Abstract
Bovine herpesvirus 1 (BHV-1), an alphaherpesvirinae subfamily member, establishes latency in sensory neurons. Elevated corticosteroid levels, due to stress, reproducibly triggers reactivation from latency in the field. A single intravenous injection of the synthetic corticosteroid dexamethasone (DEX) to latently infected calves consistently induces reactivation from latency. Lytic cycle viral gene expression is detected in sensory neurons within 6 h after DEX treatment of latently infected calves. These observations suggested that DEX stimulated expression of cellular genes leads to lytic cycle viral gene expression and productive infection. In this study, a commercially available assay—Bovine Gene Chip—was used to compare cellular gene expression in the trigeminal ganglia (TG) of calves latently infected with BHV-1 versus DEX-treated animals. Relative to TG prepared from latently infected calves, 11 cellular genes were induced more than 10-fold 3 h after DEX treatment. Pentraxin three, a regulator of innate immunity and neurodegeneration, was stimulated 35- to 63-fold after 3 or 6 h of DEX treatment. Two transcription factors, promyelocytic leukemia zinc finger (PLZF) and Slug were induced more than 15-fold 3 h after DEX treatment. PLZF or Slug stimulated productive infection 20- or 5-fold, respectively, and Slug stimulated the late glycoprotein C promoter more than 10-fold. Additional DEX-induced transcription factors also stimulated productive infection and certain viral promoters. These studies suggest that DEX-inducible cellular transcription factors and/or signaling pathways stimulate lytic cycle viral gene expression, which subsequently leads to successful reactivation from latency in a small subset of latently infected neurons.
INTRODUCTION
Bovine herpesvirus 1 (BHV-1) is an alphaherpesvirinae subfamily member that causes significant economical losses to the cattle industry (86). The ability of BHV-1 to suppress the immune system can result in life-threatening pneumonia due to secondary bacterial infections. This multifactorial disorder is referred to as bovine respiratory disease complex (reviewed in references 35 and 39). Like different alphaherpesvirinae subfamily members, the primary site for BHV-1 latency is sensory neurons within trigeminal ganglia (TG). Viral gene expression (73) and infectious virus (29) are detected in TG from 1 to 6 days after acute infection. Lytic cycle viral gene expression is subsequently extinguished in sensory neurons, and latency is established. The BHV-1 genome is stably maintained in sensory neurons, but infectious virus is not detected by standard virus isolation procedures (reviewed in references 33 and 34). The only viral gene abundantly expressed in latently infected sensory neurons is the latency-related (LR) gene (reviewed in reference 38). Stress, due to confinement, transporting cattle, restricting food and water, weaning, or increased corticosteroid levels increases the incidence of reactivation from latency (35, 39). The latency reactivation cycle of BHV-1 is crucial for virus transmission and survival in nature (33, 34).
Administration of the synthetic corticosteroid dexamethasone (DEX) to latently infected calves or rabbits consistently initiates reactivation from latency (29, 33, 34, 37, 38, 68). A single intravenous injection of DEX also leads to lytic cycle viral transcription in neurons within 6 h after treatment of latently infected calves (87, 89). DEX represses LR promoter activity (36) and reduces LR-RNA levels, which reduces the number of TG neurons that express LR-RNA at 18 to 21 h after treatment (68). Calves latently infected with wild-type (wt) BHV-1 or LR mutant virus, which does not reactivate from latency (29), frequently express bICP0 mRNA after DEX treatment, but only wt BHV-1 expresses late genes (94), which correlates with virus shedding during reactivation from latency (29). These findings are consistent with an earlier study that concluded DEX induces viral gene expression in many latently infected neurons, but only a subset of these neurons produce infectious virus (68). DEX treatment of latently infected calves also induces apoptosis of T cells that persist in TG after infection (87). Persistence of T cells in TG of humans or mice latently infected with HSV-1 also occurs (9, 25, 48, 77–79, 82), and persistent CD8+ T cells in TG produce factors that promote maintenance of latency (41, 42, 46, 47, 66). Although DEX may have many effects that promote reactivation from latency, the ability of DEX to stimulate lytic cycle viral gene expression is predicted to be crucial for successful reactivation from latency.
In the present study, cellular gene expression was examined in TG during the early stages of DEX-induced reactivation from latency using a commercially available bovine array. Within 90 min or 3 h after DEX treatment, 7 or 11 genes, respectively, were stimulated at least 10-fold in the TG of calves latently infected with BHV-1. The Pentraxin 3-like gene, which regulates innate immune responses and neurodegeneration, was stimulated more than any other gene by DEX (63-fold at 6 h after DEX treatment). Two cellular transcription factors—zinc finger and BTB domain containing 16, also referred to as promyelocytic leukemia zinc finger (PLZF), and Snail homolog 2 (also known as Slug in mammals)—were induced more than 15-fold at 3 h after DEX treatment. Additional transcription factors induced by DEX were also identified. Plasmids expressing these respective cellular transcription factors activated productive infection and certain viral promoters, suggesting that these cellular transcription factors promote lytic cycle viral gene expression during the early stages of reactivation from latency.
MATERIALS AND METHODS
Cells and virus.
Murine neuroblastoma (neuro-2A), rabbit skin (RS), and bovine kidney (CRIB) cells were grown in Earle's modified Eagle medium supplemented with 10% fetal calf serum, penicillin (10 U/ml), and streptomycin (100 μg/ml).
A BHV-1 recombinant virus (gCblue) containing the lacZ gene in place of the viral gC gene was obtained from S. Chowdhury (Baton Rouge, LA). The virus grows to similar titers as the wild-type parent virus and expresses the β-Gal gene as a true late gene.
Calf studies.
BHV-1-free crossbred calves (∼200 kg) were used for the present study. Calves were inoculated with 107 PFU of BHV-1 into each nostril and eye for a total of 4 × 107 PFU/animal as described previously (29, 30, 50, 61–63). Calves were housed under strict isolation and given antibiotics before and after BHV-1 infection to prevent secondary bacterial infection. At 60 days postinfection, calves were injected intravenously (jugular vein) with 100 mg of DEX. Calves were then transported to the Veterinary Diagnostic lab. Prior to euthanasia by electrocution, calves were heavily sedated with xylazine. After decapitation, TG were collected, and samples from each TG were formalin fixed and then paraffin embedded. The remainder of both TG was minced into small pieces and placed into a single 50-ml conical tube, and the tube was placed in a dry ice ethanol bath. TG samples were then stored at −80°C. After decapitation, it took approximately 5 min to collect the TG, mince the TG, place the pieces in a 50-ml conical tube, and then submerge the tube in a dry ice-ethanol bath. One calf was decapitated at a time, to ensure that TG was processed in a timely manner to reduce the possibility of degrading RNA. Calves were decapitated in the same order in which they were injected with DEX to ensure that the time points after DEX treatment were as close as possible to the designated time point. Three calves/time point were used for these studies. Experiments were performed in accordance with the American Association of Laboratory Animal Care guidelines, and the University of Nebraska IACUC committee.
RNA preparation for microarray analysis and reverse transcription.
TG from BHV-1-infected calves were collected at necropsy at 60 days after infection (latency) and at 1.5, 3, 6, or 24 h after DEX treatment and then stored at −80°C. TG were minced into small pieces, the tissue was solubilized with a Polytron tissue homogenizer, and the total RNA was prepared using TRIzol reagent (Life Technologies) as previously described (95).
Microarray processing.
Microarray studies were performed at the University of Nebraska Medical Center (UNMC) Microarray Core Facility in Omaha, Nebraska. Briefly stated, 200 ng of total RNA was reverse transcribed, processed using an Affymetrix 3′ IVT Express kit, and hybridized according to the manufacturer's suggested protocols for the Bovine Gene Chip assay (Affymetrix, Santa Clara, CA), which contains more than 23,000 genes. After overnight hybridization, arrays were stained and scanned with the Affymetrix 450 fluidics station and a 3000 7G high-resolution scanner. After quality control assessment was performed with respect to background and other standardized parameters, as suggested by Affymetrix, the .cel files were used for further analysis.
Microarray data analyses were conducted with BRB ArrayTools developed by Richard Simon and Amy Peng. Robust multiarray average RMA was used to do background correction, quantile normalization, and median polish summarization of probe-level data (31). A preliminary filter was applied retaining genes where at least 20% of the arrays had at least a 1.5-fold change from the median value for that gene. To determine differentially expressed genes over time, we compared control (0 h) and DEX 1.5-, 3-, 6-, and 24-h groups using a random variance F-test and then selected genes with a false discovery rate of <10%. Fold changes were calculated for each differentially expressed gene relative to the control group. Heat maps were created to display differentially expressed genes using Gene Cluster and TreeView (13, 16).
RT-PCR.
To confirm microarray data, reverse transcription-PCR (RT-PCR) was performed. One microgram of RNA was treated with amplification-grade DNase I (Invitrogen). RT was performed using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's directions. RNA was reverse transcribed using oligo(dT) primers (Invitrogen). Portions (100 ng) of the resulting cDNA were used as a template for PCR using specific primers for the cellular gene of interest. PCR was performed using GoTaq DNA polymerase (Promega) and initiated at 95°C for 5 min. This was followed by 30 cycles of 95°C for 45 s, annealing at the temperatures listed below for 45 s, and 72°C for 45 s. Final extension was performed at 72°C for 10 min. PCR products were analyzed on a 1.3% agarose gel. The following primer sequences and PCR conditions were used: GATA6, forward primer (5′-CACCAGTATCGCCCTT-3′) and reverse primer (CCTGTGGGTTAGTCACACTA), at an annealing temperature of 48°C; SPDEF, forward primer (5′-CAAGAAGGGCATCATCCG-3′) and reverse primer (5′-CCCTCTTTCCCAGCTCTG-3′), at an annealing temperature of 48°C; PLZF, forward primer (5′-TCTTTGGCATATGGGCTCAGTC-3′) and reverse primer (5′-AGGCTGACTTCTGTCTCCACA-3′), at an annealing temperature of 51°C; Slug, forward primer (5′-GACCCCAGAATGGAACAGC-3′) and reverse primer (5′-CAGGATTGCCTAACACACAGC-3′), at an annealing temperature of 51°C; KLF4, forward primer (5′-AAGACCAGAACCCCTTGAG-3′) and reverse primer (5′-GACTTACCAAGCACCATCG-3′), at an annealing temperature of 48°C; KLF6, forward primer (5′-GCCTTACAGATGCTCATGGG-3′) and reverse primer (5′-GTCTCTTCATGTGCAGGGC-3′), at an annealing temperature of 51°C; Pentraxin, forward primer (5′-AAGGAGAGAGTTGAGAT-3′) and reverse primer (5′-TTCTCCAGTCTCCCTT-3′), at an annealing temperature of 46°C; and GAPDH, forward primer (5′-CCATGGAGAAGGCTGGGG-3′) and reverse primer (5′-CAAAGTTGTCATGGATGACC-3′), at an annealing temperature of 55°C.
