K. C.; Ding H.; Lee E. L.; Steel D. G.; Gafni A. J. Mol. Biol. 2009, 386, 81–96. [DOI] [PubMed] [Google Scholar]
- Gu L. Q.; Dalla Serra M.; Vincent J. B.; Vigh G.; Cheley S.; Braha O.; Bayley H. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, 3959–3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maglia G.; Restrepo M. R.; Mikhailova E.; Bayley H. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 19720–19725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V.; Indi S.; Rao K. J. Mol. Neurosci. 2008, 34, 35–43. [DOI] [PubMed] [Google Scholar]
- Esler W. P.; Stimson E. R.; Fishman J. B.; Ghilardi J. R.; Vinters H. V.; Mantyh P. W.; Maggio J. E. Biopolymers 1999, 49, 505–514. [DOI] [PubMed] [Google Scholar]
- Ciccotosto G. D.; Tew D. J.; Drew S. C.; Smith D. G.; Johanssen T.; Lal V.; Lau T.-L.; Perez K.; Curtain C. C.; Wade J. D.; Separovic F.; Masters C. L.; Smith J. P.; Barnham K. J.; Cappai R. Neurobiol. Aging 2011, 32, 235–248. [DOI] [PubMed] [Google Scholar]
- Cribbs D. H.; Pike C. J.; Weinstein S. L.; Velazquez P.; Cotman C. W. J. Biol. Chem. 1997, 272, 7431–7436. [DOI] [PubMed] [Google Scholar]
- Soscia S. J.; Kirby J. E.; Washicosky K. J.; Tucker S. M.; Ingelsson M.; Hyman B.; Burton M. A.; Goldstein L. E.; Duong S.; Tanzi R. E.; Moir R. D. PLoS ONE 2010, 5, e9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang H.; Arce F. T.; Mustata M.; Ramachandran S.; Capone R.; Nussinov R.; Lal R. Biophys. J. 2011, 100, 1775–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade D.; Boman A.; Wahlin B.; Drain C. M.; Andreu D.; Boman H. G.; Merrifield R. B. Proc. Natl. Acad. Sci. U.S.A. 1990, 87, 4761–4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyasaki K. T.; Iofel R.; Oren A.; Huynh T.; Lehrer R. I. J. Periodontal Res. 1998, 33, 91–98. [DOI] [PubMed] [Google Scholar]
- Boman H. G.; Agerberth B.; Boman A. Infect. Immun. 1993, 61, 2978–2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Castillo F. J.; del Castillo I.; Moreno F. J. Bacteriol. 2001, 183, 2137–2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vunnam S.; Juvvadi P.; Merrifield R. B. J. Pept. Res. 1997, 49, 59–66. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.