Abstract
Hendra virus is a highly pathogenic paramyxovirus classified as a biosafety level four agent. The fusion (F) protein of Hendra virus is critical for promoting viral entry and cell-to-cell fusion. To be fusogenically active, Hendra virus F must undergo endocytic recycling and cleavage by the endosomal/lysosomal protease cathepsin L, but the route of Hendra virus F following internalization and the recycling signals involved are poorly understood. We examined the intracellular distribution of Hendra virus F following endocytosis and showed that it is primarily present in Rab5- and Rab4-positive endosomal compartments, suggesting that cathepsin L cleavage occurs in early endosomes. Hendra virus F transmembrane domain (TMD) residues S490 and Y498 were found to be important for correct Hendra virus F recycling, with the hydroxyl group of S490 and the aromatic ring of Y498 important for this process. In addition, changes in association of isolated Hendra virus F TMDs correlated with alterations to Hendra virus F recycling, suggesting that appropriate TMD interactions play an important role in endocytic trafficking.
INTRODUCTION
The Hendra and Nipah viruses are recently emerged, closely related paramyxoviruses that are highly pathogenic in humans and other animal species (10, 15, 17). They are enveloped viruses, classified as biosafety level four agents due to the absence of treatments and vaccines. Hendra and Nipah viruses are single-stranded RNA viruses that enter cells with the help of two surface glycoproteins: the attachment protein, G, and the fusion protein, F (3, 17, 84). The G protein promotes viral binding through interactions with cell surface receptor Ephrin B2 or B3 (3, 49, 50). It is thought that these interactions trigger the F protein to undergo a series of conformational rearrangements that lead to the fusion of the two membranes (15, 84). In addition to virus-cell fusion, F and G can also promote cell-cell membrane fusion after viral infection (40, 84).
The paramyxovirus F protein is synthesized as an inactive precursor, F0, which must be proteolytically processed into the fusogenically active, disulfide-linked F1+F2 form (Fig. 1A) (16, 38, 39). Cleavage places the fusion peptide at the N terminus of the newly formed F1 subunit, allowing it to be inserted into the target cell membrane when fusion is initiated (Fig. 1A). While the majority of paramyxovirus F proteins are cleaved during transport through the trans-Golgi network (39), a small number of F proteins are cleaved after they reach the cell surface by tissue-specific extracellular proteases (39, 69). Unique among viral fusion proteins, Hendra virus F and Nipah virus F are proteolytically processed by the endosomal/lysosomal protease cathepsin L in a process that requires endocytic recycling (14, 54, 55, 81) (Fig. 2A) and the low pH of endosomal compartments (56). However, a detailed map of Hendra and Nipah virus F endocytic trafficking, as well as the signals that drive F recycling, remains largely unclear.
Fig 1.
(A) Schematic of Hendra virus F protein and the HRB and TMD amino acid sequences. FP, fusion peptide; HR, heptad repeat; TMD transmembrane domain; CT, cytoplasmic tail; S-S, disulfide bond. (B) Helical wheel representation of the Hendra virus F TMD. Residues at positions a and d of the helical wheel, denoted by stars, are hypothesized to form the interface of the three TMD helices.
Fig 2.
(A) Model for proteolytic processing of the Henipavirus F proteins, with the effects of mutations noted. The Hendra and Nipah virus F proteins are synthesized in the endoplasmic reticulum (ER), transit through the secretory pathway to the plasma membrane (PM), and are subsequently endocytosed. Following interaction with cathepsin L at an undetermined point in the endocytic pathway, the cleaved protein is recycled to the plasma membrane. Steps affected by the various S490 and Y498 mutants are indicated. (B) Hendra virus F proteolytic cleavage requires delivery to the early endosomes but not the late endosomes. Vero cells expressing Hendra virus F alone or together with DN Rab5 or DN Rab7 were starved for 30 min and metabolically labeled for 4 h. Surface proteins were labeled with biotin. Hendra virus F was immunoprecipitated with an anti-Hendra virus F antibody, and the surface population was isolated by streptavidin pulldowns. (C) Expression of DN Rab5 slows the kinetics of Hendra virus F cleavage. Vero cells expressing Hendra virus F alone or together with DN Rab5 were starved for 30 min, metabolically labeled for 2 h, and chased for 0, 1, 2, 4, and 8 h. At the end of each chase interval, surface proteins were labeled with biotin. Hendra virus F was immunoprecipitated with an anti-Hendra virus F antibody, and the surface population was isolated by streptavidin pulldowns.
Hendra virus F is a 546-amino-acid type I integral membrane protein. It folds as a homotrimer and contains the typical domains of class I viral fusion proteins: a fusion peptide (FP), two heptad repeat regions (heptad repeat A [HRA] and HRB), a transmembrane domain (TMD), and a 28-amino-acid-long cytoplasmic/intraviral tail (CT) (Fig. 1A). Hendra and Nipah virus F proteins share 88% homology (29), and a YSRL endocytosis motif in the Hendra virus F and Nipah virus F cytoplasmic tails is critical for F protein internalization and proteolytic processing (46, 81). It has been suggested that YXXΦ motifs (where X represents any amino acid and Φ represents a hydrophobic amino acid) function as endocytic signals when they are positioned 10 to 40 residues from the TMD and as lysosomal targeting signals when they are 6 to 11 residues from the TMD (4, 5). Although Hendra and Nipah virus F proteins are recycled to the cell surface after cathepsin L cleavage (46, 55, 81), their CT YSRL motif is present only 6 residues from the TMD, suggesting that additional sorting signals may contribute to the Hendra and Nipah virus F protein recycling.
After internalization, plasma membrane proteins are first delivered to the early endosomes, which represent a major intracellular sorting station (24, 30, 36, 44). From here, the proteins are targeted either to the plasma membrane, the recycling endosomes, or the late endosomes (30, 44). These processes are complex as endocytic compartments are highly dynamic (44). Recent studies have identified a number of recycling motifs in the CTs of several G protein-coupled receptors and transferrin receptor (13, 27, 28), but the overall process of protein sorting and the signals influencing recycling decisions remain poorly understood. In addition to the CT signals, residues within the TMD have also been implicated in protein sorting (60, 86). Previous work on transferrin receptor, a classic model for recycling, has shown that removing its CT (31, 34) or ectodomain (61) does not affect its recycling, implicating the TMD in trafficking. Zaliauskiene et al. showed that substitutions of polar residues within the transferrin receptor TMD changed the fate of the protein from a recycled receptor to one that is rapidly downregulated (86), suggesting that TMD polar residues may modulate the sorting decisions toward downregulation. In addition, the introduction of acidic residues within the TMD of Pep12p also altered sorting (60). However, specific TMD signals have not been identified for either recycling or downregulation. The Hendra virus F TMD contains a significant number of polar residues, and in this study we investigated their potential role in endocytic trafficking.
The Hendra virus F TMD (Fig. 1) has a predicted length of approximately 30 amino acids and is thought to span the lipid bilayer as a helix. An extensive analysis of the parainfluenza virus 5 (PIV5) F TMD confirmed that the TMD forms a helical structure and that the interacting face of the three helices was likely formed primarily of leucine and isoleucine residues (2). Hydropathy plots of the Hendra virus F TMD sequence suggest that the TMD starts at leucine 488 and ends at valine 518 (Fig. 1A). Interestingly, a leucine-isoleucine zipper spans the length of the Hendra virus F HRB and TMD (Fig. 1A), suggesting that the HRB continues with TMD as a straight helix. This model also predicts that the residues present at positions a and d on the helical wheel shown in Fig. 1B will form the internal, interacting faces of the helices. Although the structures of paramyxovirus F proteins determined to date do not contain the TMD, the structure of a SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) fusion complex was determined recently, showing that the postfusion helix bundle continues as a straight helix in the lipid bilayer (72), with potential implications for membrane fusion.