Detection of viral DNA in the TG of calves.
TG slices obtained from the TG pieces collected from infected calves were suspended in cold TNE buffer (50 mM Tris [pH 7.5], 100 mM NaCl, 10 mM EDTA) plus sodium dodecyl sulfate (SDS) 0.1% and then solubilized in a Polytron homogenizer. Samples were then treated with RNase (40 μg/ml) for 1 h at 37°C, followed by proteinase K (250 μg/ml) treatment for 3 h at 37°C and then at room temperature in the dark overnight. Total DNA was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), followed by chloroform only and precipitated by 100% ethanol at −80°C. DNA was suspended in TE buffer and used as a template for PCR with primers specific for the BHV-1 glycoprotein B (gB) gene (forward, 5′-GTGGTGGCCTTTGACCGCGAC-3′; reverse, 5′-GCTCCGGCGAGTAGCTGGTGT-3′). Amplified products were electrophoresed on a 1.5% agarose gel and stained with ethidium bromide.
Quantifying β-Gal-positive infected cells.
The gCblue virus grows to similar titers as wt BHV-1 and was grown in CRIB cells. Procedures for preparing genomic DNA were described previously (95). RS cells grown in six-well plates were cotransfected with 1 μg of the gCblue viral genome and the designated amounts of plasmid expressing bICP0 or the DEX-inducible cellular transcription factor of interest using Lipofectamine 2000 (catalog no. 11668-019; Invitrogen). At 24 h after transfection, the cells were fixed (2% formaldehyde and 0.2% glutaraldehyde in phosphate-buffered saline [PBS]), stained (1% Bluo-Gal, 5 mM potassium ferricyanide, 5 mM potassium ferricyanide, and 0.5 M MgCl2 in PBS), and the number of β-galactose-positive (β-Gal+) cells was determined as described previously (95). The number of β-Gal+ cells in cultures expressing the blank vector was set to 100%. To calculate percent plaque formation, the number of blue cells in cultures transfected with the cellular transcription factor were divided by the number of blue cells in cultures transfected with the blank vector. This representation of the data minimized the differences in cell density, Lipofectamine lot variation, and transfection efficiency. The results are an average of at least three independent experiments.
Small interfering RNA (siRNA) knockdown of cellular transcription factors was accomplished by transfection of human osteosarcoma cells (U2OS) with 100 nM specific siRNA or control RNA using Lipofectamine 2000 according to the manufacturer's specifications. The Block-iT-Fluorescent oligonucleotide was used as a control siRNA (catalog no. 44-2926; Invitrogen). It is a fluorescence-conjugated control containing a scrambled sequence that does not reduce the levels of any known mammalian gene. At 24 h after transfection of the siRNA, cells were transfected with 1 μg of gCblue viral genome. At 24 h after the transfection of viral genome, the cells were fixed, stained, and β-Gal+ cells counted as described above. The siRNAs were purchased from Santa Cruz Biotechnology (Santa Cruz) and were as follows: GATA6 (sc-37907), SPDEF (sc-45845), PLZF (sc-37149), Slug (sc-38393), KLF4 (sc-35480), and KLF6 (sc-38021).
Western blot analysis.
To evaluate the efficiency of siRNA knockdown, U2OS cells were cotransfected with 0.5 μg of a plasmid encoding a DEX-inducible transcription factor and 100 nM the respective siRNA or control siRNA as described above. At 36 h after transfection, whole-cell lysate was prepared. The cells were washed with phosphate-buffered saline (PBS) and suspended in NP-40 lysis buffer (100 mM Tris [pH 8.0], 1 mM EDTA, 100 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride) and one tablet of Complete protease inhibitor (Roche Molecular Biochemicals) in 10 ml of buffer. The cell lysate was incubated on ice for 30 min, sonicated, and then clarified by centrifugation at 10,000 × g at 4°C for 15 min. The protein concentrations were quantified by the Bradford assay. For SDS-PAGE, the proteins were mixed with an equal amount of 1× sample loading buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 50 mM dithiothreitol, 0.1% bromophenol blue, 10% glycerol) and boiled for 5 min. Proteins were separated in a SDS–12% PAGE gel. After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) and blocked for 4 h in 5% nonfat dry milk with Tris-buffered saline–0.1% Tween 20 (TBS-T). Membranes were then incubated with the designated primary antibody overnight at 4°C. The primary antibody was diluted 1:1,000 in the blocking solution. An antibody directed against β-actin (Santa Cruz Biotechnology, Santa Cruz, CA) was used as a loading control. After 45 min of washing with TBS-T, the blots were incubated with donkey anti-rabbit horseradish peroxidase-conjugated immunoglobulin G (Amersham Biosciences), which was diluted 1:2,000 in 5% nonfat milk in TBS-T. Blots were washed 45 min with TBS-T and exposed to Amersham ECL reagents, and then autoradiography was performed. Primary antibodies were purchased from Santa Cruz Biotechnology: GATA6 (sc-9055), SPDEF (sc-67022), PLZF (sc-22839), KLF4 (sc-20691), and KLF6 (sc-7158). The secondary donkey anti-rabbit antibody (NA9340V) was purchased from GE Healthcare. The KLF15 antibody (sc-271675) used for the present study is a mouse monoclonal antibody purchased from Santa Cruz Biotechnology. A secondary sheep anti-mouse antibody was purchased from GE Healthcare.
RT-PCR analysis of siRNA knockdown of Slug.
Since we were unable to detect Slug using the specific antibody, RT-PCR was performed to confirm that the Slug siRNA reduced Slug mRNA levels. U2OS cells were transfected with 100 nM Slug siRNA (sc-38393; Santa Cruz Biotechnology) or a fluorescent control siRNA (catalog no. 44-2926; Invitrogen) using Lipofectamine 2000 as described above. At 24 h after transfection, total RNA was prepared from cells using TRIzol reagent (Life Technologies) as previously described (95). One microgram of RNA was treated with amplification-grade DNase I (Invitrogen). RT was performed using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's directions. RNA was reverse transcribed using oligo(dT) primers (Invitrogen). A 100-ng aliquot of the resulting cDNA was used as a template for PCR using specific primers for Slug (listed above). PCR products were analyzed on a 1.3% agarose gel.
Plasmids and promoter activity measured in transfected cells.
The construction and characteristics of the BHV-1 bICP0 E promoter-CAT constructs (EP-172, EP-143, EP-133, EP-71, EP-50, and EP-42) used in the present study were described previously (95). The numbers in the plasmid name refer to the length of the bICP0 E promoter fragment cloned into the promoterless vector, pCAT-basic (Promega). Truncations to the promoter were made from the 5′ terminus. IETu1-CAT contains 1.5 kb of upstream sequences cloned at the 5′ terminus of pSV0CAT (a promoter minus CAT expression vector). V. Misra, Saskatoon, Canada, provided the IEtu1CAT plasmid (56). Two deletion constructs, Δ1024 and Δ1391 IEtu1, have 1,024- and 1,391-bp sequences removed from the 5′ termini, respectively. The gC promoter constructs (gC-CAT, gC-PstI-CAT, and gC-XhoI-CAT) were previously described (101). The empty vector pcDNA3.1 was purchased from Invitrogen.
Neuro-2A cells grown in 60-mm dishes were cotransfected with the designated plasmids as indicated in the respective figure legends using TransIT Neural according to the manufacturer's instructions. Measurement of chloramphenicol acetyltransferase (CAT) activity in Neuro-2A cells was performed as described previously (93, 95).
The GATA 6 expression vector was obtained from Paul Herring (Indiana University School of Medicine). The KLF4 expression vector was obtained from Jonathan Katz (University of Pennsylvania). The KLF6 expression vector was obtained from Bin Guo (North Dakota State University). The SPDEF expression vector was obtained from Origene (Rockville, MD). The KLF15 expression vector was obtained from Deborah Otteson (University of Houston). The PLZF expression vector was obtained from Derek Sant'Angelo (Sloan-Kettering Cancer Center). The Slug expression vector was obtained from Paul Wade (NIEHS, Research Triangle Park, NC).
Immunohistochemistry.
Immunohistochemistry was performed essentially as previously described (54, 55, 87, 88) using an ABC kit (Vector Laboratories). In brief, TG from calves used for the microarray studies were fixed in neutral buffered formalin and then embedded in paraffin. Thin sections (4 to 5 um) were cut and mounted onto slides. Tissue sections were incubated 20 min at 65°C, followed by two incubations of 10 min in xylene and rehydrated in graded alcohols. Tissue sections were then incubated with 3% hydrogen peroxide in PBS (pH 7.4) for 20 min at room temperature to block endogenous peroxidase. After three washes in TBS (5 min each) at room temperature, tissue sections were digested with proteinase K (catalog nos. 53020[Dako]) for 20 min at 37°C. Tissue sections were then blocked with 5% normal serum diluted in TBS containing 0.25% bovine serum albumin for 45 min at room temperature in a humidified chamber.
