Abstract
The human adenovirus type 5 (HAdV5) early region 1B 55-kDa protein (E1B-55K) is a multifunctional phosphoprotein playing several critical roles during adenoviral productive infection, e.g., degradation of host cell proteins, viral late mRNA export, and inhibition of p53-mediated transcription. Many of these functions are apparently regulated at least in part by the phosphorylation of E1B-55K occurring at a stretch of amino acids resembling a potential CK2 consensus phosphorylation motif. We therefore investigated the potential role of CK2 phosphorylation upon E1B-55K during adenoviral infection. A phosphonegative E1B-55K mutant showed severely reduced virus progeny production, although viral early, late, and structural protein levels and viral DNA replication were not obviously affected. Binding studies revealed an interaction between the CK2α catalytic subunit and wild-type E1B-55K, which is severely impaired in the phosphonegative E1B mutant. In addition, in situ the α-catalytic subunit is redistributed into ring-like structures surrounding E1B-55K nuclear areas and distinct cytoplasmic accumulations, where a significant amount of CK2α colocalizes with E1B-55K. Furthermore, in in vitro phosphorylation assays, wild-type E1B-55K glutathione S-transferase fusion proteins were readily phosphorylated by the CK2α subunit but inefficiently phosphorylated by the CK2 holoenzyme. Addition of the CK2-specific inhibitors TBB (4,5,6,7-tetrabromobenzotriazole) and DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole) to infected cells confirmed that CK2α binding to E1B-55K is necessary for efficient phosphorylation of E1B-55K. In summary, our data show that CK2α interacts with and phosphorylates HAdV5 E1B-55K at residues S490/491 and T495 and that these posttranslational modifications are essential for E1B-55K lytic functions.
INTRODUCTION
The cellular CK2 protein is a serine/threonine kinase known to be ubiquitously expressed, highly conserved in eukaryotic cells, and considered to be constitutively active (36, 46, 62). Today, CK2 is known to phosphorylate more than 300 cellular and viral proteins, and yet the list is far from complete, as shown by comparative amino acid sequence screen analyses among putative CK2 phosphorylation motifs/sites (36, 53). Among these substrates are proteins involved in DNA replication (topoisomerase I [36, 44]), transcription (c-Myc [36, 44]), cell cycle control (cyclin H [55]), ribosome biogenesis (L5 [42]), apoptosis induction (Bid [14]), and cell differentiation (HOXB7 [71]), as well as numerous viral proteins (e.g., EB2 from Epstein-Barr virus [[EBV] [35], EBNA-2 from EBV [24], NS2 from hepatitis C virus [HCV] [21], and ZEBRA from EBV [17]), that comprise ca. 10% of the known CK2 substrates (36). Apart from its role in normal cellular signaling pathways and viral infections, CK2 is reported to be involved in tumorigenesis. For example, increased CK2 activity is linked to several kinds of malignancies such as breast cancer (31, 38) or colorectal carcinoma (47), while reduced activity has been associated with limited cell viability (60, 66, 68). Indeed, the large number of cellular substrates highlights the important role CK2 plays in maintaining cell homeostasis. Several studies have shown that knockout of the CK2α or β subunit is lethal at the embryonic stage in mice (10, 58, 66).
CK2α or CK2α′ can form homo- or heterodimers and assemble with a homodimer of β subunits to form the CK2 holoenzyme (18, 23, 46). Either the holoenzyme or α and α′ subunits show constitutive activity, so CK2 activity is not regulated through a “classical” signal transduction cascade, as known, for example, for mitogen-activated protein kinases (45) or cyclin-dependent kinases (CDKs [46]). It is assumed that constitutive activity and ubiquitous expression are two of the reasons why CK2 is exploited by many different viral pathogens (36). Mechanistically, CK2 can use either ATP or GTP as phosphoryl donors (39). Phosphorylation occurs specifically at serine or threonine residues on target proteins, although CK2 shows a higher propensity to phosphorylate serine residues (36). The general consensus motif for a CK2 phosphorylation site was found to be S/T-X-X-E/D (36, 46, 53).
The human adenovirus type 5 (HAdV5) early region 1B 55-kDa protein (E1B-55K) is a multifunctional phosphoprotein (33, 64, 65, 70) and regulator of adenoviral replication (6). E1B-55K in complex with E4orf6, cellular factors cullin-5, Rbx1, or elongins B and C mediates the degradation of cellular proteins such as p53, Mre11, DNA ligase IV, and integrin α3 (5, 7, 11, 13, 50). It has been shown that these functions are necessary to antagonize the DNA damage response that would eventually lead to concatenation of viral genomes (63), as well as stress responses that would initiate cellular antiviral defense mechanisms (32, 56, 69). Furthermore, E1B-55K, along with E4orf6, mediates the export of viral late mRNA transcripts (8, 16, 20, 22) and is responsible for blocking cellular mRNA export (3, 4, 43). Moreover, E1B-55K alone can induce a decrease in protein levels of Daxx (death domain-associated protein), which was shown to be a restrictive factor for adenoviral replication (56). E1B-55K is known to be phosphorylated at amino acids serine 490, serine 491, and threonine 495 (64, 65). Phenotypic analyses of a mutant virus lacking these sites point to an essential involvement of this posttranslational modification in E1B-55K functions and viral replication in general (65, 57). For example, these modifications seem to be necessary for E1B-55K's p53-repression functions, i.e., the ability to bind p53 (57), inhibit its transactivational abilities (64, 65) and induce the proteasomal degradation of p53 (50, 57). Also, localization of the E1B-55K protein is affected by mutating the phosphosites (57). However, the kinase responsible for E1B-55K phosphorylation is still unknown. Teodoro et al. already speculated on an involvement of the protein kinase CK2 (64) since E1B-55K has at least two amino acids that fit, almost perfectly, the criteria of the general CK2 phosphorylation motif (see Fig. 1B and C). However, in vitro kinase studies performed by Teodoro et al. with purified CK2 could not efficiently show an involvement of this kinase in E1B-55K phosphorylation. Hence, we set out to determine whether CK2 is responsible for the crucial modifications of E1B-55K.
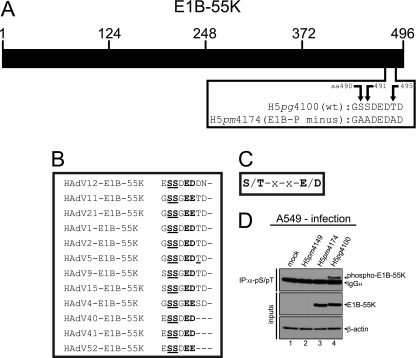
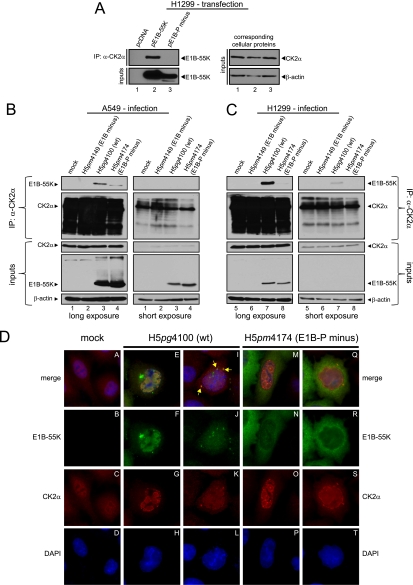
Fig 1.
CK2 phosphorylation consensus motif of E1B-55K and E1B-55K phosphorylation. (A) Schematic representation of E1B-55K showing the amino acid sequence of the C terminus of E1B-55K in H5pg4100 (wt) and the H5pm4174 (E1B-P minus) mutant. (B) Alignment of E1B-55K C-terminal amino acid sequences from different adenovirus types. Amino acids highly conserved throughout different E1B-55K proteins and known to be phosphorylated in HAdV5 are underlined. Amino acids matching the general CK2 consensus motif are in bold. (C) Simplified CK2 consensus motif according to Meggio et al. (36) and Pinna et al. (46). A serine or threonine in boldface denotes CK2-targeted amino acids in this motif. (D) A549 cells were mock infected or infected with H5pm4149 (E1B minus), the E1B-P minus mutant H5pm4174 and H5pg4100 wt virus at an MOI of 20 FFU per cell. Total cell lysates were prepared and subjected to immunoprecipitation using anti-phosphoserine/phosphothreonine (pS/pT) antibody. Proteins were separated by SDS-PAGE and detected by immunoblotting with anti-E1B antibody 2A6. Steady-state concentrations (inputs) of E1B-55K and β-actin were determined by immunoblotting of protein extracts with anti-E1B-55K (2A6) and anti-β-actin (AC-15). The Western blot represents one experiment which had been independently repeated at least four times.
We show here that E1B-55K is a substrate of the α subunit of protein kinase CK2 in vitro, but the CK2 holoenzyme does not efficiently phosphorylate E1B-55K. Moreover, it was also possible to show that a protein derivative generated by alternative splicing of E1B-mRNA, E1B-156R (1, 59) is phosphorylated by CK2α. Our data point to the importance of CK2 for adenoviral infection as the kinase regulating E1B-55K's phosphorylation state, and thus its proper functions, which in turn have global effects on virus replication.
MATERIALS AND METHODS
Cells.
The cell lines A549 (DSMZ [Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany], ACC107), HEK293 (DSMZ, ACC305), and H1299 (37) were grown in monolayers and cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 5 to 10% newborn calf serum (NCS), 100 U of penicillin, and 100 μg of streptomycin per ml in a 5% CO2 atmosphere at 37°C (48).
Plasmids and transient transfections.
The following plasmids were used in the present study: pcDNA3 vector, which contains the cytomegalovirus (CMV) immediate-early promoter (Invitrogen), or pcDNA3 harboring either HAdV5 wild-type E1B-55K (pE1B-55K) or mutant E1B-55K (pE1B-P minus). pE1B-P minus carries three alanine substitutions at positions 490, 491, and 495 (S490/491A,T495A), which were generated using the oligonucleotide primers 941 (5′-GCT GAG TTT GGC GCT GCC GAT GAA GAT ACA G-3′), 942 (5′-CTG TAT CTT CAT CGG CAG CGC CAA ACT CAG C-3′), 943 (5′-GCC GAT GAA GAT GCA GAT TGA GGT ACT G-3′), and 944 (5′-CAG TAC CTC AAT CTG CAT CTT CAT CGG C-3′).
