Griffin et al., respond:
In January 2011, we reported the unique CD20+CD27+CD43+ phenotype of human B1 cells (Griffin et al., 2011a). In December 2011, we reported that the human B1 cell pool consists of functionally distinct CD11b− and CD11b+ populations, with the former particularly proficient at antibody secretion and the latter particularly efficient in stimulating T cells (Griffin and Rothstein, 2011). We also noted that the frequency of CD11b+ B1 cells is increased in the circulation of patients with lupus. As part of this work, we included microarray data that pointed to transcriptional differences between CD11b− and CD11b+ B1 cells. We have read with great interest the letter by Reynaud and Weill, who have mined the microarray data we deposited in the National Center for Biotechnology Information (NCBI) repository and drawn inferences, incorrectly as it turns out, regarding the nature and origin of CD11b− and CD11b+ B1 cells.
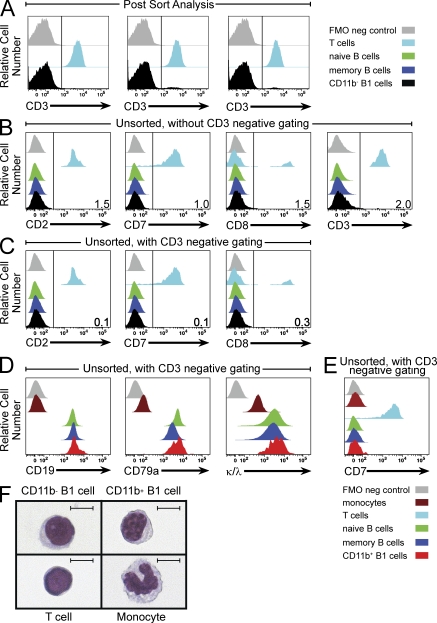
Reynaud and Weill speculated that CD11b− B1 cells represent B cell–T cell doublets because this population, in aggregate, expressed genes characteristic of T cells along with genes characteristic of B cells. The potential issue of B cell–T cell doublets, raised previously by these same authors (Descatoire et al., 2011), is a topic we have already addressed by demonstrating that our sort-purified B1 cells exist as singlets (Griffin and Rothstein, 2011; Griffin et al., 2011b). At the same time, we identified procedures that can be used to circumvent the potential problem of doublet events (Griffin et al., 2011b). In our published work, we reported that B1 cells sort-purified without CD3− gating contained an estimated 1–3% CD3+ events (Griffin and Rothstein, 2011; Griffin et al., 2011b). We have now sort-purified CD11b− B1 cells again, exactly as was done to generate our published microarray results, and have analyzed three independent postsort samples for CD3 expression, along with purified T cells (Fig. 1 A). We again find a very small proportion of CD3+ events (1.9 ± 0.66%; mean ± SEM) in the CD11b− B1 cell population. This very low level of CD3 positivity is likely attributable to a minor degree of T cell contamination that could account for the appearance of very low levels of some characteristically T cell transcripts among CD11b− B1 cells. In keeping with this, flow cytometric analysis of unsorted CD19-enriched PBMCs demonstrates a small degree of CD2, CD7, CD8, and CD3 staining among CD11b− B1 cells that is eliminated by CD3 exclusion and subsequent analysis of CD3−CD20+CD27+CD43+CD11b− B1 cells (Fig. 1, B and C). Importantly, when CD3 exclusion is combined with sort purification, expression of T cell genes such as CD7, CD8, and granzyme A is essentially eliminated (to <0.2% that of CD3+CD20− T cells from the same samples), as assessed by TaqMan QPCR (unpublished data). Collectively, these results indicate that CD11b− B1 cells represent a distinct B cell population, as we previously reported. They are not doublets and do not express multiple T cell markers. Our deposited NCBI data show minor amplification of some characteristically T cell genes caused by the presence of very few T cells in the samples used for microarray analysis.
Figure 1.
