Abstract
Postexposure treatment (PET) of wild-type rabies virus (RV)-infected mice with a live-attenuated triple-glycoprotein RV variant (TriGAS) promotes survival but does not prevent the pathogenic RV from invading and replicating in the brain. Successful PET is associated with the induction of a robust virus-neutralizing antibody response and clearance of the wild-type RV from brain tissues. Comparison of the transcriptomes of normal mouse brain with those of wild-type-RV-infected mice that had received either mock or TriGAS PET treatment revealed that many of the host genes activated in the mock-treated mice represent type I interferon (IFN) response genes. This indicates that RV infection induces an early type I IFN response that is unable to control the infection. In contrast, most of the activated genes in the brain of the RV-infected, TriGAS-treated mouse play a role in adaptive immunity, including the regulation of T cell activation, T cell differentiation, and the regulation of lymphocyte and mononuclear cell proliferation. These findings were confirmed by quantitative PCR (qPCR) array studies, which showed that 3 genes in particular, encoding chemokine ligand 3 (Ccl3), natural killer cell activator 2 (interleukin 12B [IL-12B]), and granzyme A (GzmA), were activated earlier and to a greater extent in the brains of RV-infected mice treated with TriGAS than in the brains of mock-treated mice. The activation of these genes, known to play key roles in the regulation of lymphocyte and mononuclear cell proliferation, is likely an important part of the mechanism by which TriGAS mediates its PET activity.
INTRODUCTION
Rabies is a zoonotic disease that remains an important public health issue, causing an estimated 55,000 human deaths globally each year (2). The causative agent is rabies virus (RV), a negative-strand RNA virus of the family Rhabdoviridae. The main feature of RV is neuroinvasiveness, which refers to its unique ability to invade the central nervous system (CNS) from peripheral sites, which is a major facet of its pathogenicity. Virus uptake, axonal transport, transsynaptic spread, and the rate of viral replication are key factors that determine the neuroinvasiveness of particular RV variants (7–11, 34, 35). In addition to neuroinvasiveness, the rate of viral replication appears to be one of the important arbiters of RV pathogenesis. Pathogenic RVs have considerably lower replication rates than attenuated strains. This benefits the pathogenic RV strains both by conserving the structures of neurons that are used by these viruses to reach the CNS and by limiting exposure of viral antigens to the host immune system (35). Alternatively, the more rapidly replicating attenuated recombinant RV strains induce strong innate and adaptive immune responses that pathogenic RVs tend to evade. In particular, attenuated recombinant RVs express large amounts of viral surface glycoprotein (G), the RV antigen that elicits the production of the RV-neutralizing antibodies (VNAs) (5), which play a crucial role in preventing RV invasion of the CNS (40, 42). The immunogenicity of live-attenuated recombinant RV strains is enhanced through increasing G expression by introducing up to two additional G genes into the RV genome (12, 13). The recently developed live-attenuated recombinant RV vaccine strain TriGAS, which contains three G genes, has been shown to be highly immunogenic and nonpathogenic for mice that either are developmentally immunocompromised or have inherited deficits in immune function, as well as normal adult animals, even when administered intracranially (12). Immunization with live, but not UV-inactivated, TriGAS induces immune effectors and processes that promote infiltration of T cells into the CNS tissues so that a pathogenic RV infection in the CNS may be contained and cleared, preventing an otherwise lethal outcome (12). This more desirable outcome refutes the general belief that death from an established CNS infection with pathogenic RV is virtually inevitable.
Although certain mechanisms have been implicated in the clearance of an RV infection from the CNS (12, 40, 46), the specific host-virus interactions targeted by TriGAS that result in the clearance of pathogenic RV from the CNS have not been resolved. While VNAs are a known requirement for the clearance of an established RV infection in the CNS, contributions from cell-mediated immune responses are likely to be necessary for the clearance (3, 20, 56).
Genomic technologies have facilitated studies of complex biological systems, including pathways involved in the host-pathogen response (54). In this study, we applied transcriptome analysis in combination with real-time quantitative PCR (qPCR) array analysis to identify transcriptional events that might contribute to the clearance of an RV infection from the CNS and the prevention of lethal RV encephalopathy. These analyses reveal that the safe clearance of RV from the CNS correlates with the rapid production of high levels of serum RV-neutralizing antibodies and the induction of factors that promote the activity of immune effectors in brain tissue.
MATERIALS AND METHODS
Viruses.
The recombinant RV SPBAANGAS-GAS-GAS (TriGAS), which contains three G genes, was generated as described elsewhere (12). The pathogenic RV strain DOG4 was propagated in NA cells (C1300 mouse neuroblastoma, clone NA) as described previously (12).
Determination of virus titers.
To determine virus titers, NA cells were grown for 2 days, and the monolayers were infected with virus in 10-fold serial dilutions. Forty-eight hours postinfection (p.i.), the cells were fixed with 80% acetone and stained with fluorescein isothiocyanate (FITC)-labeled RV nucleoprotein (N)-specific antibody (Fujirebio Diagnostics, Malvern, PA). Virus titers in triplicate samples were determined using a fluorescence microscope.
Infection and PET of mice.
Eight- to 10-week-old female C57BL/6 mice were purchased from the National Cancer Institute at Frederick, MD. To determine the survival after postexposure treatment (PET), groups of 10 mice were infected under anesthesia intranasally (i.n.) with 10 times the 50% lethal doses (LD50) of the highly pathogenic RV strain DOG4. The i.n. route was chosen because it is the most efficient means of rapidly promoting RV entry into the brain while avoiding the injury that induces innate immune mechanisms that accompany intracranial (i.c.) RV administration. Four hours after infection, one group was treated intramuscularly (i.m.) (in the masseter muscle) with 107 focus-forming units (FFU) of the live-attenuated TriGAS RV vaccine strain (12) in 100 μl phosphate-buffered saline (PBS), and the other group was mock treated with 100 μl PBS. Groups of healthy 10-week-old female C57BL/6 mice (uninfected and untreated mice) served as controls. The mice were observed daily for 40 days for clinical signs of rabies. Moribund mice were euthanized. All procedures were conducted in accordance with the Public Health Service Policy on Humane Care and Use of Laboratory Animals under protocols approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University.
Real-time PCR analysis.
Forty 8- to 10-week old female C57BL/6 mice were infected i.n. with DOG4, and 4 h later, 20 of the infected mice were treated with TriGAS and the other 20 mice were mock treated with PBS as described above. A group of 4 healthy 10-week-old female C57BL/6 mice (uninfected and untreated mice) served as controls. Four, 6, 8, 12, and 40 days p.i., 4 mice each from the TriGAS- and 4 mock-treated groups were euthanized. After blood collection and cardiac perfusion, the brains were removed and total RNA was isolated from the brains using the RNeasy minikit (Qiagen, Valencia, CA) according to the manufacturer's instructions. For quantification of mRNA, reverse transcription (RT) and qPCR were performed on each individual mouse brain RNA in quadruplicate as described previously (12, 40). Briefly, cDNA was synthesized by reverse transcription using oligo(dT)15 primer (Promega, Madison, WI). qPCR was performed on the iCycler iQ real-time detection system (Bio-Rad), using iQ Supermix (Bio-Rad Laboratories, Hercules, CA) and gene-specific probes and primers or iQ SYBR green Supermix (Bio-Rad) and gene-specific primers (IFN-α4 and IFN-β1). The mRNA copy numbers for a particular gene were normalized to the copy numbers of housekeeping gene L13 in each sample. The primers and probes used in quantitative PCR for testing IFN-α4, IFN-β1, and RV N of DOG4 (DOG4 N) were as follows: IFN-α4 forward (5′-ATTTTGGATTCCCCTTGGAG-3′) and reverse (5′-TGATGGAGGTCATTGCAGAA-3′); IFN-β1 forward (5′-CATCAACTATAAGCAGCTCCA-3′) and reverse (5′-TTCAAGTGGAGAGCAGTTGAG-3′); and DOG4 N forward (5′-CAGGCATGAACGCTTCCAAA-3′), reverse (5′-CGTCAGTGCGCTTTATCTCCAA-3′), and probe (6-carboxyfluorescein [FAM-5′-TGATCCCGATGATGTATGCTCCTACTTGGC-3′-6-carboxytetramethylrhodamine [TAMRA]). The sequences of the primers and probes for other genes, including TriGAS N, have been described previously (10, 40).
