Abstract
The life cycle of adenoviruses is divided by convention into early and late phases, separated by the onset of viral genome replication. Early events include virus adsorption, transport of the genome into the nucleus, and the expression of early genes. After the onset of viral DNA replication, transcription of the major late transcription unit (MLTU) and thereby synthesis of late proteins is induced. These steps are controlled by an orchestra of regulatory processes and require import of the genome and numerous viral proteins into the nucleus, as well as active transport of viral transcripts and proteins from the nucleus to the cytoplasm. The latter is achieved by exploiting the shuttling functions of cellular transport receptors, which normally stimulate the nuclear export of cellular mRNA and protein cargos. A set of adenoviral early and late proteins contains a leucine-rich nuclear export signal of the HIV-1 Rev type, known to be recognized by the cellular export receptor CRM1. However, a role for CRM1-dependent export in supporting adenoviral replication has not been established. To address this issue in detail, we investigated the impact of two different CRM1 inhibitors on several steps of the adenoviral life cycle. Inhibition of CRM1 led to a reduction in viral early and late gene expression, viral genome replication, and progeny virus production. For the first time, our findings indicate that CRM1-dependent shuttling is required for the efficient export of adenoviral early mRNA.
INTRODUCTION
The human subgroup C adenoviruses, such as adenovirus type 5 (Ad5), encode several gene products that are required for efficient synthesis of viral macromolecules and progeny production. During the early phase, the E1A gene products interact with a variety of cellular proteins to induce an optimal cellular environment that is conducive to viral gene expression and replication (reviewed in references 23, 28, and 66). The late phase is characterized by the production of large quantities of viral macromolecules and a severe inhibition of cellular protein synthesis (2, 53, 83). This highly efficient expression of viral late genes is achieved by the preferential accumulation of viral late transcripts in the cytoplasm and inhibition of nuclear export of most cellular mRNA induced by the E1B-55K and E4orf6 proteins (1, 10, 37, 49, 60). Furthermore, the selective translation of viral late mRNA is induced by VA-RNA1 and the L4-100K protein (64, 75, 79, 80). In addition, the L4-100K and pVI structural proteins are known to participate in trimerization and nuclear import of Hexon, as well as capsid assembly (14, 15, 39, 78).
As infection progresses the early E1A, E1B-55K, E4orf6, and late L4-100K and pVI proteins localize in both the cytoplasm and nucleus at different times of viral replication. However, the impact of their precise intracellular distribution on their activities is not well understood. These proteins all possess nuclear export signals (NES) of the HIV-1 Rev type, and it is clearly established that the nucleocytoplasmic export of E1A, E1B-55K, and E4orf6 can be directed through their NES by the cellular export receptor CRM1 (16, 19, 44, 46, 69, 78).
During the early stages of adenovirus infection, nuclear targeting of protein pVII and viral DNA into the nucleus is directed by a cellular import mechanism that requires binding of viral particles to the nucleoporin CAN/Nup214 at the nuclear pore complex (NPC) (34, 51, 76). Interestingly, leptomycin B (LMB), a specific inhibitor of CRM1, blocks the dissociation of incoming viral particles from microtubules. This inhibits the particles' binding to the NPC, suggesting that either CRM1 or CRM1-dependent export is required for the viral genome to reach the nucleus (74). The CRM1 export pathway is also known to be responsible for E1A export through an NES located between amino acid residues 70 and 80 in the Ad5 protein. Inactivation of the E1A-NES by mutation abrogates export of the protein in infected cells and severely reduces viral progeny production (41). A Rev-type NES is also present in L4-100K (16) and pVI (78), suggesting that CRM1 activity could also be required at later stages of viral replication for selective translation of viral late mRNA or nuclear import and assembly of viral late proteins (14, 39, 55, 64, 78–80).