Since the Hendra virus F TMD (Fig. 1) is enriched in noncharged polar residues compared to other TMDs (42), we used the unique recycling pathway of Hendra virus F to examine the role of these polar residues in endocytic trafficking. We identified two residues which are critical for Hendra virus F internalization and recycling: S490 and Y498. S490, predicted to be placed at the border with the ectodomain, is important for Hendra virus F endocytosis, and our results indicate that the hydroxyl group is necessary for correct protein trafficking. In addition, Y498 is critical for proper F recycling, and our work demonstrates that the aromatic ring, and not the hydroxyl group, is needed for correct Hendra virus F recycling. Furthermore, analytical ultracentrifugation sedimentation equilibrium analysis of isolated Hendra virus F TMD demonstrated that TMD associations are altered for the Hendra virus F Y498 mutants that fail to properly recycle, suggesting that proper interactions between Hendra virus F TMDs may play a role in modulating trafficking decisions.
MATERIALS AND METHODS
Cell lines.
Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-Invitrogen) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin.
Plasmids.
Hendra virus F and G genes were kindly provided by Lin-Fa Wang of the Australian Animal Health Laboratory, and the genes were ligated into the pCAGGS mammalian expression vector (51) as previously described (56). Primers were designed to include the described TMD mutations. Site-directed mutagenesis was performed on the plasmids pGEM4Z-Hendra virus F or pET11A SN-Hendra F TMD (pFT11A containing staphylococcal nuclease [SN] fused to Hendra virus F TMD) using a QuikChange site-directed mutagenesis system (Stratagene, La Jolla, CA). The constructs were sequenced in their entirety, and the Hendra virus F mutants were subcloned from pGEM to pCAGGS as previously described (56). Wild-type (WT) Rab5 and dominant negative (DN) Rab5 S34N cloned as green fluorescent protein (GFP) N-terminal fusions in the pGreenLantern vector (Gibco-BRL, Grand Island NY) were a kind gift from Gary Whittaker, Cornell University (68). Rab4 was cloned into pEGFP (where EGFP is enhanced GFP) and was kindly provided by Marci A. Scidmore (62). WT pEGFP-Rab7 and pEYFP-Lamp1 (where EYFP is enhanced yellow fluorescent protein) were obtained from Addgene (Addgene plasmids 12605 [9] and 1816 [67]), where they were provided by Walter Moses and Richard Pagano. The pET11A vector containing SN linked to the glycophorin TMD was kindly provided by Karen Fleming (Johns Hopkins University). The Hendra virus F TMD domain was cloned into this vector to replace the glycophorin TMD and fused to SN using the SmaI and XhoI restriction sites.
Antibodies.
Polyclonal antibodies were generated to residues 526 to 539 of the Hendra virus F cytoplasmic tail (Genemed Custom Peptide Antibody Service, San Francisco, CA). Monoclonal antibody 7F7 to the Hendra virus F ectodomain was kindly provided by Christopher Broder (Uniformed Services University). Goat anti-mouse IgG DyLight 549 secondary antibody was purchased from Jackson ImmunoResearch Laboratories (code number 115-505-146).
Time course surface biotinylation experiments.
WT Hendra virus F or the Hendra virus F TMD mutants were expressed in subconfluent monolayers of Vero cells using the pCAGGS system and Lipofectamine Plus (Invitrogen). The cells were transfected in 60-mm dishes, using 4 μg of total DNA per dish, according to the manufacturer's protocol. The next day cells were washed with phosphate-buffered saline deficient in calcium and magnesium chloride (PBS−) and starved for 30 min in cysteine-methionine-deficient DMEM. Cells were then labeled for 2 h with trans-35S (100 μCi/ml; Perkin Elmer), washed twice with PBS−, and chased (DMEM, 10% FBS, 1% penicillin-streptomycin) for 0, 1, 2, 4, and 8 h. At the end of each time point, cells were washed three times with ice-cold PBS− at pH 8 and then biotinylated using 1 ml EZ-Link sulfo-N-hydroxysuccinimide-biotin (sulfo-NHS-biotin; 1 mg/ml; Pierce, Rockford, IL) in PBS−, pH 8, by rocking gently for 35 min at 4°C. Cells were then washed again three times in cold PBS− at pH 8 and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer supplemented with protease inhibitors and 25 mM iodoacetamide (46). The lysates were centrifuged at 55,000 rpm for 12 min at 4°C, and immunoprecipitations were performed as described previously (21, 83) using Hendra virus F antipeptide antibodies and protein A-conjugated Sepharose beads (Amersham/GE Healthcare Bio-Sciences, Piscataway, NJ). To separate the surface population of Hendra virus F, protein A Sepharose beads were boiled twice in 50 μl of 10% SDS, and the SDS supernatant was transferred to a microcentrifuge tube, resulting in a total of 100 μl. From this, 15 μl was saved for analysis of the total protein population, while the surface population was isolated from the remaining 85 μl by rocking the samples for 1 h at 4°C in the presence of 35 μl of streptavidin beads and 500 μl of biotinylation buffer (20 mM Tris, pH 8, 150 mM NaCl, 5 mM EDTA, 1% Triton X-100, 0.2% bovine serum albumin [BSA]). Samples were washed twice with RIPA buffer containing 0.30 M NaCl, twice with RIPA buffer containing 0.15 M NaCl, and one time with SDS wash II buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 2.5 mM EDTA), and the total and surface fractions of the proteins were analyzed on 15% SDS-PAGE gels under reducing conditions and visualized using a Typhoon imaging system (GE Healthcare, Piscataway, NJ).
Immunofluorescence experiments.
Vero cells in subconfluent monolayers in eight-well chamber slides (BD Falcon) were transfected with 0.3 μg of WT or mutant Hendra virus F, along with 0.3 μg of GFP-expressing Rab5, Rab7, or Rab4 or YFP-Lamp1 using Lipofectamine 2000 (Invitrogen). For the antibody capture experiments, cells were washed five times with PBS−, followed by incubation for 40 min at 4°C with PBS− containing 1% normal goat serum (NGS), 1% bovine serum albumin, and the 7F7 Hendra virus F monoclonal antibody at a 1:1,000 dilution (17.5 μg/ml). Cells were then washed three times with cold PBS− and transferred to 37°C for 0, 10, 20, or 30 min, followed by another wash and fixation with 4% paraformaldehyde for 15 min. Cells were washed and permeabilized with 1% Triton X-100 (in PBS− supplemented with 0.02% NaN3), followed by a 60-min incubation at 4°C with a goat anti-mouse DyLight 549 secondary antibody (diluted 1:800 in PBS− supplemented with 0.02% NaN3, 1% NGS, and 1% BSA). The secondary antibody was removed, and cells were washed seven times with PBS− containing 0.05% Tween 20. The slides were mounted on coverslips using Vectashield and 4′,6′-diamidino-2-phenylindole ([DAPI] Vector Laboratories, Inc., Burlingame, CA), and the edges were sealed with nail polish. The samples were stored at −20°C. Confocal microscopy was performed using an Olympus FluoView 1000 inverted confocal system. Cells were visualized with a 60× oil objective (numerical aperture [NA], 1.35), and images were captured with FluoView software. Quantitation of colocalization was performed using ImageJ software.
Syncytium assay.
Subconfluent monolayers of Vero cells were transfected with WT Hendra virus F or the described Hendra virus F mutants along with Hendra virus G. The transfection was performed using Lipofectamine Plus, and the F-to-G ratio was 1:2 (0.5 μg of F and 1 μg of G). Syncytium formation was examined at 24 to 48 h posttransfection using a Nikon TS 100 inverted phase-contrast microscope. The cells were photographed at a magnification of ×100 using a Nikon Coolpix 995 digital camera.
Analytical ultracentrifugation sedimentation equilibrium.