The designated rabbit polyclonal antibodies directed against cellular transcription factors or the mouse monoclonal antibody directed against KLF15 were used at a 1:1,000 dilution, incubated overnight in a humidified chamber at 4°C, and the next day washed in TBS (pH 7.6). Biotinylated goat anti-rabbit IgG (PK-6101; Vector Labs) or biotinylated donkey anti-mouse IgG (PK-6102; Vector Labs) was then incubated with the section for 30 min at room temperature in a humidified chamber. Next, the avidin-biotinylated enzyme complex was added to slides for 30 min at room temperature in a humidified chamber. After three washes in TBS, slides were incubated with freshly prepared substrate (SK-4800; Vector Labs), rinsed with distilled water, and counterstained with hematoxylin. Thin sections from mock-infected or latently infected calves were used as a negative control.
RESULTS
Identification of DEX-regulated genes in TG.
DEX reproducibly induces reactivation from latency, as judged by virus shedding from ocular or nasal cavities (1, 8, 24, 27, 37, 40, 68, 75, 87, 88). Six hours after DEX treatment viral transcripts are detected in a subset of TG neurons by in situ hybridization using a probe that hybridizes to ribonucleotide reductase, glycoprotein C, and ICP4 (68, 87, 89). The same probe does not readily detect viral gene expression in calves latently infected with BHV-1 (60 days after infection). Based on these observations, we predicted that viral gene expression is stimulated by DEX-inducible cellular factors expressed prior to 6 h after DEX treatment. Consequently, we examined cellular gene expression in TG at 90 min or 3, 6, or 24 h after DEX treatment using a commercially available bovine GeneChip array. These results were compared to those for cellular genes expressed in TG during latency.
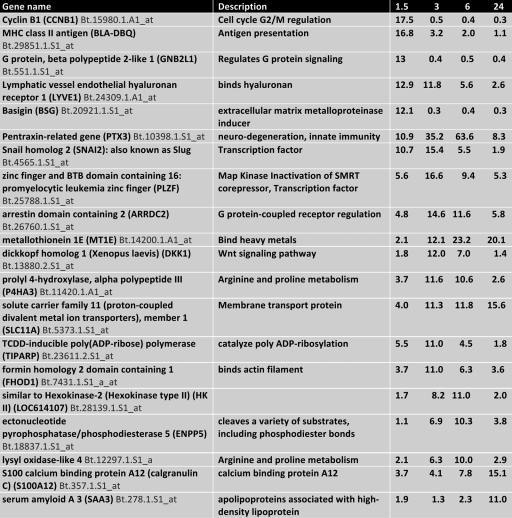
To identify cellular genes regulated by DEX that may promote reactivation from latency, calves latently infected with BHV-1 were given a single intravenous injection of DEX (100 mg). Seven genes were induced more than 10-fold within 90 min after DEX treatment, and by 3 h after DEX treatment four of these genes had levels that were less than that during latency (Table 1). At 3 or 6 h after DEX treatment, eleven or eight cellular genes were induced more than 10-fold. Only four genes were identified that had a 10-fold induction relative to latency at 24 h after DEX treatment.
Table 1.
Genes induced more than 10 fold after DEX treatmenta
Values are expressed as the fold induction at 1.5, 3, 6, and 24 h compared to TG of latently infected calves not infected with DEX.
Pentraxin-related gene three (PTX3) RNA levels were increased more than 35- and 60-fold at three and 6 h after DEX treatment, respectively (Table 1). PTX3 belongs to a family of genes that are involved with innate immune response, neurodegeneration, and PTX3 reduces mouse cytomegalovirus replication (5, 20). We predict that PTX3 may be detrimental to neuronal survival and/or may interfere with reactivation from latency in certain neurons. The RNA levels of two transcription factors, Snail homolog 2 (also referred to as Slug in mammals) and promyelocytic leukemia zinc finger (PLZF) were increased more than 15-fold at 3 h after DEX treatment. Slug and PLZF induction appear to fit the time frame for stimulating viral gene expression and/or productive infection during the early phases of reactivation from latency.
Additional transcription factors were induced more than 3-fold by 6 h after treatment with DEX (Table 2). The SPDEF (Sam-pointed domain containing Ets transcription factor) was induced 6-fold at 3 and 6 h after DEX treatment. SPDEF encodes a transcription factor belonging to the Ets (erythroblast transformation specific) family of transcription factors that regulate cell lineage specification, proliferation, differentiation, angiogenesis, and apoptosis (reviewed in references 23 and 74). The nuclear factor of kappa light polypeptide gene enhancer in B cells inhibitor, alpha (NFKB1A) encodes a 105-kDa protein that can undergo processing by the 26S proteasome to produce a 50-kDa protein. The 105-kDa protein is a Rel protein-specific transcriptional inhibitor, and the 50-kDa protein is a DNA binding subunit of the NF-κB protein complex (reviewed in reference 26).
Table 2.
Transcription factors induced more than 3-folda
Values are expressed as the fold induction at 1.5, 3, 6, and 24 h compared to TG of latently infected calves not treated with DEX.
Kruppel-like factor 15 (KLF15), which was induced 3.9-fold at 3 h after DEX treatment, belongs to a large family of transcription factors that can function as either repressors or activators. PLZF also belongs to the KLF family of transcription factors (reviewed in references 4 and 23). Expression of KLF4 and KLF6 was stimulated more than 10-fold in one calf, but not in the other calves. KLF family members may be important factors during reactivation from latency because (i) they generally bind GC-rich motifs resembling Sp1 binding sites or C-rich motifs (4), (ii) alphaherpesvirinae subfamily members are >70% GC-rich, and (iii) most viral promoters contain Sp1 binding sites. Zinc finger protein 36 homolog (ZFP36) is a protein encoded by the ZFP36 gene (14). The ZFP36 protein binds to AU-rich elements (AREs) in the 3′-untranslated regions (UTRs) of mRNAs of certain cytokines, including tumor necrosis factor alpha, and promotes their degradation (10). The transcription factor GATA6 (99) was induced 6-fold in one calf at 3 and 6 h after DEX treatment, but not in the other two calves at 3 or 6 h after DEX treatment.
The hairy and enhancer of split 6 (HES6) is a downstream transcription factor in the Notch signaling pathway, which regulates development, cell growth, and cell survival (7, 15). We recently demonstrated that a protein encoded by the LR gene (ORF2) interacts with Notch1 and Notch3 (93). Notch1 stimulated productive infection and certain viral promoters, whereas Notch3 did not stimulate productive infection and only activated the BHV-1 glycoprotein C (gC) promoter. Notch3 (93) and Notch4 (data not shown) RNA levels are also activated by DEX in TG. As judged by the Ingenuity program, the Notch signaling pathway was activated at 6 h after DEX treatment (data not shown), adding support to our conclusion that the Notch signaling pathway may regulate certain aspects of productive infection and/or reactivation from latency.
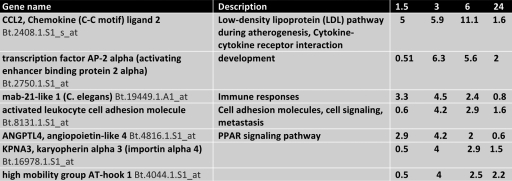
The mRNAs of cellular genes that were reduced at least 4-fold following DEX treatment are shown in Table 3. The only cellular gene that was reduced more than 10-fold was the chemokine CCL2 ligand 2. The transcription factor AP-2 alpha was also reduced more than 5-fold at 3 and 6 h after DEX treatment. In summary, these studies demonstrated that DEX had a rapid effect on cellular gene expression in TG of latently infected calves.
Table 3.
Genes repressed at least 4-fold by DEXa
Values are expressed as the fold induction at 1.5, 3, 6, and 24 h compared to TG of latently infected calves not treated with DEX.
Analysis of DEX-inducible cellular genes by RT-PCR.
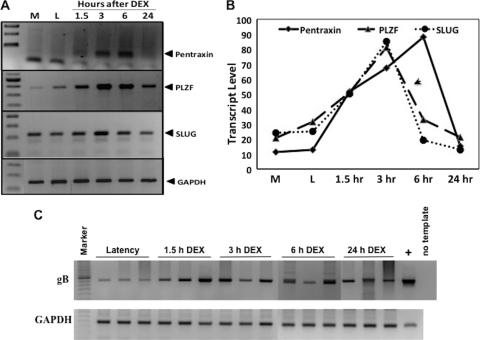
To confirm the changes in cellular gene expression identified by the microarray studies, RT-PCR was performed. The genes examined were PTX3, PLZF, and Slug, the three genes stimulated most by DEX. Bovine GAPDH was used as a loading control. PCR products were separated by gel electrophoresis (Fig. 1A), and the relative amounts of the respective PCR products measured by densitometry as described in Materials and Methods (Fig. 1B). The graph represents the average density of three TG samples from three calves at each time point. PTX3 RNA levels were readily detected at 3 or 6 h after DEX treatment, but not prior to DEX treatment. PLZF and SLUG RNA levels were highest at 3 h after DEX treatment, but the levels decreased as a function of time after DEX treatment. In summary, the RT-PCR results confirmed that PTX3, PLZF, and Slug RNA levels were higher in TG of calves latently infected with BHV-1 after DEX treatment.
Fig 1.
RT-PCR confirmation of DEX-inducible cellular genes. (A) RT-PCR using specific primers was performed to analyze expression of cellular genes identified by microarray analysis. Three latently infected calves were treated with DEX for 1.5, 3, 6, or 24 h. In addition, three mock (M)-infected calves as well as three latently infected calves (i.e., no DEX; L) were used as controls for baseline expression values. TG was collected at necropsy, and total RNA was isolated. RT-PCR was performed using specific primers as described in Materials and Methods. Products were separated by agarose gel electrophoresis and visualized using ethidium bromide. (B) Graphical representation of RT-PCR results. The amount of PCR products was quantified using a biomolecular imager (Bio-Rad), and density values were normalized to a GAPDH control. Values represent the average of TG from three calves at each time point. (C) Total DNA was prepared from the respective TG. Viral DNA was amplified from 1 μg of DNA using gB-specific primers. Genomic viral DNA extracted from BHV-1-infected CRIB cells served as a template for the positive control PCR (+). GAPDH amplification was used as a loading control. The details of the PCR are described in Materials and Methods.