The contribution made by the NES sequences within the E1B-55K and E4orf6 proteins to viral replication is not completely clear. To address the role of CRM1 in E1B-55K and E4orf6-mediated viral mRNA transport, the CRM1 inhibitor LMB was used, which associates covalently to cysteine 528 in the NES binding region of CRM1 and thereby irreversibly blocks its interaction with NES containing proteins (26, 47). These experiments showed that the functional inhibition of CRM1 by LMB had only a minor effect on viral late protein synthesis (an indirect measurement of viral late mRNA export), leading to the conclusion that LMB treatment during the late phase of adenoviral replication does not abrogate viral late mRNA accumulation in the cytoplasm (11, 62). A different experimental approach, in which a specific peptide inhibitor of CRM1 was used to make direct measurements of viral late mRNA export, supported these findings. Even though shuttling of E1B-55K was drastically reduced, viral late mRNA transport was unaffected by the peptide inhibitor (25). We recently generated virus mutants carrying amino acid substitutions in the NES sequences of either or both the E1B-55K and E4orf6 to evaluate the role of CRM1-dependent nuclear export of both proteins in efficient viral replication in the context of Ad5-infected cells (44, 69).
Consistent with previous observations (11, 25, 62), our findings demonstrated that inhibition of CRM1-mediated export of E1B-55K or E4orf6 has no effect on viral late mRNA export. Rather, the NES present in E1B-55K and E4orf6 are differentially required for the export of p53 and Mre11, two important cellular targets of the adenovirus-dependent cell-specific E3 ubiquitin ligase, indicating that although dispensable for viral late mRNA export, CRM1 is required for other activities that depend on E1B-55K/E4orf6 shuttling (44, 69).
Furthermore, since the adenoviral E1A, L4-100K, and pVI proteins are known to possess leucine-rich NES sequences, the CRM1 pathway is likely to contribute to a variety of both early and late events during viral replication (16, 41, 78). However, the contribution of CRM1-dependent export to the activities of these adenoviral proteins is not completely understood (16, 19, 41, 44, 69, 78).
To address this issue, we designed our experiments to analyze how inhibition of the CRM1 transport pathway affects viral replication at different stages. We developed a system that exploits the modular nature of the nucleoporin 214, also known as CAN. The C terminus of CAN (CANc) contains the FG (phenylalanine, glycine) repeats that associate with CRM1 during its translocation through the NPC (reviewed in reference 40). Whereas treatment with LMB blocks CRM1 binding to proteins containing a leucine-rich NES (26), overexpression of CANc leads to a dominant-negative inhibition of CRM1-dependent nuclear export by selectively binding to CRM1. This prevents the association of CRM1 with the NPC (6, 27, 42, 84). In contrast to CRM1, TAP does not interact with the C terminus of CAN; hence, TAP-dependent export, predominantly bulk cellular mRNA transport, is unaffected (6, 77). In this system, the production of CANc can be induced for the period of time required, allowing a detailed analysis of the role of CRM1 in both early and late phenotypes.
Using controlled expression of CANc in parallel with LMB treatment of Ad-infected cells, we evaluated whether CRM1 was required for protein production, early mRNA synthesis and export, viral DNA replication, or virus yield. While, as expected, the inhibition of CRM1 by either CANc or LMB did not impair the export of viral late transcripts, we observed for the first time a reduced cytoplasmic accumulation of viral early mRNA. Furthermore, the abrogation of CRM1 activity reduced viral protein and DNA synthesis, as well as virus growth, indicating that a functional CRM1-dependent pathway is required for early activities of the adenoviral replication cycle.
MATERIALS AND METHODS
Cell culture.
A549 (DSMZ ACC 107; Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH, Braunschweig, Germany), 2E2 (13), HEK293 (33), and HepaRG (35) cells were grown as monolayer cultures in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum (FCS), 100 U of penicillin per ml, and 100 μg of streptomycin per ml in a 5% CO2 atmosphere at 37°C. For HepaRG cells, the medium was supplemented with 5 μg of bovine insulin per ml and 0.5 μM hydrocortisone.