Subconfluent H1299 cells were transfected with 2 μg of pE1B-55K encoding plasmids (for pE1B-P minus, 6 μg) or empty control vector, pcDNA3, using PEI (polyethyleneimine; Polysciences). Transfection mixtures were prepared by incubating 2 μg of DNA with 20 μg of PEI (6 μg of DNA with 60 μg of PEI, respectively) and 100 μl of DMEM for 10 min at room temperature. Before the transfection mixture was added to the cells, the medium was removed and changed to DMEM with 5% NCS without antibiotics. Transfection mixtures were incubated for 6 to 8 h before being removed and replaced by DMEM containing 5% NCS, 100 U of penicillin, and 100 μg of streptomycin per ml. Cells were harvested 24 to 48 h posttransfection (p.t.).
Viruses.
The following viruses were used in the present study: H5pg4100 (wt), H5pm4149 (E1B minus), H5pm4174 (E1B-P minus), and H5pm4230 (ΔE1B-55K/ΔE4orf3). H5pg4100 is an HAdV5-derived virus with a deletion in the E3 region and contains an additional BstBI restriction site (25, 27). Several nucleotides were exchanged creating four stop codons at amino acid positions 3, 8, 86, and 88 in the E1B-55K open reading frame of H5pm4149, rendering this virus unable to express E1B-55K (25, 28). H5pm4174 was created as described previously (25). Here, mutagenesis of the E1B-55K gene via site-directed mutagenesis PCR was carried out with the primers 1020 (5′-CGC GCT GAG TTT GGC GCT GCC GAT GAA GAT GCA GAT TGA GGT ACT G-3′) and 1021 (5′-CAG TAC CTC AAT CTG CAT CTT CAT CGG CAG CGC CAA ACT CAG CGC G-3′) to create the mutated E1B open reading frame, which was further cloned as described previously (25). H5pm4230 was created using the genomic backbone of H5pm4149 as described in reference 25. An insertion of a thymidine at genome position 34592 (5′-3′) results in a frameshift with subsequent three stop codons in the E4orf3 open reading frame, making this virus unable to express E4orf3 and E1B-55K (due to the H5pm4149 backbone) at the same time. Infection of cells was carried out with virus dilutions in DMEM at indicated multiplicities of infection (MOI). At 2 h postinfection (p.i.), virus-containing medium was removed and replaced by DMEM containing 5 to 10% NCS, 100 U of penicillin, and 100 μg of streptomycin per ml. Cells were harvested at the indicated time points. To determine virus growth, infected cells were harvested 24 or 48 h p.i. and lysed by three cycles of freeze-thawing. The cell lysates were serially diluted in DMEM for infection of HEK293 cells, and the virus yield was determined by using quantitative E2A-72K immunofluorescence staining 22 h after infection.
Protein analysis.
Cell pellets of infected or transfected cells were lysed in ice-cold radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-chloride [pH 8.0], 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40 [NP-40], 0.1% sodium dodecyl sulfate [SDS], and 0.1% Triton X-100 [Sigma-Aldrich]) with freshly added protease inhibitors (1% [vol/vol] PMSF [phenylmethylsulfonyl fluoride], 0.1% [vol/vol] aprotinin, 1 μg of leupeptin/ml, and 1 μg of pepstatin/ml) for 30 min on ice, following sonication and subsequent centrifugation at 15,000 × g and 4°C to pellet the cell debris. After normalization for protein concentration using a Bio-Rad protein assay, whole-cell extracts were used for immunoprecipitation or directly in Western blotting assays.
For immunoblotting, equal amounts of total protein were separated by SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to nitrocellulose membranes (Whatman). Membranes were incubated for 45 min or 1 h at room temperature in phosphate-buffered saline (PBS) containing 5% nonfat dry milk and then for 1 to 3 h in PBS containing 0.05% Tween 20 and 1% nonfat dry milk plus the appropriate primary antibody. Membranes were washed three times in PBS containing 0.05% Tween 20, incubated with a secondary antibody linked to horseradish peroxidase (GE Healthcare) in PBS containing 0.05% Tween 20 overnight at 4°C, and washed once in PBS containing 0.05% Tween 20 and twice in PBS. The bands were visualized by enhanced chemiluminescence as recommended by the manufacturer (Pierce) on X-ray films (CEA RP). Autoradiograms were scanned, cropped, and prepared by using Adobe Photoshop CS4 and Adobe Illustrator CS4 software. For Western blot band intensity quantification ImageJ software was used.
Analysis of viral DNA synthesis.
Adenoviral DNA replication was determined by PCR. At the indicated time points, infected cells were harvested and lysed in ice-cold lysis buffer containing protease inhibitors as described above. Then, 5-μg portions of total protein lysates were treated with 5 μg of proteinase K (Sigma) and Tween 20 (0.5%; Applichem). Next, 18 cycles of PCR (30 s at 95°C, 1 min at 55°C, and 2 min at 72°C) were performed with 24.5-μl portions of the lysates and 1.25 U of DreamTaq polymerase (Fermentas) in a 50-μl reaction volume. Two synthetic oligonucleotides—64 (5′-CGC GGG ATC CAT GGA GCG AAG AAA CCC ATC TGA GC-3′) and 110 (5′-CGG TGT CTG GTC ATT AAG CTA AAA-3′)—were used to amplify a specific 399-bp DNA fragment from the E1B gene. As an internal loading control, the primers 1447 (5′-CCTG CAC CAC CAA CTG CTT A-3′) and 1448 (5′-GCC ATG CCA GTG AGC TTC CCG-3′) were used to amplify specific GAPDH (glyceraldehyde-3-phosphate dehydrogenase) DNA fragments. The reaction products were analyzed on 1% agarose gels containing 0.66 μg of ethidium bromide/ml.
Antibodies.
The following primary antibodies were used in the present study: mouse monoclonal antibody (MAb) anti-E1A (M73 [26]), mouse MAb anti-E2A-72K (B6-8 [51]), mouse MAb anti-E1B-55K (2A6 [54]), rat MAb anti-E1B-55K (7C11 [26]), rabbit polyclonal antibody (PAb) anti-E4orf6 (1807 [9]), rat MAb anti-L4-100K (6B10 [30]), rabbit PAb anti-HAdV5-capsid (L133 [27]), mouse MAb anti-β-actin (AC-15; Sigma-Aldrich), mouse MAb anti-p53 (DO-1; Santa Cruz Biotechnology, Inc.), rabbit PAb anti-Mre11 (pNB 100-142; Novus Biologicals, Inc.), rabbit PAb anti-DNA ligase IV (NB110-57379; Novus Biologicals, Inc.), rabbit PAb anti-CK2α (ab13410; Abcam), mouse MAb anti-CK2β (catalog no. 51; Santa Cruz Biotechnology, Inc.), mouse MAb anti-CK2β (6D5; Sigma), and mouse MAb anti-phosphoserine/phosphothreonine (22a; BD Transduction). The following secondary antibodies were used in the present study: donkey anti-rabbit immunoglobulin G (IgG), sheep anti-mouse IgG, and goat anti-rat IgG (GE Healthcare).
Immunoprecipitation.
A549 or H1299 cells were infected at an MOI of 20 or 50 fluorescence-forming units (FFU)/cell and harvested 24 h p.i., or H1299 cells were transfected and harvested between 24 and 48 h p.t. Cells were lysed in ice-cold RIPA or NP-40 lysis buffer (50 mM Tris-chloride [pH 8.0], 150 mM NaCl, 1% NP-40, 1 mM PMSF, 2 mM dithiothreitol [DTT]) with freshly added protease inhibitors (as described above). Next, 2 mg of protein A-Sepharose coupled to 0.5 to 2 μg of antibody was added to 600 to 1,000 μg of protein A-Sepharose-precleared or Pansorbin (Calbiochem)-precleared protein lysates. The immune complexes were washed three times with lysis buffer, separated by SDS-PAGE, and analyzed by immunoblotting. For CK2β immunoprecipitation studies, 0.8 μg of anti-CK2β antibody (mouse MAb 51; Santa Cruz Biotechnology, Inc.) was added to 800 to 1,100 μg of Pansorbin (Calbiochem)-precleared protein lysates and incubated overnight at 4°C. On the next day, 2 to 3 mg of protein A-Sepharose was added, followed by incubation for 2 h (while rotating). The immune complexes were washed three times, eluted from the Sepharose beads using 0.1 M triethylamine (pH 11.5), separated by SDS-PAGE, and analyzed by immunoblotting.
For inhibitor treatment, DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole; Calbiochem) or TBB (4,5,6,7-tetrabromobenzotriazole; Sigma-RBI) was added to the cell culture at 4 h p.i. to a final concentration of 2.5 or 12.5 μM, respectively.
Indirect immunofluorescence.
Cells were grown on glass coverslips, mock infected or infected at an MOI of 20, and methanol fixed/permeabilized at 24 h p.i. To block unspecific binding, coverslips were incubated with TBS-BG (20 mM Tris-chloride [pH 7.6], 137 mM NaCl, 3 mM KCl, 1.5 mM MgCl2, 5 g of glycine/liter, 5 g of bovine serum albumin fraction V [PAA]/liter, Tween 20 [0.05%], 0.5 g of sodium azide/liter) for 30 min at room temperature while shaking, followed by two washing steps with 0.05% TBS-Tween 20 (TBS-T). Next, the cells were stained directly with hybridoma supernatant of 2A6 or 7C11 or in double-label in situ staining together with anti-CK2α or anti-CK2β (6D5) antibody for 1 h. Furthermore, double-label in situ staining was also performed for CK2α (ab13410; Abcam) and CK2β (6D5). After three washing steps with TBS-T or PBS-Tween 20 (PBS-T; 0.05%), cells were incubated for 1 h with fluorescein isothiocyanate-conjugated anti-mouse secondary antibody and/or Texas Red-conjugated anti-rabbit secondary antibody (Invitrogen). Nuclear DNA content was detected by DAPI (4′,6′-diamidino-2-phenylindole) staining. After three subsequent washing steps with TBS-T or PBS-T, coverslips were mounted in Glow medium (Energene). Digital immunofluorescence images were acquired on a DMRB fluorescence microscope (Leica) with a charge-coupled device camera (Diagnostic Instruments).
Expression and purification of recombinant fusion proteins.