PCR analysis was also performed to determine whether the respective samples contained similar levels of viral DNA. This was a concern because pieces of bovine TG were used for RNA preparation. To reduce the risk of choosing samples that did not contain similar levels of latently infected neurons, the TG were minced into small pieces at the time of necropsy. Thin slices were then obtained from the individual frozen TG pieces, and this was used to prepare total RNA or high-molecular-weight DNA. It was not possible to examine LR-RNA expression for these studies because it is well established that during DEX-induced reactivation from latency LR-RNA levels are reduced dramatically (32, 68). Using gB primers, we were able to readily detect viral DNA in all of the TG of latently infected calves (Fig. 1C). As expected, there was some variability in the levels of viral DNA detected in TG of the respective samples.
Analysis of DEX-induced cellular transcription factors by IHC.
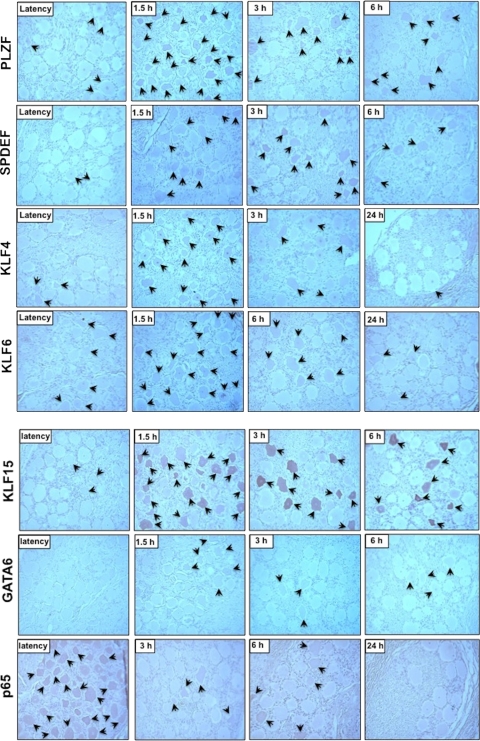
As another confirmation of the results obtained by the microarray analysis, immunohistochemistry (IHC) was performed to determine whether the cellular transcription factors were expressed in neurons, satellite cells, or infiltrating lymphocytes after DEX treatment. A PLZF-specific antibody recognized a subset of TG neurons at 1.5 h after DEX treatment. In general, we observed areas where many neurons were recognized by PLZF at 1.5 h after DEX (Fig. 2; arrows denote PLZF-positive neurons). Conversely, other areas of TG sections contained few PLZF positive neurons. At 3 or 6 h after DEX treatment, the number of PLZF-positive neurons was reduced. The PLZF antibody recognized few neurons in TG sections from latently infected calves (Fig. 2) or at 24 h after DEX treatment (data not shown). The commercially available Slug antibody we tested did not specifically recognize a protein in TG sections prepared from calves or when cells were transfected with the Slug expression vector.
Fig 2.
Immunohistochemical (IHC) analysis of DEX-inducible transcription factors. Thin sections were cut from formalin fixed and paraffin embedded TG at the designated times after calves latently infected with BHV-1 were treated with DEX. The sections designated as latency were at least 60 days after infection. IHC was performed as described in Materials and Methods with antiserum directed against the designated DEX-inducible transcription factor. Arrows denote neurons that were recognized by the respective antibody. Magnification, ×400.
The SPDEF specific antibody weakly recognized a few TG neurons in latently infected calves (Fig. 2) or uninfected calves (data not shown). At 1.5, 3, or 6 h after DEX treatment, there were more neurons recognized by SPDEF relative to TG from latently infected calves. At 3 h after DEX treatment, more neurons were recognized by the SPDEF antibody compared to TG from 6 h after DEX treatment. At 24 h after DEX treatment, the staining was similar to that in TG of latently infected calves (data not shown).
KLF4 and KLF6 antibodies yielded similar results. In general, there were areas of TG from latently infected calves that were recognized by the KLF4 or KLF6 antibodies (Fig. 2), and this was similar to mock-infected calves (data not shown). At 1.5 h after DEX treatment, there were areas in TG that contained high numbers of KLF4 or KLF6 positive neurons. After 1.5 h after DEX treatment, the number of neurons that were recognized by the respective antibodies decreased. By 24 h after DEX treatment, there were only a few neurons recognized by either KLF4 or KLF6 antibodies. The KLF15 specific antibody strongly reacted with neurons in certain areas of TG sections at 1.5, 3, or 6 h after DEX treatment (Fig. 2). Although KLF15-positive neurons were detected in TG of latently infected neurons, the number of neurons was fewer and the intensity of staining was low. At 24 h after DEX treatment, we observed a staining pattern similar to that observed during latency (data not shown).
It was difficult to identify areas of TG neurons during latency or after DEX treatment that were stained strongly by the GATA6 antibody. At 1.5, 3, or 6 h after DEX treatment, a few neurons were weakly stained by the GATA6 antibody. With the exception of the GATA6 antibody, antibodies directed against the DEX-inducible transcription factors recognized more neurons at 1.5 h after DEX treatment than the other time points examined. This may imply that DEX induction of these specific transcription factors occurred prior to 1.5 h of treatment.
DEX is an anti-inflammatory hormone, in part because it inactivates NF-κB signaling (reviewed in references 3 and 67). Consequently, we examined expression of p65, a component of the NF-κB specific transcription factor that is necessary for activating transcription, after DEX treatment. In contrast to the other cellular transcription factors examined, the number of neurons recognized by the p65 specific antibody was high in TG neurons of latently infected calves (Fig. 2). Although the p65 antibody did not stain all neurons, it was relatively easy to identify clusters of neurons recognized by the p65 antibody. In mock-infected TG, the results were similar to latently infected calves (data not shown). In contrast, the number of neurons recognized by the p65 antibody was much lower after DEX treatment.
DEX-inducible transcription factors stimulate productive infection.
The data presented above indicated that DEX has a rapid and direct effect on certain cellular transcription factors in TG neurons of latently infected calves. However, these data do not prove these factors promote reactivation from latency. Since we predict that only a subset of latently infected neurons actually produce infectious virus following DEX treatment, identifying these neurons is difficult with the technology available today. To determine whether these respective transcription factors play a role in stimulating reactivation from latency, we tested whether the respective DEX-inducible cellular transcription factors can stimulate productive infection.
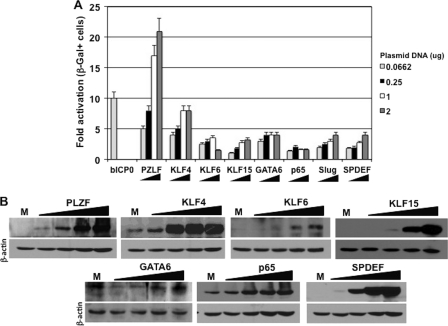
To test whether plasmids expressing a DEX-inducible cellular transcription factor stimulated productive infection in cultured cells, BHV-1 genomic DNA was cotransfected with a plasmid expressing one of the DEX-inducible cellular transcription factors, and the efficiency of productive infection was measured. The BHV-1 virus used for these studies is gCblue, which contains the lacZ gene downstream of the gC promoter. This allows one to measure productive infection by counting β-Gal+ cells. The gCblue virus grows to similar titers as wt BHV-1 in bovine cells, and the number of β-Gal+ cells directly correlates with plaque formation (21, 22, 30, 55). As expected, bICP0 increased the levels of β-Gal+ cells by ∼10-fold (Fig. 3A). PLZF increased the number of β-Gal+ cells 20-fold relative to the empty expression vector (Fig. 3A). To achieve this level of activation, the amount of the PLZF plasmid had to be higher than bICP0. KLF4 increased the number of β-Gal+ cells 9-fold. GATA6, KLF6, KLF15, SLUG, and SPDEF increased the number of β-Gal+ cells ∼4-fold relative to an empty expression vector. The plasmid expressing p65 increased the number of β-Gal+ cells ∼2-fold. Additional studies were performed to test whether the cellular transcription factors had a synergistic effect on productive infection. Although certain combinations had an additive effect, there was no evidence of synergism (data not shown). When RS cells were transfected with increasing amounts of a plasmid encoding a DEX-inducible transcription factor, higher levels of the protein were detected (Fig. 3B). The commercially available Slug antibody we used did not specifically recognize a protein. In summary, these studies indicated that the DEX-inducible transcription factors stimulated productive infection in RS cells.
Fig 3.
DEX-inducible transcription factors stimulate productive infection. (A) Rabbit skin (RS) cells were cotransfected with 62 ng of a plasmid encoding bICP0 or increasing concentrations of the respective plasmids (62 ng, 250 ng, 1 μg, or 2 μg) encoding DEX-inducible transcription factors, along with 1 μg of gCblue BHV-1 DNA, which contains the lacZ gene inserted into the gC locus of BHV-1. At 24 h after infection, cells were fixed, stained, and β-Gal+ cells were counted. The results are the average of three independent experiments. (B) As described for panel A, RS cells were cotransfected with increasing concentrations of plasmids that express the designated DEX-inducible transcription factor (0.0662, 0.25, 1, or 2 μg of DNA). To maintain equivalent amounts of plasmid in the transfection mixture, an empty expression plasmid (pcDNA3.1) was included. At 40 h after transfection, cell lysate was prepared and Western blotting was performed as described in Materials and Methods. For each lane, 100 μg of protein was loaded. The lane M denotes mock-transfected cells to allow determination of the amount of the designated DEX-inducible transcription factor that was in normal cells. β-Actin was used as a loading control.