Measurement of serum antibody titers.
Mouse sera were tested for the presence of VNA by the rapid fluorescent focus inhibition test (RFFIT) as described previously (53). In brief, RV CVS-11 was treated with 3-fold serially diluted mouse sera and then added to 2-day-old 90% confluent NA cells. Thirty hours after infection, the cells were fixed with 80% acetone and stained with FITC-labeled RV N-specific antibody (Fujirebio Diagnostics). Neutralization titers, defined as the inverse of the highest serum dilution that neutralizes 50% of the infected virus, were normalized to international units (IU) using the World Health Organization anti-RV antibody standard. Levels of RV-specific IgG1, IgG2a, IgG2b, and IgM in sera were assessed by enzyme-linked immunosorbent assay (ELISA) as described previously (20). Briefly, 96-well plates coated with purified RV G antibodies captured from serially diluted sera were detected with alkaline phosphatase-conjugated anti-mouse IgG1 (Pharmingen), IgG2a (Cappel), IgG2b (Cappel), and IgM (Cappel), and p-nitrophenyl phosphate (pNNP) (Sigma) was used as a substrate. Absorbance was measured at 405 nm for phosphatase activity in a microplate spectrophotometer (Biotek, Winooski, VT).
Statistical analyses.
The statistical significance of differences in the expression of specific target genes was assessed using one-way analysis of variance (ANOVA) and Tukey's multiple-comparison test. The graphs were plotted and analyzed using Graph PadPrism.
Whole-transcriptome analysis of mouse brain RNA.
Total brain RNAs isolated from the brains of single mice representing the groups of healthy mice and TriGAS- or mock-treated DOG4-infected mice at 6 days p.i. were used for transcriptome analysis. Single mice were selected based on the expression levels of mRNAs for RV N and protein markers of innate and adaptive immune responses described above. Whole-transcriptome analysis was performed using the Applied Biosystems SOLiD 4 platform (Applied Biosystems, Foster City, CA).
Mapping of RNA-seq fragments to the mouse genome.
Mouse whole-transcriptome RNA sequence reads were mapped to the mm9 genome using Life Technologies/Applied Biosystems BioScope software, version 1.3. Default parameters for 50 base reads were used to perform the mapping. Transcript expression values were then estimated using the “count known exons” analysis pipeline in Bioscope, which produced read per kilobase of exon per million mapped reads (RPKM) values for each exon.
Differential gene expression.
Differential comparisons included DOG4 RV infection plus mock treatment versus normal, DOG4 RV infection plus TriGAS treatment versus DOG4 RV infection plus mock treatment, and DOG4 RV infection plus TriGAS treatment versus normal. Preliminary differential expression analysis was performed using Student's t test on RPKM values. Because replicates were not used for RNA-seq, each exon in multiexon genes was treated as a replicate value for that gene, and differential expression P values were computed from the t statistic of the log2 RPKM expression ratio across a gene's constituent exons. This was followed by a more comprehensive analysis of differential expression using the Cuffdiff program available in the Cufflinks software package (http://cufflinks.cbcb.umd.edu/manual.html) (49). The set of transcripts analyzed by Cuffdiff included the mouse refGene database, obtained from the UCSC genome browser (version dated 20 August 2009), in addition to mouse-specific noncoding RNAs downloaded from the functional RNA database (version dated 23 February 2010) at http://www.ncrna.org. Noncoding RNAs annotated as C_D_box_snoRNA, H_ACA_box_snoRNA, antisense_RNA, lincRNA, mature_transcript, nc_conserved_region, pre_miRNA, snRNA, snoRNA, and tRNA, were evaluated for differential expression, but molecules annotated as simply “ncRNA” were excluded due to their large numbers and lack of specific annotation. Cuffdiff was run on the mapping results produced by Bioscope and produced its own normalized expression values as fragments per kilobase of exon per million fragments mapped (FPKM), in addition to differential expression P values for both protein-coding and noncoding transcripts. The resulting P values were adjusted for multiple testing using the Benjamini-Hochberg correction (58). Significant genes were defined as having an adjusted P value of ≤0.05 and absolute fold change greater than 1.5.
Functional annotation.
Gene ontology enrichment analysis was performed on sets of significant differentially regulated transcripts using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) version 6.7. Hierarchical clustering was performed among the transcripts differentially expressed in at least one of these three comparisons in order to subdivide them into groups based on patterns of up- and downregulation. Gene groups resulting from the hierarchical-clustering analysis were also analyzed for gene ontology term enrichment.
RT-PCR array analysis.
A subset of genes with differential-expression t test P values below 0.01 was used for validation on RT-PCR arrays. Total RNA was DNase I treated and extracted with the RNeasy Mini Kit (Qiagen) from the brains of mouse groups as described above. RNA (2.5 μg) was reverse transcribed using an RT2 First-Strand Kit (Qiagen) and mixed with RT2 SYBR green qPCR MasterMix (Qiagen). Each sample was loaded onto a custom RT2 Profiler PCR 96-well array plate (SABiosciences), and qRT-PCR was performed on an ABI Prism 7900. Gene expression was calculated from three samples per time point using the ΔΔCT method, and the array data analysis was performed as described in the manufacturer's instructions. The significance of differences between the treatment and control groups was assessed using scatter plot analysis with a fold change boundary greater than 2, as described on the SABiosciences web portal. Heat maps were constructed from the RT-PCR array data using a Microsoft Excel Visual Basic macro modified from the version online.
RESULTS
PET with TriGAS induces high levels of circulating VNA and the expression of markers of innate and adaptive immunity in the brain, promoting the clearance of pathogenic RV from mouse brains.