The Hendra virus F SN-TMD proteins (WT and Y498A, Y498F, and Y498S mutants) were purified and exchanged into C14SB [3-(N,N-dimethylmyristyl-ammonio) propane sulfonate] as previously described (75). Sedimentation equilibrium analytical ultracentrifugation experiments were performed in a Beckman XL-A analytical ultracentrifuge, using three protein concentrations and three rotor speeds (20,000, 25,000, and 30,000 rpm), as previously presented (18, 20). The equilibrium profiles were analyzed with KaleidaGraph, using the following equation:
where A represents the total absorbance at a radial position, r, while α0 represents the monomer absorbance at a reference position, r0. The monomer molecular mass, M, and the partial specific volumes, v̄, were calculated using SEDNTERP; ρ represents the solvent density, ω is the angular velocity, R is the universal gas constant, T is temperature, and ζ is the offset, representing nonsedimenting material (20). For the monomer–N-mer fits, the molecular mass of the monomer was kept constant, and the association constants were allowed to vary (absorbance units). The best fit was chosen based on the smallest square root of the variance (SRV) at the three concentrations and three speeds tested. Protein concentrations were determined by spectrophotometry, using E280 = 17,420 M−1 cm−1.
RESULTS
Hendra virus F proteolytic cleavage requires delivery to the sorting endosomes.
Previous work has shown that Hendra and Nipah virus F protein endocytosis and cathepsin L processing are necessary for proper cleavage and function (14, 46, 55, 81). In addition, Diederich et al. showed the presence of Nipah virus F in the early and recycling endosomes through colocalization experiments with transferrin. However, as cathepsin L has primarily been thought to localize to the late endosomes and lysosomes (45, 79), it is unclear what compartment must be reached to facilitate cathepsin L cleavage. To determine if trafficking to the early or late endosomes is needed, we examined Hendra virus F cell surface expression and cleavage in the presence of the dominant negative Rab5 S34N (DN Rab5), which inhibits early endosomal fusion (73), or the dominant negative Rab7 T22N (DN Rab7), which inhibits late endosomal fusion. Hendra virus F was expressed in Vero cells alone or in combination with DN Rab5 or DN Rab7. Following a 30-min label and 4-h chase, biotinylation of the surface proteins was performed (Fig. 2B). The surface population of Hendra virus F was then isolated by immunoprecipitation and streptavidin pulldown. The presence of the cleaved F1+F2 form on the cell surface indicates that the protein was internalized, processed by cathepsin L, and recycled to the surface. Expression of DN Rab5 dramatically decreased the processing of Hendra virus F, indicating that trafficking to the early endosome is important for F protein cleavage. Surprisingly, no significant reduction in processing was seen in the presence of DN Rab7 (Fig. 2B), indicating that trafficking to the late endosome is not necessary for cathepsin L to process Hendra virus F. To verify the DN Rab5 block and determine the kinetics of the cleavage process, cells transfected to express Hendra virus F with or without DN Rab5 were metabolically labeled and chased for 0, 1, 2, 4, and 8 h, and biotinylation of the surface proteins was performed (Fig. 2C). When F was expressed alone, some of the cleaved form (F1) was present on the cell surface after the 2-h label (0 min time point); by the 4- and 8-h time points the active F1 form was the predominant form on the cell surface (Fig. 2C). However, in the presence of DN Rab5 there was a significant alteration in the kinetics of F processing as F0 was the predominant surface-expressed form at all time points (Fig. 2C). By the 4- and 8-h time points, some F1 was apparent, suggesting that inhibition by DN Rab5 was not complete. However, the overall levels of processing were much lower than those seen without DN Rab5. Taken together, these data suggest that trafficking to the early endosomes, but not the late endosomes, is needed for proper Hendra virus F processing.
Hendra virus F is recycled through the early endosomal compartments.
To further evaluate the intracellular identity of the compartment where the Hendra virus F-cathepsin L interaction occurs, we examined Hendra virus F endosomal distribution at steady state using immunofluorescence experiments and analysis of Hendra virus F colocalization with Rab5, Rab7, and Rab4 (Fig. 3). Rab5 plays important roles in the fusion of the incoming endocytosed vesicles with the sorting endosomes and is considered a marker of the sorting endosomes (70, 74). Rab4 is involved in membrane recycling from the sorting endosomes and is considered a marker for endocytic recycling compartments (53, 80), while Rab7 plays roles in membrane trafficking to late endosomes and lysosomes and is considered a late endosomal marker (8, 44, 59). Vero cells were transfected with Hendra virus F and GFP-tagged Rab5, Rab7, or Rab4. The next day cells were fixed, permeabilized, and incubated for 40 min with a monoclonal antibody to the Hendra virus F ectodomain, kindly provided by Christopher Broder (Uniformed Services University of the Health Sciences). Hendra virus F was detected by labeling with a secondary DyLight 549 fluorescent antibody (Fig. 3). Hendra virus F showed moderate colocalization with Rab5- and Rab4-positive compartments, corresponding to sorting and recycling endosomes (Pearson's coefficients 0.15 to 0.3), but very little colocalization (negative Pearson's coefficients) of Hendra virus F was observed with the late endosomal marker Rab7 (Fig. 3). These results suggest that Hendra virus F is not present at a significant level within late endosomes, which have previously been shown to contain cathepsin L (45, 79).
Fig 3.
At steady state, Hendra virus F colocalizes with Rab5- and Rab4-positive endosomes. Vero cells were transfected with Hendra virus F and the WT form of GFP-tagged Rab5, Rab7, or Rab4, and Hendra virus F steady-state distribution was analyzed by single confocal sections of fixed cells. Insets show enlargements of the boxed areas. Scale bar, 5 μm. Experiments were repeated two times, and representative pictures are shown.
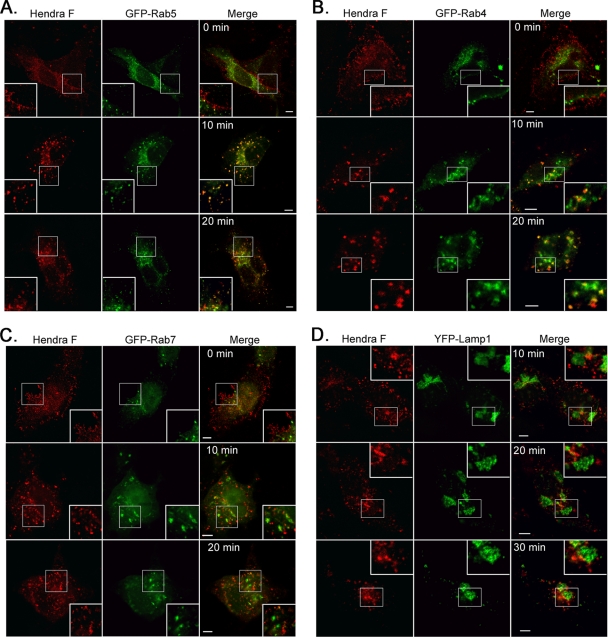
To further investigate and map the route of Hendra virus F after internalization, we performed antibody capture experiments and followed F colocalization with specific endosomal/lysosomal markers during endocytosis (Fig. 4). Briefly, Vero cells were transfected with Hendra virus F and one of the GFP-Rabs. The next day, cells were washed and incubated for 40 min at 4°C with a monoclonal antibody to the Hendra virus F ectodomain. Cells were then incubated at 37°C for 0, 10, 20, or 30 min, fixed, permeabilized, and labeled with a secondary fluorescent antibody. We have previously shown that cleavage of Hendra virus F occurs rapidly after endocytosis, with the vast majority of internalized protein undergoing processing within 30 min (46). As expected, at the 0-min time point Hendra virus F had a plasma membrane distribution (Fig. 4A to C) as cells were maintained at 4°C and thus did not undergo appreciable levels of endocytosis. However, after 10 min of incubation at 37°C, the majority of F was internalized (Fig. 4A to D) and showed moderate colocalization (Pearson's coefficients of 0.2 to 0.4) with Rab5 (Fig. 4A) and Rab4 (Fig. 4B) but almost no colocalization (negative Pearson's coefficients) with Rab7 and the lysosome-associated membrane protein 1 (Lamp1), which are markers of late endosomes/lysosomes (Fig. 4C and D). By 20 min Hendra virus F was still predominantly present in Rab5- and Rab4-positive compartments (Fig. 4A and B), with almost no colocalization with late endosomal/lysosomal markers (Fig. 4C and D), suggesting that Hendra virus F processing uses the classical recycling pathway for its processing. In addition, these results suggest that cathepsin L is present and is functionally active within the early and/or the recycling endosomes.