The glutathione S-transferase (GST) bacterial expression vectors used in the present study were as follows (see Fig. 7 and 8): GST-wt-E1B (E1B-55K amino acids [aa] 448 to 496), GST-E1B-P minus (E1B-S490/1A/T495A; contains 48 aa of the C terminus like GST-AAT, GST-DDT, and GST-wt-E1B), GST-AAT (E1B-S490/1A), GST-DDT (E1B-S490/1D), GST-E1B-156R, and GST-April (kindly provided by Joachim Hauber). GST-wt-E1B was generated via amplification of the carboxy terminus of wild-type (wt) HAdV5 E1B-55K using the oligonucleotide primers 687 (5′-GCC AGG ATC CTG TGG CAA CTG CGA GG-3′) and 688 (5′-GCC AGA ATT CTC AAT CTG TAT CTT C-3′) and subsequent cloning into the expression vector pGEX-2T (PL-Pharmacia) using BamHI/EcoRI restriction enzyme sites. GST-AAT was generated by introducing point mutations using site-directed mutagenesis (Stratagene) into GST-wt-E1B with the primers 2179 (5′-CGC GCT GAG TTT GGC GCT GCC GAT GAA GAT ACA GAT TGA GAA TTC ATC-3′) and 2180 (5′-GAT GAA TTC TCA ATC TGT ATC TTC ATC GGC AGC GCC AAA CTC AGC GCG-3′). GST-DDT was generated as GST-AAT but using the primers 2181 (5′-CGC GCT GAG TTT GGC GAT GAC GAT GAA GAT ACA GAT TGA GAA TTC ATC-3′) and 2182 (5′-GAT GAA TTC TCA ATC TGT ATC TTC ATC GTC ATC GCC AAA CTC AGC GCG-3′). Expression of GST-fusion proteins in Escherichia coli (TOPP3 or TOPP6) was induced for 2 to 4 h by adding IPTG (isopropyl-β-d-thiogalactopyranoside; PEQLAB) to a final concentration of 1 mM. The bacterial cells were centrifuged, and the pellets were washed with STE buffer (10 mM Tris-chloride [pH 8.0], 150 mM NaCl, 1 mM EDTA) and lysed by the addition of 1 mg of lysozyme (15 min on ice; Sigma-Aldrich), followed by the addition of DTT (5 mM) and N-lauroylsarcosine (10%; Sigma-Aldrich) and sonication twice with ultrasound (30 pulses, output 40, 0.5 impulse; Branson Sonifier 450). To precipitate insoluble cell parts, the lysate was centrifuged, and the supernatant transferred to a new 15-ml Falcon tube. Triton X-100 was then added (end concentration 1%), and the supernatant was vortexed and filtered using a 0.45-μm-pore-size filter. The samples were batch purified, and 100 μl of a 50% slurry (0.5 bed volume) of glutathione-Sepharose 4B beads (Amersham Biosciences) in TBS was added to the filtered lysate supernatant. The mixture was incubated for 1 h at 4°C in an overhead incubator (GFL; Society for Laboratory Technology), and the beads were pelleted and washed six times with TBS. To analyze the protein content, the beads were boiled in Laemmli buffer and analyzed by SDS-PAGE. Proteins were visualized by safe stain (Coomassie) staining (Invitrogen).
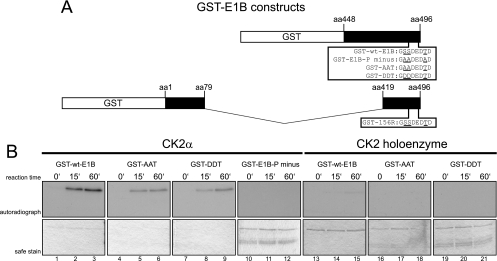
Fig 7.
CK2α, but not the CK2 holoenzyme, phosphorylates E1B-55K's C terminus in vitro. (A) C-terminal amino acid sequences of different GST-E1B fusion proteins used in the phosphorylation assays. Underlined are amino acids known to be phosphorylated and/or changed to the indicated amino acids. Numbers above the schematic drawing represent amino acid (aa) positions of E1B-55K from which the GST fragments are derived. (B) The indicated GST-E1B fusion proteins were incubated either with recombinant CK2α or with CK2 holoenzyme together with radioactive [γ-32P]ATP for the indicated reaction times at 30°C. Kinase assays performed with the holoenzyme contained double the amount of substrate (GST-wt-E1B, GST-E1B-P minus, GST-AAT, and GST-DDT) and holoenzyme units compared to CK2α assays. After extensive washing, kinase assay samples were separated by SDS-PAGE and Coomassie stained (“safe stain,” lower panels), and the gels were vacuum dried for autoradiographic detection with X-ray films (upper panels). Autoradiographs represent one of at least four independent experiments.
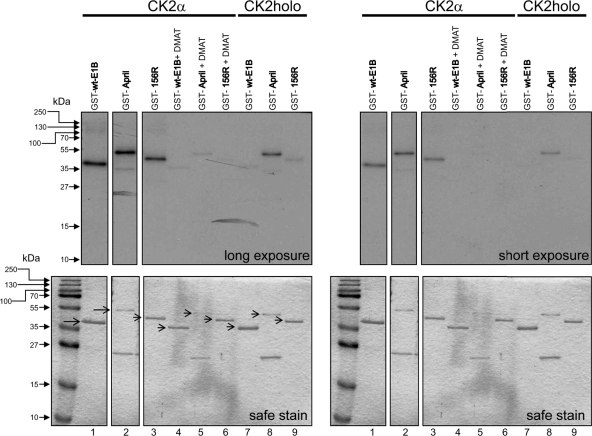
Fig 8.
The 156R splice product of E1B-55K is also phosphorylated by CK2α but not by the holoenzyme. The indicated GST-E1B and GST-April fusion proteins were incubated either with recombinant CK2α or CK2 holoenzyme together with radioactive [γ-32P]ATP for 30 min at 30°C. Where indicated, CK2α batches were incubated for 15 min in 20 μM DMAT together with substrates at room temperature prior to starting the in vitro kinase reaction. Kinase assay products were analyzed by SDS-PAGE, and the gels were Coomassie stained (“safe stain,” lower panels) and then vacuum dried for autoradiography on X-ray films (top panels). Additional control lanes were omitted due to clarity. Segments are from the same autoradiograph. The autoradiograph represents one of at least three independent experiments.
Kinase assays.
For in vitro phosphorylation, GST-fusion proteins (1 μg) were incubated in kinase reaction mixture (0.1 mM Na3VO4, 2 mM DTT, 1 mM Pefabloc SC [AEBSF; Roche], 1 mM phenylmethylsulfonyl fluoride [PMSF], 1.2% aprotinin, 200 μM ATP) together with recombinant CK2α (170 U) or CK2 holoenzyme (340 U; both from proteinkinase.de) in the presence of [γ-32P]ATP (1 μCi). CK2 holoenzyme assays were performed with 340 U of kinase and 2 μg of substrate protein. Samples were incubated at 30°C for 0, 15, 30, or 60 min. In vitro phosphorylation was stopped by adding ice-cold TBS supplemented with fresh protease inhibitors (1% [vol/vol] PMSF, 0.1% [vol/vol] aprotinin, 1 μg of leupeptin/ml, and 1 μg of pepstatin/ml) to reaction mixtures. GST fusion proteins were washed three times with ice-cold TBS (as described above), resuspended in Laemmli loading buffer, and boiled at 95°C. The samples were separated via SDS-PAGE and SimplyBlue safe stained according to the manufacturer's instructions (Invitrogen). Phosphorylation signals were detected from dried gels via autoradiography. For inhibitor treatment in this setting, GST fusion proteins were incubated with 20 μM DMAT in a kinase reaction mixture for 30 min at room temperature before adding [γ-32P]ATP (1 μCi) to the samples.
Pulsed-field gel electrophoresis.
Samples were prepared as follows. First, 1.2 × 107 A549 cells, mock-infected or infected with the indicated viruses, were treated with trypsin at 30 h p.i., and cell pellets were resuspended in 400 μl of PBS. Subsequently, 400 μl of molten low-melting-point agarose (LMP agarose [Lonza]) was added. The mixtures were pipetted into mold plugs and incubated for 30 min at 4°C. Solid agarose plugs were incubated in proteinase K solution (2% N-lauroylsarcosine [Sigma-Aldrich], 0.4 M EDTA [pH 8.0], and 2 mg of proteinase K [Sigma-Aldrich]/ml) for 24 h at 50°C. After renewing the proteinase K solution, plugs were incubated additional 24 h, rinsed in double-distilled H2O several times, and incubated for 24 h in TE50 buffer (10 mM Tris-HCl [pH 8.0], 50 mM EDTA) at 4°C. Afterward, the TE50 buffer was renewed, and PMSF was added (0.1 mM), followed by incubation at 4°C for 4 h with TE50/PMSF renewal after 2 h. Plugs were incubated in new TE50 buffer for an additional 24 h and transferred into storage buffer (0.5 M EDTA [pH 8.0]).
The DNA in agarose blocks was analyzed by electrophoresis with a Biometra Rotaphor system through 1.5% agarose gel at 150 V with ramped pulse times from 10 to 90 s for 39 h at 15°C in 0.5× TBE (0.045 M Tris base, 0.045 M sodium borate, 0.005 M EDTA). The gel was stained with ethidium bromide for 30 min at room temperature to visualize viral DNA.
RESULTS
E1B-55K is phosphorylated at highly conserved residues at the C terminus, which has similarity to the CK2 consensus phosphorylation motif.
Previous studies by Branton and coworkers have shown that the HAdV5 E1B-55K protein is phosphorylated at serine and threonine residues near the C terminus (S490/491, T495) within sequences characteristic of CK2 substrates (Fig. 1A, B, and C) (64, 65). To investigate the role of this cellular protein kinase in phosphorylating the adenoviral protein, we generated a mutant virus, H5pm4174 (E1B-P minus), which contains three alanine substitutions at positions 490, 491, and 495 within the 496-amino-acid residue E1B-55K polypeptide (Fig. 1A). To test whether the amino acid changes abolish phosphorylation of E1B-55K, we performed combined immunoprecipitation-immunoblotting assays using total cell lysates from wt and mutant virus-infected A549 cells (Fig. 1D). As expected, wt E1B-55K (phospho-E1B-55K) was precipitated with an antibody reactive for phosphoserine and phosphothreonine residues from H5pg4100 wt-infected cells (Fig. 1D, lane 4). In contrast, no E1B-55K protein was detected in the same immunoblots from cells infected with E1B mutants H5pm4149 lacking E1B-55K or H5pm4174 lacking the phosphorylation sites (S490/491A, T495A, and E1B-P minus; Fig. 1D, lanes 2 and 3).
Alanine substitution of E1B-55K's phosphosites results in profound negative effects.
We next analyzed the effect of mutations on the ability of the Ad protein to promote viral early and late protein production. In effect, H5pg4100 (wt) and H5pm4174 (E1B-P minus) infection revealed almost identical adenoviral protein production (Fig. 2A, E1A, E2A, E1B-55K, E4orf6, L4-100K, and HAdV5 capsid protein stainings). In contrast, Mre11 steady-state reduction was delayed and p53 degradation was impaired during E1B-P minus virus infection (Fig. 2A). To examine the binding capacity of the mutated E1B-55K from E1B-P minus toward Mre11 and p53, coimmunoprecipitation studies from infected lysates were performed. Figure 2B lanes 4 and 8, illustrates reduced binding of the phosphonegative E1B-55K toward Mre11 (∼80 to 90% reduction [ImageJ quantification]) and p53 (not quantifiable due to immunoglobulin heavy-chain band), which in turn might explain the delayed steady-state reduction of Mre11 or the increase in p53 protein levels during H5pm4174 infection.
