Abstract
Influenza A virus NS1 protein has multiple functions in the infected cell during the virus life cycle. Identification of novel cellular factors that interact with NS1 and understanding their functions in virus infection are of great interest. Recombinant viruses carrying a tagged NS1 are valuable for investigation of interactions between NS1 and cellular factors in the context of virus infection. Here, we report the generation of replication-competent recombinant influenza A viruses bearing a Strep tag in the NS1 protein. Purification of a protein complex associated with Strep-tagged NS1 from virus-infected cells followed by mass spectrometry revealed a number of attractive host factors. Among them, we focused our study on RNA helicase A (RHA) in this report. Through biomedical and functional analyses, we demonstrated that RHA interacts with NS1 in an RNA-dependent manner. Knockdown of RHA resulted in a significant reduction on virus yield and polymerase activity in a minigenome assay. Our cell-free viral genome replication assay showed that viral RNA replication and transcription can be enhanced by addition of RHA, and the enhanced effect of RHA required its ATP-dependent helicase activity. In summary, we established a system to identify cellular factors that interact with NS1 protein during virus infection and furthermore demonstrated that RHA interacts with NS1 and enhances viral replication and transcription.
INTRODUCTION
Influenza A viruses are important pathogens that cause acute respiratory disease in humans and different animal species. The genome of influenza A virus contains eight single-stranded RNA segments of negative polarity that encode 11 viral proteins (35). Like all viruses, influenza A viruses require host proteins and pathways to carry out many of the phases of their life cycles. Thus, cellular factors that are essential for efficient virus infection might be the target for anti-influenza interventions. Recently, many groups of investigators have made great efforts in searching for new host factors that are involved in the influenza virus life cycle by using different strategies (4, 13, 20, 23, 41). The most popular strategy is genome-wide RNA interference (RNAi) screening. Although it is a powerful tool for identifying novel host factors that modulate the influenza virus life cycle and although these studies provided fundamental information in this regard, the application of RNAi screening is limited by the coverage of human genes in the small interfering RNA (siRNA) library, the efficiency of knockdown, and cell viability after gene knockdown (48). Therefore, it is believed that there are many host factors involved in regulating influenza virus infection that remain undiscovered.
Another approach to identify host factors regulating virus infection is through mass spectrometry (MS) analysis of the protein complex that interacts with viral proteins. By using proteomics-based approaches, Mayer et al. identified 41 host factors that associate with viral RNPs (vRNPs) (31); Jorba et al. reported a catalogue of cellular factors that interact with the viral RNA polymerase (19). These cellular factors are candidates that potentially are involved in regulating the virus life cycle.
The RNA segment 8 encodes two proteins: nonstructural protein 1 (NS1) and nuclear export protein (NEP). NS1 is translated from the full-length unspliced mRNA and can be divided into two functional domains, the RNA-binding domain (RBD) near the N terminus and the effector domain (ED) at the C terminus (37). NS1 is a virulence factor and has multiple functions during virus infection, such as regulation of virus replication, viral protein synthesis, host innate and adaptive immune responses, and cellular signaling pathways (12). The various functions of NS1 are fulfilled by its interaction with many cellular factors. Although some NS1 interaction partners have been identified, such as the cleavage and polyadenylation specificity factor (CPSF) and the poly(A)-binding protein II (PABII) (28), the p85β subunit of phosphatidylinositol 3-kinases (PI3K) (11, 42, 44), TRIM25 (10), and heterogeneous nuclear RNA-F (hnRNA-F) (26), most of them were identified by either yeast two-hybrid screening (26, 34) or immunoprecipitation using antibodies directed against a suspected cellular interaction partner (10, 42, 44). Thus, it is imperative to establish a comprehensive list of proteins that interact with NS1 protein, with the aim of finding novel cellular proteins that are pivotal during influenza virus infection.
Here, we report the establishment of a novel system, where a Strep-Tag II (WSHPQFEK) (45) is inserted into the NS1 gene between the RBD and ED. The rescued recombinant virus has similar growth properties to those of the wild-type (WT) virus during single-cycle replication, which enabled us to purify NS1 protein and its associated proteins from infected cells by Strep-Tactin beads. Purified protein complex was then subjected to multidimensional liquid chromatography (LC) and tandem mass spectrometry (MS/MS) analysis. We found that NS1 pulled down several RNA-binding proteins, in particular, a member of the DEXD/H box family of proteins, RNA helicase A (RHA). Through biomedical and functional analyses, we verified that RHA interacts with NS1 in an RNA-dependent manner. Knockdown of RHA resulted in a significant reduction in virus yield and polymerase activity in a minigenome assay. Furthermore, an in vitro cell-free viral genome replication assay showed that viral RNA replication and transcription can be enhanced by the addition of RHA, and the enhanced effect of RHA requires its ATP-dependent helicase activity. Thus, we demonstrated the important role of RHA in the influenza A virus life cycle.
MATERIALS AND METHODS
Cells and viruses.
A549 and 293T cells were maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma) containing 10% fetal bovine serum (FBS; Invitrogen). Madin-Darby canine kidney (MDCK) cells were cultivated in minimal essential medium (MEM) (Invitrogen) supplemented with 10% FBS. Influenza A/Puerto Rico/8/34 (H1N1) (PR8), A/Texas/36/91 (Tx91), and A/Halifax/210/2009 (H1N1) (Halifax210) viruses were propagated in 11-day-old embryonated chicken eggs as described previously (42).
Antibodies.
Rabbit polyclonal NS1 and NP antibodies were generated in our laboratory as previously described (43). The other antibodies were purchased from different sources as follows: monoclonal anti-Flag antibody (Sigma), strepMAB-Classic antibody conjugated to horseradish peroxidase ([HRP] IBA), monoclonal anti-RHA (Abcam), monoclonal anti-β-actin (Santa Cruz), normal mouse IgG (Santa Cruz), alkaline phosphatase-conjugated anti-rabbit IgG (Jackson ImmunoResearch), and IRDye 800-conjugated donkey polyclonal anti-mouse IgG (Li-Cor Biosciences).
Plasmid construction and generation of mutant viruses.
Strep-Tag II (WSHPQFEK)-encoding sequence (TGGTCACACCCACAGTTCGAAAAA) was introduced into pHW198-NS (14) by standard overlapping PCR. Plasmid 427 encodes PR8 NS1 with a Strep tag inserted after amino acid (aa) 79; plasmid 442 encodes PR8 NS1 with a Strep tag inserted after aa 52; plasmid 443 encodes PR8 NS1 with aa 79 to 84 replaced by a Strep tag.
An eight-plasmid reverse genetics system (15) was utilized for rescuing the viruses. To generate PR8 NS1 mutant viruses, the plasmids pHW191-PB2, pHW192-PB1, pHW193-PA, pHW194-HA, pHW195-NP, pHW196-NA, and pHW197-M and one of the Strep tag-encoding plasmids (plasmid 427, plasmid 442, or plasmid 443) were used for transfection. The rescued viruses, designated PR8-427, PR8-442, and PR8-443, were propagated in 10- to 11-day-old embryonated chicken eggs and characterized by sequencing of the reverse transcription-PCR (RT-PCR) product.
Plasmid pCR-XL-TOPO-DHX9 containing full-length RHA was purchased from Open Biosystems. Full-length RHA (aa 1 to 1269) was amplified from pCR-XL-TOPO-DHX9 by PCR and cloned into pCMV-3×Flag (where CMV is cytomegalovirus and the plasmid carries three copies of the Flag tag) (27) at NotI/KpnI sites, generating pCMV-3×Flag-RHA. pCMV-3×Flag-RHA-K417R was generated by site-directed mutagenesis, where aa 417 of RHA was mutated from Lys to Arg. pCMV-3×Flag-p85β and pCMV-3×Flag-p85α encode Flag-tagged PI3K regulatory subunits p85β and p85α, respectively (27). pcDNA3-NS1-R38AK41A, encoding NS1 with mutations at aa 38 and 41, where the original Arg and Lys were both replaced by Ala residues, was constructed from pcDNA3-NS1 (42) by site-directed mutagenesis. The coding regions of all plasmids were verified by DNA sequencing to ensure that no unwanted mutation was introduced during PCR amplification. All PCR primers are available upon request.
Purification of Strep-tagged NS1 protein complex from infected cells and identification of cellular interaction proteins.