Fig 4.
After internalization, Hendra virus F colocalizes with Rab5- and Rab4-positive endosomes. Antibody capture experiments were performed as described. Hendra virus F colocalization with specific endosomal markers was examined at 0, 10, 20, and 30 min after transfer to 37°C, by single confocal sections on fixed cells. Insets represent enlargements of the boxed areas. Scale bar, 5 μm.
The S490 hydroxyl group modulates Hendra virus F endocytosis and recycling.
The signals involved in protein recycling remain poorly understood. The Hendra and Nipah virus F protein TMDs contain a high number of polar residues, and several reports have implicated polar residues in protein sorting (43, 60, 86). To investigate the potential role of polar TMD residues in Hendra virus F recycling, we performed alanine mutagenesis on Hendra virus F TMD polar residues and identified two amino acids that are critical for Hendra virus F endocytic recycling: S490 and Y498 (Fig. 1A).
To examine Hendra virus F S490A trafficking and surface expression over time, we performed time course surface biotinylation experiments, as described above. WT Hendra virus F and the S490A mutant were expressed in Vero cells and metabolically labeled for 2 h, and a chase was performed for the times indicated on Fig. 5. Surface F was biotinylated and separated by streptavidin pulldown (Fig. 5A). The vast majority of the Hendra virus F S490A mutant was present on the cell surface as the uncleaved F0 form throughout the 8-h time course, with very little of the cleaved form present on the cell surface (Fig. 5A). The Hendra virus F S490A mutant was also present primarily in the F0 form in the “total” fraction, which includes the intracellular F population (Fig. 5B), suggesting that the S490A mutation either made the cathepsin L cleavage site inaccessible or disrupted endocytosis, preventing F from reaching a cathepsin L positive compartment.
Fig 5.
Hendra virus F S490 TMD residue plays critical roles in Hendra virus F trafficking. Vero cells expressing WT or S490 Hendra virus F mutants were starved for 30 min, metabolically labeled for 2 h, and chased for 0, 1, 2, 4, and 8 h. At the end of each chase interval, surface proteins were labeled with biotin. Proteins were immunoprecipitated with an anti-Hendra virus F antibody. Samples were separated into a surface and total fraction, as indicated. Surface proteins were then separated using streptavidin-agarose beads and analyzed by SDS-PAGE. (A) Time course of surface expression of WT or S490A, S490T, or S490V Hendra virus F. (B) Total expression of WT and S490A Hendra virus F. (C) Surface expression of WT or S490A Hendra virus F after trypsin digestion. Wild-type or S490A Hendra virus F was expressed in Vero cells, labeled for 2 h, and chased for 1.5 h. TPCK-trypsin (1.0 μg/ml) was added to the chase medium, and cells were incubated for an additional 30 min. Hendra virus F proteins were biotinylated, isolated using streptavidin beads, resolved on a 15% polyacrylamide gel, and analyzed using a Typhoon imaging system. (D) Time course of surface expression of S490E and S490K Hendra virus F, in the presence or absence of DN Rab5. The proteins were transiently expressed in Vero cells alone or along with DN Rab5, and they were labeled, chased, and immunoprecipitated as described above. Surface proteins were biotinylated, isolated using streptavidin beads, resolved on a 15% polyacrylamide gel, and analyzed using a Typhoon imaging system. Experiments shown are representative of at least three repetitions.
To determine if the cathepsin L cleavage site was accessible in the F S490A mutant, we examined cleavage of Hendra virus F S490A when extracellular tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK)-trypsin was added to the medium as addition of exogenous trypsin has been shown to cleave Hendra virus F at the same site (12). Briefly, cells expressing the WT or S490A mutant were labeled for 2 h, followed by a 30-min incubation with 1 μg/ml TPCK-trypsin. As seen in Fig. 5C, Hendra virus F S490A was cleaved, generating F1 proteins of similar sizes to the proteins of the WT Hendra virus F, indicating that the S490A mutation does not prevent accessibility of trypsin to the Hendra virus F cleavage site. Additionally, the trypsin-cleaved S490A Hendra virus F was fusogenically active (data not shown), suggesting that the S490A mutation does not cause additional changes in the structure or function of the protein.
To examine if the S490A mutation alters the rate of Hendra virus F internalization, antibody capture experiments were performed, as described above. Hendra virus F S490A was coexpressed in Vero cells along with WT GFP-Rab5; cells were washed and incubated for 40 min at 4°C with an anti-Hendra virus F monoclonal antibody, followed by their transfer to 37°C for 0, 10, 20, or 30 min. As seen in Fig. 6, Hendra virus F S490A displayed a strong plasma membrane distribution even after 20 and 30 min at 37°C, while most of WT Hendra virus F was internalized by 10 min (Fig. 4), indicating that the S490A substitution alters Hendra virus F internalization (Fig. 6).
Fig 6.
Hendra virus F S490A is deficient in endocytosis. Hendra virus F S490A and WT GFP-Rab5 were expressed in Vero cells, and antibody capture experiments were performed as described in the text. Single confocal sections were acquired and analyzed. Insets represent enlargements of the boxed areas. Scale bar, 5 μm. Experiments were repeated twice, and the images are representative.
To further delineate the characteristics of the S490 residue that are needed for Hendra virus F endocytosis, S490T and S490V were created, replacing amino acids whose side chains are similar in size, but with only the S490T mutant containing the hydroxyl group to mimic S490. Expression and recycling of these mutants were examined by time course surface biotinylation experiments, as described above. Interestingly, Hendra virus F S490T was cleaved and recycled back to the cell surface at rates similar to those of the WT Hendra virus F (Fig. 5A). In contrast, the S490V mutant initially appeared on the cell surface in the F0 uncleaved form but disappeared by the 2-h time point without returning to the cell surface in the active F1 form at later time points (Fig. 5A). These results suggest that the hydroxyl group of S490 is important for proper trafficking of Hendra virus F. In contrast, other small side chains caused either an endocytic defect (S490A) or a recycling defect (S490V), indicating that subtle amino acid changes at the TMD-ectodomain border can have major effects on protein trafficking.
The fusion activity of these mutants was examined using a syncytium assay, as previously described (58). Membrane fusion between neighboring cells, promoted by Hendra virus F and G proteins, leads to the formation of giant multinucleated cells called syncytia. WT or the S490 Hendra virus F mutants were expressed in Vero cells along with the attachment protein, G, which is required for fusion activity. At 24 h posttransfection syncytium formation was examined (Fig. 7). As expected, only the WT and the properly cleaved and trafficked S490T mutant formed syncytia, while the F mutants S490A and S490V were fusion dead (Fig. 7 and data not shown). As cleavage of Hendra virus F is needed for fusion promotion, these results substantiate the idea that recycling of the cleaved form to the cell surface is critical for the fusion process.
Fig 7.
S490T and Y498F Hendra virus F proteins promote syncytium formation, while S490A and Y498A TMD mutants are fusogenically inactive. Vero cells were transfected with WT, S490, or Y498 Hendra virus F TMD mutants, along with Hendra virus G at a ratio of 1:2 (F/G). At 24 h posttransfection multinucleated cells were photographed.