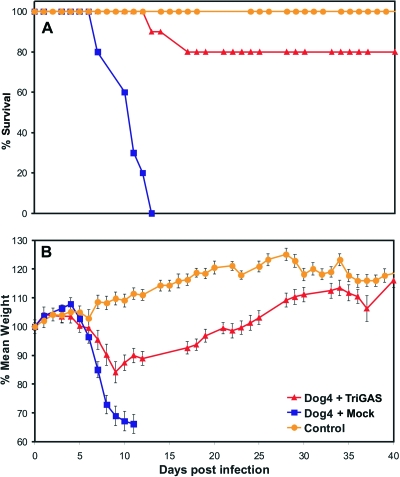
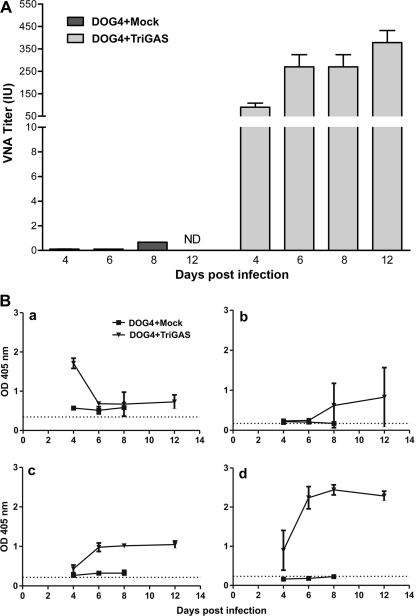
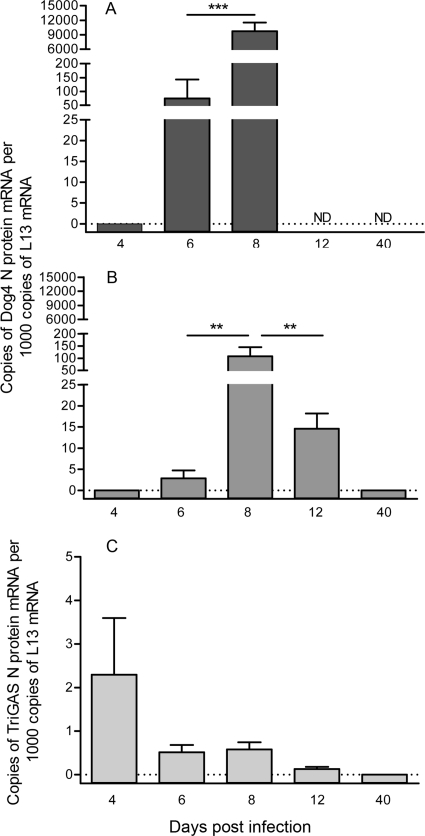
To identify elements of the innate and adaptive immune responses that play essential roles in the clearance of pathogenic RV from the CNS, we used a mouse rabies PET model in which 8- to 10-week-old female C57BL/6 mice were infected i.n. with 10 LD50 of DOG4 RV and then treated 4 h later i.m. with either PBS (mock treatment) or 107 FFU of live-attenuated TriGAS vaccine. Whereas all mock-treated mice succumbed to the DOG4 RV challenge infection, 80% of the TriGAS-treated mice survived the infection. The TriGAS-treated mice that survived showed only subtle clinical signs and loss of body weight (∼10 to 12%) between 8 and 17 days p.i. but completely recovered by 40 days after infection (Fig. 1). Comparison of serum VNA titers at 4, 6, 8, and 12 days after challenge infection revealed that TriGAS-treated DOG4-infected mice develop RV-specific immunity considerably more rapidly than mock-treated DOG4-infected mice (Fig. 2). While low levels of VNA become detectable in mock-treated mice only by day 8 (<0.5 IU) after challenge, TriGAS-treated mice produce high VNA titers (>50 IU) as early as 4 days after challenge (Fig. 2A). Serum VNA levels continued to increase in the TriGAS-treated animals, reaching titers of 350 IU at 12 days p.i. In addition, isotype switching from the IgM that predominates at day 4 p.i. to the IgG isotypes, which are known to be effective against an RV infection in vivo (47), rapidly occurred in the TriGAS-treated animals (Fig. 2B). It is generally considered that the principal contribution of circulating VNA to the anti-RV response is through blocking virus spread to the CNS. To provide insight into the possibility that the early onset of VNA in TriGAS-treated animals may prevent DOG4 RV from spreading into the CNS, we used qPCR analysis to compare transcription of the mRNAs for the N proteins of DOG4 and TriGAS in CNS tissues at different times after challenge. No TriGAS N mRNA was detected with primer-specific DOG4 N mRNA probe and vice versa, indicating that the detection of these two RV N mRNAs is mutually exclusive (see Fig. S1 in the supplemental material). As shown in Fig. 3, DOG4 N mRNA is readily detected in the brains of both mock- and TriGAS-treated mice at 6 days after infection, albeit at a considerably lower level in the latter. DOG4 N mRNA levels increased in the brains of both groups of mice between days 6 and 8 p.i., indicating that virus replication likely occurs in the CNS, but levels in the brains of TriGAS-treated animals remained lower and declined significantly by day 12 p.i., by which time the mock-treated mice had died. No DOG4 N mRNA could be detected in the brains of surviving TriGAS-treated mice at day 40 p.i. TriGAS N mRNA copies were also detected in the brains of the treated mice at low levels, which were highest at day 4 p.i. and undetectable by 12 days p.i.
Fig 1.
Postexposure treatment of mice with TriGAS promotes survival of infection with DOG4 RV. Groups of 10 adult C57BL/6 mice were infected i.n. with 105 FFU of DOG4 RV and 4 h later treated i.m. (in the masseter muscle) with PBS (mock treatment) or 107 FFU of TriGAS as described in Materials and Methods. Ten uninfected, untreated mice served as controls. The mice were observed for 4 weeks, and survival (A) and body weight (B) are presented. The error bars indicate standard errors (±SE) .
Fig 2.
Postexposure TriGAS treatment induces the rapid development of RV-specific antibodies in mice intranasally infected with DOG4 RV. (A) Groups of C57BL6 mice were infected i.n. with 105 FFU of DOG4 RV and treated with107 FFU of TriGAS or mock treated with PBS 4 h later. Blood samples were obtained 4, 6, 8, and 12 days afterward, and VNA titers were determined as described in Materials and Methods, normalized to international units using the World Health Organization standard, and presented as mean titers (plus or minus standard deviations [±SD]). (B) RV G-specific antibody isotypes were determined using ELISA as described in Materials and Methods and are presented in graphs a (IgM), b (IgG1), c (IgG2a), and d (IgG2b) as means ± SD of the absorbance at a serum dilution of 1:10.
Fig 3.
Postexposure treatment of DOG4 RV-infected mice with TriGAS rapidly reduces the accumulation of RV N mRNA in the brain. Mice were infected i.n. with 105 FFU of DOG4 RV and either mock treated with PBS (A) or treated with 107 FFU of TriGAS (B and C) 4 h later. Four mice per group were euthanized at each indicated time point, and the number of DOG4 RV N mRNA (A and B) or TriGAS RV N mRNA (C) copies in brain tissue samples was quantified by qRT-PCR as described in Materials and Methods. The results are presented as the mean N mRNA copy numbers (±SE) per 1,000 copies of L13 mRNA. The bars indicate significant differences in copy numbers between different time points determined by one-way ANOVA and Tukey's multiple-comparison test. *** and **, P < 0.001 and P < 0.01, respectively. ND, not determined.
Despite the rapid production of VNA shortly after TriGAS treatment, DOG4 RV was able to invade the brain and replicate prior to clearance, which occurred several days after circulating VNA levels had become extensive. These findings are consistent with the considerable evidence that immune mediators other than VNA, including type I interferons and T cell products, are likely to be essential in the clearance of the virus from the CNS (20, 21, 40).