293T cells were infected with PR8-427 at multiplicity of infection (MOI) of 5. At 10 h postinfection (hpi), cells were harvested and lysed in cell lysis buffer (Cell Signaling Technology) with supplementation of phenylmethylsulfonyl fluoride (PMSF). After brief sonication, cell lysates were clarified by centrifugation at 15,000 × g for 10 min at 4°C, and the supernatants were subjected to purification using Strep-Tactin Sepharose (IBA) according to the manufacturer's protocol with minor modifications. Briefly, the Strep-Tactin Sepharose was washed extensively and saturated with cell lysis buffer. Thereafter, the clarified cell supernatants were incubated with the Sepharose at 4°C overnight. Subsequently, the lysate-Sepharose mixture was loaded onto a polypropylene column (Qiagen). The Sepharose was then washed with 5 column volumes (CV) of washing buffer (100 mM Tris-Cl, pH 8.0, 150 mM NaCl, 1 mM EDTA), and proteins bound to the Sepharose were then eluted with 3 CV of elution buffer (washing buffer containing 2.5 mM desthiobiotin). The eluted proteins were concentrated with a 10-kDa-pore-size centrifugal filter (Millipore), and the protein complex was subjected directly to protein identification by LC-MS/MS at the University of Victoria-Genome BC Proteomics Centre.
Transfection, immunoprecipitation, and Western blotting.
To examine whether endogenous RHA interacts with NS1, A549 cells were infected with PR8 at an MOI of 1. Cell lysates were prepared at 10 hpi, precleared by mouse IgG immobilized on protein G; an immunoprecipitation assay was performed as described previously (42) with the exception that RHA monoclonal antibody was used.
To examine whether RHA expressed from plasmid transfection interacts with NS1, 293T cells were seeded in six-well plates at a density of 1 × 106 cells/well. Two micrograms of DNA constructs, specifically pCMV-3×Flag-RHA, pCMV-3×Flag-p85β, pCMV-3×Flag-p85α, or empty vector pCMV-3×Flag, was transfected using TransIT-LT1 (Mirus) per the manufacturer's protocol. Twenty-four hours later, cell lysates were prepared and were incubated with anti-Flag M2 affinity gel (Sigma) for 4 h at 4°C. After samples were washed with a buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100, the freshly prepared Flag resin conjugated with Flag-tagged RHA, p85β, or p85α was incubated with the cell lysates of 293T cells either transfected with pcDNA3-NS1 for 24 h or infected with PR8, Tx91, or Halifax210 at an MOI of 1 for 10 h. After incubation at 4°C for 4 h, resins were spun down, washed, and eluted with 1× Tris-buffered saline ([TBS] 0.1 M Tris, pH 7.6, 0.9% NaCl) containing 15 μg/ml Flag peptide (Sigma). The eluted samples were subjected to SDS-PAGE using antibodies specific for Flag or NS1.
Western blotting was performed as described previously (43) with minor modifications. Briefly, 30 μg of total protein or a portion of precipitated protein was resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto nitrocellulose membranes (Bio-Rad). Membranes were blocked for nonspecific binding with Odyssey blocking buffer (Li-Cor Biosciences) for 1 h at room temperature and then incubated with a primary antibody diluted in Odyssey blocking buffer overnight at 4°C. Infrared dye-linked secondary antibody (1:15,000) was then added, and membranes were incubated at room temperature for 1 h. The immunoblots were visualized using an Odyssey infrared imaging system (Li-Cor Biosciences). To examine Strep-tagged NS1, membrane was probed with strepMAB-Classic-HRP conjugate (1:15,000), and visualized with an enhanced chemiluminescence reagent (ECL Advance Western Blotting Detection Kit; GE Healthcare).
Knockdown of RHA expression and viral infection.
Four siRNA constructs targeted to different regions of the human RHA gene, designated RHA-6, RHA-9, RHA-10, and RHA-11, were purchased from Qiagen. Additionally, nonsense siRNA negative-control constructs were obtained from Invitrogen. The combination of the four RHA siRNAs was found to be the most effective at knocking down the endogenous expression of RHA in A549 cells and was therefore used throughout the experiments. A549 cells (7 × 104/well of a 24-well plate) were transfected with siRNAs at a concentration of 40 nM (10 nM each siRNA) using X-tremeGene siRNA transfection reagent (Roche) according to the manufacturer's protocol with minor modifications. Briefly, siRNA and X-tremeGene siRNA transfection reagent were diluted in OptiMEM in separate vials. The diluents were mixed immediately, and the mixture was further incubated at room temperature (RT) for 20 min before addition to cells at 30 to 40% confluence. At 48 h posttransfection, one set of transfected cells was harvested, and the efficiency of RHA knockdown was confirmed by Western blotting with antibody specific for RHA. The other set of siRNA-transfected cells was infected with WT PR8 or Halifax210 virus. Culture supernatant and cells were collected at predetermined times. Virus titers, viral protein expression, and viral RNA levels were determined by various assays.
Knockdown of RHA and minigenome assay.
Since cells would be transfected twice in this assay, a reverse transfection method was used to knock down RHA. Briefly, 24 pmol of RHA siRNAs or off-target siRNA was diluted in 100 μl of OptiMEM medium in 24-well plates. Two microliters of Lipofectamine RNAiMAX (Invitrogen) was added to each well containing diluted siRNA and mixed gently, and cells were incubated at room temperature for 20 min. A549 cells were trypsinized and diluted at a density of 6 × 104 cells/ml in OptiMEM. To each well, 500 μl of the diluted A549 cells was added to obtain a final siRNA concentration of 40 nM in a final volume of 600 μl. Cells were incubated at 37°C for 12 h; the medium was then replaced by OptiMEM. At 48 h after siRNA transfection, the cells were further transfected with phPolI-LUC-NP, which contains a firefly luciferase open reading frame flanked by noncoding regions of NP under the control of human RNA polymerase I protein (36), together with the plasmids pcDNA3-PB2, pcDNA3-PB1, pcDNA3-PA, and pcDNA-NP, which encode PB2, PB1, PA, and NP, respectively, and Renilla luciferase expression plasmid pTK-rLuc, using Lipofectamine LTX with Plus reagents per the manufacturer's protocol (Invitrogen). At 24 h posttransfection, the luciferase activity was measured using a Dual-Luciferase Reporter Assay kit (Promega) according to the manufacturer's protocol. Relative luciferase activities were calculated as the ratio of firefly to Renilla luciferase light units. Each luciferase activity value is the average of three independent experiments.
Cell-free viral genome replication assay.
Virion-derived vRNP (vvRNP) was prepared from purified PR8 virus as described by Vreede and Brownlee (46). The cell-free viral genome replication assay was performed as described previously (21, 46) with modifications. Briefly, the following components were added to a 25-μl reaction mixture: 50 mM Tris (pH 8.0), 5 mM MgCl2, 1.5 mM dithiothreitol, 1 mM each nucleoside triphosphate (NTP), 8 U of RNase inhibitor, 50 ng of rabbit globin mRNA (Sigma), and vvRNP (30 ng of NP equivalents), with or without addition of various amount of purified proteins (Flag-RHA, Flag-RHA-K417R, and NS1) as indicated on Fig. 7 and 8. The reaction mixture was incubated at 30°C for 12 h. Synthesized RNA was extracted with TRIzol and precipitated by addition of 5 μg of glycogen (Roche) and 1 volume of isopropanol, followed by washing with 75% ethanol. The dried RNA was dissolved in 20 μl of 50 mM Tris buffer (pH 8.0) and was subjected to a primer extension assay and a real-time RT-PCR assay as described in the following sections.
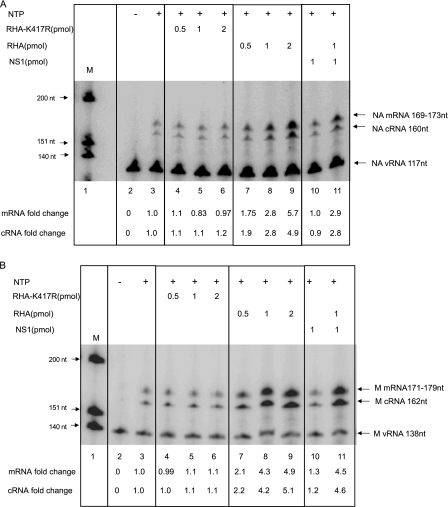
Fig 7.