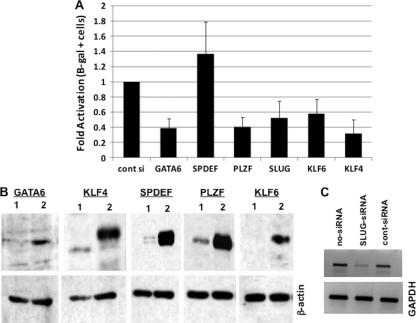
Additional studies were performed to examine the effect of a siRNA specific for certain DEX-inducible transcription factors on BHV-1 productive infection. These studies were performed in a human osteosarcoma (U2OS) cell line that is permissive for BHV-1 infection (69) because commercially available siRNAs directed against the bovine transcription factors are not available. U2OS cells were transfected with 100 nM concentrations of the respective siRNAs or a scrambled control siRNA 24 h prior to transfection with the gCblue viral genome. At 24 h after transfection with the viral genome, the cells were fixed, stained, and β-Gal+ cells counted. With the exception of SPDEF, silencing of a DEX-inducible cellular transcription factor reduced the number of β-Gal+ cells ∼2-fold (Fig. 4A). In RS cells, overexpression of SPDEF enhanced productive infection, whereas silencing SPDEF in U2OS cells increased productive infection. One explanation for this difference may be that SPDEF has cell type-specific effects on productive infection or in the highly transformed human tumor cell line (U2OS) SPDEF was not important for productive infection. Western blot analysis confirmed that the siRNA specifically reduced the respective transcription factor levels in U2OS cells (Fig. 4B). Since the SLUG antibody did not recognize a specific band, RT-PCR was performed to confirm that endogenous SLUG mRNA levels were reduced (Fig. 4C).
Fig 4.
siRNA directed against the DEX-inducible genes reduces the efficiency of productive infection. (A) Human osteosarcoma cells (U2OS) were transfected with 100 nM siRNA targeting the designated cellular transcription factor or a control siRNA, which does not reduce the expression of any known mammalian gene. After 24 h, the cells were transfected with gCblue viral genome and efficiency of productive infection was measured as described in Materials and Methods. The results are the average of three independent experiments. (B) To confirm that the siRNAs reduced the levels of the expected proteins, U2OS cells were cotransfected with 100 nM siRNA and 0.5 μg of the designated plasmid expressing the DEX-inducible transcription factor. At 24 h after transfection, the cells were collected and processed for Western blot analysis. β-Actin was assayed as a loading control. Lane 1 was transfected with the control siRNA, and lane 2 was transfected with the gene-specific siRNA. (C) The Slug antibody used for the present study did not recognize a specific protein in U2OS cells. Therefore, U2OS cells were transfected with 100 nM Slug siRNA, control siRNA, or no siRNA, and RT-PCR was performed as described in Materials and Methods. PCR products were run on a 1.3% agarose gel and visualized with ethidium bromide.
DEX-induced cellular transcription factors activate certain viral promoters.
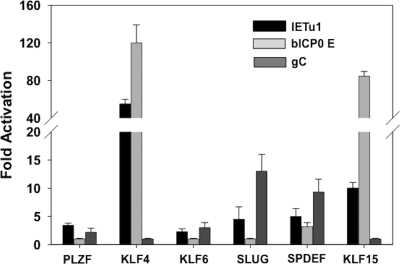
Additional studies were performed to test whether the DEX-inducible cellular transcription factors transactivated certain BHV-1 promoters. For these studies, we examined the immediate-early transcription unit 1 promoter (IEtu1), bICP0 early (E) promoter, or the gC late (L) promoter. The IEtu1 promoter activates expression of bICP0 and bICP4, the major transcriptional regulatory proteins encoded by BHV-1 (90, 92). The bICP0 E promoter also activates bICP0 expression, and previous studies indicated that it is preferentially stimulated during reactivation from latency (94). Neuro-2A cells were cotransfected with the designated promoter construct and DEX-inducible gene, and the CAT activity was measured at 48 h after transfection. Neuro-2A cells were used for these studies because they are neuron-like cells and are readily transfected. PLZF, which activated productive infection 21-fold, was unable to strongly transactivate any of the three viral promoters examined (Fig. 5). KLF4 activated the IEtu1 and bICP0 E promoter more than 50- and 100-fold, respectively, but not the gC promoter. KLF15 transactivated the bICP0 E promoter ∼80-fold and the IEtu1 promoter ∼8-fold, but it had no effect on the gC promoter. KLF6 did not have a dramatic effect on any of the three promoters examined. Slug activated the IEtu1 promoter 5-fold and the gC promoter 13-fold but had no effect on the bICP0 E promoter. SPDEF activated the gC promoter ∼7-fold but activated the IEtu1 or bICP0 E promoter <5-fold. GATA6 stimulated the promoter minus control; thus, reliable data were not obtained for the three BHV-1 promoters. In summary, these studies suggested that DEX-inducible transcription factors enhanced productive infection by directly and/or indirectly regulating viral promoter activity.
Fig 5.
DEX-inducible cellular transcription factors stimulate specific viral promoters. Mouse neuroblastoma cells (Neuro-2A) were cotransfected with 1 μg of a CAT reporter plasmid with the IETu1 promoter (IETu1Δ1024), the bICP0 E promoter (EP-638), or the gC L promoter and 1 μg of pcDNA3.1 empty vector or 1 μg of a plasmid expressing the designated DEX-inducible transcription factor. At 48 h after transfection, the CAT reporter activity was measured. The numbers represent the fold induction divided by the empty vector control. The results are the average of three independent experiments.
Localization of KLF4 and KLF15 responsive sequences in the IEtu1 promoter.
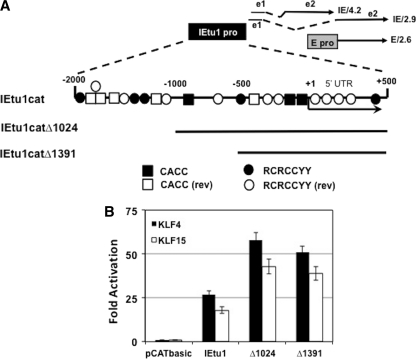
The results presented in Fig. 5 demonstrated that KLF4 and KLF15 activated the IETu1 and bICP0 E promoter more than 50- and 8-fold, respectively, in transient-transfection assays. The KLF4 consensus binding sites are CACC or RCRCCYY (2, 12). KLF15 consensus binding sites are similar to KLF4 (59). Three consensus CACC and five RCRCCYY sites were identified in the IEtu1 promoter sequences (Fig. 6A). Five reverse CACC motifs and nine reverse RCRCCYY motifs were detected in the IEtu1 promoter.
Fig 6.
Localization of KLF4 and KLF15 responsive sequences in the IETu1. (A) Positions of transcripts that encode bICP4 and bICP0. The immediate-early (IE) transcription unit 1 (IEtu1) encodes bICP4 (IE/4.2) and bICP0 (IE/2.9) (91, 92). The IEtu1 promoter (denoted by the black rectangle) activates IE expression of IE/4.2 and IE/2.9. E/2.6 is the early transcript that encodes bICP0, and an early promoter (denoted by the gray rectangle) activates expression of the early bICP0 transcript (E/2.6) (90). Exon 2 (e2) of bICP0 contains all of the protein coding sequences of bICP0. The dashed lines are intron sequences. KLF4 consensus sequences were located in the IETu1 promoter using the free online program fuzznuc (http://emboss.open-bio.org/wiki/Appdocs). The KLF4 consensus binding sites are CACC or RCRCCYY. The location of consensus KLF4 sequences in the IETu1 promoter and 5′UTR included in CAT constructs is shown. The reverse (rev) sequences of CACCC and RCRCCYY are also shown. (B) Neuro-2A cells were cotransfected with the designated IETu1 promoter-CAT reporter plasmid (1 μg) and 1 μg of pcDNA3.1 empty vector or 1 μg of KLF4 or KLF15. At 48 h after transfection, the CAT activity was measured. The numbers represent the fold induction over the empty vector control. The results are the averages of three independent experiments.
To localize sequences in the IEtu1 promoter responsive to KLF4 and KLF15, two deletion mutants were used (Fig. 6A). The basal promoter activity of IEtu1catΔ1024 is ∼4-fold less than IEtu1cat and IEtu1 catΔ1391 is 25-fold less than IEtu1cat (93). Neuro-2A cells were cotransfected with the designated promoter construct, along with KLF4 or KLF15, and CAT reporter activity was measured at 48 h after transfection. KLF4 activated the full-length IEtu1 25-fold and IEtu1Δ1024 and IEtu1Δ1391 ∼50-fold (Fig. 6B). Although KLF15 transactivated the IEtu1 promoters less efficiently than KLF4, both transcription factors exhibited a similar trend for transactivating the three IEtu1 promoter constructs. IEtu1Δ1391 contains two CACC motifs, two RCRCCYY motifs, one reverse CACC motif, and six reverse RCRCCYY motifs, suggesting these motifs at the distal region of the IEtu1 promoter were not necessary for transactivation by KLF4 or KLF15.
Localization of KLF4 and KLF15 responsive sequences in the bICP0 E promoter.
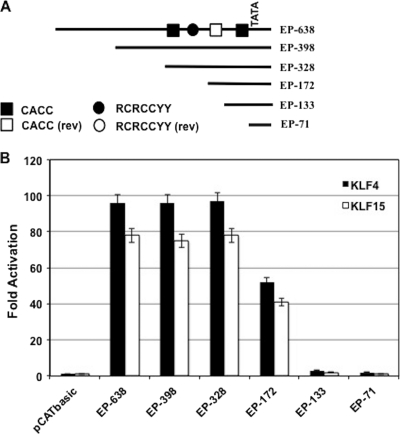
To localize bICP0 E promoter sequences necessary for KLF4- or KLF15-mediated transactivation, six deletion mutants previously described (94, 95) were examined (Fig. 7A). The basal promoter activity of EP-638, EP-398, EP-328, and EP-172 does not vary more than 2-fold (94, 95). Relative to EP-638, EP-133, and EP-71 have ∼3-fold reduced basal promoter activity. EP-638, EP-398, and EP-328 promoter activity were stimulated ∼100-fold by the KLF4 expression plasmid and ∼75-fold by the KLF15 expression plasmid (Fig. 7B). EP-172 was activated ∼50-fold by KLF4 and >40-fold by KLF15. In contrast, EP-133 and EP-71 promoter constructs were activated <5-fold by KLF4 and KLF15 (Fig. 7B). Two consensus KLF4/KLF15 binding sites (CACCC) in the forward orientation plus one in the reverse orientation were identified in EP-328. In addition, there was one RCRCCYY motif in the forward orientation. The reduction of KLF4 and KLF15 responsiveness between EP-328 and EP-172 correlated with the loss of one CACCC consensus in the forward orientation plus a RCRCCYY motif. From EP-172 to EP-133, there was a dramatic reduction in promoter responsiveness to KLF4 and KLF15, which correlated with the loss of 1 CACCC consensus in the reverse orientation. EP-133 retained one CACCC binding site: however, this single motif does not appear to be sufficient for KLF4 or KLF15 responsiveness. EP-71 contained no binding sites, which correlated with its inability to be stimulated by KLF4 or KLF15.