It is unclear how a hydroxyl group at position 490, predicted to be near the TMD-ectodomain border, mediates Hendra virus F recycling. Previous work from our lab has not detected the phosphorylation of Hendra virus F (data not shown). However, transient phosphorylation could occur but go undetected by anti-phospho antibodies. To test whether the negative charge associated with possible phosphorylation events influenced Hendra virus F trafficking, we created an S490E substitution, which serves as a mimic of phosphorylation due to the negative charge of the glutamic acid. In addition, an S490K mutation was introduced as positive charges are often found at the membrane-water interface (23). Hendra virus F S490E and S490K surface expression and proteolytic cleavage were analyzed by pulse-chase surface biotinylation experiments (Fig. 5D). In contrast to results with the Hendra virus F S490A and S490V mutants, a small fraction of the proteins was trafficked back to the cell surface, suggesting that either a positive or negative charge at this position can facilitate the recycling process (Fig. 5D). However, the surface levels of S490E and S490K (S490E/S490K) mutants were much lower than those of WT F, suggesting that either less protein was produced or a greater fraction was targeted for degradation after internalization (Fig. 5D). To differentiate these possibilities, we performed surface biotinylation experiments in the presence of DN Rab5, which inhibits endocytosis. When endocytosis was blocked, much higher levels of F0 S490E/S490K were detected on the cell surface, suggesting that after internalization, a fraction of S490E/S490K Hendra virus F was mistrafficked toward degradation (Fig. 5D). Taken together, the data suggest that Hendra virus F endocytosis and recycling are finely tuned by residues at the Hendra virus F TMD-ectodomain border.
The aromatic ring of Y498 is important for Hendra virus F recycling.
Our analysis of TMD polar residues also found that alanine substitution of Hendra virus F TMD residue Y498 resulted in a protein that reached the cell surface in the F0 uncleaved form at early time points but disappeared from the surface without returning in the cleaved, processed form (Fig. 8A). Analysis of total expression showed that the protein was properly cleaved (Fig. 8B), suggesting that this mutant was targeted toward degradation after cathepsin L cleavage. To further confirm that this mutant was mistrafficked after internalization and was not degraded on the cell surface by extracellular proteases, we expressed it in Vero cells along with DN Rab5, which inhibits endocytosis (73) (Fig. 8C). As seen in Fig. 8C, the protein was present on the cell surface in the F0 uncleaved form until the 4- and 8-h time points, indicating that Hendra virus F Y498A is degraded only after internalization.
Fig 8.
Hendra virus F Y498 TMD residue plays a critical role in Hendra virus F recycling. Surface biotinylation experiments were performed as described above. (A) Time course of surface expression of WT, Y498A, Y498F, Y498S, and Y498L. (B) Total expression of the proteins shown in panel A. (C) Time course of surface expression of WT and Y498A Hendra virus F in the presence or absence of DN Rab5. (D) The surface expression of WT and Y498A Hendra virus F in the presence of E64D. The cathepsin L inhibitor E64D (10 μM) was added to the medium during starvation, label, and chase. Each experiment was repeated at least three times with similar results.
To further delineate the characteristics of the Y498 residue important for Hendra virus F recycling, three additional substitutions were made: Y498F, Y498S, and Y498L. The phenylalanine residue also contains an aromatic ring; the serine presents a hydroxyl group, while the leucine is similar in size with the tyrosine but lacks the hydroxyl or aromatic elements. Time course surface biotinylations were performed, and the results indicated that only the Y498F substitution promoted proper Hendra virus F recycling (Fig. 8A), while Y498S and Y498L mutants were targeted toward degradation after cathepsin L cleavage (Fig. 8A and B).
As with other class I viral fusion proteins, proteolytic processing of Hendra virus F generates a metastable, prefusion form. At the right time and place, this cleaved form can be triggered to undergo the conformational changes needed for fusion. Since Hendra virus F Y498A, Y498S, and Y498L mutants are targeted toward degradation after cathepsin L cleavage (Fig. 8A and B), it is possible that the cleaved form of these mutants is unstable and rapidly folds to the postfusion conformation. These major conformational changes could result in the sorting toward degradation of these mutants. To determine if cathepsin L processing affects trafficking of these mutants, we performed the time course biotinylation of WT or Y498A Hendra virus F in the presence of E64d, which is a membrane-permeable inhibitor of cysteine proteases (Fig. 8D). The WT Hendra virus F maintained surface expression throughout the 8-h time course, suggesting that it was recycled back, but since cathepsin L was inactivated, only the F0 form was present. Hendra virus F Y498A was still targeted toward degradation, suggesting that premature triggering following cathepsin L cleavage is not the cause of the mistrafficking seen with these mutations. In addition, these experiments suggest that cathepsin L processing is likely not involved in the trafficking decisions of WT Hendra virus F.
The fusion activity of Hendra virus F Y498 mutants was also examined using a syncytium assay. Only the correctly cleaved and recycled Hendra virus F Y498F mutant formed syncytia (Fig. 7), while Hendra virus F Y498A, Y498S, and Y498L mutants, which were not recycled in the cleaved form, were fusogenically inactive (Fig. 7 and data not shown).
The Y498A substitution alters Hendra virus F TMD-TMD association.
Recent work from our lab has shown by analytical ultracentrifugation sedimentation equilibrium that the Hendra virus F TMDs, separate from the rest of the protein, interact and associate in a monomer-trimer equilibrium (E. C. Smith et al., submitted for publication) suggesting that TMDs may play roles in the folding and function of Hendra virus F. Previous studies of the Hendra virus F protein have shown a strong correlation between loss of surface expression and trimerization defects within the whole protein (21, 22). The S490 or Y498 mutants were transported efficiently from the endoplasmic reticulum (ER) to the cell surface, suggesting that these mutations do not significantly impact overall protein folding and trimerization. However, as our Hendra virus F model with a continuous leucine-isoleucine zipper places the Y498 residue at the helical interface, we tested if Y498 substitutions alter the association of Hendra virus F isolated TMDs. For our study we used a chimeric protein containing the staphylococcal nuclease (SN) protein linked to the Hendra virus F WT TMD (residues 485 to 520) or the TMD containing the Y498A, Y498F, or Y498S mutations. SN is a monomeric protein under the conditions used for analytical ultracentrifugation (20), allowing us to analyze the association and oligomeric status of the transmembrane domains.
The SN-Hendra virus F TMD constructs were expressed in the E. coli strain Rosetta-Gami, and the protein was purified by FPLC and exchanged into 3-(N,N-dimethylmyristyl-ammonio) propane sulfonate (C14SB) detergent (7). Samples at three different protein concentrations were brought to sedimentation equilibrium in a Beckman XL-A analytical ultracentrifuge, and radial absorbance data were obtained at 20,000, 25,000, and 30,000 rpm. The data sets were analyzed using KaleidaGraph, and the best fits for the 25,000- and 30,000-rpm speeds at one concentration are illustrated in Fig. 9. The residuals for each mutant are shown above each data set. After analyzing the data at all the three speeds and concentrations examined, we found that the overall best fit was that of monomer-trimer for the WT and Y498 TMD mutants tested. However, while Y498F Hendra virus F TMDs presented association constants similar to those of the WT F (Table 1), the association constants of Y498A were only 34% to 50% of those of the wild type (Table 1), suggesting that the Y498A mutation decreases the interactions between the TMDs. In addition, the association constants of Y498S were about 70% of that of the WT, suggesting that altered TMD interactions may contribute to the trafficking defect seen for the Hendra virus F Y498A and Y498S mutants.
Fig 9.
Sedimentation equilibrium analysis of the Hendra virus F Y498 SN-TMD proteins demonstrates a monomer-trimer equilibrium for WT F and Y498 mutants, with lower association constants for Y498A and Y498S mutants. The Hendra virus F SN-TMD was expressed in the E. coli strain Rosetta-Gami, purified, and exchanged in C14SB detergent. Samples were brought to sedimentation equilibrium in a Beckman XL-A analytical ultracentrifuge, and radial absorbance data were obtained at 20,000, 25,000, and 30,000 rpm. The best fit was determined by analyzing the square root of the variance for the three speeds and concentrations tested. The data points at 25,000 and 30,000 rpm and one concentration are presented. The residuals for each mutant are shown above each data set.
Table 1.