RHA enhanced viral transcription and replication in a cell-free viral genome replication system measured by primer extension assay. NA segment-specific (A) and M segment-specific (B) primer extension assays were performed in the absence or presence of different doses of purified WT RHA, RHA-K417R, NS1, or WT RHA plus NS1. mRNA or cDNA was first normalized to the level of vRNA in each sample and then compared to that in the sample without addition of any form of RHA. This number is indicated as mRNA or cRNA fold change.
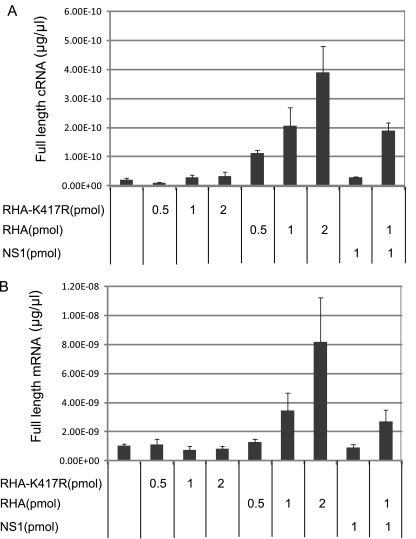
Fig 8.
RHA enhanced viral transcription and replication in cell-free viral genome replication system measured by strand-specific real-time RT-PCR. Full-length cRNA (A) and mRNA (B) levels in the absence or presence of different doses of purified WT RHA, RHA-K417R, NS1, or WT RHA plus NS1 were measured by NA strand-specific real-time PCR assay.
Primer extension assay.
Total RNAs were extracted from virus-infected cells or from an in vitro cell-free viral genome replication assay. Levels of matrix (M) segment- or neuraminidase (NA) segment-specific mRNA, cRNA, and vRNA were determined by primer extension assay as described previously (8). The primers used in this study are the following: M-vRNA, 5′-GAA AGG AGG GCC TTC TAC GG-3′; M-m/cRNA, 5′-AGC CAT TCC ATG AGA ACC TC-3′; NA-vRNA, 5′-TGG ACT AGT GCG AGC AGC AT-3′; NA-m/cRNA, 5′-TCC AGT ATG GTT TTG ACT TCC A-3′; 5s rRNA, 5′-TCC CAG GCG GTC TCC CAT CC-3′. The primer extension products were subjected to 6% PAGE in the presence of 8 M urea. The gel was visualized by phosphorimaging using a personal FX phosphorimager (Bio-Rad). The band density was quantified using Quantity One software (Bio-Rad). As a marker [γ-32P]ATP-labeled ϕ174 DNA/HinfI dephosphorylated marker (Promega) was loaded.
Real-time RT-PCR.
The full-length viral RNAs (cRNA, mRNA, and vRNA) were quantified by a novel strand-specific real-time RT-PCR method recently developed by Kawakami et al. (22). This method is based on introducing a tag sequence, which is unrelated to influenza virus, at the 5′ end of the reverse transcription primer. In this study, we designed primers specific for the NA segment. cRNA was reverse transcribed by 5′-GCTAGCTTCAGCTAGGCATCAGTAGAAACAAGGAGTTTTTTGAAC-3′, followed by PCR using 5′-GCTAGCTTCAGCTAGGCATC-3′/5′-CTGTATGAGGCCGTGCTTCTG-3′. mRNA was reverse transcribed by 5′-CCAGATCGTTCGAGTCGTTTTTTTTTTTTTTTTTGAACAGACTAC-3′, followed by PCR using 5′-CTGTATGAGGCCGTGCTTCTG-3′/5′-CCAGATCGTTCGAGTCGT-3′. vRNA was reverse transcribed by 5′-GGCCGTCATGGTGGCGAATCTATAATGACTGATGGCCCGAGT, followed by PCR using 5′-GGCCGTCATGGTGGCGAAT-3′/5′-TTCGAACCATGCCAATTGTC-3′. The RNA quantification was conducted in two steps using a SuperScript III Platinum two-step quantitative RT-PCR kit with SYBR green from Invitrogen, per the manufacturer's instructions, in an iCycler iQ-Multicolor real-time PCR detection system (Bio-Rad). The concentrations of viral RNAs were obtained by comparison with a serially diluted plasmid, pHW195-NA, of known concentration (14). All RNA determinations were assayed in triplicate and repeated three times.
RESULTS
Construction and characterization of Strep-tagged NS1 mutants.
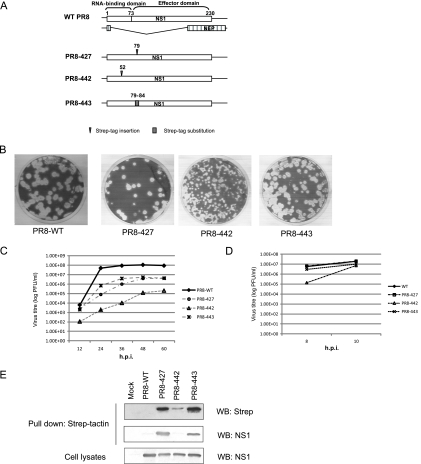
We had previously identified two flexible linker regions in the NS1 gene that were able to accommodate insertion or substitution with a tetracysteine (TC) tag (CCPGCC), namely, a loop region connecting two α-helixes at an RBD (aa 52) and a highly variable linker region between the NS1 RBD and ED (aa 79 to 84). These rescued mutant viruses possessing the TC tag allowed us to monitor the NS1 movement in live cells (29). Motivated by these results, we extended these findings to rescue mutant viruses possessing the optimized Strep-Tag II motif (WSHPQFEK) in NS1, which would enable us to pull down the proteins associated with NS1 in the context of virus infection. Three mutant PR8 viruses were rescued: PR8-427 that encodes NS1 with a Strep tag inserted after aa 79, PR8-442 that encodes NS1 with a Strep tag inserted after aa 52, and PR8-443 that encodes NS1 with a Strep tag replacement of aa 79 to 84 (Fig. 1A).
Fig 1.
Construction and characterization of mutant viruses bearing Strep tag in NS1. (A) Schematic representation of the gene structure of influenza A virus segment 8 and of Strep-tagged NS1 constructs. (B) Plaques formed by the WT, PR8-427, PR8-442, and PR8-443 viruses on MDCK cells. (C) Multiple-cycle growth curves of WT, PR8-427, PR8-442, and PR8-443 on MDCK cells. Cells were infected in triplicate with each virus at an MOI of 0.001. Medium was collected at the indicated time points, and titers were determined by plaque assays on MDCK cells. Mean titer values at each time point were plotted. (D) Virus yield of WT, PR8-427, PR8-442, and PR8-443 after a single-cycle infection. MDCK cells were infected at an MOI of 5, and virus titers were determined at 8 and 10 hpi by plaque assay. (E) NS1 can be pulled down by Strep-Tactin. MDCK cells were infected by the WT or mutant viruses at an MOI of 1. At 8 hpi, cell lysates were prepared and subjected to the pulldown assay with Strep-Tactin Sepharose, followed by Western blotting (WB) with antibodies specific for NS1 and Strep tag. NS1 levels in the crude cell lysates were monitored by Western blotting with NS1 antibody.
The replication potential of the mutant viruses was first assessed by monitoring virus plaque size. As shown in Fig. 1B, while PR8-427 and PR8-443 formed similarly sized plaques relative to the WT virus, PR8-442 formed smaller plaques, suggesting that PR8-442 is the most attenuated virus. To assess the degree of virus attenuation, we compared the multiple-cycle growth kinetics of the mutant viruses to that of the WT virus in MDCK cells. MDCK cells were infected at an MOI of 0.001, supernatant was harvested every 12 h until 60 hpi, and virus titers were determined by plaque assay. As shown in Fig. 1C, both PR8-427 and PR8-443 were attenuated in growth to approximately 1.5 logs lower than WT virus by 60 hpi. Consistent with the smaller-plaque phenotype, PR8-442 grew much more slowly than the WT virus. A previous study with TC-tagged viruses showed that although the TC-tagged virus was attenuated in multiple-cycle replication, the mutant virus was not significantly attenuated during single-cycle growth assays (29). Therefore, we were interested in assessing the replication potential of the Strep-tagged viruses in a single-cycle growth assay. MDCK cells were infected with the respective virus at an MOI of 5, and virus titers in the supernatant were determined by plaque assay at 8 and 10 hpi. As shown in Fig. 1D, PR8-427 and PR8-443 exhibited growth rates similar to the growth rate of the WT virus, whereas PR8-442 grew more slowly than the WT virus.