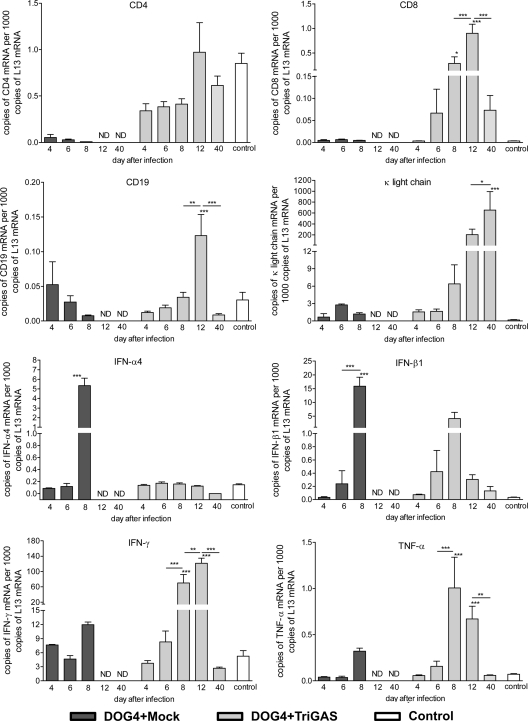
Since TriGAS, in addition to DOG4, invaded the brain in animals that received PET, we speculated that TriGAS may confer protection by enhancing the development of innate and specific antiviral immunity in brain tissues. To test the hypothesis, we used qPCR analysis to compare the expression of several adaptive and innate immune effectors in the brains of mock- or TriGAS-treated DOG4-infected mice at different times after challenge (Fig. 4). IFN-α4 and IFN-β1 mRNAs were expressed at the highest levels in the brains of the mock-treated, DOG4-infected mice at day 8 p.i., correlating with the expression levels of DOG4 RV N mRNA. In the brains of TriGAS-treated mice, IFN-β1 mRNA was significantly increased, peaking at day 8 p.i., again correlating with the highest levels of DOG4 N mRNA. However, IFN-α4 levels were not elevated in these animals. Levels of mRNA specific for the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) were significantly increased in only the TriGAS-treated mice at days 8 and 12 postchallenge, a pattern that was also evident for gamma interferon (IFN-γ) expression. Evidence of immune cell infiltration was also restricted to the TriGAS-treated, DOG4-challenged mice. mRNAs specific for CD8 and CD19, a marker found on B cell subsets but not plasma cells, were both present at their highest levels in TriGAS-treated mice at 12 days postchallenge but were not significantly elevated at any point in the mock-treated animals. Brain tissues from mock-treated, DOG4-infected mice showed substantially reduced levels of CD4 transcript. TriGAS treatment of DOG4 RV-infected mice limited the reduction in brain CD4 transcripts, which showed a trend toward becoming elevated at 12 days p.i. Finally, κ light-chain mRNA, indicative of antibody production, was elevated to high levels only in the TriGAS-treated mice, being initially detected at day 8 postchallenge and continuing to rise over the 12- and 40-day time points. Notably, the increased expression of the mRNAs relevant to innate and adaptive immunity seen in the TriGAS-treated, DOG4 RV-infected mice is due to combined infection with both viruses. The administration of TriGAS i.m. in the absence of a DOG4 RV challenge, like the treated animals, results in a low TriGAS N mRNA copy number in CNS tissues but does not significantly increase the CNS expression of mRNAs for any of the adaptive and innate immune markers studied here at any time point (see Fig. S2 in the supplemental material).
Fig 4.
The accumulation of mRNAs specific for markers of adaptive and innate immunity in mouse brains after DOG4 RV i.n. infection differs between mock and TriGAS treatments. Four mice per group were euthanized at each indicated time point, and the number of mRNA copies present in brain tissue samples was quantified by qRT-PCR as detailed in Materials and Methods. The data are presented as the mean (±SD) mRNA copy numbers per 1,000 copies of L13 mRNA. The asterisks indicate significant differences in the copy numbers between different time points determined by one-way ANOVA and Tukey's multiple-comparison test: ***, **, and * represent P values of less than 0.001, 0.01, and 0.05, respectively.
Comparison of the kinetics of RV replication in CNS tissues with those of markers of immunity suggests that the activity of antigen-specific T and B cells in the CNS tissues may ultimately be responsible for the clearance of DOG4. However, the initial reduction in DOG4 N mRNA accumulation in TriGAS-treated animals manifests at 6 days postchallenge, before mRNAs for either the innate or adaptive markers of immunity studied have become significantly elevated in CNS tissues. This suggests that additional processes induced by TriGAS that are not reflected by these markers may contribute to the early inhibition of DOG4 replication.
The induction of early responses in the brain that might play a decisive role in the protection against lethal encephalopathy is revealed by transcriptome analysis.
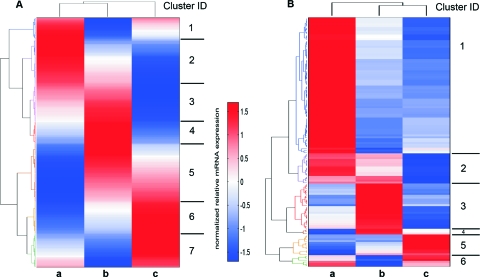
To obtain insight into the early host responses in the CNS that could influence the outcome of the infection, we performed comparative whole-transcriptome analysis of normal mouse brain with the brains of mock an-d TriGAS-treated, DOG4 RV-infected mouse brains that were collected at day 6 p.i. Differential expression analysis initially identified 411 protein-coding mRNAs that were differentially expressed in the brains of these mice (Fig. 5A). Hierarchical clustering of the genes differentially expressed between treatment groups produced seven distinct clusters (Fig. 5A; see Table S1 in the supplemental material). Cluster 1, consisting of genes that in comparison with normal mouse brain are overexpressed in DOG4 RV-infected, mock-treated brain but downregulated in TriGAS-treated mouse brain, contains 35 mRNAs involved in nucleotide and nucleic acid metabolic processes, transcription, and chromosome organization. Cluster 2, in which gene expression is increased in mock-treated and to a lesser extent in TriGAS-treated mouse brain, has 73 members, including mRNAs implicated in the regulation of virus growth, antigen processing, and nervous system development. The 63 mRNAs of cluster 3 are also upregulated in infected mouse brain regardless of treatment but to a greater extent with TriGAS treatment and include genes that regulate defense responses to virus, including innate immune responses and immune effector processes. Cluster 4 consists of genes that are strongly upregulated in TriGAS-treated mouse brain but remain almost unchanged in mock-treated mouse brain. Within these 37 mRNAs are those with roles in adaptive immunity, including the regulation of lymphocyte and mononuclear cell proliferation and T cell differentiation, as well as the regulation of antigen receptor signaling pathways, calcium ion sequestration, and programmed cell death. Genes that are expressed at higher levels in TriGAS-treated but lower levels in mock-treated brain than in normal brain comprise cluster 5. Members of the 95 mRNAs of this cluster have been implicated in the control of programmed cell death, the indol metabolic process, the ephrin receptor signaling pathway, vessel morphogenesis, and the regulation of cell adhesion. Cluster 6 consists of 53 mRNAs that in comparison to normal brain are downregulated in both mock-treated and TriGAS-treated mouse brain, but to a much lesser extent in the latter. Included are genes involved in the regulation of cation and metal ion transport, transmembrane transport, intracellular signaling cascades, neuropeptide signaling pathways, and establishment of tissue polarity, among other functions. Finally, the 55 mRNAs that are primarily downregulated in TriGAS-treated mouse brain are grouped in cluster 7. Among these are genes with a role in the positive regulation of biosynthetic and macromolecular metabolic processes and the metabotropic glutamate receptor pathway.
Fig 5.
Clusters of genes are differentially expressed in the transcriptomes of whole brains from DOG4-infected, mock-treated (a), DOG4-infected, TriGAS-treated (b), and healthy, uninfected, and untreated (c) mice at 6 days p.i. Hierarchical-clustering heat maps depict differential expression of protein-coding RNA transcripts (A) and noncoding RNAs (B) assessed by RNA sequencing. The transcript profiles are mean centered and scaled to unitary standard deviation, so that values above the mean are shown in red and values below the mean are shown in blue.