Fig 7.
Localization of KLF4 and KLF15 responsive sequences in the bICP0 E promoter. (A) Schematic of bICP0 E promoter constructs. KLF4 and KLF15 consensus sequences were located in the bICP0 E promoter using the free online program fuzznuc (http://emboss.open-bio.org/wiki/Appdocs). These consensus-binding sites (CACC or RCRCCYY) are shown in the bICP0 E promoter. The reverse (rev) sequences of CACCC and RCRCCYY are also shown. (B) Mouse neuroblastoma cells (neuro-2A) were cotransfected with the designated bICP0 E promoter-CAT reporter plasmid (1 μg) and 1 μg of pcDNA3.1 empty vector, 1 μg of KLF4, or 1 μg of KLF15. At 48 h after transfection, the CAT activity was measured. The numbers represent the fold induction over the empty vector control. The results are the averages of three independent experiments.
DISCUSSION
In this study, the effect of DEX on cellular gene expression in TG of calves latently infected with BHV-1 was examined. As discussed above, DEX consistently induces BHV-1 reactivation from cattle (33, 34, 38, 39, 84). DEX is a synthetic corticosteroid that mimics the effects of stress: consequently, DEX is a biologically relevant stimulus to examine the steps that occur during BHV-1 reactivation from latency. Apart from the BHV-1 LR gene, other viral transcripts are not abundantly transcribed during latency (reviewed in references 33, 34, 38, and 64). Since we have no evidence that LR gene products stimulate viral transcription and LR-RNA levels are reduced by DEX (68), cellular transcription factors induced by DEX appear to be important for initiating lytic cycle viral gene expression during the early phase of reactivation from latency.
The cellular transcription factor, PLZF, was strongly induced by DEX within 3 h after DEX treatment and stimulated productive infection by 20-fold, suggesting that it promotes reactivation from latency. PLZF is also activated by glucocorticoid and progesterone in human endometrial stromal cells and myometrial smooth muscle cells (17). Although several studies concluded that PLZF represses transcription (19, 51, 53), phosphorylated PLZF activates expression of interferon-stimulated genes (96). The inability of PLZF to activate any of the BHV-1 promoters tested suggested that PLZF (i) was not phosphorylated by the proper kinase in transiently transfected cells, (ii) does not directly activate viral transcription, (iii) activates a promoter that was not tested, (iv) has neuron-specific transcriptional activity, and/or (v) “represses a repressor” that is expressed during latency. PLZF is a member of the KLF family of transcription factors (4). Several other KLF family members (KLF4, KLF6, and KLF15) were also activated by DEX in TG, suggesting that certain KLF transcription factors stimulate lytic viral gene expression during reactivation from latency.
KLF4 and KLF15 transactivated both the bICP0 E promoter and IEtu1 promoter: thus, it was not surprising to find KLF4 and/or KLF15 consensus binding sites in both promoters. KLF4 interacts with CACCC sequences (2, 12) and GAGGTCC or GGGTGT motifs (57), which are necessary for Oct3/4 and KLF4 to cooperate with Sox2 to activate Lefty1 (left-right determination factor 1) expression. KLF4 also interacts with the NF-κB family member p65 (RelA) to activate the iNOS (inducible nitric oxide synthase) promoter (18), suggesting that interactions between KLF4 and p65 are important for activating lytic cycle viral gene expression. KLF4 is stimulated by heat stress (49) and, under certain circumstances, promotes apoptosis (100). Both heat stress and apoptosis can increase the frequency of HSV-1 reactivation from latency in vivo (72) or in cultured neurons (28). KLF4 also stimulates the noncanonical Notch signaling pathway (71). KLF15 encodes a protein that interacts with a CG/TCCCC motif, G-rich motifs, or a CACCC site (59), and its expression is stimulated by glucocorticoids (52, 80). KLF15 interacts with the Sp1 transcription factor (97) and, like most KLF factors, activates certain promoters while repressing others. These studies provide evidence that KLF4 and KLF15 have the potential to stimulate lytic cycle viral gene expression by several distinct mechanisms.
The finding that Slug and SPDEF transactivated the gC promoter more efficiently than the bICP0 E or IEtu1 promoter suggested that the normal cascade of viral gene expression was altered during the early phases of latency. Slug was the only transcription factor induced by >10-fold at 90 min after DEX treatment, implying that transactivation of the gC promoter was an early event during reactivation from latency. Several independent studies concluded that the normal cascade of viral gene expression during productive infection is different during reactivation from latency. For example, the bICP0 E promoter, but not the bICP0 IE promoter (IEtu1), is consistently stimulated during reactivation from latency (94). HSV-1 E gene expression and DNA replication is proposed to occur prior to IE gene expression during reactivation from latency (43, 58, 65, 81). Finally, expression of a late HSV-1 gene (VP16), which activates expression of IE genes (70), was proposed to stimulate reactivation from latency (83). Slug family members, including Snail1 and Snail2, also reduce expression of at least two proapoptotic proteins (Bid and p53) and consequently inhibit apoptosis (44, 45). Perhaps increased levels of Slug promote successful reactivation from latency by enhancing the survival of latently infected neurons after DEX treatment.
None of the DEX-inducible transcription factors that we identified stimulated a BHV-1 VP16 promoter CAT construct by >2-fold (data not shown). However, when the same BHV-1 VP16 promoter CAT construct was transfected into bovine cells and then infected with BHV-1, CAT activity was stimulated >10-fold. We had anticipated that the BHV-1 VP16 promoter would be activated by DEX-inducible transcription factors because a recent report concluded that expression of the HSV-1 VP16 promotes the exit from latency (83). In general, our results suggested that the BHV-1 VP16 promoter was not transactivated during the early phases of reactivation from latency. It is also possible that cellular transcription factors stimulated by DEX can transactivate the BHV-1 VP16 promoter during the early stages of reactivation from latency, but these are only expressed in a minor population of latently infected neurons. If this scenario were true, these transcription factors would be difficult to identify by microarray studies. It is also possible that BHV-1 VP16, unlike HSV-1 VP16, is not required for inducing reactivation from latency because bICP0 is the trigger for BHV-1 reactivation from latency. Evidence supporting a prominent role for bICP0 during BHV-1 reactivation from latency comes from the finding that the bICP0 gene contains two promoters: an early promoter and an immediate-early promoter (IEtu1) (90, 91). The bICP0 early promoter, but not IEtu1, is consistently stimulated by DEX-induced reactivation from latency (94).
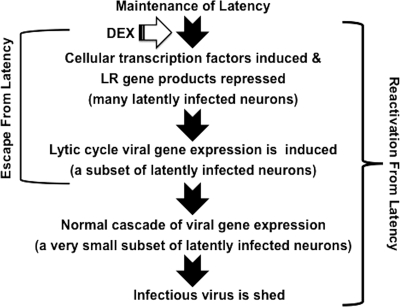
Our results provided evidence that several distinct steps occur when the transition from maintenance of latency is disrupted by DEX and reactivation from latency occurs (see Fig. 8 for a schematic of these putative steps). During the maintenance of latency, products encoded by the LR gene interfere with productive infection (6, 22, 32, 93), enhance neuronal survival (11, 50, 76), and stabilize normal neuronal functions (60). Stressful stimuli, in this case DEX, have several important effects on sensory neurons within TG. The early events that occur in TG after DEX treatment are operationally defined as the escape from latency. Important events that occur during the escape from latency include: reduced levels of LR gene products (68), apoptosis of infiltrating lymphocytes in TG (87), and induction of cellular gene expression. Lytic cycle viral gene expression can be detected by in situ hybridization within 6 h after DEX treatment (87). That study also indicated that only a small subset of neurons in TG expressed lytic cycle viral genes, which agrees with an independent study by Rock et al. (68). Activation of any viral gene during the escape from latency would seem to favor extensive viral transcription. Many latently infected neurons that escape latency are likely to reestablish or return to latency (68) and thus would not produce infectious virus.
Fig 8.
Working model for steps involved in the reactivation from latency. For details, see the Discussion.
Increased expression of BHV-1 regulatory proteins—bICP0, bICP4, or perhaps VP16—during the escape from latency may lead to the normal cascade of viral gene expression in certain latently infected neurons. Consequently, all viral proteins would be expressed, and infectious virus would be produced. Interestingly, the LR mutant virus, which does not reactivate from latency, consistently expresses bICP0 transcripts in TG after DEX treatment (94), suggesting that merely activating bICP0 expression is not sufficient for reactivation from latency. Conversely, gC or VP16 transcripts (both late genes) were not detected when calves latently infected with the LR mutant virus were treated with DEX. There are several subtypes of murine sensory neurons in TG (98), suggesting that certain subtypes of latently infected neurons possess the necessary factors to support production of infectious virus. None of the transcription factors identified in the present study were induced when murine TG are explanted to induce reactivation from latency (85), suggesting that stimulus-specific events mediate reactivation from latency or explant-induced reactivation from latency does not reflect the events that occur during DEX-induced BHV-1 reactivation from latency in vivo.
ACKNOWLEDGMENTS
This research was supported by a grant from the U.S. Department of Agriculture (09-01653). A grant to the Nebraska Center for Virology (1P20RR15635) also supported certain aspects of these studies. A.W. and D.S. were partially supported by a fellowship from a Ruth L. Kirschstein National Research Service Award 1 T32 AIO60547 (National Institute of Allergy and Infectious Diseases). The UNMC Microarray Core Facility receives support from the Nebraska Research Network in Functional Genomics NE-INBRE P20 RR16469, the Molecular Biology of Neurosensory Systems CoBRE 2P20RR018788-06, and the Nebraska Research Initiative.