Expression level of NS1 and the accessibility of the Strep tag to Strep-Tactin, a derivative of streptavidin which binds to the Strep tag specifically with high affinity (40), were evaluated. MDCK cells were infected by the WT or mutant viruses at an MOI of 1. At 8 hpi, cell lysates were prepared and subjected to a pulldown assay with Strep-Tactin Sepharose followed by Western blotting with antibodies specific for NS1 and the Strep tag. As shown in Fig. 1E, while levels of NS1 expression in the mutant viruses were similar to the level of the WT virus (lower panel), NS1 was able to be pulled down efficiently in PR8-427- and PR8-443-infected cells with Strep-Actin, as detected by Western blotting using Strep tag antibody (upper panel) and NS1 antibody (middle panel). In contrast, in PR8-442-infected cells, small amounts of NS1 were present in the pulldown complex, which was barely detected by Strep tag antibody (upper panel) and insufficient to be detected by NS1 antibody (middle panel). These data indicate that the Strep tag inserted after aa 52 was not accessible to its ligand, thus indicating that the PR8-442 mutant virus was not suitable for the purpose of this study.
Identification of viral and host proteins associated with NS1 during virus infection.
The foregoing results showed that both PR8-427 and PR8-443 mutants were suitable for use in experiments to identify the cellular proteins that associate with NS1 during virus infection. We chose PR8-427 to infect cells and performed a pulldown assay, the rationale being that PR8-427 carrying a Strep tag insertion rather than a substitution would minimize the possibility of missing any NS1 function related to a deletion of aa 79 to 84. At 10 hpi, infected cells were harvested and subjected to a pulldown assay with Strep-Tactin Sepharose. The purified protein complexes were subjected directly to protein identification by LC-MS/MS.
The LC-MS/MS analysis identified viral NS1, NP, and PA proteins. Additionally, 25 cellular proteins, which fall into different functional categories, were also identified (Table 1). The most abundant proteins are RNA-binding proteins (such as ATP-dependent RNA helicase A, RNA-binding protein Raly, insulin-like growth factor 2 mRNA-binding protein 1, interleukin enhancer-binding factor 3, and heterogeneous nuclear ribonucleoproteins C, R, H, U, etc.) that are involved in regulation of RNA processing, RNA transcription, mRNA localization, and double-stranded RNA-regulated gene expression (9, 18). Heat shock protein, other chaperone proteins, and tubulin are also presented in the protein complex. Among the identified cellular factors, we were particularly interested in RNA-binding proteins. By analyzing the LC-MS/MS data of RHA, we found that five queries matched RHA protein. Considering the ion scores for each matching peptide, three of them were identified as being homologous to RHA, and two of them are identical to RHA. Given the important role of RHA in modulating RNA processing and RNA-protein remodeling and the fact that influenza virus is an RNA virus, we decided to focus on investigating the potential role of RHA in the influenza virus life cycle.
Table 1.
Cellular proteins associated with Strep-tagged NS1
| Accession code | Protein name | Mass (Da) | Scorea | emPAIb |
|---|---|---|---|---|
| P17042 | Nonstructural protein 1 of influenza A virus | 25,964 | 1,075 | 22.14 |
| ABD62785 | Nucleoprotein of influenza A virus | 56,430 | 1,320 | 5 |
| P11498 | Pyruvate carboxylase, mitochondrial | 130,293 | 601 | 0.56 |
| P05166 | Propionyl-CoA carboxylase beta chain, mitochondrial | 58,806 | 475 | 1.09 |
| Q67256 | Acetyl-CoA carboxylase 1 | 26,260 | 359 | 1.25 |
| Q00839 | Heterogeneous nuclear ribonucleoprotein U | 91,198 | 336 | 0.27 |
| Q9HCC0 | Methylcrotonoyl-CoA carboxylase beta chain, mitochondrial | 61,808 | 319 | 0.51 |
| Q12906 | Interleukin enhancer-binding factor 3 | 95,678 | 301 | 0.21 |
| Q5RA82 | Heterogeneous nuclear ribonucleoprotein C | 33,707 | 195 | 0.53 |
| O42254 | Insulin-like growth factor 2 mRNA-binding protein 1 | 63,574 | 178 | 0.19 |
| Q43390 | Heterogeneous nuclear ribonucleoprotein R | 71,184 | 120 | 0.23 |
| Q9D0E1 | Heterogeneous nuclear ribonucleoprotein M | 77,940 | 87 | 0.10 |
| Q8VHV7 | Heterogeneous nuclear ribonucleoprotein H | 49,442 | 68 | 0.08 |
| Q5R874 | ATP-dependent RNA helicase A | 142,140 | 118 | 0.08 |
| Q4U0F3 | Heat shock 70-kDa protein 1B | 70,495 | 115 | 0.11 |
| P37108 | Signal recognition particle 14-kDa protein | 14,675 | 101 | 0.25 |
| Q96RQ3 | Methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial | 80,935 | 98 | 0.14 |
| Q28618 | Nuclease-sensitive element-binding protein 1 | 35,803 | 81 | 0.10 |
| Q9UKM9 | RNA-binding protein Raly | 32,494 | 75 | 0.12 |
| Q9IQ47 | Polymerase acidic protein of influenza A virus | 83,364 | 64 | 0.09 |
| Q8WN55 | Polypyrimidine tract-binding protein 1 | 57,241 | 49 | 0.07 |
| Q9V1E8 | Signal recognition 54-kDa protein | 49,750 | 42 | 0.08 |
| A2Q0Z1 | Chaperone protein dnaK | 71,082 | 86 | 0.17 |
| Q2KJD0 | Tubulin beta-5 chain | 50,095 | 72 | 0.24 |
| Q8BP67 | 60S ribosomal protein L24 | 17,882 | 71 | 0.22 |
| Q7L2J0 | 7SK snRNA methylphosphate capping enzyme | 74,937 | 66 | 0.05 |
| Q0D9S3 | Cation transporter HKT1 | 59,542 | 65 | 0.06 |
| O95696 | Bromodomain-containing protein 1 | 121,554 | 42 | 0.03 |
The sum of the unique peptide scores.
emPAI, exponentially modified protein abundance index.
RHA interacts with the NS1 protein of influenza A virus and is mediated by RNA.
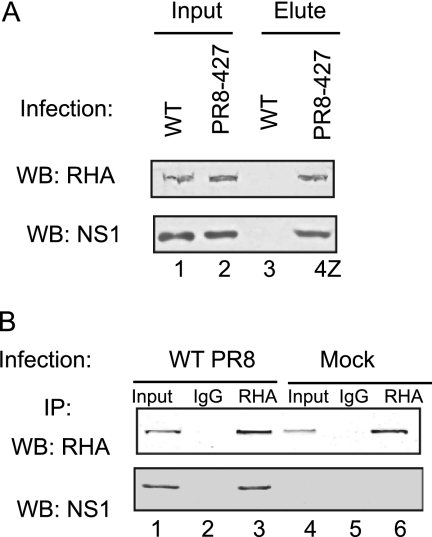
It is possible that some of the protein candidates represent nonspecific interactions. In order to validate the specific interaction of RHA with NS1, we performed the following experiments. First, we infected A549 cells with WT and PR8-427 virus at an MOI of 1. At 10 hpi, cell lysates were prepared and subjected to a pulldown assay with Strep-Tactin Sepharose. Proteins eluted from the Sepharose were subjected to Western blotting using RHA and NS1 antibodies. As seen in Fig. 2A, RHA and NS1 could be detected in the input cell lysates prepared from WT- and PR8-427-infected cells (lanes 1 and 2). While neither RHA nor NS1 was present in the Strep-Tactin Sepharose-eluted samples prepared from WT virus-infected cell lysates (lane 3), RHA and NS1 were readily detected in the eluted samples from PR8-427-infected cells (lane 4). These data demonstrated that Strep-Tactin specifically binds to the Strep tag and furthermore that RHA was associated with Strep-tagged NS1. Next, to exclude the possibility that RHA interacts with the Strep tag, i.e., to validate that endogenous RHA interacts with authentic NS1 protein, we infected A549 cells with WT PR8 virus at an MOI of 1. At 10 hpi, cell lysates were precleared with mouse IgG and incubated with RHA monoclonal antibody-protein or normal mouse IgG-protein G complexes. Immunoprecipitated proteins were subjected to Western blotting by RHA or NS1 antibody. As shown in Fig. 2B, endogenous RHA was present in the input samples in either mock-infected or virus-infected cells (lanes 4 and 1). As expected, NS1 was coimmunoprecipitated in complex with RHA using an RHA antibody in WT virus-infected cells (lane 3). No NS1 was detected in samples that were immunoprecipitated using a control mouse IgG (lane 2). Thus, we confirmed that RHA interacts with NS1 during WT virus infection.