Subsequent differential-expression analysis of noncoding transcripts identified 132 RNAs with significant differences among the brains of DOG4-infected, mock-treated, and TriGAS RV-treated mice (Fig. 5B). Similar to protein-coding mRNAs, the 132 differentially expressed noncoding RNAs (ncRNAs) in the mouse brains can be mapped into six clusters as follows: cluster 1, high expression in mock-treated brain with roughly equivalent lower expression in normal mouse brain and TriGAS-treated mouse brain; cluster 2, relatively high expression in mock-treated and TriGAS-treated mouse brains compared to normal mouse brain; cluster 3, strongly elevated in TriGAS-treated mouse brain, with comparably low levels in normal and mock-treated mouse brains; cluster 4, medial expression in normal mouse brain with high expression in TriGAS-treated and low expression in mock-treated mouse brains; cluster 5, expressed at the highest levels in normal mouse brain, downregulated in TriGAS-treated and more so in mock-treated mouse brains; and cluster 6, downregulated in the TriGAS-treated mouse brain relative to both normal and mock-treated mouse brains (see Table S2 in the supplemental material). Long intergenic ncRNAs (lincRNAs), which are suggested to play a role in cell regulation, also appear in several clusters. The majority of the ncRNAs of clusters 1 and 2 are small nucleolar RNAs (snoRNAs), which have been associated with pseudouridylation of other RNAs but may also function as microRNAs (25). Several of these differentially regulated snoRNAs were identified as H/ACA box and C/D box RNAs. These are small RNA molecules that primarily guide chemical modifications of other RNAs, mainly ribosomal RNAs, transfer RNAs, and small nuclear RNAs (26). Interestingly, the majority of ncRNAs in clusters 5 and 6, downregulated in the brains of the mock-treated and TriGAS-treated mouse, respectively, consist mainly of noncoding antisense RNAs.
TriGAS treatment induces the early expression in the brain of a subset of mRNAs associated with lymphocyte activation and inflammatory responses.
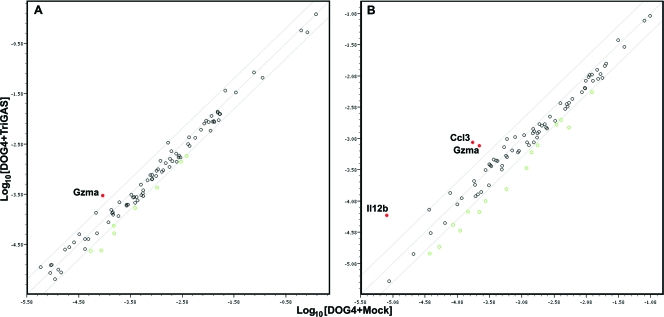
To validate the results of the transcriptome analysis and to obtain information on the kinetics of expression of particular host genes in the brain during disease progression or clearance of DOG4 RV infection, we used PCR array analysis. The expression of 90 genes identified by the ontology analysis as having an absolute fold change greater than 2 with an adjusted P value of ≤0.01 was assessed. The PCR array technique used is generally considered to be less sensitive than the deep sequencing used for the ontology analysis. Nevertheless, the results confirmed that the expression of 63 of these genes is significantly altered in the brains of mock- and TriGAS-treated mice between 4 and 8 days p.i. in comparison to normal brain (see Table S3 in the supplemental material). As illustrated in the heat map in Fig. 6, many of the upregulated genes are located in clusters 2 to 4 and include genes associated with innate immunity, such as the Mx1, Mx2, Gbp10, and Ifih1 type I IFN response genes. It is noteworthy that the expression levels of these genes are in certain cases higher in the brains of mock-treated than those of TriGAS-treated mice at 6 and 8 days p.i. At the earliest stage of infection studied, day 4 p.i., when DOG4 N protein mRNA is virtually undetected and only a few copies of TriGAS N protein mRNA are seen (see Fig. 3), the expression of 16 mRNAs was already increased in the brains of the mock-treated mice. However, at this time, only 3 mRNAs (Irf7, Usp18, and GzmA) were expressed at significantly higher levels in the brains of the TriGAS-treated mice. When expression levels of these genes were compared between the brains of mock- and TriGAS-treated animals at day 4 p.i., only GzmA mRNA was higher in the latter (Fig. 7A). By day 6 p.i, both the number of upregulated genes and their expression levels had increased in the two experimental groups of mice. For example, in comparison to normal brain, 39 and 23 mRNAs were found to be expressed at higher levels in the brains of mock- or TriGAs-treated mice, respectively. At this time point, 3 mRNAs known to be associated with lymphocyte activation and inflammatory responses, interleukin 12B (IL-12B), Ccl3, and GzmA, were increased in the brains of the TriGAS-treated mice compared to the brains of the mock-treated animals (Fig. 7B).
Fig 6.
Quantitative RT-PCR arrays reveal temporal changes in gene expression between the brains of DOG4 RV-infected mice receiving mock and TriGAS treatments. Groups of mice were either left untreated or infected with DOG4 and subjected to mock or TriGAS treatment 4 h later. The brains from 3 uninfected controls, as well as 3 mice from each treatment group, were collected at 4, 6, and 8 days (d) p.i., and RNA was isolated for qRT-PCR array analysis as detailed in Materials and Methods. The relative fold changes between healthy control mice and infected mice that were mock treated or treated with Tri-GAS are expressed as heat maps, with color coding performed with Microsoft Excel.
Fig 7.
A limited number of genes are expressed at higher levels in TriGAS-treated than in mock-treated mice at days 4 (A) and 6 (B) after DOG4 RV infection. Scatter plots depicting the normalized gene expression levels in the brains of the mock-treated (x axes) versus TriGAS-treated (y axes) mice determined as described in the legend to Fig. 6 are shown for 4 (A) and 6 (B) days postinfection with DOG4 RV. The red symbols identify genes that are expressed at higher levels in the brains of TriGAS- versus mock-treated mice.
DISCUSSION
The recently developed live-attenuated recombinant RV strain TriGAS not only has a far superior safety profile, but is also more effective than conventional rabies vaccines in inducing RV-specific immunity, as well as in promoting immune effector delivery to CNS tissues (12). Based on these characteristics and its capacity to prevent lethal rabies when administered after exposure to a pathogenic RV (12), we theorized that this vaccine may be effective in clearing pathogenic RV from CNS tissues. Indeed, unlike their mock-treated counterparts, mice treated i.m. with TriGAS 4 h after i.n. infection with the highly pathogenic DOG4 RV strain generally survive, despite clear evidence of DOG4 replication in brain tissue. Although significant protection was seen when PET with TriGAS was performed as late as 16 h p.i. (see Fig. S3 in the supplemental material), the 4-h PET time point was chosen for transcriptome and qPCR analyses so that the effect of the more rapidly spreading DOG4 on host gene expression would be minimized. Comparative analyses of peripheral and CNS host responses in mock- and TriGAS-treated mice were performed to identify potential markers associated with the protective antiviral response and virus clearance from CNS tissues. Striking differences in measures of adaptive immunity are evident between TriGAS- and mock-treated mice both in the induction of circulating RV-neutralizing antibodies and in the delivery of immune effector cells into the CNS. As early as 4 days after DOG4 infection, TriGAS-treated mice exhibited high titers of circulating VNA of the IgM isotype, which switched to IgG isotypes within 2 days. This contrasts with mock-treated mice, in which serum VNAs are virtually undetectable until 8 days p.i., when their levels are less than 0.5% of those of the TriGAS-treated animals. Messages specific for CD8 and CD19, markers of cytotoxic T cells and B cells, respectively, and IFN-γ appear in the CNS tissues of TriGAS- but not mock-treated, DOG4-infected mice beginning around 6 to 8 days p.i., and antibody production, reflected by increasing levels of κ light-chain mRNA, begins in the CNS shortly thereafter. It is noteworthy that the level of CD4 transcription is greatly reduced in the brains of mock-treated, DOG4-infected mice. Although CD4 is the prototypic phenotype marker for T helper cells, truncated CD4 transcripts with no known function are expressed by CNS-resident cells (16, 29). Since the CD4 mRNA-specific primers and probes used in our study detect both conventional and truncated CD4 transcripts, our analysis cannot determine whether the higher level of CD4 expression in the brain tissues of TriGAS-treated compared to mock-treated mice is due to CD4 T cell infiltration or the maintained expression of the transcripts by CNS resident cells. Nevertheless, we consider it very likely that CD4 T cells accumulate in the brain tissues of DOG4-infected, TriGAS-treated mice due to the elevated expression of mRNAs specific for IFN-γ and antibody light chain.