To generate monoclonal HA-CANc cells, the nontransformed HepaRG cells were first transduced with the lentiviral vector, pLKOneo.CMV.EGFPnlsTetR, expressing the EGFPnlsTetR repressor, as described previously (21, 22). Transduced cells were selected and maintained in medium containing neomycin (250 μg/ml). Once established, the HA-TetR cell line was transfected with pLKO.DCMV.TetO.CANc-HA and selected and maintained in medium also containing puromycin (1 μg/ml). To determine the cell viability, the cells were collected at the indicated time points and stained with trypan blue, and viable cells were counted using a hemocytometer.
Plasmids and transient transfections.
To generate a tetracycline-inducible plasmid expressing a CANc-HA protein downstream of the Tet On promoter, we inserted restriction sites for both NheI and EcoRV at the ends of pLKO.DCMV.TetO (21, 22) by PCR using the primers 1560 and 1561 (Table 1). In addition, we obtained a CANc-HA gene fusion with attached NheI and EcoRV restriction sites, by PCR amplification of pcDNA3-CANc (65, 71) using the oligonucleotide primers 1556 and 1557 (Table 1). After NheI/EcoRV digestion, CANc-HA was ligated with the backbone of pLKO.DCMV.TetO, resulting in pLKO.DCMV.TetO.CANc-HA.
Table 1.
Primers used in this study
| Primer | Sequence (5′–3′) |
|---|---|
| 1204 | CATCCTGGGCTACACTGA |
| 1205 | TTGACAAAGTGGTCGTTG |
| 1556 | GCGCTAGCATGCAGCAATCATCC |
| 1557 | GCGATATCTCAAGCGTAGTCTGG |
| 1560 | GCGATATCGAATTCTCGACCTCGAGACAAATGG |
| 1561 | GCGCTAGCGGCTCACGAGCGAAGCTTGATCTCTATCACTG |
| 1569 | GAGGGTAACTCCAGGGTGCG |
| 1570 | TTTCACTAGCATGAAGCAACCACA |
| 1686 | GTGCCCCATTAAACCAGTTG |
| 1687 | GGCGTTTACAGCTCAAGTCC |
| 1688 | GGTCTGGGCGTTAGGATACA |
| 1689 | CAATCAGTTTTCCGGCAAGT |
| 1767 | GCTGGTTTAGGATGGTGGTG |
| 1768 | CCCTCATAAACACGCTGGAC |
| 2183 | GAAACCAGAGGGCGAAGACC |
| 2184 | AGTCTGGGTCACGGTGAAGG |
The expression vector p3_eGFP-CD83complete is a derivative of pcDNA3 (Invitrogen) that contains the enhanced green fluorescent protein (eGFP) coding sequence, followed by the human CD83 cDNA and its flanking 5′- and 3′-untranslated regions (61).
For transient transfection of HA-TetR cells with pLKO.DCMV.TetO.CANc-HA, the cells were treated as described previously (70). After transfection, medium was replaced with DMEM supplemented with 10% FCS, 100 U of penicillin per ml, 100 μg of streptomycin per ml, 5 μg of bovine insulin per ml, and 0.5 μM hydrocortisone.
Viruses.
H5pg4100 served as the wild-type (wt) Ad5 parent virus in these studies (44). H5pm4149 has been described recently (18) and carries four stop codons in the 55K reading frame. H5pm4154 expresses normal E4orf6/7 protein but an unstable or nonfunctional product of E4orf6 containing only the first 66 residues (5). H5pg4100, H5pm4149, and H5pm4154 were propagated in 2E2 monolayer cultures. All virus titers were determined by fluorescent focus assays (36). To measure virus growth, infected cells were harvested at the indicated time postinfection (p.i.). The cell lysates were serially diluted in DMEM for infection of HEK293 cells, and the virus yield was determined by quantitative E2A-72K immunofluorescence staining at 24 h p.i (36). To measure the viral DNA replication, viral DNA was isolated, and quantitative PCR was performed as described previously (70).
Antibodies, protein analysis, and inhibitors.