Fig 2.
Validation of RHA-NS1 interaction during virus infection. (A) A549 cells were infected with WT or PR8-427 at MOI of 1. At 10 hpi, cell lysates were prepared and subjected to the pulldown assay with Strep-Tactin Sepharose. Proteins eluted from the Sepharose were subjected to Western blotting (WB) using RHA and NS1 antibodies. (B) A549 cells were mock or WT virus infected at an MOI of 1. Cell lysates were prepared at 10 hpi, precleared by mouse IgG-protein G, and incubated with either RHA antibody-protein G or mouse IgG-protein G. Immunoprecipitated (IP) proteins were subjected to Western blotting (WB) with RHA and NS1 antibodies. Ten percent of input was loaded as a control.
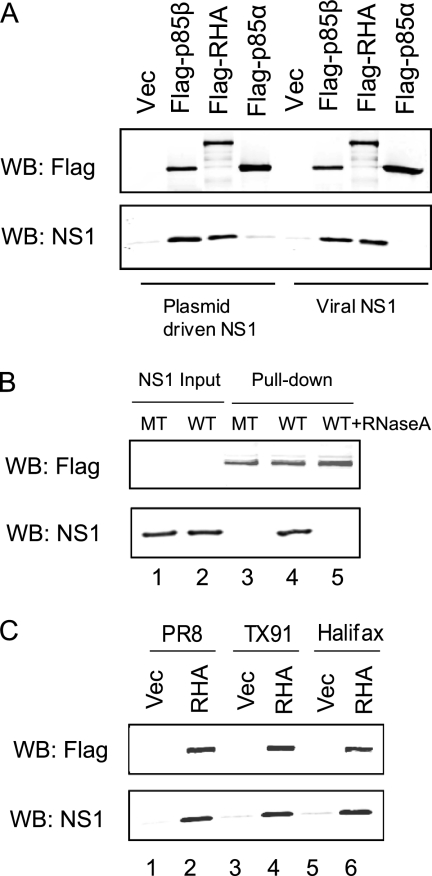
To further characterize RHA and NS1 interaction, we constructed pCMV-3×Flag-RHA, where Flag-tagged RHA is under the control of the cytomegalovirus promoter, and transfected this plasmid into 293T cells. We had previously demonstrated that NS1 interacts with PI3K regulatory subunit p85β but not p85α independent of other viral components; thus cells were also transfected with pCMV-3×Flag-p85β, pCMV-3×Flag-p85α, or pFlag vector (27). Flag-tagged proteins immobilized on an anti-Flag M2 affinity gel were incubated with lysates obtained from 293T cells either transfected with pcDNA3-NS1 (42) or infected with WT PR8. Precipitated proteins were analyzed by Western blotting with anti-Flag or -NS1 antibody. Figure 3A shows that RHA interacted with NS1 as efficiently as p85β. Flag tag alone and p85α did not interact with NS1. Similar amounts of the Flag-p85β, Flag-RHA, and Flag-p85α proteins were used, as demonstrated by Western blotting using Flag antibody (Fig. 3A, upper panel).
Fig 3.
Characterization of RHA and NS1 interaction. (A) Flag-tagged p85β, RHA, or p85α was purified from transfected 293T cells by M2 Flag resin and incubated with cell lysates prepared from pcDNA3-NS1-transfected cells or PR8 WT virus-infected cells. After 4 h of incubation at 4°C, precipitated proteins were eluted by Flag peptide and subjected to Western blotting (WB) using antibodies specific for Flag or NS1. (B) 293T cells were transfected with either pcDNA3-NS1 (WT) or pcDNA3-NS1-R38AK41A (MT). Cell lysates were prepared at 24 h posttransfection and incubated with Flag-tagged RHA immobilized to the M2 resin in the absence or presence of RNase A (10 μg/ml). The precipitated protein complex was then subjected to Western blotting using antibodies specific for Flag or NS1. (C) MDCK cells were infected with PR8, Tx91 or Halifax210 virus at an MOI of 1. At 8 hpi, cell lysates were prepared and subjected to a precipitation assay with Flag-tagged RHA or vector immobilized on M2 resin as described above. Vec, vector.
Since both RHA and NS1 proteins have RNA-binding activity, we were interested in investigating whether RHA and NS1 interaction was mediated by RNA molecules. It has been previously found that the two basic amino acids R38 and K41 on NS1 are essential for RNA-binding activity (6, 47); we therefore constructed pcDNA3-NS1-R38AK41A, where R38 and K41 are replaced by Ala residues. 293T cells were transfected with either pcDNA3-NS1 or pcDNA3-NS1-R38AK41A. Cell lysates were prepared at 48 h posttransfection and were incubated with Flag-RHA immobilized on M2 resin. Figure 3B shows that while similar levels of NS1 expression were obtained (lanes 1 and 2), NS1 with mutations on R38 and K41 failed to interact with RHA (lane 3), suggesting that RNA-binding activity on NS1 is required for interaction with RHA. Moreover, RNase A could completely eliminate the interaction between RHA and WT NS1 (lane 5), further confirming that RHA-NS1 interaction requires RNA.
To examine whether RHA interacts with NS1 derived from other strains of influenza A viruses, we infected MDCK cells with two contemporary strains, Tx91 and Halifax210. Cell lysates were incubated with immobilized Flag-RHA. Figure 3C shows that NS1 proteins derived from Tx91 and Halifax210 could interact with RHA (lanes 4 and 6), as did the NS1 derived from PR8 (lane 2), suggesting that interaction between RHA and NS1 does not seem to be strain specific.
Knockdown of endogenous RHA led to reduction of virus replication.
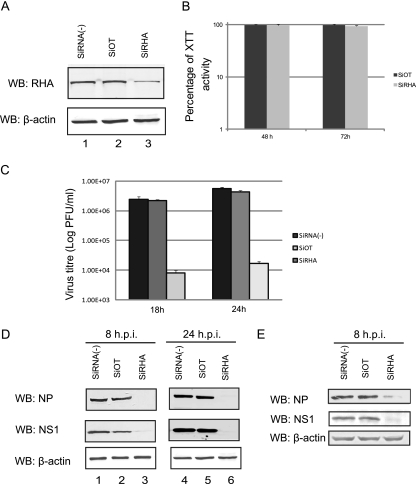
To examine the biological function of RHA during influenza A virus infection, we assessed the gene knockdown effect on virus yield and viral protein expression. A549 cells were transfected with an siRNA mixture that targeted four different regions of RHA. After 48 h, one set of cells was harvested, and the knockdown effect was determined by Western blotting with RHA antibody. As shown in Fig. 4A, off-target siRNA treatment did not alter the expression level of RHA compared to that in untreated cells (lane 2 versus lane 1). However, transfection of RHA siRNA led to significant inhibition of RHA expression (lane 3). siRNA transfection did not alter other cellular protein expression levels dramatically, as measured by β-actin expression levels. The effect of RHA knockdown on cell viability was further determined by XTT (2,3-bis [2-methoxy-4-nitro-5-sulfophenyl]-5-[(phenylamino) carbonyl]-2H-tetrazolium hydroxide) assay. XTT activity was calculated as a percentage of that in off-target siRNA-transfected cells. Figure 4B shows that at both 48 and 72 h after siRNA transfection, knockdown of RHA did not induce detectable cell toxicity compared with off-target siRNA-transfected cells.
Fig 4.