The association constants of Y498A, Y498F, and Y498S TMDs relative to the WT F TMD at 25,000 rpm
| F protein or mutant | Relative association constant at:a |
|
|---|---|---|
| 25,000 rpm | 30,000 rpmb | |
| WT | 1 | 0.70 ± 0.26 |
| Y498A | 0.34 ± 0.03 | 0.35 ± 0.07 |
| Y498F | 0.97 ± 0.33 | 0.80 ± 0.20 |
| Y498S | 0.67 ± 0.07 | 0.49 ± 0.18 |
The data represent averages of three or four independent protein concentrations, spun at 25,000 rpm or 30,000 rpm, relative to the WT value. The errors represent the standard error of the mean.
Relative to WT F at 25,000 rpm, (1.001 ± 0.33) × 109 M−2.
DISCUSSION
While a number of signals involved in endocytic recycling have been identified, the larger picture of what drives this important process remains unclear. Viral proteins have been outstanding models for dissecting cellular secretory trafficking pathways (11, 26, 78) because they lack an endogenous counterpart and use host cell mechanisms. During endocytotic trafficking, Hendra virus F and Nipah virus F are proteolytically processed by cathepsin L, generating the fusion-active form of the protein and providing an excellent biochemical assay to assess trafficking.
Our antibody capture experiments show that Hendra virus F is recycled to the plasma membrane through Rab5- and Rab4-positive endosomes (Fig. 4A and B), with little protein detected in late endosomes or lysosomes (Fig. 4C and D), which are classically considered the main location of cathepsin L (59, 79). These results are in agreement with a previous study which found Nipah virus F colocalization with transferrin receptor at 5 and 30 min after internalization (14). As Rab4 is involved in vesicular transport both directly to the plasma membrane and to the recycling endosomes, it is possible that both pathways may be utilized. Interestingly, a CT Myc-tagged form of Hendra virus F which contains a dileucine motif associated with slower recycling and longer retention in the endosomal recycling compartment (32, 33, 41) displayed a much lower recycling rate than WT Hendra virus F (58).
Our results suggest that cathepsin L is present and functionally active in early endosomal compartments, fitting with the finding that Hendra virus F cleavage is observed within 15 min after internalization, with the majority of Hendra virus F cleaved by 30 min (46). Recent studies have shown that cathepsin L can cleave nanoparticle-associated peptides shortly after internalization (65) and that cathepsin L proteolysis in lysosomal-like environments resulted in extensive degradation, while proteolysis under endosomal-like conditions resulted in few fragments of higher molecular weight, suggesting that the biochemical environment plays an important role in the cleavage specificity of cathepsin L (35). The lower concentration of proteases and the variation in pH present in endosomes have been proposed to promote controlled and selective proteolytic processing of proteins (1, 57). Selective early endosomal activity of cathepsin L is compatible with the specific proteolytic processing observed for Hendra virus F.
The Hendra virus F TMD is critical for F endocytosis and recycling, with TMD residues S490 and Y498 specifically modulating these processes. TMDs are often viewed as simple membrane anchors. However, recent studies have shown that TMDs play roles in protein folding (43), oligomerization (25, 52), receptor activation (76, 77), and trafficking through both secretory (47, 48, 63, 66, 71, 85) and endocytic pathways (60, 86). Plasma membrane proteins were shown to have smaller mean residue volumes in the outer leaflet of the TMD than Golgi proteins (66), suggesting that residues within the outer half of the bilayer may contribute to protein sorting and localization. The residues identified in our study, S490 and Y498, are likely situated in the outer half of the bilayer. However, our data show that characteristics beyond side chain volume are important for correct Hendra virus F trafficking and sorting.
S490 is located near the start of the predicted TMD (Fig. 1), likely situated in close proximity to the hydrophilic head groups. Correct trafficking was observed only with the Hendra virus F S490T mutation, suggesting that hydrogen bond interactions with the polar head group of the bilayer or with other plasma membrane proteins promoted by the serine or threonine side chains may be important. Phosphorylation of S490 is likely not involved in Hendra virus F recycling, but a charge at this position, either positive (S490K) or negative (S490E), allows partial recycling of Hendra virus F to the cell surface, albeit at much lower levels than WT F (Fig. 5D), potentially by anchoring the Hendra virus F TMD into the bilayer at a position similar to that of the WT Hendra virus F. Interestingly, the Hendra virus F S490A and S490V mutants display very different trafficking pathways (Fig. 5A), indicating that very small TMD changes can have dramatic effects on protein trafficking and sorting.
Residue Y498 is also critical for Hendra virus F recycling. Y498A, Y498S, and Y498L Hendra virus F substitutions are endocytosed but are sorted toward lysosomal degradation after cathepsin L processing (Fig. 8A and B). Only the Y498F mutation is tolerated, indicating that an aromatic ring at this position plays an important role in endocytic recycling (Fig. 8 A). This residue is predicted to be positioned within the hydrophobic environment of the bilayer at the interacting face of the helices (Fig. 1A and B). Our results suggest that targeting to degradation is not directed by a specific residue or signal but, rather, by the absence of the aromatic ring at this position (Fig. 8A and B). Interestingly, transferrin receptor also contains two phenylalanine residues toward the middle of its TMD, but to our knowledge their function in recycling has not been determined.
Why would an aromatic residue in the center of the TMD be important for Hendra virus F recycling? Analytical ultracentrifugation sedimentation equilibrium analysis found that isolated WT and mutant F protein TMDs associate as monomer-trimers, with association constants for the Y498F mutant similar to those of the WT TMD (Fig. 9 and Table 1). In contrast, the association constants for Y498A TMDs were much lower (Fig. 9), suggesting a decreased stability of the Y498A TMD trimer. Changes in TMD associations may lead to changes in the TMD conformation or interaction with the bilayer in the context of the whole protein, which could potentially influence trafficking decisions. As the S490 and Y498 residues are also present in the Nipah virus F TMD at the same positions, it is very likely that they are also important for Nipah virus F endocytic trafficking.
Recent work (19, 82) showed that Nipah virus F and G have a basolateral distribution in polarized cells. Since previous studies have shown that the TMDs of respiratory syncytial virus (RSV) F (6) and influenza virus hemagglutinin (43) and neuraminidase (37) are necessary for apical sorting in polarized cells, it is possible that the Hendra and Nipah virus F TMDs contribute to the basolateral trafficking of F, but further studies are needed to examine this function. In addition, the basolateral location of Nipah virus F and G suggests that the assembly process of henipaviruses occurs in the basolateral region, in agreement with the systemic infection generated by these viruses (17, 64). This contrasts with the apical budding and limited infection of the majority of paramyxoviruses, suggesting that the unique recycling pathway of F is important for the pathogenesis of henipaviruses. Taken together, our data indicate that Hendra virus F represents an excellent model for studying endocytic recycling and that the F protein TMD is important in this process, with S490 and Y498 residues playing critical roles.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01AI051517 and NIH grant U54 AI057157 from the Southeastern Regional Center of Excellence for Emerging Infections and Biodefense to R.E.D., R01GM070662 to M.G.F, and NIH grant 2P20 RR020171 from the National Center for Research Resources to R.E.D and M.G.F.
We are thankful to Lin-Fa Wang of the Australian Animal Health Laboratory for the Hendra virus F and G plasmids and to Christopher Broder for kindly providing the 7F7 monoclonal antibody to the Hendra virus F ectodomain. We also thank Gary Whittaker, Marci A. Scidmore, Walther Moses, and Richard Pagano for providing the Rabs and Lamp1 plasmids. Karen Fleming kindly provided us the pET11A SN-glycophorin TMD plasmid. We are grateful to Carole Moncman and Emilia Galperin for offering assistance with the confocal microscopy experiments and image analysis and to Jia Jianhang for access to the confocal microscopy facilities. We also thank Clint Smith for assistance with the analytical ultracentrifugation Kaleidagraph data analysis, and the members of the Dutch lab for critical reviews of the manuscript.