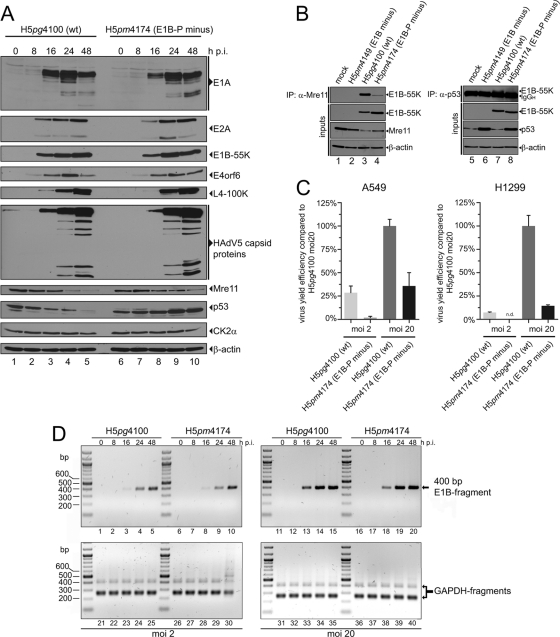
Fig 2.
Phenotypic characterization of H5pg4100 (wt) and H5pm4174 (E1B-P minus) viruses. (A) Viral and cellular protein expression analyses. A549 cells were mock infected or infected with the indicated viruses at an MOI of 20 FFU per cell and harvested at the indicated time points. Total cell extracts were prepared, separated by SDS-PAGE, and immunoblotted for the indicated proteins. (B) Coimmunoprecipitation with Mre11 or p53. A549 cells were mock infected or infected with wt and mutant viruses at an MOI of 20 FFU per cell. Whole-cell lysates were subjected to immunoprecipitation with anti-Mre11 (pNB 100-142) or anti-p53 (DO-1) antibodies. Proteins separated by SDS-PAGE were detected by immunoblotting with anti-E1B antibody 2A6. Steady-state concentrations (inputs) of E1B-55K (2A6), Mre11 (pNB 100-142), β-actin (AC-15), and p53 (DO-1) were determined by immunoblotting of protein extracts with the appropriate antibodies. (C) Virus growth. A549 and H1299 cells were infected with wt and E1B-P minus virus at an MOI of 2 or 20 FFU per cell and harvested at 48 h p.i. The virus yield was determined by quantitative E2A-72K immunofluorescence staining (B6-8) on HEK293 cells. The results represent the average of at least three independent experiments. Bars indicate the standard error of the mean values. Virus yield efficiency is represented as a percentage of H5pg4100 (wt; MOI = 20) yield efficiency. (D) Viral DNA synthesis. A549 cells were infected with H5pg4100 (wt) or H5pm4174 (E1B-P minus) virus at an MOI of 2 or 20 FFU per cell. Total DNA was isolated at the indicated time points and subjected to PCR. PCR products were analyzed by agarose gel electrophoresis. PCR amplification of GAPDH (lanes 21 to 40) served as an internal control.
In past experiments, Teodoro et al. observed a severe defect in viral replication efficiency using a virus mutant lacking two of the three phosphosites in E1B-55K (pmS490/91A [65]). To test whether our mutant, lacking all three phosphosites (S490/491/T495A, E1B-P minus), also shows a defect in virus progeny production, the virus yield was determined in A549 and H1299 cells at 48 h p.i. (Fig. 2C).
At an MOI of 2, we detected up to 93% less viral particles in H5pm4174 (E1B-P minus) infected A549 cells compared to wt-infected cells. To try to rescue this defect, we performed similar experiments with an MOI of 20 FFU per cell. However, the reduction was still a significant 65%, so the defect could not be compensated completely (Fig. 2C, A549). A549 infection with H5pm4174 (E1B-P minus) resulted in increased p53 protein levels, which might be responsible for the detrimental effect on virus growth. Interestingly, H5pm4174 infection in H1299 p53-negative cells yielded undetectable virus progeny amounts at an MOI of 2 and 85% less viral particles at an MOI of 20 FFU per cell (Fig. 2C, H1299). These results indicate that factors other than, for example, p53 activation, are responsible for reduced progeny virion numbers.
Since defects in viral progeny production cannot be traced back to lower viral protein levels in general (Fig. 2A), we investigated the ability of the H5pm4174 (E1B-P minus) virus to synthesize DNA. Viral DNA replication efficiency during E1B-P minus infection was comparable to wt infection at MOIs of 2 or 20 FFU per cell (Fig. 2D). These results suggest that decreased adenoviral progeny production is not due to reduced viral DNA synthesis and cannot be fully explained by our results.
Consistent with previous work from different groups (57, 64, 65), our data show that E1B-55K is phosphorylated at residues S490, S491, and T495 in virus-infected cells and confirm that mutations that convert these amino acids to alanines have profound negative effects on binding to and ubiquitin-dependent degradation of Mre11 and p53 and on virus growth but, interestingly, neither affect viral early and late protein production nor viral DNA synthesis (Fig. 2). In addition, it appears that phosphorylation at these carboxy-terminal sites regulates the subcellular distribution of E1B-55K since the mutant protein exhibits a substantially different localization pattern compared to the wt product (Fig. 3D and Fig. 4B and C).
Fig 3.
Coimmunoprecipitation and indirect immunofluorescence to analyze E1B-55K and its interaction with CK2α in infected cells. (A) CK2α interaction with E1B-55K after transfection. H1299 cells transfected with empty plasmid vector (pcDNA) and vectors encoding E1B-55K (pE1B-55K) or mutant E1B-55K (pE1B-P minus) were subjected to immunoprecipitation with an anti-CK2α antibody (ab13410). Proteins separated by SDS-PAGE were detected by Western blots with anti-E1B antibody (2A6). Western blot analyses of protein input levels are shown below for E1B-55K (2A6) and to the right for CK2α (ab13410) and β-actin (AC-15). (B and C) CK2α interaction with E1B-55K during infection. A549 and H1299 cells either mock infected or infected (MOI = 20 FFU/cell) with H5pm4149 (E1B minus), H5pg4100 (wt), and H5pm4174 (E1B-P minus) virus were analyzed by immunoprecipitation assays using protein A-Sepharose-coupled anti-CK2α antibody (ab13410). Proteins separated by SDS-PAGE were detected by immunoblotting with anti-E1B (2A6) and anti-CK2α (ab13410) antibodies. Immunoblot analyses of protein input levels are shown below for CK2α, E1B-55K (2A6), and β-actin (AC-15). (D) CK2α is relocalized during adenoviral infection. A549 cells either mock infected or infected with the indicated viruses (MOI = 20 FFU/cell) were analyzed by in situ immunofluorescence staining for E1B-55K (2A6), CK2α (ab13410), and DNA content (DAPI). Examples of the two major CK2α relocalization patterns observed in this cell line are shown (n = 168).
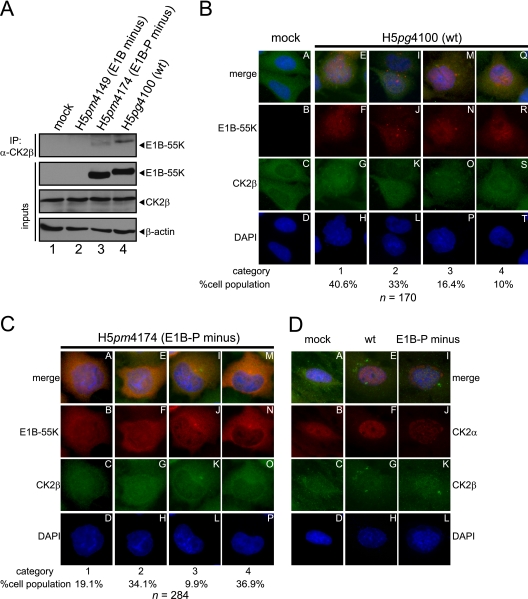
Fig 4.
Coimmunoprecipitation and indirect immunofluorescence to analyze E1B-55K and its interaction with CK2β in infected cells. (A) CK2β interaction with E1B-55K during infection. A549 cells either mock infected or infected (MOI = 20 FFU/cell) with H5pm4149 (E1B minus), H5pg4100 (wt), or H5pm4174 (E1B-P minus) virus were analyzed by immunoprecipitation assays using protein A-Sepharose-coupled anti-CK2β antibody (Santa Cruz, “51”). Proteins separated by SDS-PAGE were detected by immunoblotting with anti-E1B (2A6) antibody. Immunoblot analyses of protein input levels are shown below for CK2β (6D5), E1B-55K (2A6), and β-actin (AC-15). (B and C) CK2β is relocalized during adenoviral infection. A549 cells either mock infected or infected with the indicated viruses (MOI = 20 FFU/cell) were analyzed by in situ immunofluorescence staining for E1B-55K (7C11), CK2β (6D5), and DNA content (DAPI). Examples of the four major CK2β relocalization patterns observed in this cell line are shown (B, n = 170; C, n = 284). (D) Comparison of CK2α and CK2β localization during adenoviral infection. A549 cells either mock infected or infected with the indicated viruses (MOI = 20 FFU/cell) were analyzed by in situ immunofluorescence staining for CK2α (ab13410), CK2β (6D5), and DNA content (DAPI). Presented are blots and immunofluorescence data which show results that have been reproduced in at least three independent experiments.
CK2α interacts with E1B-55K and is relocalized during adenovirus infection.
As mentioned above, E1B-55K phosphorylation sites S490 and S491 lie within a protein kinase CK2 phosphorylation consensus sequence, suggesting that this kinase is responsible for E1B-55K phosphorylation (Fig. 1B and C). Therefore, we investigated whether E1B-55K interacts with the cellular protein kinase CK2. For this, we first focused on the α subunit which confers kinase activity either alone or in complex with the β subunit as a holoenzyme (36, 39). In a first attempt to show interaction between E1B-55K and CK2α, endogenous CK2α was immunoprecipitated from p53-negative H1299 cells transfected with wt pE1B-55K or mutant pE1B-55K (pE1B-P minus) encoding plasmids (Fig. 3A). By using this approach, we could exclude that either p53 or another adenoviral protein might mediate an interaction between E1B-55K and CK2 since both proteins are also p53 interaction partners (36, 57). Next, A549 and H1299 cells were infected with H5pm4149 (E1B minus), H5pg4100 (wt), and H5pm4174 (E1B-P minus) viruses (Fig. 3B and C), and immunoprecipitation experiments were performed as described above. Immunoblotting revealed that wt E1B-55K coprecipitated with CK2α after transfection or infection of either cell line (Fig. 3A, lane 2, and Fig. 3B, lane 3, 3C lane 7). In contrast, mutant E1B-55K from E1B-P minus did not coprecipitate in the transient-transfection assay and only did so inefficiently after infection of either cell line (Fig. 3A, lane 3, Fig. 3B, lane 4, and Fig. 3C, lane 8).