CD11b− and CD11b+ B1 cells are B cells, not T cells or monocytes. (A) Adult PBMCs from three separate donors were CD19 enriched and immunofluorescently stained for CD11b, CD20, CD27, and CD43. CD11b− B1 cells (CD20+CD27+CD43+CD11b−) were sort-purified (Influx; BD) and evaluated postsort for expression of CD3 by flow cytometric analysis (LSRII; BD), and then compared with separately purified T cells, as indicated. (B and C) Adult PBMCs were CD19-enriched and immunofluorescently stained for CD11b, CD20, CD27, and CD43, as well as CD2, CD3, CD7, and CD8. The expression of T cell markers on naive (CD20+CD27−CD43−), memory (CD20+CD27+CD43−), and CD11b− B1 (CD20+CD27+CD43+CD11b−) cells was evaluated by flow cytometric analysis and compared with T cells (CD3+CD20−), as indicated (B). B cells were gated to exclude CD3+ events, and the expression of T cells markers was evaluated on CD11b− B1 cells by flow cytometric analysis and compared with T cells (CD3+CD20−), as indicated (C). (D) Adult PBMCs were immunofluorescently stained for CD11b, CD20, CD27, and CD43, as well as CD19, CD79a, and κ and λ light chains (combined). B cells were gated to exclude CD3+ events, and naive, memory, and CD11b+ B1 cells were then evaluated for expression of B cell markers by flow cytometric analysis and compared with T cells (CD3+CD20−) and monocytes (CD11b+CD3−CD20−), as indicated. (E) Adult PBMCs were CD19 enriched and immunofluorescently stained for CD11b, CD20, CD27, and CD43, as well as CD7. B cells were gated to exclude CD3+ events, and naive, memory, and CD11b+ B1 cells were then evaluated for CD7 expression by flow cytometric analysis and compared with T cells and monocytes, as indicated. (F) Sort-purified CD11b− B1 cells, CD11b+ B1 cells, T cells, and monocytes were cytocentrifuged onto glass slides, stained with Wright-Giemsa, and examined by light microscopy. Representative images for each cell type are shown. Bars, 5 μm. Results shown in B–F represent one of three separate donors.
Reynaud and Weill also contend that CD11b+ B1 cells are not B cells, but are monocytes, because this population underexpressed some characteristically B cell genes and overexpressed some characteristically monocyte genes by microarray. Although some B cell genes were not well expressed by CD11b+ B1 cells, the corresponding proteins are expressed normally, as shown by immunofluorescent staining. For example, CD19 and CD79a (Fig. 1 D), as well as CD21, CD22, and BAFFR (not depicted), are expressed on CD11b+ B1 cells at levels similar to those on naive and memory B cells. These staining results do not represent nonspecific binding, as CD11b+ B1 cells are negative for staining with CD7 (Fig. 1 E), CD2, CD3, and CD8 (not depicted), as well as CD80 and CD71 (Griffin and Rothstein, 2011). Importantly, CD11b+ B1 cells all express surface immunoglobulin, as demonstrated by positive staining with anti-κ and anti-λ Ig light chain antibodies conjugated to the same fluorophore (Fig. 1 D). Consistent with this, additional evidence that CD11b+ B1 cells belong to the B cell lineage is provided by single-cell PCR amplification of rearranged immunoglobulin (Tiller et al., 2008; Griffin et al., 2011a), which we find to be even more efficient in CD11b+ B1 cells than naive B cells from the same samples (unpublished data). On the basis of immunoglobulin gene rearrangement, B cell receptor expression, and expression of characteristically B-cell surface antigens, CD11b+ B1 cells are B cells.
The notion that CD11b+ B1 cells express monocyte genes is entirely consistent with what is known about mouse B1 cells. Expression of monocyte genes by some human B1 cells fits well with an extensive literature indicating a close relationship between B1 cells and monocytes, extending even to B1 cell phagocytic activity (Cumano et al., 1992; Borrello and Phipps, 1996; Almeida et al., 2001; Montecino-Rodriguez et al., 2001; Ghosn et al., 2006; Kawamoto, 2006; Popi et al., 2009; Parra et al., 2011). This does not mean, however, that CD11b+ B1 cells are monocytes, as can be readily seen by the morphology of CD11b+ B1 cells, which is quite different from the distinctive features of monocytes (Fig. 1 F).