Footnotes
Published ahead of print 11 January 2012
REFERENCES
- 1. Authier F, Posner BI, Bergeron JJ. 1996. Endosomal proteolysis of internalized proteins. FEBS Lett. 389:55–60 [DOI] [PubMed] [Google Scholar]
- 2. Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. 2009. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J. Mol. Biol. 386:14–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bonaparte MI, et al. 2005. Ephrin-B2 ligand is a functional receptor for Hendra virus and Nipah virus. Proc. Natl. Acad. Sci. U. S. A. 102:10652–10657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonifacino JS, Traub LM. 2003. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72:395–447 [DOI] [PubMed] [Google Scholar]
- 5. Braulke T, Bonifacino JS. 2009. Sorting of lysosomal proteins. Biochim. Biophys. Acta 1793:605–614 [DOI] [PubMed] [Google Scholar]
- 6. Brock SC, Heck JM, McGraw PA, Crowe JE., Jr 2005. The transmembrane domain of the respiratory syncytial virus F protein is an orientation-independent apical plasma membrane sorting sequence. J. Virol. 79:12528–12535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Burgess NK, Stanley AM, Fleming KG. 2008. Determination of membrane protein molecular weights and association equilibrium constants using sedimentation equilibrium and sedimentation velocity. Methods Cell Biol. 84:181–211 [DOI] [PubMed] [Google Scholar]
- 8. Cabrera M, Ungermann C. 2010. Guiding endosomal maturation. Cell 141:404–406 [DOI] [PubMed] [Google Scholar]
- 9. Choudhury A, et al. 2002. Rab proteins mediate Golgi transport of caveola-internalized glycosphingolipids and correct lipid trafficking in Niemann-Pick C cells. J. Clin. Invest. 109:1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chua KB, et al. 1999. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 354:1257–1259 [DOI] [PubMed] [Google Scholar]
- 11. Copeland CS, Doms RW, Bolzau EM, Webster RG, Helenius A. 1986. Assembly of influenza hemagglutinin trimers and its role in intracellular transport. J. Cell Biol. 103:1179–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Craft WW, Jr, Dutch RE. 2005. Sequence motif upstream of the Hendra virus fusion protein cleavage site is not sufficient to promote efficient proteolytic processing. Virology 341:130–140 [DOI] [PubMed] [Google Scholar]
- 13. Dai J, et al. 2004. ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev. Cell 7:771–776 [DOI] [PubMed] [Google Scholar]
- 14. Diederich S, Moll M, Klenk HD, Maisner A. 2005. The nipah virus fusion protein is cleaved within the endosomal compartment. J. Biol. Chem. 280:29899–29903 [DOI] [PubMed] [Google Scholar]
- 15. Dutch RE. 2010. Entry and fusion of emerging paramyxoviruses. PLoS Pathog. 6:e1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dutch RE, Jardetsky TS, Lamb RA. 2000. Virus membrane fusion proteins: biological machines that undergo a metamorphosis. Biosci. Rep. 20:597–612 [DOI] [PubMed] [Google Scholar]
- 17. Eaton BT, Broder CC, Middleton D, Wang LF. 2006. Hendra and Nipah viruses: different and dangerous. Nat. Rev. Microbiol. 4:23–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ebie AZ, Fleming KG. 2007. Dimerization of the erythropoietin receptor transmembrane domain in micelles. J. Mol. Biol. 366:517–524 [DOI] [PubMed] [Google Scholar]
- 19. Erbar S, Maisner A. 2010. Nipah virus infection and glycoprotein targeting in endothelial cells. Virol. J. 7:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fleming KG, Ackerman AL, Engelman DM. 1997. The effect of point mutations on the free energy of transmembrane alpha-helix dimerization. J. Mol. Biol. 272:266–275 [DOI] [PubMed] [Google Scholar]
- 21. Gardner AE, Dutch RE. 2007. A conserved region in the F2 subunit of paramyxovirus fusion proteins is involved in fusion regulation. J. Virol. 81:8303–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gardner AE, Martin KL, Dutch RE. 2007. A conserved region between the heptad repeats of paramyxovirus fusion proteins is critical for proper F protein folding. Biochemistry 46:5094–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Granseth E, von Heijne G, Elofsson A. 2005. A study of the membrane-water interface region of membrane proteins. J. Mol. Biol. 346:377–385 [DOI] [PubMed] [Google Scholar]
- 24. Grant BD, Donaldson JG. 2009. Pathways and mechanisms of endocytic recycling. Nat. Rev. Mol. Cell Biol. 10:597–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gurezka R, Laage R, Brosig B, Langosch D. 1999. A heptad motif of leucine residues found in membrane proteins can drive self-assembly of artificial transmembrane segments. J. Biol. Chem. 274:9265–9270 [DOI] [PubMed] [Google Scholar]
- 26. Hammond C, Helenius A. 1994. Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J. Cell Biol. 126:41–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanyaloglu AC, von Zastrow M. 2007. A novel sorting sequence in the β2-adrenergic receptor switches recycling from default to the Hrs-dependent mechanism. J. Biol. Chem. 282:3095–3104 [DOI] [PubMed] [Google Scholar]
- 28. Hanyaloglu AC, von Zastrow M. 2008. Regulation of GPCRs by endocytic membrane trafficking and its potential implications. Annu. Rev. Pharmacol. Toxicol. 48:537–568 [DOI] [PubMed] [Google Scholar]
- 29. Harcourt BH, et al. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334–349 [DOI] [PubMed] [Google Scholar]
- 30. Hsu VW, Prekeris R. 2010. Transport at the recycling endosome. Curr. Opin. Cell Biol. 22:528–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jing S, Spencer T, Miller K, Hopkins C, Trowbridge IA. 1990. Role of the human transferrin receptor cytoplasmic domain in endocytosis: Localization of a specific signal sequence for internalization. J. Cell Biol. 110:283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Johnson AO, Lampson MA, McGraw TE. 2001. A di-leucine sequence and a cluster of acidic amino acids are required for dynamic retention in the endosomal recycling compartment of fibroblasts. Mol. Biol. Cell 12:367–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Johnson AO, et al. 1998. Identification of an insulin-responsive, slow endocytic recycling mechanism in Chinese hamster ovary cells. J. Biol. Chem. 273:17968–17977 [DOI] [PubMed] [Google Scholar]
- 34. Johnson LS, Dunn KW, Pytowski B, McGraw TE. 1993. Endosome acidification and receptor trafficking: bafilomycin A1 slows receptor externalization by a mechanism involving the receptor's internalization motif. Mol. Biol. Cell 4:1251–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jordans S, et al. 2009. Monitoring compartment-specific substrate cleavage by cathepsins B, K, L, and S at physiological pH and redox conditions. BMC Biochem. 10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jovic M, Sharma M, Rahajeng J, Caplan S. 2010. The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 25:99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kundu A, Avalos RT, Sanderson CM, Nayak DP. 1996. Transmembrane domain of influenza virus neuraminidase, a type II protein, possesses an apical sorting signal in polarized MDCK cells. J. Virol. 70:6508–6515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lamb RA, Jardetzky TS. 2007. Structural basis of viral invasion: lessons from paramyxovirus F. Curr. Opin. Struct. Biol. 17:427–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lamb RA, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication, p 1305–1340 In Knipe DM, et al. (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 40. Lamb RA, Parks GD. 2007. Paramyxoviridae: the viruses and their replication, p 1449–1496 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 41. Lampson MA, Schmoranzer J, Zeigerer A, Simon SM, McGraw TE. 2001. Insulin-regulated release from the endosomal recycling compartment is regulated by budding of specialized vesicles. Mol. Biol. Cell 12:3489–3501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RA. 1993. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J. Mol. Biol. 229:602–608 [DOI] [PubMed] [Google Scholar]
- 43. Lin S, Naim HY, Rodriguez AC, Roth MG. 1998. Mutations in the middle of the transmembrane domain reverse the polarity of transport of the influenza virus hemagglutinin in MDCK epithelial cells. J. Cell Biol. 142:51–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Maxfield FR, McGraw TE. 2004. Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5:121–132 [DOI] [PubMed] [Google Scholar]