Souquere-Besse et al. previously showed that CK2α and β subunits are separated and relocalized during adenoviral infection (61). To investigate the relative localization of CK2α and E1B-55K in A549 cells, we performed double-label immunofluorescence analyses of cells infected with wt H5pg4100 or E1B-P minus H5pm4174 virus. We observed similar CK2α redistribution patterns in both wt and E1B-P minus-infected cells. Nearly 53% of wt-infected A549 cells (n = 168) showed relocalization of CK2α into nuclear ring-like structures, which showed specific CK2α-positive foci at the periphery of these ring-like structures (Fig. 3D, panels E to H). Intriguingly, these ring-like structures form specifically around E1B-55K-positive nuclear regions (Fig. 3D, panels E to H), implicating that interaction between CK2α and E1B-55K might take place in these areas of the nucleus. In contrast, the remaining ∼47% of cells largely revealed a CK2α-staining with neither nuclear ring-like structures nor nuclear CK2α foci (Fig. 3D, panels I to L). Here, CK2α is found evenly distributed in the nucleus with some aggregates in the cytoplasm or directly at the periphery of the nucleus. Interestingly, a significant number of the cells showing this phenotype (almost 15%) also displayed colocalization between CK2α and E1B-55K in cytoplasmic aggregates (Fig. 3D, indicated by yellow arrows, panels I to L). Intriguingly, the relocalization pattern of CK2α in E1B-P minus H5pm4174-infected cells was very similar qualitatively and quantitatively to that displayed during wt infection (Fig. 3D, panels M to P [55.2%] and panels Q to T [44.8%]), which suggests that CK2α relocalization is independent of E1B-55K's phosphorylation status or even E1B-55K interaction at all as indirectly derivable from our coimmunoprecipitation studies (Fig. 3A, B, and C).
Taken together, our results clearly demonstrate that wt E1B-55K can interact with CK2α. In contrast, the E1B-55K mutant from E1B-P minus exhibits significantly reduced or no binding to CK2α in virus-infected A549 or H1299 cells, respectively (Fig. 3B and C) or in single wt E1B-55K/mutant E1B-55K H1299 transfection experiments (Fig. 3A). This indicates that the phosphorylatable amino acids S490, S491, and T495 contribute significantly to the CK2α binding site on E1B-55K. Furthermore, interaction between E1B-55K and CK2α is apparently independent of p53 and other viral proteins, since both proteins can interact in pE1B-55K plasmid-transfected p53-negative H1299 cells (Fig. 3A). Moreover, our data from immunofluorescence analyses suggest that the location of interaction (if interaction takes place) between CK2α and E1B-55K occurs mainly at the periphery of infection-induced CK2α-positive ring-like structures in the nucleus and to a lesser extent in cytoplasmic accumulations (Fig. 3D).
CK2β binds to E1B-55K and is relocalized during adenoviral infection.
Association of CK2α with CK2β can result in different functional outcomes. For example, association of the CK2 subunits into a stable holoenzyme reduces the amount of CK2β in the nucleus (19). Furthermore, substrate specificity can be altered by holoenzyme association (36). Interestingly, our results from infected A549 cells analyzed at 24 h p.i. demonstrate that CK2α is redistributed upon H5pg4100 (wt) and H5pm4174 (E1B-P minus) infection into nuclear areas, as well as cytoplasmic accumulations (Fig. 3D). To test whether CK2β also interacts with E1B-55K and to investigate its relative localization during wt and E1B-P minus virus infection in comparison to E1B-55K and CK2α, coimmunoprecipitation and double-label immunofluorescence analyses were performed in A549 cells. Our results clearly show that wt E1B-55K interacts with endogenous CK2β and that this interaction also occurs with the phosphonegative E1B-55K, although this interaction is greatly weakened (Fig. 4A, lanes 3 and 4). Interestingly, we could only detect in very rare cases a specific overlap in the staining of E1B-55K and CK2β accumulations (not quantifiable). Nevertheless, we observed several different redistribution patterns of CK2β during adenoviral infection, which strongly depend on the phosphorylation status of E1B-55K (Fig. 4B, C, and D). We tried to categorize those phenotypes and defined four general categories (Fig. 4B and C). During wt infection (H5pg4100), we observed that 40.6% of CK2β relocalized into small, mainly nuclear accumulations (Fig. 4B, panels E to H; category 1), whereas in the second largest cohort (33%, category 2) CK2β is more evenly distributed and concentrated in the nucleus (compare Fig. 4B, panels A to D and panels I to L, and compare the upper and lower cell in panel K, category 2). A total of 16.4% of the wt-infected cells showed a relocalization of CK2β into mainly cytoplasmic bodies at the periphery of the nucleus (Fig. 4B, panels M to P, category 3), and 10% displayed no redistribution at all upon H5pg4100 infection (Fig. 4B, panels Q to T, category 4).
In contrast, CK2β redistribution during H5pm4174 (E1B-P minus) infection revealed the same phenotypes but with mainly two different quantitative outcomes. Here, only 19.1% showed a CK2β relocalization into small, nuclear accumulations (Fig. 4C, panels A to D, category 1). In addition, the largest cohort of H5pm4174-infected cells displayed no CK2β redistribution (Fig. 4C, panels M to P, category 4). However, no major differences could be observed between wt and mutant virus-infected cells in the amount of cells showing redistributed CK2β with phenotypes of categories 2 and 3 (compare Fig. 4B, panel K [33%] to Fig. 4C, panel G [34.1%] and Fig. 4B, panel O [16.4%] to Fig. 4C, panel K [9.9%]). These data suggest that phosphorylation of E1B-55K is important for proper CK2β relocalization, although CK2α reorganization displayed no major differences between wt and E1B-P minus infection (Fig. 3D). As illustrated in Fig. 4B and C, CK2β redistribution does not follow the same pattern as CK2α relocalization (compare to Fig. 3D). Costaining of CK2α and CK2β in mock-infected, wt (H5pg4100)-infected, or E1B-P minus (H5pm4174)-infected A549 cells revealed no significant overlap between both cellular proteins (Fig. 4D). However, infection with wt and E1B-P minus viruses resulted in redistribution of both CK2 subunits into the nucleus (Fig. 4D, panels E to H and panels I to L), indirectly supporting observations from Souquerre-Besse et al. (61) and Filhol et al. (19).
In conclusion, E1B-55K also binds to the β subunit of CK2, which is relocalized in a different manner than CK2α during adenoviral infection. Moreover, this redistribution is probably dependent on the phosphorylation status of E1B-55K indicated by reduced binding of the phosphonegative E1B-55K and the increased number of cells showing no CK2β relocalization during H5pm4174 (E1B-P minus) infection.
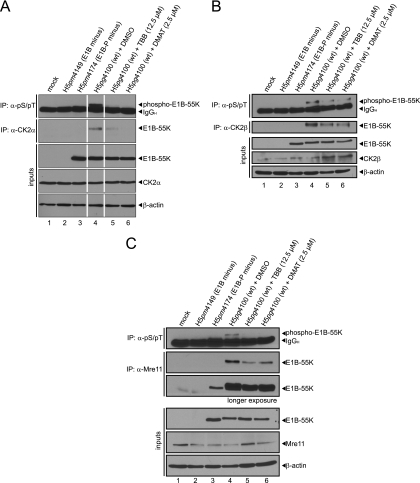
Inhibition of CK2 during adenovirus infection abolishes E1B-55K phosphorylation and interaction with CK2α, CK2β, and Mre11.
To test whether specific CK2 inhibition leads to reduced or completely abolished detection of phospho-E1B-55K, we determined the phosphorylation status of E1B-55K and its interaction with CK2α, CK2β, and Mre11 after treatment with two specific CK2 inhibitors (DMAT and TBB). Resembling the results with mutant E1B-55K (E1B-P minus) from H5pm4174 and E1B minus from H5pm4149 infection (Fig. 1, top panel, lanes 3 and 4), we either could not detect or only detected severely reduced phosphorylation of E1B-55K after treatment with the DMAT or TBB inhibitors (Fig. 5A, B, and C, top panels, lanes 4 to 6). We also observed either reduced or no coprecipitation of E1B-55K with endogenous CK2α, CK2β, and Mre11 (Fig. 5A, B, and C, second panels, lanes 4 to 6, respectively) after inhibitor treatment. We conclude from our results that CK2α must bind to E1B-55K in order to phosphorylate the viral protein efficiently. Moreover, these data suggest that CK2α-mediated phosphorylation of E1B-55K determines the affinity of protein-protein interaction or substrate specificity toward CK2β and Mre11, too (Fig. 5B and C, respectively).
Fig 5.
Inhibitors TBB or DMAT reduce or abolish CK2α, CK2β, and Mre11 binding and phosphorylation of E1B-55K. (A, B, and C) A549 cells were mock infected or infected with H5pm4149 (E1B minus), H5pg4100 (wt), and H5pm4174 (E1B-P minus) virus at an MOI of 20 FFU/cell. As indicated, cells were also treated with DMSO, TBB, or DMAT 4 h p.i. until harvesting the cells at 24 h p.i. Immunoprecipitation assays were performed with anti-phosphoserine/phosphothreonine (pS/pT, 22a, A, B, and C), anti-CK2α (ab13410 [A]; segments originate from one blot, and double control lanes were omitted due to clarity), anti-CK2β (“51” [B]) and Mre11 (pNB 100-142 [C]) antibodies. Proteins separated by SDS-PAGE were immunoblotted with anti-E1B antibody (2A6). Protein steady-state levels were detected by immunoblotting with antibodies specific for E1B-55K (2A6), CK2α (ab13410 [A]), CK2β (6D5 [B]), Mre11 (pNB 100-142 [C]), and β-actin (AC-15 [A, B, and C]). The Western blots represent one of at least three independent experiments.