While the production of antibody in CNS tissues may be essential to clearing the virus, it is evident that the replication of DOG4 in the CNS is reduced in TriGAS-treated animals prior to the accumulation of substantial numbers of immune cells in brain tissues. Reduced in these animals, DOG4 replication increases from days 6 to 8 p.i. despite the presence of high levels of circulating VNA. These observations suggest that the innate response of brain tissues to TriGAS infection may play an important role in the control of DOG4 replication. Interestingly, significant upregulation of the expression of the IFN-α and IFN-β mRNAs was seen only in the brains of the mock-treated mice at 8 days p.i., where virus loads had reached maximum, strongly suggesting that type I IFNs by themselves cannot prevent the lethal outcome of an RV infection. In addition, expression of the important proinflammatory cytokine TNF-α, while higher in TriGAS-treated mice, is also a relatively late event, occurring after the difference in DOG4 replication between TriGAS- and mock-treated mice is evident. With the analysis of conventional mediators of innate and adaptive immunity failing to provide clear insight into the early reduction of DOG4 replication in TriGAS-treated mice, we used comparative transcriptome analysis to identify host genes expressed in the CNS that may contribute to this process. Array analysis has previously been used to profile gene expression in the CNS of mice infected with either pathogenic or attenuated RV variants, identifying a number of genes relevant to immune function that are expressed at enhanced levels regardless of the nature of the infecting virus (43, 51). The availability of a live-attenuated vaccine, TriGAS, that can induce the clearance of wild-type RV from CNS tissues has allowed us to profile the changes in gene expression unique to the treatment paradigm. Substantial differences in the expression profiles of brain RNAs were seen between healthy mice and TriGAS-treated and mock-treated, DOG4-infected mice at day 6 p.i. The important general conclusion reached from these data is that DOG4 infection causes alterations in expression in the brain of a wide variety of both coding and noncoding RNAs and that superinfection with TriGAS results in a pattern of expression that is largely different from that of healthy mice or the DOG4-infected, mock-treated brain.
Many of the genes activated in the brain of the mock-treated mouse are involved in nucleic acid metabolism, RNA transcription, and other processes that may contribute to both the positive and negative regulation of virus infection. A number, such as those encoding 2′-5′-oligoadenylate synthetase (Oas), interferon-induced helicase C domain-containing protein 1 (Ifih1), and guanylate binding protein (Gbp), represent type I IFN response genes, suggesting that the DOG4 RV induces an early type I IFN response that by itself is unable to control the infection and may even exacerbate the disease process. In this respect, it has recently been shown that the induction of type I IFNs in the CNS interferes with the elimination of RV from the brain by slowing down T cell infiltration (4). TriGAS treatment of the DOG4 infection results in enhanced expression of a number of genes with established roles in the regulation of different aspects of adaptive immunity, including γ/δ T cell activation, T cell differentiation, and lymphocyte and mononuclear cell proliferation. In addition, the expression of genes involved in a variety of other functions relevant to immunity, such as programmed cell death, regulation of cell adhesion, and vessel morphogenesis, is higher after TriGAS treatment than after mock treatment. Together, these findings suggest that TriGAS induces processes in the brain tissues that facilitate immune cell access to and modulate immune activity in the sites of infection. In addition to coding mRNAs, DOG4 infection induces substantial changes in the expression of ncRNAs in the CNS that were generally not seen in conjunction with TriGAS treatment. Although the functions of these annotated ncRNAs are not precisely known, it is possible that a number of them act as enhancers of gene expression (37). In this case, the increased ncRNA expression specific to the DOG4-infected, mock-treated brain may be part of the global response to virus replication. On the other hand, immunologically relevant roles for ncRNAs have been suggested in respiratory syndrome coronavirus and influenza virus infections (38), and it would be interesting if those specifically induced in the brain of the TriGAS-treated mouse may contribute to the protective response.
To examine the kinetics of the changes in gene expression detected by transcriptome analysis, we used a PCR array approach, examining brain samples from TriGAS- and mock-treated, DOG4-infected mice at 4, 6, and 8 days p.i., as well as tissues from uninfected animals. Of the 90 genes identified by ontology analysis as differentially expressed at day 6 p.i. that were chosen for the less sensitive PCR array analysis, 63 were found to be expressed at different levels in the brains of the mock- or TriGAS-treated, DOG4-infected mice than in control brain. At day 4 p.i., more of these genes, including several involved in nucleic acid metabolism, RNA transcription, regulation of virus infection, and innate immune responses, were overexpressed in mock-treated, DOG4-infected brain than in similar tissues from DOG4-infected mice treated with TriGAS, confirming the results of transcriptome analysis. However, by day 8 p.i., most of the genes were expressed at high levels in the brains of both cohorts. Only 3 genes, encoding granzyme A (GzmA), IL-12B, and chemokine ligand 3 (Ccl3), were found to be expressed at higher levels in the brains of TriGAS-treated mice than in those of mock-treated mice between days 4 and 6 p.i.
Survival of a rabies virus infection requires the rapid induction of RV-specific immunity (2) and, when the virus has reached CNS tissues, the induction of processes that facilitate immune effector cell entry into the brain (45). The finding that TriGAS superinfection rapidly induces the expression in the brain of genes involved in immune activation and cell recruitment has implications for its mode of action in containing and clearing DOG4 infection. GzmA is a serine protease originally associated with perforin-dependent cytotoxicity (18, 48). Recent work, however, suggests that GzmA may not be cytolytic under physiological conditions but that it contributes to the induction of proinflammatory cytokines (33) and interferes with cellular components necessary for viral replication (reviewed in reference 1). The latter mechanisms are consistent with the possibility that GzmA has a protective role in TriGAS-treated mice and that RV infection is characterized by neuronal dysfunction rather than the death of infected neurons (23, 57). Based on the fact that elevated GzmA mRNA appears in TriGAS-treated CNS tissues prior to CD8, we speculate that natural killer (NK) cells, known to be the first line of defense against virus-infected cells and to produce GzmA (reviewed in reference 50), may be its source at early stages of the CNS antiviral response.