Knockdown of RHA led to a significant reduction in virus yield and viral protein expression. A549 cells were mock transfected or transfected with off-target siRNA (siOT) or siRNAs targeting RHA (siRHA). At 48 h posttransfection, cells were subjected analysis. (A)Western blotting with RHA or β-actin antibody to determine the knockdown effect. (B) XTT assay to determine cell viability after siRNA transfection (a set of cells was also tested at 72 h after siRNA knockdown). (C) siRNA-transfected cells were infected by WT PR8 virus at an MOI of 0.1; virus titers at indicated times were determined by plaque assay. (D) Western blotting to determine the viral protein synthesis with NP and NS1 antibodies. (E) RHA siRNA-transfected cells were infected by Halifax210 at an MOI of 1; at 8 hpi, NP and NS1 protein levels were determined by Western blotting.
Having confirmed the RHA knockdown effect, the siRNA-transfected cells were then infected by WT PR8 virus at an MOI of 0.1. At 18 and 24 hpi, virus titers in the supernatant were determined by plaque assay. As seen in Fig. 4C, the virus titer in off-target siRNA-treated cells was similar to that in untreated cells (2.2 × 106 versus 2.4 × 106 PFU/ml at 18 hpi and 4.4 × 106 versus 5.6 × 106 PFU/ml at 24 hpi). However, siRNA knockdown of RHA in A549 cells caused a more than 300-fold reduction in virus yield (8 × 103 versus 2.4 × 106 PFU/ml at 18 hpi and 1.67 × 104 versus 5.6 × 106 PFU/ml at 24 hpi). Furthermore, inhibition of RHA expression caused a significant reduction in viral protein synthesis. As shown in Fig. 4D, at 8 and 24 hpi, while NS1 and NP protein levels were readily detected in untreated and off-target siRNA-treated cells (lanes 1, 2, 4, and 5), they were barely detectable in RHA siRNA-treated cells (lanes 3 and 6). Levels of β-actin were determined as loading controls. In order to test whether RHA plays a role in other influenza A virus strains, A549 cells were transfected by RHA siRNA or off-target siRNA and were infected 48 h later with Halifax210 at an MOI of 1. At 8 hpi, viral protein synthesis was determined by Western blotting (Fig. 4E). In agreement with the results obtained by using PR8 virus, RHA knockdown resulted in a significant reduction in viral NP protein and NS1 protein synthesis. Cellular β-actin levels were not altered by siRNA transfection and virus infection.
Knockdown of endogenous RHA led to reduction of vRNA, cRNA, and mRNA synthesis during virus infection.
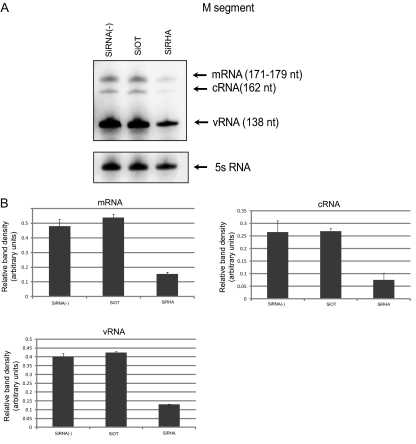
The DEXD/H family of RNA helicases is known to play an important role in all aspects of RNA synthesis and function. Since we observed that RHA knockdown significantly decreased virus propagation, we wished to understand whether RHA would have any effect on viral RNA synthesis. Thus, A549 cells transfected by respective siRNAs were infected by WT PR8 virus. At 8 hpi, total RNA was isolated from cells and subjected to primer extension assay. As shown in Fig. 5A, in off-target siRNA-treated cells, viral RNA levels were not markedly changed compared to those in untreated cells. However, knockdown of RHA gene expression led to a dramatic reduction of mRNA, cRNA, and vRNA levels of the M segment. Band densities of mRNA, cRNA, and vRNA were normalized to that of 5S RNA in the same sample (Fig. 5B). Similar results were obtained by using primers specific for NP and NS1 segments (data not shown).
Fig 5.
Knockdown of RHA led to a significant reduction in virus RNA synthesis. (A) Primer extension assay of mRNA, cRNA, and vRNA of the M segment in RHA knocked down cells. One representative of three independent experiments is shown here. (B) Quantification of band density by Quantity One software. mRNA, cRNA, and vRNA levels were normalized to 5S RNA levels in the same sample.
RHA enhances viral transcription and replication in vitro.
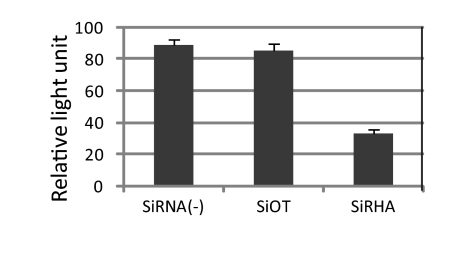
Influenza virus viral RNA exists in the form of RNP, where the viral RNA is associated with NP and forms a helical hairpin that is bound on one end by the heterotrimeric polymerase complex consisting of PB2, PB1, and PA proteins (35). Once in the nucleus, the incoming negative-sense vRNA will be used as a template to synthesize mRNA and cRNA. Considering that DEXD/H box proteins have the ability to unwind RNA structure or dissociate RNA-protein complex and the forgoing results that knockdown of RHA led to a reduction of cRNA and mRNA levels during virus infection, it is reasonable to examine whether RHA plays a regulatory role during viral transcription and replication. Thus, we performed a minigenome assay in RHA-depleted A549 cells by cotransfection of pPol1-NP-Luc and pTK-rLuc together with plasmids expressing PB2, PB1, PA, and NP proteins (36). As seen in Fig. 6, polymerase activity in RHA knockdown cells was approximately 2.6- and 2.7-fold less than activity in off-target siRNA-transfected cells and untreated cells, respectively.
Fig 6.
Knockdown of RHA led to a reduced RNA polymerase activity in the minigenome assay. A549 cells were transfected by the respective siRNA for 48 h. Cells were then transfected with plasmids expressing polymerase subunits and NP together with phPolI-LUC-NP and pTK-rLuc. Luciferase activity was measured at 24 h posttransfection and normalized to that of Renilla luciferase. Each luciferase activity value is the average of three independent experiments.
To further confirm the role of RHA in regulating RNA synthesis, we employed a cell-free viral genome replication system with virion-derived vRNP (vvRNP). In this assay, purified vvRNP is able to synthesize primer-independent cRNA and primer-dependent mRNA (when globin mRNA is provided), whose level can be determined by gene-specific primer extension assay (46). We first purified RHA from 293T cells and added it to the in vitro reaction mixture. All reaction mixtures contained the ATP required for RHA activity. A primer extension assay was performed using primers specific for NA (Fig. 7A) as well as for M (Fig. 7B). Figure 7A shows that in the absence of NTP, neither mRNA nor cRNA was detected (lane 2), indicating that there was no contamination of mRNA and cRNA molecules in the vvRNP preparation. The level of vRNA in this sample represents the input vRNA in the vvRNP. In the presence of NTP, newly synthesized cRNA and mRNA were evident (lane 3). These levels were set as the reference levels to which all the others were normalized. Addition of RHA at 0.5, 1, and 2 pmol per reaction volume led to an increase of mRNA and cRNA synthesis in a dose-dependent manner by 1.75-, 2.8-, and 5.7-fold for mRNA and by 1.9-, 2.8-, and 4.9-fold for cRNA, respectively (lanes 7 to 9). This increase was not due to additional vvRNP in the reaction mixture since the vRNA levels in all samples were similar.
Next, we were interested in determining whether RHA helicase activity is required for enhancing viral transcription and replication. It has been shown that ATP is the general energy source for RHA's helicase activity. Mutation of Lys to Arg at aa 417 results in a reduced ATP binding capability and thus abolishes its helicase activity (1, 33). We made a construct which expresses RHA with a K417R mutation (RHA-K417R) and purified this protein from 293T cells. Figure 7A shows that when increased doses of RHA-K417R were added to the in vitro reaction mixture, similar levels of mRNA and cRNA were synthesized in each sample (lanes 4 to 6), and all levels were comparable to the basal level (lane 3). Specifically, when the basal level was set as the reference, addition of RHA-K417R at 0.5, 1, or 2 pmol to the reaction mixture resulted in a 1.1-, 0.83-, or 0.9-fold change in mRNA synthesis and in a 1.1-, 1.1-, or 1.2-fold change in cRNA level, respectively. These results suggest that the helicase activity of RHA is critical in enhancing influenza virus viral RNA transcription and replication.