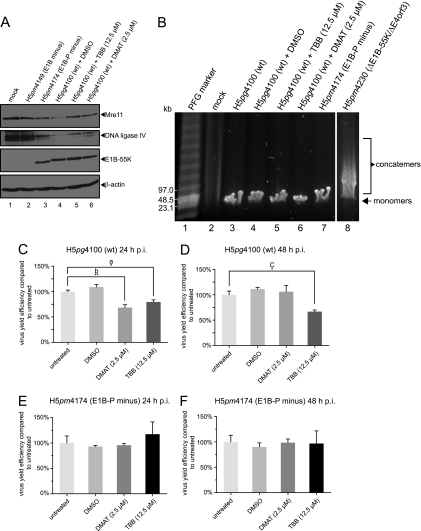
Inhibition of CK2 during adenovirus infection limits downregulation of Mre11 and DNA ligase IV protein levels but does not induce adenoviral genome concatemerization, although the virus yield is significantly reduced.
The phospho mutant of E1B-55K displays several defects such as delayed degradation of Mre11 and reduced viral progeny production (Fig. 2A and C). Moreover, specific inhibition of CK2 decreases binding efficiency of E1B-55K toward Mre11 (Fig. 5C). To evaluate the effect of CK2 inhibition during adenoviral infection more in detail, we used further approaches. First, degradation of Mre11 and DNA ligase IV was investigated by combined infection and CK2 inhibitor treatment in A549 cells. We observed a phospho-E1B-55K-dependency on Mre11 and DNA ligase IV degradation. Obviously, the phosphonegative E1B-55K mutant and CK2 inhibition after wt virus infection resulted in higher Mre11 and DNA ligase IV steady-state protein levels than after H5pg4100 (wt) infection with dimethyl sulfoxide (DMSO) control treatment (Fig. 6A, lanes 3 to 6). However, since degradation of Mre11 and (mainly) DNA ligase IV is mandatory to circumvent adenoviral genome concatemerization (57), we used pulsed-field gel electrophoresis to visualize adenoviral genomes in the context of H5pg4100 (wt) and H5pm4174 (E1B-P minus) infection and CK2 inhibitor treatment (Fig. 6B). Here, we could detect no assembly of adenoviral genome concatemers (Fig. 6B, lanes 3 to 7), only in the positive control after infection with H5pm4230 virus (ΔE1B-55K/ΔE4orf3, a similar virus mutant, was described previously [63]). Therefore, lower virus yield cannot be explained by accumulation of viral DNA concatemers after H5pm4174 (E1B-P minus) infection. Intriguingly, and as expected, CK2 inhibition led to a modest reduction in virus yield after wt (H5pg4100) infection in A549 cells at 24 and 48 h p.i. (Fig. 6C, 32% DMAT and 21% TBB; Fig. 6D, 33% TBB), whereas CK2 inhibitor treatment had no effect upon H5pm4174 (E1B-P minus) virus yield at either 24 or 48 h p.i. (Fig. 6E and F).
Fig 6.
Inhibitors TBB and DMAT reduce Mre11 and DNA ligase IV degradation efficiency but have no effect on inducing viral genome concatemerization although virus yield is significantly decreased. (A) Mre11 and DNA ligase IV degradation upon CK2 inhibitor treatment. A549 cells were mock infected or infected with H5pm4149 (E1B minus), H5pg4100 (wt), and H5pm4174 (E1B-P minus) virus at an MOI of 20 FFU/cell. As indicated, cells were also treated with DMSO, TBB, or DMAT at 4 h p.i. until harvesting the cells at 24 h p.i. Total cell extracts were prepared, separated by SDS-PAGE, and immunoblotted for Mre11(pNB 100-142), DNA ligase IV (NB110-57379), E1B-55K (2A6), and β-actin (AC-15). (B) Analysis of adenoviral genome concatemerization. A549 cells were mock infected or infected with the indicated viruses at an MOI of 30 FFU/cell. H5pm4230 (ΔE1B-55K/ΔE4orf3) infection was performed at an MOI of 100 FFU/cell. Cells were also treated with inhibitors as in panel A, but the cells were harvested for isolating total DNA at 30 h p.i. Adenoviral genome monomers and concatemers were visualized after pulsed-field gel electrophoresis by ethidium bromide staining (segments originate from one agarose gel, and triple control lanes were omitted due to clarity). (C to F) Virus growth upon CK2 inhibitor treatment. A549 cells were infected with wt and E1B-P minus virus at an MOI of 20 FFU/cell and harvested at 24 or 48 h p.i. In addition, the cells were also treated with DMSO or CK2 inhibitors as described above (in panel A). Virus yield was determined by quantitative E2A-72K immunofluorescence staining (B6-8) on HEK293 cells. The results represent the average of at least three independent experiments. P values of unpaired, two-tailed t tests (GraphPad Prism 5) were determined. Single and double asterisks indicate significant differences (a, P = 0.0158; b, P = 0.0082; c, P = 0.0135). Bars indicate the standard error of the mean values. Virus yield efficiency is represented as a percentage of “untreated” virus yield efficiency.
Taken together, cellular substrates for adenovirus-mediated degradation such as Mre11 and DNA ligase IV are inefficiently degraded in the context of a phosphonegative E1B-55K virus infection (H5pm4174) and upon CK2 inhibition. In addition, other reasons than viral genome concatemerization are responsible for lower virus yield after H5pm4174 (E1B-P minus) infection or CK2 inhibitor treatment. Strikingly, the reduced production of progeny virions upon inhibitor treatment could not be further reduced in the E1B-P minus virus (H5pm4174) infection, stressing the specific positive effect of CK2 upon E1B-55K.
CK2α, but not the CK2 holoenzyme, phosphorylates E1B-55K in vitro.
Our results clearly demonstrate that CK2 is interacting with E1B-55K and that phosphorylation of the viral protein by the cellular kinase is important for viral progeny production. We next determined whether this kinase can phosphorylate E1B-55K in vitro. GST-E1B fusion proteins (Fig. 7A) were incubated with recombinant CK2α or the holoenzyme in the presence of [γ-32P]ATP, and the reaction products were analyzed by SDS-PAGE and autoradiography (Fig. 7B). Efficient phosphorylation was observed with GST-wt-E1B (Fig. 7B, lanes 1 to 3) but not with the triple mutant GST-E1B-P minus (Fig. 7B, lanes 10 to 12). Less efficient phosphorylation by CK2α was also observed with mutant GST-E1B fusion proteins where both serine residues were changed to alanines (GST-E1B AAT) or aspartic acids (GST-E1B DDT) (Fig. 7B, lanes 4 to 6 and lanes 7 to 9, respectively), strongly indicating that T495 is also phosphorylated by CK2α. Since at least one report (2) defined a regulatory role for the β subunit of CK2, in vitro phosphorylation assays were also performed with recombinant CK2 (double α plus double β) holoenzyme (Fig. 7B, lanes 13 to 21). Interestingly, however, no signals were detected with either GST-wt-E1B (Fig. 7B, lanes 13 to 15) or the mutant constructs (Fig. 7B, lanes 16 to 21). Because double amounts of both kinase and substrate were included in the CK2 holoenzyme assays (Fig. 7B, lanes 13 to 21), one can exclude the possibility that insufficient kinase and substrate amounts were responsible for the undetectable phosphorylation. Collectively, these results suggest not only that CK2α interacts with E1B-55K and phosphorylates this protein at the C terminus but that the β subunit may mediate inhibitory effects on the phosphorylation of E1B-55K.
The 156R splice product of E1B-55K is also targeted by CK2α but not by the holoenzyme.
A splice product of the full-length E1B mRNA, called E1B-156R (156R), shares 77 identical C-terminal amino acids (and 79 N-terminal amino acids) with the full-length protein (59) and has been shown to be phosphorylated (Fig. 7A) (65). To examine whether 156R is also a target for phosphorylation via protein kinase CK2, a GST-156R fusion protein was used in a similar in vitro phosphorylation assay as described above. Recombinant CK2α efficiently phosphorylated GST-156R, GST-wt-E1B, and GST-April, an essential protein for HuR-mediated nucleocytoplasmic translocation of the CD83 mRNA, which has been shown to be a CK2 target (12) and served as a positive control (Fig. 8, lanes 1 to 3). Moreover, the CK2 holoenzyme also phosphorylated GST-April efficiently (Fig. 8, lane 8) but was not capable of phosphorylating GST-156R or GST-wt-E1B efficiently (Fig. 8, lanes 7 and 9). Preincubation with the CK2 inhibitor DMAT (40, 41) almost completely abrogated CK2α-mediated phosphorylation, (Fig. 8, lanes 4 to 6). Our data reveal that E1B-156R is also targeted by CK2α but not by the CK2 holoenzyme in vitro.
DISCUSSION
It has long been speculated that E1B-55K might be phosphorylated by the protein kinase CK2. However, until now experiments have only concentrated on the properties of an E1B-55K phosphorylation-deficient mutant, which showed that phosphorylation regulates many E1B-55K functions, and therefore viral behavior in general, and is thus one of the most critical posttranslational modifications of E1B-55K (57, 64, 65). However, the kinase responsible was not identified, so our main goal was to clarify this question.
It is known that many viruses and their gene products exploit the functions of CK2 for their own benefit, such as EBV protein ZEBRA (17), Kaposi's sarcoma-associated herpesvirus (KSHV) protein ORF57 (34), or herpesvirus protein ICP27 (29). EBV ZEBRA is reported to require phosphorylation by CK2 to turn the viral protein into a transcriptional repressor or activator and that this posttranslational modification contributes to ZEBRA's function in controlling viral lytic cycle gene expression (17). CK2-mediated phosphorylation of ORF57 from KSHV is also necessary for controlling gene expression during the lytic infectious cycle (34). As another example, ICP27 requires CK2-mediated phosphorylation for correct subcellular localization and proper binding to several interaction partners (29, 52).
In light of these reports and the fact that E1B-55K contains a highly conserved CK2 consensus motif (CKM-490SSDEDTD496; compare Fig. 1B and C), it is not surprising that the long suspected protein kinase CK2 is also involved in regulating E1B-55K activity. Indeed, we demonstrate here for the first time that E1B-55K binds to the catalytic α and the regulatory β subunit of CK2 and is a substrate for CK2α phosphorylation in vitro and in cell culture (Fig. 3, 4, 5, 7, and 8). Our coimmunoprecipitation experiments in H1299 p53-negative cells transfected with E1B-55K alone demonstrated an interaction independent of the presence of p53 and other viral proteins. Furthermore, according to our in situ stainings, redistribution of CK2α seems to be necessary for interaction with E1B-55K but not vice versa. In fact, although mutant E1B-55K from E1B-P minus does not efficiently bind to, and is not phosphorylated by CK2α, we still observed almost the same CK2α protein relocalization during E1B-P minus infection, indicating that an as-yet-unidentified adenoviral protein may also participate in CK2α redistribution (Fig. 3D). This is further supported by our observation that CK2α redistribution does not occur in cells transfected with wt pE1B-55K or pE1B-P minus alone (data not shown).