- 45. McGrath ME. 1999. The lysosomal cysteine proteases. Annu. Rev. Biophys. Biomol. Struct. 28:181–204 [DOI] [PubMed] [Google Scholar]
- 46. Meulendyke KA, Wurth MA, McCann RO, Dutch RE. 2005. Endocytosis plays a critical role in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12643–12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Miyauchi K, et al. 2010. The membrane-spanning domain of gp41 plays a critical role in intracellular trafficking of the HIV envelope protein. Retrovirology. 7:95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Munro S. 1995. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 14:4695–4704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Negrete OA, et al. 2005. Ephrin B2 is the entry receptor for Nipah virus, an emergent deadly paramyxovirus. Nature 436:401–405 [DOI] [PubMed] [Google Scholar]
- 50. Negrete OA, et al. 2006. Two key residues in Ephrin B3 are critical for its use as an alternative receptor for Nipah virus. PLoS Pathog. 2:e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Niwa H, Yamamura K, Miyazaki J. 1991. Efficient selection for high-expression transfectants by a novel eukaryotic vector. Gene 108:193–200 [DOI] [PubMed] [Google Scholar]
- 52. Oates J, Hicks M, Dafforn TR, DiMaio D, Dixon AM. 2008. In vitro dimerization of the bovine papillomavirus E5 protein transmembrane domain. Biochemistry 47:8985–8992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pagano A, Crottet P, Prescianotto-Baschong C, Spiess M. 2004. In vitro formation of recycling vesicles from endosomes requires adaptor protein-1/clathrin and is regulated by rab4 and the connector rabaptin-5. Mol. Biol. Cell 15:4990–5000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pager CT, Craft WW, Jr, Patch J, Dutch RE. 2006. A mature and fusogenic form of the Nipah virus fusion protein requires proteolytic processing by cathepsin L. Virology 346:251–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pager CT, Dutch RE. 2005. Cathepsin L is involved in proteolytic processing of the Hendra virus fusion protein. J. Virol. 79:12714–12720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pager CT, Wurth MA, Dutch RE. 2004. Subcellular localization and calcium and pH requirements for proteolytic processing of the Hendra virus fusion protein. J. Virol. 78:9154–9163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pillay CS, Elliott E, Dennison C. 2002. Endolysosomal proteolysis and its regulation. Biochem. J. 363:417–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Popa A, Pager CT, Dutch RE. 2011. C-terminal tyrosine residues modulate the fusion activity of the Hendra virus fusion protein. Biochemistry 50:945–952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Press B, Feng Y, Hoflack B, Wandinger-Ness A. 1998. Mutant Rab7 causes the accumulation of cathepsin D and cation-independent mannose 6-phosphate receptor in an early endocytic compartment. J. Cell Biol. 140:1075–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reggiori F, Black MW, Pelham HR. 2000. Polar transmembrane domains target proteins to the interior of the yeast vacuole. Mol. Biol. Cell 11:3737–3749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Rutledge EA, Mikoryak CA, Draper RK. 1991. Turnover of the transferrin receptor is not influenced by removing most of the extracellular domain. J. Biol. Chem. 266:21125–21130 [PubMed] [Google Scholar]
- 62. Rzomp KA, Scholtes LD, Briggs BJ, Whittaker GR, Scidmore MA. 2003. Rab GTPases are recruited to chlamydial inclusions in both a species-dependent and species-independent manner. Infect. Immun. 71:5855–5870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schaecher SR, Diamond MS, Pekosz A. 2008. The transmembrane domain of the severe acute respiratory syndrome coronavirus ORF7b protein is necessary and sufficient for its retention in the Golgi complex. J. Virol. 82:9477–9491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schmitt AP, Lamb RA. 2004. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 283:145–196 [DOI] [PubMed] [Google Scholar]
- 65. See V, et al. 2009. Cathepsin L digestion of nanobioconjugates upon endocytosis. ACS Nano 3:2461–2468 [DOI] [PubMed] [Google Scholar]
- 66. Sharpe HJ, Stevens TJ, Munro S. 2010. A comprehensive comparison of transmembrane domains reveals organelle-specific properties. Cell 142:158–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sherer NM, et al. 2003. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic 4:785–801 [DOI] [PubMed] [Google Scholar]
- 68. Sieczkarski SB, Whittaker GR. 2003. Differential requirements of Rab5 and Rab7 for endocytosis of influenza and other enveloped viruses. Traffic 4:333–343 [DOI] [PubMed] [Google Scholar]
- 69. Smith EC, Popa A, Chang A, Masante C, Dutch RE. 2009. Viral entry mechanisms: the increasing diversity of paramyxovirus entry. FEBS J. 276:7217–7227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Somsel Rodman J, Wandinger-Ness A. 2000. Rab GTPases coordinate endocytosis. J. Cell Sci. 113:183–192 [DOI] [PubMed] [Google Scholar]
- 71. Sousa VL, et al. 2003. Importance of Cys, Gln, and Tyr from the transmembrane domain of human alpha 3/4 fucosyltransferase III for its localization and sorting in the Golgi of baby hamster kidney cells. J. Biol. Chem. 278:7624–7629 [DOI] [PubMed] [Google Scholar]
- 72. Stein A, Weber G, Wahl MC, Jahn R. 2009. Helical extension of the neuronal SNARE complex into the membrane. Nature 460:525–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stenmark H, et al. 1994. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 13:1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stenmark H, et al. 1994. Distinct structural elements of rab5 define its functional specificity. EMBO J. 13:575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sulistijo ES, Jaszewski TM, MacKenzie KR. 2003. Sequence-specific dimerization of the transmembrane domain of the “BH3-only” protein BNIP3 in membranes and detergent. J. Biol. Chem. 278:51950–51956 [DOI] [PubMed] [Google Scholar]
- 76. Talbert-Slagle K, DiMaio D. 2009. The bovine papillomavirus E5 protein and the PDGF beta receptor: it takes two to tango. Virology 384:345–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Talbert-Slagle K, et al. 2009. Artificial transmembrane oncoproteins smaller than the bovine papillomavirus E5 protein redefine sequence requirements for activation of the platelet-derived growth factor beta receptor. J. Virol. 83:9773–9785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Thor F, Gautschi M, Geiger R, Helenius A. 2009. Bulk flow revisited: transport of a soluble protein in the secretory pathway. Traffic 10:1819–1830 [DOI] [PubMed] [Google Scholar]
- 79. Turk D, Guncar G. 2003. Lysosomal cysteine proteases (cathepsins): promising drug targets. Acta Crystallogr. D Biol. Crystallogr. 59:203–213 [DOI] [PubMed] [Google Scholar]
- 80. van der Sluijs P, et al. 1992. The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70:729–740 [DOI] [PubMed] [Google Scholar]
- 81. Vogt C, Eickmann M, Diederich S, Moll M, Maisner A. 2005. Endocytosis of the Nipah virus glycoproteins. J. Virol. 79:3865–3872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Weise C, et al. 2010. Tyrosine residues in the cytoplasmic domains affect sorting and fusion activity of the Nipah virus glycoproteins in polarized epithelial cells. J. Virol. 84:7634–7641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Whitman SD, Dutch RE. 2007. Surface density of the Hendra G protein modulates Hendra virus F protein-promoted membrane fusion: role for Hendra G protein trafficking and degradation. Virology 363:419–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Whitman SD, Smith EC, Dutch RE. 2009. Differential rates of protein folding and cellular trafficking for the Hendra virus F and G proteins: implications for F-G complex formation. J. Virol. 83:8998–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yang M, Ellenberg J, Bonifacino JS, Weissman AM. 1997. The transmembrane domain of a carboxyl-terminal anchored protein determines localization to the endoplasmic reticulum. J. Biol. Chem. 272:1970–1975 [DOI] [PubMed] [Google Scholar]
- 86. Zaliauskiene L, et al. 2000. Down-regulation of cell surface receptors is modulated by polar residues within the transmembrane domain. Mol. Biol. Cell 11:2643–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]