The role of NS1 in RHA-mediated regulation of virus transcription and replication was also tested. There were no apparent changes in mRNA and cRNA levels in the presence of NS1 compared to those in the control reaction products (lane 10 versus lane 3). Addition of RHA increased transcription/replication by 2.8-fold (lane 8); simultaneous addition of NS1 did not further increase transcription/replication (lane 11).
In order to test whether this stimulation is virus genomic RNA segment specific, the synthesized RNAs were detected with primers specific for the M segment. As shown in Fig. 7B, the synthesis patterns of M cRNA and M mRNA were similar to those of NA cRNA and NA mRNA. Therefore, it is reasonable to believe that the stimulation effect of RHA was not viral RNA segment specific.
The primer extension assay detects only the portion close to the 5′ end of the nascent cRNA and mRNA with a length of around 200 nucleotides (nt). In order to investigate whether RHA is able to stimulate full-length viral transcription and replication, total RNA was isolated from the above experiment. All three types of full-length influenza virus NA RNAs were quantified by a novel strand-specific real-time RT-PCR (22). In agreement with the results obtained from primer extension assays, in the presence of 0.5, 1, and 2 pmol of RHA, cDNA levels increased by 5.7-, 10.56-, and 19.94-fold, respectively (Fig. 8A), and mRNA levels increased by 1.27-, 3.33-, and 7.86-fold (Fig. 8B). Addition of RHA-K417R did not alter full-length cRNA and mRNA significantly, further confirming that the helicase activity in RHA is essential for viral transcription and replication. NS1 did not have a significant influence on cRNA and mRNA synthesis in this system. In the presence of 1 pmol of NS1 and 1 pmol of RHA, the cRNA and mRNA levels increased by 9.6- and 2.71-fold, respectively, changes which are slightly lower than the levels with 1 pmol of RHA alone (10.56- and 3.33-fold, respectively). vRNA levels in all samples were very similar, as detected by real-time RT-PCR (data not shown).
DISCUSSION
Influenza virus NS1 protein is a multifunctional protein and a virulence factor. In order to exert various functions during the virus life cycle, NS1 functions to modulate the activity of cellular machinery by interacting with many cellular factors. Although some cellular factors associated with NS1 have been identified, it is believed that many factors remain undiscovered. To identify these factors in the context of virus infection, one approach is to perform a coimmunoprecipitation assay using antibody specific for NS1; however, this requires a highly specific monoclonal antibody against NS1, and such antibody is not always available. The other approach is to fuse a tag in the protein of interest and perform affinity purification. The challenge of this approach is to generate replication-competent viruses bearing a tag in the NS1 protein. Here, we have extended our previous study that identified regions in the NS1 gene that could accommodate a small tag insertion/substitution (29); we inserted Strep Tag II, a small affinity peptide, in NS1 protein and generated some mutant viruses. Among those viruses, PR8-427 and PR8-443 grew to titers similar those of the WT virus during a single cycle of infection though they were attenuated in multicycle growth analysis (Fig. 1), indicating that these two viruses possess similar properties to the WT virus during single-cycle infection. Furthermore, the Strep tag inserted in these two viruses is exposed on the surface of NS1, which is accessible to its ligand Strep-Tactin, allowing the affinity purification of Strep-tagged NS1 protein from virus-infected cells (Fig. 2).
Mass spectrometry of the protein complex associated with NS1 protein revealed two other viral proteins, NP and PA. This suggested that NS1 may associate with RNP complex during infection. Since we did not find other components of RNP complex by LC-MS/MS, we performed Western blotting of the protein complex associated with Strep-tagged NS1 by using antibodies specific for PB2, PB1, PA, and NP. Our results showed that all the RNP components were present in the complex pulled down with NS1 (data not shown). These results are in agreement with the recent study of Robb et al., who reported the interaction of NS1 with NP protein of viral RNP complex (38), thus validating our system for isolation of Strep-tagged NS1 protein and its binding partners in virus-infected cells. However, we did not find other known cellular factors that interact with NS1, such as CPSF and RIG-I (32). This might be due to reasons such as time of harvesting after virus infection or cell line and virus strain used in this study. Indeed, NS1 derived from PR8 virus does not bind CPSF (5). Interestingly, we found some proteins, such as acetyl-coenzyme A (CoA) carboxylase 1, interleukin enhancer-binding factor 3, heat shock 70-kDa protein, and heterogeneous nuclear ribonucleoprotein M, that were previously identified as interaction partners of the influenza virus RNP complex using Strep-tagged NP as bait (19, 31). Since NS1 interacts with NP, whether these proteins represent components of a large protein complex bridged through NP or directly interact with NS1 remains to be elucidated in the future study.
In this report, we focused on the characterization and functional analysis of RHA, a novel cellular factor, identified by the system we established. RHA, also known as DHX9, is a 130-kDa protein which belongs to the DEXD/H box family of proteins. Members of this family act as ATP-driven motors and can unwind both double-stranded RNA and DNA (50, 51) or control switches at very specific points in processes such as pre-mRNA splicing or during ribosome biogenesis (17). In addition to its helicase activity, with the energy derived from ATP, RHA is able to bind and remodel RNA or RNA-protein complexes and subsequently regulates many aspects of cellular RNA metabolism, such as transcription, RNA nuclear export, and translation initiation (9, 17).
In addition to its multiple cellular functions, RHA, as demonstrated by a wealth of data, is also involved in the modulation of replication of various viruses, including human immunodeficiency virus type 1 (HIV-1), lymphotropic retrovirus, hepatitis C virus, and foot-and-mouth disease virus (3, 16, 25, 39). RHA has been reported to be associated with the Gag protein of HIV, which mediates the incorporation of RHA into HIV-1 particles and coordinates with Gag protein to promote the annealing of tRNA to HIV RNA (39, 49). RHA is necessary for efficient HIV-1 RNA translation that requires ATPase-dependent helicase function (2).
The interaction of RHA with NS1 was validated by a coimmunoprecipitation assay with RHA monoclonal antibody followed by Western blot analysis with NS1 antibody in WT virus-infected cells (Fig. 2). Furthermore, Flag-tagged RHA could interact with NS1 derived not only from virus infection but also from plasmid transfection (Fig. 3A), indicating that RHA-NS1 interaction does not require the presence of other viral proteins. The NS1-RHA interaction was eliminated by the presence of RNase A, and the mutant form of NS1 did not interact with RHA (Fig. 3B), suggesting that the interaction requires RNA molecules. The biological function of RHA in the influenza virus life cycle was further assessed. Knockdown of RHA gene expression did not significantly alter cell viability; however, it considerably reduced virus yield to a magnitude of 300-fold (Fig. 4), indicating that RHA plays a pivotal role in the virus life cycle. The reduction of mRNA and cRNA levels in RHA-depleted cells (Fig. 5) indicated that RHA might be involved in viral RNA transcription and replication.
Our in vitro viral genome replication assay showed that in the presence of RHA, cRNA and mRNA synthesis increased in a dose-dependent manner (Fig. 7 and 8). Addition of NS1 alone to the polymerase reaction mixture did not alter cRNA and mRNA synthesis. The addition of NS1 and RHA led to increased cRNA and mRNA synthesis of equal levels such as with RHA alone. These data seem to suggest that RHA regulation of viral transcription and replication is independent of interaction with NS1. One explanation would be that in an in vitro system, when RHA is added alone, it can easily find vRNP and then is involved in regulation; therefore, it does not need NS1. However, this would not exclude the possibility that in the infected cells, RHA might be recruited to the vRNP via NS1 through RNA-mediated interaction. This notion was supported by the results that RNP could be pulled down along with NS1 (Table 1) and by several reports that the interaction of NS1 with RNP plays an important role in regulating viral replication (7, 24, 30). Considering that DEXD/H box proteins have the ability to unwind RNA structure or dissociate RNA-protein complex, a possible mechanism by which RHA regulates influenza virus RNA synthesis might be by remodeling of RNA-NP or RNA secondary structure to enhance RNA synthesis or by directly enhancing viral RNA polymerase activity. The exact mechanism by which RHA regulates influenza A virus replication remains to be elucidated.