It has been reported that a substrate's phosphosites make a significant contribution to the overall binding energy between a kinase and its substrate (67). Thus, it might be expected that mutating the phosphosites of E1B-55K, or blocking the active site of CK2 by its specific inhibitors DMAT or TBB, leads to inefficient or completely abolished binding. Indeed, DMAT or TBB treatment of adenovirus-infected A549 cells led to lower amounts of phospho-E1B-55K, which is probably due to the reduction of CK2α binding toward E1B-55K (Fig. 5A).
In addition, it is possible that structural proteins such as the adenoviral fiber proteins play a role in CK2 subunit redistribution. Souquerre-Besse et al. reported that infection with an adenovirus deleted of the fiber gene in its genome resulted in relocalization of both α and β CK2 subunits into the same viral nuclear structures, in contrast to wt HAdV5 infection, where both subunits separate into two morphologically distinct virus-induced nuclear areas (61). We also observed relocalization of CK2α in the cytoplasm and not only in the nucleus. Furthermore, we could also show that E1B-55K not only interacts with the α but also with the β subunit of CK2 and that phosphorylation of E1B-55K strongly determines the binding efficiency toward that subunit like for CK2α (Fig. 4A and Fig. 5B). Moreover, the phosphorylation status of E1B-55K is obviously also important for the outcome of CK2β redistribution (Fig. 4B and C). Importantly, in H5pm4174 infection (E1B-P minus) the amount of cells showing small nuclear CK2β accumulations decreases from 40.6 to 19.1% (Fig. 4C, category 1) and cell numbers showing no relocalization at all rise from 10 to 36.9% (Fig. 4C, category 4).
From our data we deduce that interactions between E1B-55K and CK2α can take place partly in cytoplasmic accumulations or at the borders of specific ring-like structures precisely surrounding areas of nuclear E1B-55K regions after adenoviral infection in A549 cells (Fig. 3D), whereas deducing where CK2β and E1B-55K interaction takes place is more complicated since CK2β shows many different phenotypes with no significant overlap of E1B-55K and CK2β staining (Fig. 4B).
The fact that Souquerre-Besse et al. observed a separation of CK2 subunits also fits very well with our in vitro observation that the holoenzyme phosphorylates the wt E1B-55K C terminus inefficiently. When combined with our in vitro phosphorylation and immunofluorescence data, a picture emerges suggesting that separation of the CK2α and β subunits is necessary during the course of infection for efficient E1B-55K phosphorylation. Furthermore, though at this point only speculative but not improbable, we propose that β subunits exert inhibitory effects upon E1B-55K C-terminal phosphorylation. There is one example where such an unusual regulatory mechanism for CK2 substrates is indeed described. Arrigoni et al. (2) reported that calmodulin is a target for CK2 phosphorylation, which in turn is inhibited by CK2β. In light of these observations and knowledge owing to another study describing that dissociation of CK2α and CK2β into separate subunits favors increased amounts of nuclear CK2β (19), we assume that E1B-55K is directly involved in CK2β redistribution in order to support its own phosphorylation by CK2α. This hypothesis explains why E1B-55K binds to CK2β, although we assume at the same time an inhibitory effect of that subunit upon E1B-55K phosphorylation.
Since E1B-55K is reported to have several splice products, and at least one is also shown to be phosphorylated (65), we examined E1B-156R, which contains 77 amino acids of the full-length carboxy terminus. We found that E1B-156R is also a target of CK2α that similarly cannot be efficiently phosphorylated by the CK2 holoenzyme in vitro. This implies that CK2α may regulate the functions of the full-length, as well as the E1B-156R splice product, and that the phenotypes observed during E1B-P minus infection can also be attributed at least in part to disturbed functioning of the similarly mutated E1B-156R peptide. Investigations into the role of E1B-156R phosphorylation are under way.
In the present study wt and E1B-negative viruses originated from an HAdV5 genomic backbone. Thus, we wanted to analyze E1B-55K phosphorylation with an E1B-55K phosphorylation-deficient virus in the same genomic HAdV5 backbone. To confirm that the phenotype conferred by such a virus mutant located in this genomic backbone is comparable to previously reported viruses carrying the same mutations (57, 64), we conducted several phenotypic analyses. Carried out in part previously, we confirmed that under lytic infection conditions the newly generated virus mutant has similar characteristics, such as deficiency in p53 degradation and mislocalization of the mutated E1B-55K. Therefore, data obtained in the present study are to a great extent consistent and comparable to earlier data (49, 57, 64, 65). Moreover, we could substantiate our data by CK2 inhibitor treatment of infected cells where we observed a similar effect upon binding to and degradation of Mre11 (Fig. 5C and Fig. 6A).
Very intriguing in this context, we obtained significantly reduced levels of E1B-P minus virus progeny, although we neither detected a negative effect on viral DNA replication efficiency, nor on early or late viral protein expression (Fig. 2A, C, and D). Hence, a defect in viral mRNA synthesis or transport can most probably be excluded. To support our observations concerning the phosphonegative E1B-55K virus mutant, we used CK2 inhibitor studies and achieved a reduction of viral progeny at 24 h p.i., but at 48 h p.i. only with TBB treatment. It is possible that the DMAT inhibitor is not stable for a longer incubation period than 20 h and exerted no inhibitory effect anymore. Nevertheless, we were able to achieve up to 33% reduction of viral progeny virions upon CK2 inhibitor treatment (Fig. 6D, TBB). Probably, the remaining CK2 activity, as shown in Fig. 5A to C, is sufficient to support virus growth leading to this modest virus growth defect in comparison to virus growth defect after E1B-P minus virus infection. Strikingly, the CK2 inhibitors added no further negative effect on production of progeny virions upon E1B-P minus (H5pm4174) infection, clearly supporting the notion that specifically the phosphorylation of E1B-55K by CK2 is crucial for proper virus growth. Furthermore, since p53 protein levels increase upon infection with E1B-P minus virus (Fig. 2A), it cannot be excluded that virus growth is decreased due to increased p53 activation with subsequent negative effects for the virus such as cell cycle arrest. Interestingly, we observed an even more pronounced defect in virus growth upon H5pm4174 infection in H1299 p53-negative cells (Fig. 2C, H1299), strongly arguing for p53 not being involved in the virus growth defect of H5pm4174 (E1B-P minus).
One possible explanation for the reduced production of progeny virions can be deduced from the observed delay in Mre11 degradation during E1B-P minus infection (Fig. 2A) or the inhibited degradation of DNA ligase IV (Fig. 6A). Mre11, which forms with Rad50 and Nbs1 the MRN complex, is responsible, among others, for DNA double-strand break repair and has been shown to mediate adenoviral genome concatenation, like DNA ligase IV, if not disturbed in its functions (63). Thus, it cannot be excluded that a delayed Mre11 or DNA ligase IV degradation during E1B-P minus infection might give the MRN complex more time to exert negative effects such as viral genome concatenation (63) and in effect hinder the appropriate assembly of virions with genome content. However, analyses on viral genome concatemerization during E1B-P minus infection or during wt infection with subsequent CK2 inhibitor treatment, revealed no production of viral concatemers (Fig. 6B). Thus, the reason for reduced viral progeny production during E1B-P minus (H5pm4174) infection or CK2 inhibitor treatment remains obscure and can only be explained by a non- or misfunctioning of one or more viral factors or by a loss of regulatory functions exerted on an unknown cellular factor/s.
Of note, considering the fact that E1B-55K's phosphosites at the C terminus also roughly fit the criteria for glycogen synthase kinase-3 (S-X-X-X-pS [15, 36]) and CK1 (pS-X-X-S/T [36]) consensus phosphorylation sites, it is far too early to say that CK2 is the sole kinase phosphorylating E1B-55K. Experiments are under way to clarify potential specific roles of these other two kinases in E1B-55K regulation by phosphorylation. Nevertheless, our data demonstrate for the first time that CK2α plays a significant part in phosphorylation of E1B-55K and is also involved in threonine 495 phosphorylation in vitro.
ACKNOWLEDGMENTS
We thank Ruediger Meyer and Georg Mayr for assisting with the in vitro phosphorylation assays. We thank Joachim Hauber for providing the plasmid GST-April. We thank Ramon A. Gonzalez for critically reading the manuscript and helpful discussions.
This study was supported by grants from the Leibniz Center Infection and the Wilhelm-Sander Stiftung, Munich, Germany. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit (BMG).