The IL-12B gene encodes the p40 subunit of two heterodimeric cytokines, the proinflammatory IL-12, which plays a central role in the induction and development of Th1 cells (22, 31), and the Th17-inducing IL-23 (36). IL-12 p40 is typically produced in gross excess of the IL-12 p35 subunit (6, 55), but it remains the limiting factor for functional IL-12 availability, since p35 is constitutively expressed (6). Primarily produced by phagocytes and dendritic cells (DCs) (6, 30), but also by astrocytes, IL-12 is involved in T cell differentiation and the activation of NK cells and T cells, leading to IFN-γ production by both cell types (27). Prior studies have demonstrated that immune effector entry into the brain and RV clearance require CD4+ T cells and are associated with the expression of IFN-γ, the signature Th1 cytokine, in the brain (39, 40). IL-12 p40 homodimers have been discovered in mouse studies and found to act as IL-12 antagonists (15, 19), as well as to induce iNOS expression in primary mouse microglia (24). Conceivably, p40 homodimers may contribute to the iNOS expression and iNOS-associated radical-mediated formation of nitrotyrosine seen in the CNS of mice clearing apathogenic RV (57).
Chemokine (C-C motif) ligand 3 (CCL3/MIP-1α) promotes leukocyte recruitment and activation and can lead to induction of other proinflammatory cytokines (32). Elaborated in the periphery by a number of cells, including monocytes and dendritic cells, CCL3 can also be produced by brain-resident cells, such as vascular endothelial cells, neurons, and astrocytes (17, 28), and has previously been found to be expressed in the CNS during attenuated-RV clearance (40, 41). A recombinant RV engineered to express CCL3 has been demonstrated to have enhanced immunogenicity, which was accredited to increased recruitment and activation of DCs and B cells (52, 59). However, CCL3 overexpression tends to promote the Th1 arm of a response (32), which has been implicated in RV clearance from the CNS (39).
Neither the mechanisms that trigger the expression of these chemoattractant and immunomodulatory genes nor their cell source in the brains of TriGAS-treated mice in the early phase of infection has been established. Since certain RV strains can replicate in astrocytes (14, 44), it is possible TriGAS might be able to infect and directly stimulate astrocytes to produce these factors. Alternatively, the infected neurons may be directly or indirectly involved. Regardless, the early production of these and other factors induced by TriGAS treatment evidently promotes the containment of DOG4 infection and its eventual clearance from the CNS. Current PET is very effective when initiated shortly after exposure before the virus infects the brain (2). Unfortunately, ignorance of the risk of an RV infection or, particularly in the developing world, lack of accessibility to medical resources often prevents timely PET, leading to tens of thousands of rabies-related deaths each year. Our findings confirm in a mouse model that an established CNS infection with pathogenic RV can be cleared by PET with live-attenuated RV vaccine TriGAS, resulting in the survival of the large majority of infected animals. Virus clearance from the brain requires the rapid induction of robust innate and adaptive antiviral immunity in the periphery and CNS. We have identified several TriGAS-induced genes that are likely to contribute to this process. Characterization of the upstream and downstream pathways associated with the induction and activity of these gene products will ultimately offer further insight into the mechanisms involved in RV clearance from the CNS and, eventually, help to extend the window of PET from several days after exposure until after RV has reached the CNS.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grants R01AI093666 (to M.F.), R01AI060686 and 2R56AI060686-06 (to B.D.), and UO1AI083045 and R01AI093369 (to D.C.H.).
Footnotes
Published ahead of print 11 January 2012
Supplemental material for this article may be found at http://jvi.asm.org.
REFERENCES
- 1. Andrade F. 2010. Non-cytotoxic antiviral activities of granzymes in the context of the immune antiviral state. Immunol. Rev. 235:128–146 [DOI] [PubMed] [Google Scholar]
- 2. Anonymous 2010. Rabies vaccines: WHO position paper. Wkly. Epidemiol. Rec. 85:309–320 [Google Scholar]
- 3. Celis E, Wiktor TJ, Dietzschold B, Koprowski H. 1985. Amplification of rabies virus-induced stimulation of human T-cell lines and clones by antigen-specific antibodies. J. Virol. 56:426–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chopy D, et al. 2011. Ambivalent role of the innate immune response in rabies virus pathogenesis. J. Virol. 85:6657–6668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cox JH, Dietzschold B, Schneider LG. 1977. Rabies virus glycoprotein. II. Biological and serological characterization. Infect. Immun. 16:754–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. D'Andrea A, et al. 1992. Production of natural killer cell stimulatory factor (interleukin 12) by peripheral blood mononuclear cells. J. Exp. Med. 176:1387–1398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dietzschold B, Li J, Faber M, Schnell M. 2008. Concepts in the pathogenesis of rabies. Future Virol. 3:481–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dietzschold B, et al. 1987. Induction of protective immunity against rabies by immunization with rabies virus ribonucleoprotein. Proc. Natl. Acad. Sci. U. S. A. 84:9165–9169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dietzschold B, et al. 1983. Characterization of an antigenic determinant of the glycoprotein that correlates with pathogenicity of rabies virus. Proc. Natl. Acad. Sci. U. S. A. 80:70–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faber M, et al. 2007. Dominance of a nonpathogenic glycoprotein gene over a pathogenic glycoprotein gene in rabies virus. J. Virol. 81:7041–7047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faber M, et al. 2005. A single amino acid change in rabies virus glycoprotein increases virus spread and enhances virus pathogenicity. J. Virol. 79:14141–14148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Faber M, et al. 2009. Effective preexposure and postexposure prophylaxis of rabies with a highly attenuated recombinant rabies virus. Proc. Natl. Acad. Sci. U. S. A. 106:11300–11305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Faber M, et al. 2002. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 76:3374–3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fekadu M, Chandler FW, Harrison AK. 1982. Pathogenesis of rabies in dogs inoculated with an Ethiopian rabies virus strain. Immunofluorescence, histologic and ultrastructural studies of the central nervous system. Arch. Virol. 71:109–126 [DOI] [PubMed] [Google Scholar]
- 15. Gillessen S, et al. 1995. Mouse interleukin-12 (IL-12) p40 homodimer: a potent IL-12 antagonist. Eur. J. Immunol. 25:200–206 [DOI] [PubMed] [Google Scholar]
- 16. Gorman SD, Tourvieille B, Parnes JR. 1987. Structure of the mouse gene encoding CD4 and an unusual transcript in brain. Proc. Natl. Acad. Sci. U. S. A. 84:7644–7648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Guo H, et al. 1998. Regulation of beta-chemokine mRNA expression in adult rat astrocytes by lipopolysaccharide, proinflammatory and immunoregulatory cytokines. Scand. J. Immunol. 48:502–508 [DOI] [PubMed] [Google Scholar]
- 18. Hayes MP, Berrebi GA, Henkart PA. 1989. Induction of target cell DNA release by the cytotoxic T lymphocyte granule protease granzyme A. J. Exp. Med. 170:933–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Heinzel FP, Hujer AM, Ahmed FN, Rerko RM. 1997. In vivo production and function of IL-12 p40 homodimers. J. Immunol. 158:4381–4388 [PubMed] [Google Scholar]