In sum, the mining of our microarray data by Reynaud and Weill has led them to speculative inferences that are firmly contradicted by an extensive body of evidence. Our work has established the overall phenotype of human B1 cells, as well as the nature and characteristics of two functionally distinct human B1 cell subpopulations, one of which is increased in frequency in the blood of lupus patients.
References
- Almeida S.R., Aroeira L.S., Frymuller E., Dias M.A., Bogsan C.S., Lopes J.D., Mariano M.. 2001. Mouse B-1 cell-derived mononuclear phagocyte, a novel cellular component of acute non-specific inflammatory exudate. Int. Immunol. 13:1193–1201. 10.1093/intimm/13.9.1193 [DOI] [PubMed] [Google Scholar]
- Borrello M.A., Phipps R.P.. 1996. The B/macrophage cell: an elusive link between CD5+ B lymphocytes and macrophages. Immunol. Today. 17:471–475. 10.1016/0167-5699(96)20031-B [DOI] [PubMed] [Google Scholar]
- Cumano A., Paige C.J., Iscove N.N., Brady G.. 1992. Bipotential precursors of B cells and macrophages in murine fetal liver. Nature. 356:612–615. 10.1038/356612a0 [DOI] [PubMed] [Google Scholar]
- Descatoire M., Weill J.C., Reynaud C.A., Weller S.. 2011. A human equivalent of mouse B-1 cells? J. Exp. Med. 208:2563–2564. 10.1084/jem.20112232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosn E.E., Russo M., Almeida S.R.. 2006. Nitric oxide-dependent killing of Cryptococcus neoformans by B-1-derived mononuclear phagocyte. J. Leukoc. Biol. 80:36–44. 10.1189/jlb.1005603 [DOI] [PubMed] [Google Scholar]
- Griffin D.O., Rothstein T.L.. 2011. A small CD11b(+) human B1 cell subpopulation stimulates T cells and is expanded in lupus. J. Exp. Med. 208:2591–2598. 10.1084/jem.20110978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.O., Holodick N.E., Rothstein T.L.. 2011a. Human B1 cells in umbilical cord and adult peripheral blood express the novel phenotype CD20+ CD27+ CD43+ CD70−. J. Exp. Med. 208:67–80. 10.1084/jem.20101499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.O., Holodick N.E., Rothstein T.L.. 2011b. Human B1 cells are CD3-: A reply to “A human equivalent of mouse B-1 cells?” and “The nature of circulating CD27+CD43+ B cells.” J. Exp. Med. 208:2566–2569. 10.1084/jem.20111761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto H. 2006. A close developmental relationship between the lymphoid and myeloid lineages. Trends Immunol. 27:169–175. 10.1016/j.it.2006.02.004 [DOI] [PubMed] [Google Scholar]
- Montecino-Rodriguez E., Leathers H., Dorshkind K.. 2001. Bipotential B-macrophage progenitors are present in adult bone marrow. Nat. Immunol. 2:83–88. 10.1038/83210 [DOI] [PubMed] [Google Scholar]
- Parra D., Rieger A.M., Li J., Zhang Y.A., Randall L.M., Hunter C.A., Barreda D.R., Sunyer J.O.. 2011. Pivotal advance: peritoneal cavity B-1 B cells have phagocytic and microbicidal capacities and present phagocytosed antigen to CD4+ T cells. J. Leukoc. Biol. 10.1189/jlb.0711372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popi A.F., Motta F.L., Mortara R.A., Schenkman S., Lopes J.D., Mariano M.. 2009. Co-ordinated expression of lymphoid and myeloid specific transcription factors during B-1b cell differentiation into mononuclear phagocytes in vitro. Immunology. 126:114–122. 10.1111/j.1365-2567.2008.02883.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud C.-A., Weill J.-C.. 2012. Gene profiling of CD11b+ and CD11b− B1 cell subsets reveals potential cell sorting artifacts. J. Exp. Med. 209:433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiller T., Meffre E., Yurasov S., Tsuiji M., Nussenzweig M.C., Wardemann H.. 2008. Efficient generation of monoclonal antibodies from single human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods. 329:112–124. 10.1016/j.jim.2007.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]