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Anderson CW, Schmitt RC, Smart JE, Lewis JB. 1984. Early region 1B of adenovirus 2 encodes two coterminal proteins of 495 and 155 amino acid residues. J. Virol. 50:387–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arrigoni G, et al. 2004. Phosphorylation of calmodulin fragments by protein kinase CK2: mechanistic aspects and structural consequences. Biochemistry 43:12788–12798 [DOI] [PubMed] [Google Scholar]
- 3. Babiss LE, Ginsberg HS. 1984. Adenovirus type 5 early region 1b gene product is required for efficient shutoff of host protein synthesis. J. Virol. 50:202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Babiss LE, Ginsberg HS, Darnell JE., Jr 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baker A, Rohleder KJ, Hanakahi LA, Ketner G. 2007. Adenovirus E4 34k and E1b 55k oncoproteins target host DNA ligase IV for proteasomal degradation. J. Virol. 81:7034–7040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blackford AN, Grand RJ. 2009. Adenovirus E1B 55-kilodalton protein: multiple roles in viral infection and cell transformation. J. Virol. 83:4000–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Blanchette P, et al. 2004. Both BC-box motifs of adenovirus protein E4orf6 are required to efficiently assemble an E3 ligase complex that degrades p53. Mol. Cell. Biol. 24:9619–9629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blanchette P, et al. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boivin D, Morrison MR, Marcellus RC, Querido E, Branton PE. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Buchou T, et al. 2003. Disruption of the regulatory beta subunit of protein kinase CK2 in mice leads to a cell-autonomous defect and early embryonic lethality. Mol. Cell. Biol. 23:908–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cathomen T, Weitzman MD. 2000. A functional complex of adenovirus proteins E1B-55kDa and E4orf6 is necessary to modulate the expression level of p53 but not its transcriptional activity. J. Virol. 74:11407–11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chemnitz J, Pieper D, Gruttner C, Hauber J. 2009. Phosphorylation of the HuR ligand APRIL by casein kinase 2 regulates CD83 expression. Eur. J. Immunol. 39:267–279 [DOI] [PubMed] [Google Scholar]
- 13. Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. 2009. Identification of integrin α3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Desagher S, et al. 2001. Phosphorylation of bid by casein kinases I and II regulates its cleavage by caspase 8. Mol. Cell 8:601–611 [DOI] [PubMed] [Google Scholar]
- 15. Doble BW, Woodgett JR. 2003. GSK-3: tricks of the trade for a multi-tasking kinase. J. Cell Sci. 116:1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dobner T, Kzhyshkowska J. 2001. Nuclear export of adenovirus RNA. Curr. Top. Microbiol. Immunol. 259:25–54 [DOI] [PubMed] [Google Scholar]
- 17. El-Guindy AS, Miller G. 2004. Phosphorylation of Epstein-Barr virus ZEBRA protein at its casein kinase 2 sites mediates its ability to repress activation of a viral lytic cycle late gene by Rta. J. Virol. 78:7634–7644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Filhol O, Cochet C, Wedegaertner P, Gill GN, Chambaz EM. 1991. Coexpression of both alpha and beta subunits is required for assembly of regulated casein kinase II. Biochemistry 30:11133–11140 [DOI] [PubMed] [Google Scholar]
- 19. Filhol O, et al. 2003. Live-cell fluorescence imaging reveals the dynamics of protein kinase CK2 individual subunits. Mol. Cell. Biol. 23:975–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Flint SJ, Gonzalez RA. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287–330 [DOI] [PubMed] [Google Scholar]
- 21. Franck N, Le Seyec J, Guguen-Guillouzo C, Erdtmann L. 2005. Hepatitis C virus NS2 protein is phosphorylated by the protein kinase CK2 and targeted for degradation to the proteasome. J. Virol. 79:2700–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gonzalez RA, Flint SJ. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graham KC, Litchfield DW. 2000. The regulatory beta subunit of protein kinase CK2 mediates formation of tetrameric CK2 complexes. J. Biol. Chem. 275:5003–5010 [DOI] [PubMed] [Google Scholar]
- 24. Grasser FA, et al. 1992. Phosphorylation of the Epstein-Barr virus nuclear antigen 2. Biochem. Biophys. Res. Commun. 186:1694–1701 [DOI] [PubMed] [Google Scholar]
- 25. Groitl P, Dobner T. 2006. Construction of adenovirus type 5 early region 1 and 4 virus mutants, p 29–39 In Wold WS, Tollefson AE. (ed), Adenovirus methods and protocols, 2nd ed, vol 1 Humana Press, Inc, Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- 26. Harlow E, Franza BR, Jr, Schley C. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kindsmuller K, et al. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kindsmuller K, et al. 2009. A 49-kilodalton isoform of the adenovirus type 5 early region 1B 55-kilodalton protein is sufficient to support virus replication. J. Virol. 83:9045–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Koffa MD, Kean J, Zachos G, Rice SA, Clements JB. 2003. CK2 protein kinase is stimulated and redistributed by functional herpes simplex virus ICP27 protein. J. Virol. 77:4315–4325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kzhyshkowska J, Kremmer E, Hofmann M, Wolf H, Dobner T. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383:259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Landesman-Bollag E, et al. 2001. Protein kinase CK2: signaling and tumorigenesis in the mammary gland. Mol. Cell. Biochem. 227:153–165 [PubMed] [Google Scholar]
- 32. Lilley CE, Schwartz RA, Weitzman MD. 2007. Using or abusing: viruses and the cellular DNA damage response. Trends Microbiol. 15:119–126 [DOI] [PubMed] [Google Scholar]
- 33. Malette P, Yee SP, Branton PE. 1983. Studies on the phosphorylation of the 58,000 Dalton early region 1B protein of human adenovirus type 5. J. Gen. Virol. 64:1069–1078 [DOI] [PubMed] [Google Scholar]
- 34. Malik P, Clements JB. 2004. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi's sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 32:5553–5569 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35. Medina-Palazon C, et al. 2007. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J. Virol. 81:11850–11860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meggio F, Pinna LA. 2003. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17:349–368 [DOI] [PubMed] [Google Scholar]
- 37. Mitsudomi T, et al. 1992. p53 gene mutations in non-small-cell lung cancer cell lines and their correlation with the presence of ras mutations and clinical features. Oncogene 7:171–180 [PubMed] [Google Scholar]
- 38. Munstermann U, et al. 1990. Casein kinase II is elevated in solid human tumours and rapidly proliferating non-neoplastic tissue. Eur. J. Biochem. 189:251–257 [DOI] [PubMed] [Google Scholar]
- 39. Niefind K, Putter M, Guerra B, Issinger OG, Schomburg D. 1999. GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6:1100–1103 [DOI] [PubMed] [Google Scholar]
- 40. Pagano MA, et al. 2008. The selectivity of inhibitors of protein kinase CK2: an update. Biochem. J. 415:353–365 [DOI] [PubMed] [Google Scholar]
- 41. Pagano MA, et al. 2004. 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole: a novel powerful and selective inhibitor of protein kinase CK2. Biochem. Biophys. Res. Commun. 321:1040–1044 [DOI] [PubMed] [Google Scholar]
- 42. Park JW, Bae YS. 1999. Phosphorylation of ribosomal protein L5 by protein kinase CKII decreases its 5S rRNA binding activity. Biochem. Biophys. Res. Commun. 263:475–481 [DOI] [PubMed] [Google Scholar]
- 43. Pilder S, Moore M, Logan J, Shenk T. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pinna LA. 1990. Casein kinase 2: an “eminence grise” in cellular regulation? Biochim. Biophys. Acta 1054:267–284 [DOI] [PubMed] [Google Scholar]
- 45. Pinna LA. 1997. Protein kinase CK2. Int. J. Biochem. Cell Biol. 29:551–554 [DOI] [PubMed] [Google Scholar]
- 46. Pinna LA. 2002. Protein kinase CK2: a challenge to canons. J. Cell Sci. 115:3873–3878 [DOI] [PubMed] [Google Scholar]
- 47. Pistorius K, Seitz G, Remberger K, Issinger O-G. 1991. Differential CKII activities in human colorectal mucosa, adenomas, and carcinomas. Onkologie 14:255–260 [Google Scholar]
- 48. Ponten J, Saksela E. 1967. Two established in vitro cell lines from human mesenchymal tumours. Int. J. Cancer 2:434–447 [DOI] [PubMed] [Google Scholar]
- 49. Querido E, et al. 1997. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J. Virol. 71:3788–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Querido E, et al. 2001. Identification of three functions of the adenovirus e4orf6 protein that mediate p53 degradation by the E4orf6-E1B55K complex. J. Virol. 75:699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Reich NC, Sarnow P, Duprey E, Levine AJ. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480–484 [DOI] [PubMed] [Google Scholar]
- 52. Rojas S, Corbin-Lickfett KA, Escudero-Paunetto L, Sandri-Goldin RM. 2010. ICP27 phosphorylation site mutants are defective in herpes simplex virus 1 replication and gene expression. J. Virol. 84:2200–2211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Salvi M, Sarno S, Cesaro L, Nakamura H, Pinna LA. 2009. Extraordinary pleiotropy of protein kinase CK2 revealed by weblogo phosphoproteome analysis. Biochim. Biophys. Acta 1793:847–859 [DOI] [PubMed] [Google Scholar]
- 54. Sarnow P, Sullivan CA, Levine AJ. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517 [DOI] [PubMed] [Google Scholar]
- 55. Schneider E, Kartarius S, Schuster N, Montenarh M. 2002. The cyclin H/cdk7/Mat1 kinase activity is regulated by CK2 phosphorylation of cyclin H. Oncogene 21:5031–5037 [DOI] [PubMed] [Google Scholar]
- 56. Schreiner S, et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schwartz RA, et al. 2008. Distinct requirements of adenovirus E1b55K protein for degradation of cellular substrates. J. Virol. 82:9043–9055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seldin DC, Lou DY, Toselli P, Landesman-Bollag E, Dominguez I. 2008. Gene targeting of CK2 catalytic subunits. Mol. Cell. Biochem. 316:141–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sieber T, Dobner T. 2007. Adenovirus type 5 early region 1B 156R protein promotes cell transformation independently of repression of p53-stimulated transcription. J. Virol. 81:95–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Slaton JW, Unger GM, Sloper DT, Davis AT, Ahmed K. 2004. Induction of apoptosis by antisense CK2 in human prostate cancer xenograft model. Mol. Cancer Res. 2:712–721 [PubMed] [Google Scholar]
- 61. Souquere-Besse S, et al. 2002. Adenovirus infection targets the cellular protein kinase CK2 and RNA-activated protein kinase (PKR) into viral inclusions of the cell nucleus. Microsc. Res. Technol. 56:465–478 [DOI] [PubMed] [Google Scholar]
- 62. St-Denis NA, Litchfield DW. 2009. Protein kinase CK2 in health and disease: from birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell. Mol. Life Sci. 66:1817–1829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stracker TH, Carson CT, Weitzman MD. 2002. Adenovirus oncoproteins inactivate the Mre11-Rad50-NBS1 DNA repair complex. Nature 418:348–352 [DOI] [PubMed] [Google Scholar]
- 64. Teodoro JG, Branton PE. 1997. Regulation of p53-dependent apoptosis, transcriptional repression, and cell transformation by phosphorylation of the 55-kilodalton E1B protein of human adenovirus type 5. J. Virol. 71:3620–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Teodoro JG, et al. 1994. Phosphorylation at the carboxy terminus of the 55-kilodalton adenovirus type 5 E1B protein regulates transforming activity. J. Virol. 68:776–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. 2009. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell. Mol. Life Sci. 66:1858–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ubersax JA, Ferrell JE., Jr 2007. Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell. Biol. 8:530–541 [DOI] [PubMed] [Google Scholar]
- 68. Wang H, Davis A, Yu S, Ahmed K. 2001. Response of cancer cells to molecular interruption of the CK2 signal. Mol. Cell. Biochem. 227:167–174 [PubMed] [Google Scholar]
- 69. Weitzman MD, Ornelles DA. 2005. Inactivating intracellular antiviral responses during adenovirus infection. Oncogene 24:7686–7696 [DOI] [PubMed] [Google Scholar]
- 70. Wimmer P, et al. 2010. SUMO modification of E1B-55K oncoprotein regulates isoform-specific binding to the tumour suppressor protein PML. Oncogene 29:5511–5522 [DOI] [PubMed] [Google Scholar]
- 71. Yaron Y, McAdara JK, Lynch M, Hughes E, Gasson JC. 2001. Identification of novel functional regions important for the activity of HOXB7 in mammalian cells. J. Immunol. 166:5058–5067 [DOI] [PubMed] [Google Scholar]