In summary, here we report that by employing a novel NS1-tagging approach, we identified several novel cellular factors that interact with NS1 protein during influenza A virus infection. We further characterized the interaction of RHA and NS1 and investigated the possible mechanism by which RHA regulates influenza A virus infection. The recombinant viruses carrying a Strep-tagged NS1 are valuable tools for the identification of novel cellular factors that play fundamental roles in the influenza virus replication cycle.
ACKNOWLEDGMENTS
We thank Nathalie Berube for technical assistance.
Y.Z. is a recipient of Canadian Institutes of Health Research New Investigator Award. X.L. was supported by postdoctoral fellowships from the Saskatchewan Health Research Foundation. This work was supported by a grant from CIHR.
Footnotes
Published ahead of print 14 December 2011
This article is VIDO manuscript series number 600.
REFERENCES
- 1. Aratani S, et al. 2001. Dual roles of RNA helicase A in CREB-dependent transcription. Mol. Cell. Biol. 21:4460–4469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bolinger C, Sharma A, Singh D, Yu L, Boris-Lawrie K. 2010. RNA helicase A modulates translation of HIV-1 and infectivity of progeny virions. Nucleic Acids Res. 38:1686–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bolinger C, et al. 2007. RNA helicase A interacts with divergent lymphotropic retroviruses and promotes translation of human T-cell leukemia virus type 1. Nucleic Acids Res. 35:2629–2642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brass AL, et al. 2009. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 139:1243–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Das K, et al. 2008. Structural basis for suppression of a host antiviral response by influenza A virus. Proc. Natl. Acad. Sci. U. S. A. 105:13093–13098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Donelan NR, Basler CF, Garcia-Sastre A. 2003. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J. Virol. 77:13257–13266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Falcon AM, et al. 2004. Defective RNA replication and late gene expression in temperature-sensitive influenza viruses expressing deleted forms of the NS1 protein. J. Virol. 78:3880–3888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fodor E, et al. 2002. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J. Virol. 76:8989–9001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fuller-Pace FV. 2006. DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res. 34:4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gack MU, et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host. Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hale BG, Jackson D, Chen YH, Lamb RA, Randall RE. 2006. Influenza A virus NS1 protein binds p85β and activates phosphatidylinositol-3-kinase signaling. Proc. Natl. Acad. Sci. U. S. A. 103:14194–14199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hale BG, Randall RE, Ortin J, Jackson D. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376 [DOI] [PubMed] [Google Scholar]
- 13. Hao L, et al. 2008. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454:890–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoffmann E, Krauss S, Perez D, Webby R, Webster RG. 2002. Eight-plasmid system for rapid generation of influenza virus vaccines. Vaccine 20:3165–3170 [DOI] [PubMed] [Google Scholar]
- 15. Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. U. S. A. 97:6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Isken O, et al. 2007. Nuclear factors are involved in hepatitis C virus RNA replication. RNA 13:1675–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jankowsky E, Bowers H. 2006. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 34:4181–4188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jonson L, et al. 2007. Molecular composition of IMP1 ribonucleoprotein granules. Mol. Cell Proteomics 6:798–811 [DOI] [PubMed] [Google Scholar]
- 19. Jorba N, et al. 2008. Analysis of the interaction of influenza virus polymerase complex with human cell factors. Proteomics 8:2077–2088 [DOI] [PubMed] [Google Scholar]
- 20. Karlas A, et al. 2010. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 463:818–822 [DOI] [PubMed] [Google Scholar]
- 21. Kawaguchi A, Momose F, Nagata K. 2011. Replication-coupled and host factor-mediated encapsidation of the influenza virus genome by viral nucleoprotein. J. Virol. 85:6197–6204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kawakami E, et al. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods 173:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Konig R, et al. 2010. Human host factors required for influenza virus replication. Nature 463:813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuo RL, Krug RM. 2009. Influenza a virus polymerase is an integral component of the CPSF30-NS1A protein complex in infected cells. J. Virol. 83:1611–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lawrence P, Rieder E. 2009. Identification of RNA helicase A as a new host factor in the replication cycle of foot-and-mouth disease virus. J. Virol. 83:11356–11366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee JH, et al. 2010. Direct interaction of cellular hnRNP-F and NS1 of influenza A virus accelerates viral replication by modulation of viral transcriptional activity and host gene expression. Virology 397:89–99 [DOI] [PubMed] [Google Scholar]
- 27. Li Y, Anderson DH, Liu Q, Zhou Y. 2008. Mechanism of influenza A virus NS1 protein interaction with the p85beta, but not the p85α, subunit of phosphatidylinositol 3-kinase (PI3K) and up-regulation of PI3K activity. J. Biol. Chem. 283:23397–23409 [DOI] [PubMed] [Google Scholar]
- 28. Li Y, Chen ZY, Wang W, Baker CC, Krug RM. 2001. The 3′-end-processing factor CPSF is required for the splicing of single-intron pre-mRNAs in vivo. RNA. 7:920–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li Y, et al. 2010. Genetically engineered, biarsenically labeled influenza virus allows visualization of viral NS1 protein in living cells. J. Virol. 84:7204–7213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marion RM, Zurcher T, LSde la, Ortin J. 1997. Influenza virus NS1 protein interacts with viral transcription-replication complexes in vivo. J. Gen. Virol. 78:2447–2451 [DOI] [PubMed] [Google Scholar]
- 31. Mayer D, et al. 2007. Identification of cellular interaction partners of the influenza virus ribonucleoprotein complex and polymerase complex using proteomic-based approaches. J. Proteome Res. 6:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mibayashi M, et al. 2007. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 81:514–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakajima T, et al. 1997. RNA helicase A mediates association of CBP with RNA polymerase II. Cell 90:1107–1112 [DOI] [PubMed] [Google Scholar]
- 34. Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. 1998. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol. Cell 1:991–1000 [DOI] [PubMed] [Google Scholar]
- 35. Palese P, Shaw ML. 2007. Orthomyxoviridae: the viruses and their replication, p 1648–1689 In Knipe DM, et al. (ed), Fields virology, 5th ed Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 36. Ping J, et al. 2010. PB2 and hemagglutinin mutations are major determinants of host range and virulence in mouse-adapted influenza A virus. J. Virol. 84:10606–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Qian XY, Alonso-Caplen F, Krug RM. 1994. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J. Virol. 68:2433–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robb NC, et al. 2011. The influenza A virus NS1 protein interacts with the nucleoprotein of viral ribonucleoprotein complexes. J. Virol. 85:5228–5231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Roy BB, et al. 2006. Association of RNA helicase a with human immunodeficiency virus type 1 particles. J. Biol. Chem. 281:12625–12635 [DOI] [PubMed] [Google Scholar]
- 40. Schmidt TG, Skerra A. 2007. The Strep-tag system for one-step purification and high-affinity detection or capturing of proteins. Nat. Protoc. 2:1528–1535 [DOI] [PubMed] [Google Scholar]
- 41. Shapira SD, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139:1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Shin YK, et al. 2007. SH3 binding motif 1 in influenza A virus NS1 protein is essential for PI3K/Akt signaling pathway activation. J. Virol. 81:12730–12739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. J. Gen. Virol. 88:942–950 [DOI] [PubMed] [Google Scholar]
- 44. Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. 2007. Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J. Gen. Virol. 88:13–18 [DOI] [PubMed] [Google Scholar]
- 45. Skerra A, Schmidt TG. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271–304 [DOI] [PubMed] [Google Scholar]
- 46. Vreede FT, Brownlee GG. 2007. Influenza virion-derived viral ribonucleoproteins synthesize both mRNA and cRNA in vitro. J. Virol. 81:2196–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang W, et al. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Watanabe T, Watanabe S, Kawaoka Y. 2010. Cellular networks involved in the influenza virus life cycle. Cell Host Microbe 7:427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Xing L, Liang C, Kleiman L. 2011. Coordinate roles of Gag and RNA helicase A in promoting the annealing of tRNALys3 to HIV-1 RNA. J. Virol. 85:1847–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhang S, Grosse F. 1997. Domain structure of human nuclear DNA helicase II (RNA helicase A). J. Biol. Chem. 272:11487–11494 [DOI] [PubMed] [Google Scholar]
- 51. Zhou K, et al. 2003. RNA helicase A interacts with dsDNA and topoisomerase IIα. Nucleic Acids Res. 31:2253–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]