- 20. Hooper DC, et al. 1998. Collaboration of antibody and inflammation in clearance of rabies virus from the central nervous system. J. Virol. 72:3711–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hornung V, et al. 2004. Replication-dependent potent IFN-alpha induction in human plasmacytoid dendritic cells by a single-stranded RNA virus. J. Immunol. 173:5935–5943 [DOI] [PubMed] [Google Scholar]
- 22. Hsieh CS, et al. 1993. Development of TH1 CD4+ T cells through IL-12 produced by Listeria-induced macrophages. Science 260:547–549 [DOI] [PubMed] [Google Scholar]
- 23. Jackson AC, Randle E, Lawrance G, Rossiter JP. 2008. Neuronal apoptosis does not play an important role in human rabies encephalitis. J. Neurovirol. 14:368–375 [DOI] [PubMed] [Google Scholar]
- 24. Jana M, Dasgupta S, Pal U, Pahan K. 2009. IL-12 p40 homodimer, the so-called biologically inactive molecule, induces nitric oxide synthase in microglia via IL-12R beta 1. Glia 57:1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kandhavelu M, et al. 2009. Existence of snoRNA, microRNA, piRNA characteristics in a novel non-coding RNA: x-ncRNA and its biological implication in Homo sapiens. J. Bioinform. Sequence Anal. 1:31–40 [Google Scholar]
- 26. Kiss AM, Jady BE, Bertrand E, Kiss T. 2004. Human box H/ACA pseudouridylation guide RNA machinery. Mol. Cell. Biol. 24:5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kobayashi M, et al. 1989. Identification and purification of natural killer cell stimulatory factor (NKSF), a cytokine with multiple biologic effects on human lymphocytes. J. Exp. Med. 170:827–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y, Fu L, Gonzales DM, Lavi E. 2004. Coronavirus neurovirulence correlates with the ability of the virus to induce proinflammatory cytokine signals from astrocytes and microglia. J. Virol. 78:3398–3406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lonberg N, Gettner SN, Lacy E, Littman DR. 1988. Mouse brain CD4 transcripts encode only the COOH-terminal half of the protein. Mol. Cell. Biol. 8:2224–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Macatonia SE, et al. 1995. Dendritic cells produce IL-12 and direct the development of Th1 cells from naive CD4+ T cells. J. Immunol. 154:5071–5079 [PubMed] [Google Scholar]
- 31. Manetti R, et al. 1993. Natural killer cell stimulatory factor (interleukin 12 [IL-12]) induces T helper type 1 (Th1)-specific immune responses and inhibits the development of IL-4-producing Th cells. J. Exp. Med 177:1199–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maurer M, von Stebut E. 2004. Macrophage inflammatory protein-1. Int. J. Biochem. Cell Biol. 36:1882–1886 [DOI] [PubMed] [Google Scholar]
- 33. Metkar SS, et al. 2008. Human and mouse granzyme A induce a proinflammatory cytokine response. Immunity 29:720–733 [DOI] [PubMed] [Google Scholar]
- 34. Morimoto K, Foley HD, McGettigan JP, Schnell MJ, Dietzschold B. 2000. Reinvestigation of the role of the rabies virus glycoprotein in viral pathogenesis using a reverse genetics approach. J. Neurovirol. 6:373–381 [DOI] [PubMed] [Google Scholar]
- 35. Morimoto K, Hooper DC, Spitsin S, Koprowski H, Dietzschold B. 1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73:510–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oppmann B, et al. 2000. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity 13:715–725 [DOI] [PubMed] [Google Scholar]
- 37. Orom UA, et al. 2010. Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Peng X, et al. 2010. Unique signatures of long noncoding RNA expression in response to virus infection and altered innate immune signaling. MBio 1:e00206–10 doi:10.1128/mBio.00206-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Phares TW, Fabis MJ, Brimer CM, Kean RB, Hooper DC. 2007. A peroxynitrite-dependent pathway is responsible for blood-brain barrier permeability changes during a central nervous system inflammatory response: TNF-alpha is neither necessary nor sufficient. J. Immunol. 178:7334–7343 [DOI] [PubMed] [Google Scholar]
- 40. Phares TW, Kean RB, Mikheeva T, Hooper DC. 2006. Regional differences in blood-brain barrier permeability changes and inflammation in the apathogenic clearance of virus from the central nervous system. J. Immunol. 176:7666–7675 [DOI] [PubMed] [Google Scholar]
- 41. Pinto AR, Reyes-Sandoval A, Ertl HC. 2003. Chemokines and TRANCE as genetic adjuvants for a DNA vaccine to rabies virus. Cell Immunol. 224:106–113 [DOI] [PubMed] [Google Scholar]
- 42. Prosniak M, et al. 2003. Development of a cocktail of recombinant-expressed human rabies virus-neutralizing monoclonal antibodies for postexposure prophylaxis of rabies. J. Infect. Dis. 188:53–56 [DOI] [PubMed] [Google Scholar]
- 43. Prosniak M, Hooper DC, Dietzschold B, Koprowski H. 2001. Effect of rabies virus infection on gene expression in mouse brain. Proc. Natl. Acad. Sci. U. S. A. 98:2758–2763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ray NB, Power C, Lynch WP, Ewalt LC, Lodmell DL. 1997. Rabies viruses infect primary cultures of murine, feline, and human microglia and astrocytes. Arch. Virol. 142:1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Roy A, Hooper DC. 2007. Lethal silver-haired bat rabies virus infection can be prevented by opening the blood-brain barrier. J. Virol. 81:7993–7998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Roy A, Phares TW, Koprowski H, Hooper DC. 2007. Failure to open the blood-brain barrier and deliver immune effectors to central nervous system tissues leads to the lethal outcome of silver-haired bat rabies virus infection. J. Virol. 81:1110–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schumacher CL, et al. 1989. Use of mouse anti-rabies monoclonal antibodies in postexposure treatment of rabies. J. Clin. Invest. 84:971–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shi L, Kraut RP, Aebersold R, Greenberg AH. 1992. A natural killer cell granule protein that induces DNA fragmentation and apoptosis. J. Exp. Med. 175:553–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Trapnell C, et al. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vivier E, et al. 2011. Innate or adaptive immunity? The example of natural killer cells. Science 331:44–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wang ZW, et al. 2005. Attenuated rabies virus activates, while pathogenic rabies virus evades, the host innate immune responses in the central nervous system. J. Virol. 79:12554–12565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wen Y, et al. 2011. Rabies virus expressing dendritic cell-activating molecules enhances the innate and adaptive immune response to vaccination. J. Virol. 85:1634–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wiktor TJ, MacFarlan RI, Foggin CM, Koprowski H. 1984. Antigenic analysis of rabies and Mokola virus from Zimbabwe using monoclonal antibodies. Dev. Biol. Stand. 57:199–211 [PubMed] [Google Scholar]
- 54. Wise RP, Moscou MJ, Bogdanove AJ, Whitham SA. 2007. Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 45:329–369 [DOI] [PubMed] [Google Scholar]
- 55. Wysocka M, et al. 1995. Interleukin-12 is required for interferon-gamma production and lethality in lipopolysaccharide-induced shock in mice. Eur. J. Immunol. 25:672–676 [DOI] [PubMed] [Google Scholar]
- 56. Xiang ZQ, et al. 1994. Vaccination with a plasmid vector carrying the rabies virus glycoprotein gene induces protective immunity against rabies virus. Virology 199:132–140 [DOI] [PubMed] [Google Scholar]
- 57. Yan X, et al. 2001. Silver-haired bat rabies virus variant does not induce apoptosis in the brain of experimentally infected mice. J. Neurovirol. 7:518–527 [DOI] [PubMed] [Google Scholar]
- 58. Yekutieli D, Benjamini Y. 1999. Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics. J. Stat. Planning Inference 82:171–196 [Google Scholar]
- 59. Zhao L, et al. 2010. Expression of MIP-1alpha (CCL3) by a recombinant rabies virus enhances its immunogenicity by inducing innate immunity and recruiting dendritic cells and B cells. J. Virol. 84:9642–9648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.