Footnotes
Published ahead of print 21 December 2011
REFERENCES
- 1. Ackermann M, Peterhans E, Wyler R. 1982. DNA of bovine herpesvirus type 1 in the trigeminal ganglia of latently infected calves. Am. J. Vet. Res. 43:36–40 [PubMed] [Google Scholar]
- 2. Adam PJ, Reagan CP, Hautmann MB, Owens GK. 2000. Positive- and negative-acting Kruppel-like transcription factors bind a transforming growth factor beta control element required for expression of the smooth muscle cell differentiation marker SM22-alpha in vivo. J. Biol. Chem. 275:37798–37806 [DOI] [PubMed] [Google Scholar]
- 3. Barnes PJ. 1998. Anti-inflammatory actions of glucocorticoids: molecular mechanisms. Clin. Sci. 94:557–572 [DOI] [PubMed] [Google Scholar]
- 4. Bieker JJ. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355–34358 [DOI] [PubMed] [Google Scholar]
- 5. Bozza S, et al. 2006. Pentraxin 3 protects from MCMV infection and reactivation through TLR sensing pathways leading to IRF3 activation. Blood 108:3387–3396 [DOI] [PubMed] [Google Scholar]
- 6. Bratanich AC, Hanson ND, Jones CJ. 1992. The latency-related gene of bovine herpesvirus 1 inhibits the activity of immediate-early transcription unit 1. Virology 191:988–991 [DOI] [PubMed] [Google Scholar]
- 7. Bray SJ. 2006. Notch signaling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678–689 [DOI] [PubMed] [Google Scholar]
- 8. Brown GA, Field HJ. 1990. Experimental reactivation of bovine herpesvirus 1 (BHV-1) by means of corticosteroids in an intranasal rabbit model. Arch. Virol. 112:81–101 [DOI] [PubMed] [Google Scholar]
- 9. Cantin EM, Hinton DR, Chen J, Openshaw H. 1995. Gamma interferon expression during acute and latent nervous system infection by herpes simplex virus type 1. J. Virol. 69:4898–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carballo E WSL, Blackshear PJ. 1988. Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281:1001–1005 [DOI] [PubMed] [Google Scholar]
- 11. Ciacci-Zanella J, Stone M, Henderson G, Jones C. 1999. The latency-related gene of bovine herpesvirus 1 inhibits programmed cell death. J. Virol. 73:9734–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dang DT, Zhao W, Mahatan CS, Geiman DE, Yang VW. 2002. Opposing effects of Kruppel-like factor 4 (gut-enriched Kruppel-like factor) and Kruppel-like factor 5 (intestinal-enriched Kruppel-like factor) on the promoter of the Kruppel-like factor 4 gene. Nucleic Acids Res. 30:2736–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Hoon M, Imoto S, Miyano S. 2002. Cluster 3.0. University of Tokyo, Human Genome Center, Tokyo, Japan [Google Scholar]
- 14. DuBois RN, Ryder MWMK, Lau LF, Nathans DD. 2000. A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. J. Biol. Chem. 265:19185–19191 [PubMed] [Google Scholar]
- 15. Ehebauer M, Penelope P, Arias AM. 2006. Notch, a universal arbiter of cell fate decisions. Science 314:1414–1415 [DOI] [PubMed] [Google Scholar]
- 16. Eisen M. 2002. Gene cluster 3.0. Stanford University, Stanford, CA [Google Scholar]
- 17. Fahenenstich J, et al. 2003. Promyelocytic leukaemia zinc finger protein (PLZF) is a glucocorticoid- and progesterone-induced transcription factor in human endometrial stromal cells and myometrial smooth muscle cells. Mol. Hum. Reprod. 9:611–623 [DOI] [PubMed] [Google Scholar]
- 18. Feinberg MW, et al. 2005. Kruppel-like factor 4 is a mediator of proinflammatory signaling in macrophages. J. Biol. Chem. 280:38247–38268 [DOI] [PubMed] [Google Scholar]
- 19. Fillipponi D, et al. 2007. Repression of kit expression by Plzf in germ cells. Mol. Cell Biol. 27:6770–6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garlanda C, Bottazzi B, Bastone A, Mantovani A. 2005. Pentraxins at the crossroads between innate immunity, inflammation, matrix deposition, and female fertility. Annu. Rev. Immunol. 23:337–366 [DOI] [PubMed] [Google Scholar]
- 21. Geiser V, Jones C. 2003. Stimulation of bovine herpesvirus 1 productive infection by the adenovirus E1A gene and a cell cycle regulatory gene, E2F-4. J. Gen. Virol. 84:929–938 [DOI] [PubMed] [Google Scholar]
- 22. Geiser V, Inman M, Zhang Y, Jones C. 2002. The latency related (LR) gene of bovine herpesvirus 1 (BHV-1) can inhibit the ability of bICP0 to activate productive infection. J. Gen. Virol. 83:2965–2971 [DOI] [PubMed] [Google Scholar]
- 23. Gutierrez-Hartmann A, Duval DL, Bradford AP. 2007. ETS transcription factors in endocrine systems. Trends Endocrinol. Metab. 18:150–158 [DOI] [PubMed] [Google Scholar]
- 24. Hage JJ, et al. 1998. Reactivation of latent bovine herpesvirus 1 in cattle seronegative to glycoproteins gB and gE. Vet. Microbiol. 60:87–98 [DOI] [PubMed] [Google Scholar]
- 25. Halford WP, Gebhardt BM, Carr DJ. 1996. Persistent cytokine expression in trigeminal ganglion latently infected with herpes simplex virus type 1. J. Immunol. 157:3542–3549 [PubMed] [Google Scholar]
- 26. Hayden MS, Ghosh S. 2008. Shared principles in NF-κB signaling. Cell 132:344–362 [DOI] [PubMed] [Google Scholar]
- 27. Homan EJ, Easterday BC. 1983. Experimental latent and recrudescent bovine herpesvirus-1 infections in calves. Am. J. Vet. Res. 44:309–313 [PubMed] [Google Scholar]
- 28. Hunsperger EA, Wilcox CL. 2003. Caspase-3-dependent reactivation of latent herpes simplex virus type 1 in sensory neuronal cultures. J. Neurovirol. 9:390–398 [DOI] [PubMed] [Google Scholar]
- 29. Inman M, Lovato L, Doster A, Jones C. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 interferes with the latency reactivation cycle of latency in calves. J. Virol. 76:6771–6779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Inman M, Lovato L, Doster A, Jones C. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 75:8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Irizarry RA, Gautier LBM, Cope L. 2002. An r package for analyses of Affymetrix oligonucleotide arrays, p 102–119 In Parmigiani ESGG, Irizarry RA, Zeger SL. (ed), Analysis of gene expression of gene expression data: methods and software. Springer, New York, NY [Google Scholar]
- 32. Jaber T, Workman A, Jones C. 2010. Small noncoding RNAs encoded within the bovine herpesvirus 1 latency-related gene can reduce steady-state levels of infected cell protein 0 (bICP0). J. Virol. 84:6297–6307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81–133 [DOI] [PubMed] [Google Scholar]
- 34. Jones C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 16:79–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jones C. 2009. Regulation of innate immune responses by bovine herpesvirus 1 and infected cell protein 0. Viruses 1:255–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jones C, Delhon G, Bratanich A, Kutish G, Rock D. 1990. Analysis of the transcriptional promoter which regulates the latency-related transcript of bovine herpesvirus 1. J. Virol. 64:1164–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jones C, et al. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 18:3185–3195 [DOI] [PubMed] [Google Scholar]
- 38. Jones C, et al. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113:199–210 [DOI] [PubMed] [Google Scholar]
- 39. Jones C, Chowdhury S. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex, and development of improved vaccines. Adv. Anim. Health 8:187–205 [DOI] [PubMed] [Google Scholar]
- 40. Kaashoek MJ, Rijsewijk FA, Oirschot JT. 1996. Persistence of antibodies against bovine herpesvirus 1 and virus reactivation two to three years after infection. Vet. Microbiol. 53:103–110 [DOI] [PubMed] [Google Scholar]
- 41. Khanna KM, Bonneau RH, Kinchington PR, Hendricks RL. 2003. Herpes simplex virus-specific memory CD*+ T cells are selectively activated and retained in latently infected sensory ganglia. Immunity 18:593–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Knickelbein JE, et al. 2008. Noncytotoxic lytic granule-mediated CD8+ T cell inhibition of HSV-1 reactivation from neuronal latency. Science 322:268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. KosZ-Vnenchak JJ, Coen DM, Knipe DM. 1993. Evidence for a novel regulatory pathway for herpes simplex virus gene expression in trigeminal ganglion neurons. J. Virol. 67:5383–5393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee K, Gjorevski N, Boghaert E, Radisky DC, Nelson CM. 2011. Snail1, Snail2, and E46 promote mammary epithelial branching morphogenesis. EMBO J. 30:2662–2674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leroy P, Mostov KE. 2007. Slug is required for cell survival during partial epithelial-mesenchymal transition of HGF-induced tubulogenesis. Mol. Biol. Cell 18:1943–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liu T, Khanna KM, Carriere BN, Hendricks RL. 2001. Gamma interferon can prevent herpes simplex virus type 1 reactivation from latency in sensory neurons. J. Virol. 75:11178–11184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu T, Khanna KM, Chen X, Fink DJ, Hendricks RL. 2000. CD8+ T cells can block herpes simplex virus type 1 (HSV-1) reactivation from latency in sensory neurons. J. Exp. Med. 191:1459–1466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Liu T, Tang Q, Hendricks RL. 1996. Inflammatory infiltration of the trigeminal ganglion after herpes simplex virus type 1 corneal infection. J. Virol. 70:264–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Liu Y, et al. 2006. Induction of KLF4 in response to heat stress. Cell Stress Chaperones 11:379–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lovato L, Inman M, Henderson G, Doster A, Jones C. 2003. Infection of cattle with a bovine herpesvirus 1 (BHV-1) strain that contains a mutation in the latency related gene leads to increased apoptosis in trigeminal ganglia during the transition from acute infection to latency. J. Virol. 77:4848–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin JP, Delmotte M-H, Formstecher P, Lefebvre P. 2003. PLZF is a negative regulator of retinoic acid receptor transcriptional activity. Nuclear Recept. 1:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Masuno K, et al. 2011. Expression profiling identifies Klf15 as a glucocorticoid target that regulates airway hyperresponsiveness. Am. J. Respir. Cell Mol. Biol. 45:642–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. McConnell MJ, et al. 2003. Growth suppression by acute promyelocytic leukemia-associated protein PLZF is mediated by repression of c-myc expression. Mol. Cell Biol. 23:9375–9388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Meyer F, et al. 2007. Identification of a novel protein encoded by the latency-related gene of bovine herpesvirus 1. J. Neurovirol. 13:569–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Meyer F, et al. 2007. A protein encoded by the bovine herpesvirus 1 (BHV-1) latency related gene interacts with specific cellular regulatory proteins, including the CCAAT enhancer binding protein alpha (C/EBP-α). J. Virol. 81:59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Misra V, Bratanich AC, Carpenter D, O'Hare P. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV alpha gene trans-inducing factor. J. Virol. 68:4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakatake Y, et al. 2006. Klf4 cooperates with Oct3/4 and Sox2 to activate the Lefty1 core promoter in embryonic stem cells. Mol. Cell Biol. 26:7772–7782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nichol PF, Chang JY, Johnson EM, Jr, Olivo PD. 1996. Herpes simplex virus gene expression in neurons: viral DNA synthesis is a critical regulatory event in the branch point between lytic and latent pathways. J. Virol. 5476-5486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Otteson DC, Lai H, Liu Y, Zack DJ. 2005. Zinc-finger domains of the transcriptional repressor KLF15 binds multiple sites in rhodopsin and IRBP promoters including the CRS-1 and G-rich elements. BMC Mol. Biol. 6:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Perez S, et al. 2007. A protein encoded by the bovine herpesvirus 1 ORF E gene induces neurite-like morphological changes in mouse neuroblastoma cells and is expressed in trigeminal ganglionic neurons. J. Neurovirol. 13:139–149 [DOI] [PubMed] [Google Scholar]
- 61. Perez S, Meyer F, Saira K, Doster A, Jones C. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J. Gen. Virol. 89:1338–1345 [DOI] [PubMed] [Google Scholar]
- 62. Perez S, Lovato L, Zhou J, Doster A, Jones C. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild type BHV-1 versus a virus strain containing a mutation in the LR (latency-related) gene. J. Neurovirol. 12:392–397 [DOI] [PubMed] [Google Scholar]
- 63. Perez S, Inman M, Doster A, Jones C. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J. Clin. Microbiol. 43:393–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Perng G-C, Jones C. 2010. Towards an understanding of the herpes simplex virus type 1 latency reactivation cycle. Interdisc. Persp. Infect. Dis. 2010:1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pesola JM, Zhu J, Knipe DM, Coen DM. 2005. Herpes simplex virus 1 immediate-early and early gene expression during reactivation from latency under conditions that prevent infectious virus production. J. Virol. 79:14516–14525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Prbhakaran K, et al. 2005. Sensory neurons regulate the effector functions of CD8+ T cells in controlling HSV-1 latency ex vivo. Immunity 23:515–523 [DOI] [PubMed] [Google Scholar]
- 67. Rhen T, Cidlowski JA. 2005. Anti-inflammatory action of glucocorticoids: new mechanisms of old drugs. N. Engl. J. Med. 353:1711–1723 [DOI] [PubMed] [Google Scholar]
- 68. Rock D, Lokensgard J, Lewis T, Kutish G. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 66:2484–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodrigues R, Cuddington B, Mossman K. 2010. Bovine herpesvirus type 1 as a novel oncolytic virus. Cancer Gene Ther. 17:344–345 [DOI] [PubMed] [Google Scholar]
- 70. Roizman B, Sears AE. 1996. Herpes simplex viruses and their replication, p 2231–2295 In Fields BE, et al. (ed), Fields virology, 3rd ed Lippincott-Raven Publishers, New York, NY [Google Scholar]
- 71. Sanalkumar R, Dhanesh SB, James J. 2010. Non-canonical activation of Notch signaling/target genes in vertebrates. Cell. Mol. Life Sci. 67:2957–2968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sawtell NM, Thompson RL. 1992. Rapid in vivo reactivation of herpes simplex virus in latently infected murine ganglionic neurons after transient hyperthermia. J. Virol. 66:2150–2156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Schang L, Jones C. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 71:6786–6795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Sharrocks AD. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell. Biol. 2:827–837 [DOI] [PubMed] [Google Scholar]
- 75. Sheffy BE, Davies DH. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140:974–976 [DOI] [PubMed] [Google Scholar]
- 76. Shen W, Jones C. 2008. Open reading frame 2 encoded by the latency related gene of bovine herpesvirus 1 has anti-apoptosis activity in transiently transfected neuroblastoma cells. J. Virol. 82:10940–10945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Shimeld C, et al. 1995. Immune cell infiltration and persistence in the mouse trigeminal ganglion after infection of the cornea with herpes simplex virus type 1. J. Neuroimmunol. 61:7–16 [DOI] [PubMed] [Google Scholar]
- 78. Shimeld C, Whiteland JL, Williams NA, Easty DL, Hill TJ. 1997. Cytokine production in the nervous system of mice during acute and latent infection with herpes simplex virus type 1. J. Gen. Virol. 78:3317–3325 [DOI] [PubMed] [Google Scholar]
- 79. Shimeld C, Whiteland JL, Williams NA, Easty DL, Hill TJ. 1996. Reactivation of herpes simplex virus type 1 in the mouse trigeminal ganglion: an in vivo study of virus antigen and immune cell infiltration. J. Gen. Virol. 77:2583–2590 [DOI] [PubMed] [Google Scholar]
- 80. Shimizu N, et al. 2011. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 13:170–182 [DOI] [PubMed] [Google Scholar]
- 81. Tal-Singer R, et al. 1997. Gene expression during reactivation of herpes simplex virus type 1 from latency in the peripheral nervous system is different from that during lytic infection of tissue cultures. J. Virol. 71:5268–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Theil D, et al. 2003. Latent herpesvirus infection in human trigeminal ganglia causes chronic immune response. Am. J. Pathol. 163:2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Thompson RL, Preston CM, Sawtell NM. 2009. De novo synthesis of VP16 coordinates the exit form HSV latency in vivo. PLoS Pathog. 5:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Tikoo SK, Campos M, Babiuk LA. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191–223 [DOI] [PubMed] [Google Scholar]
- 85. Tsavachidou D, Podrzucki W, Seykora J, Berger SL. 2001. Gene array analysis reveals changes in peripheral nervous system gene expression following stimuli that result in reactivation of latent herpes simplex virus type 1: induction of transcription factor Bcl-3. J. Virol. 75:9909–9917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Turin L, Russo S, Poli G. 1999. BHV-1: new molecular approaches to control a common and widespread infection. Mol. Med. 5:261–284 [PMC free article] [PubMed] [Google Scholar]
- 87. Winkler MT, Doster A, Sur JH, Jones C. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86:139–155 [DOI] [PubMed] [Google Scholar]
- 88. Winkler MT, Schang LS, Doster A, Holt T, Jones C. 2000. Analysis of cyclins in trigeminal ganglia of calves infected with bovine herpesvirus 1. J. Gen. Virol. 81:2993–2998 [DOI] [PubMed] [Google Scholar]
- 89. Winkler MTC, Doster A, Jones C. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsil of latently infected calves. J. Virol. 74:5337–5346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wirth UV, et al. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 66:2763–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wirth UV, Gunkel K, Engels M, Schwyzer M. 1989. Spatial and temporal distribution of bovine herpesvirus 1 transcripts. J. Virol. 63:4882–4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wirth UV, Vogt B, Schwyzer M. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Workman A, Sinani D, Pittayakhajonwut D, Jones C. 2011. A protein (ORF2) encoded by the latency-related gene of bovine herpesvirus 1 interacts with Notch1 and Notch3. J. Virol. 85:2536–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Workman A, Perez S, Doster A, Jones C. 2009. Dexamethasone treatment of calves latently infected with bovine herpesvirus 1 (BHV-1) leads to activation of the bICP0 early promoter, in part by the cellular transcription factor C/EBP-alpha. J. Virol. 83:8800–8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Workman A, Jones C. 2010. Bovine herpesvirus 1 productive infection and bICP0 early promoter activity are stimulated by E2F1. J. Virol. 84:6308–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Xu D, et al. 2009. Promyelocytic Leukemia zinc finger protein regulates interferon-mediated innate immunity. Immunity 30:802–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Yamamoto J, et al. 2004. A Kruppel-like factor KLF15 contributes fasting-induced transcriptional activation of mitochondrial acetyl-CoA synthase gene AceCS2. J. Biol. Chem. 279:16954–16962 [DOI] [PubMed] [Google Scholar]
- 98. Yang L, Voytek CC, Margolis TP. 2000. Immunohistochemical analysis of primary sensory neurons latently infected with herpes simplex virus type 1. J. Virol. 74:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yin F, Herring BP. 2005. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J. Biol. Chem. 280:4745–4752 [DOI] [PubMed] [Google Scholar]
- 100. Li Z, et al. 2010. KLF4 promotes hydrogen-peroxide-induced apoptosis of chronic myeloid leukemia cells involving the bcl-2/bax pathway. Cell Stress and Chaperones 15:905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Zhang Y, Jiang Y, Zhou J, Jones C. 2006. The bovine herpesvirus 1 (BHV-1) immediate-early protein (bICP0) interacts with the histone acetyltransferase p300, and these interactions correlate with stimulation of gC promoter activity. J. Gen. Virol. 87:1843–1851 [DOI] [PubMed] [Google Scholar]