Primary antibodies specific for Ad proteins used in the present study included E1A mouse monoclonal antibody (MAb) M73 (38), E1B-55K mouse MAb 2A6 (67), E2A-72K (DBP) mouse MAb B6-8 (63), E4orf3 rat MAb 6A11 (57), E4orf6 rabbit polyclonal antibody 1807 (7), E4orf6 mouse MAb RSA3 (52), L4-100K rat MAb 6B10 (48), and Ad5 rabbit polyclonal serum L133 (69). Primary antibodies specific for cellular proteins included β-actin mouse MAb AC-15 (Sigma-Aldrich, Inc.) and CRM1 mouse MAb exportin-1 (BD Transduction). Primary antibodies specific for tag proteins included HA rat MAb 3F10 (Roche Applied Science, Inc.) and GFP rabbit polyclonal antibody GFP (FL) (Santa Cruz Biotechnology, Inc.).
Preparation of cell lysates for protein analysis and immunoblotting, as well as preparation of cells for indirect immunofluorescence, has been described previously (45).
LMB was purchased from Enzo Life Sciences and used at a concentration of 20 nM to block CRM1-dependent nuclear export.
Quantitative PCR analysis.
Preparation of total, cytoplasmic, and nuclear RNA from Ad-infected cells for quantitative PCR analysis was conducted exactly as described previously (69). Quantitative PCR was performed in triplicate for each sample by denaturing at 95°C for 10 min, followed by 40 cycles of 95°C for 15 s, 60°C (E1A, E2A, E4orf6 and GAPDH) or 62°C (E1B and IVa2) for 30 s, and 72°C for 15 s. Raw numbers were normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) as an internal control. The relative RNA concentrations were determined by the standard curve method. The subcellular distribution of adenoviral transcripts is presented as the ratio of cytoplasmic to nuclear mRNAs. Standard deviations were calculated by the best value of sum or quotient. For amplification of viral early RNA, oligonucleotide primers 1686/1687 (E1A), 1688/1689 (E2A), 1569/1570 (E1B), and 1767/1768 (E4orf6) (Table 1) located in the E1A/E2A/E1B/E4orf6 regions were used to generate products with lengths of 106 bp (E1A), 113 bp (E2A), 63 bp (E1B), and 120 bp (E4orf6). A 119-bp product of the IVa2 cDNA was amplified by quantitative PCR using the oligonucleotide primers 2183 and 2184 (Table 1). The oligonucleotide primers 1204 and 1205 (Table 1) were used to amplify cellular GAPDH, generating a product with a length of 111 bp.
RESULTS
Generation of an inducible cell line expressing CANc, an inhibitor of CRM1-mediated transport.
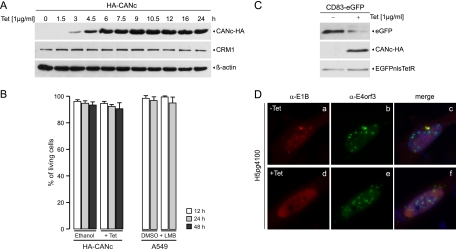
To study the function of CRM1 on different steps of viral replication, at clearly defined times postinfection, we generated a tetracycline-inducible cell line expressing the C terminus of CAN (CANc), also known as nucleoporin 214 (Nup214). Therefore, the CANc coding sequence containing the amino acid residues 1864 to 2090 of the nucleoporin with an adjacent hemagglutinin (HA) tag (CANc-HA) (61) was inserted in a plasmid harboring a tetracycline-inducible Tet On promoter (pLKO.DCMV.TetO) (21, 22), as described in Materials and Methods. Briefly, we generated restriction sites for both NheI and EcoRV at the ends of the pLKO.DCMV.TetO by PCR using the primers 1560 and 1561 (Table 1). In addition, we obtained a CANc-HA gene fusion with attached NheI and EcoRV restriction sites by PCR amplification of pcDNA3-CANc (71) using the oligonucleotide primers 1556 and 1557 (Table 1). After NheI/EcoRV digestion, CANc-HA was ligated with the backbone of pLKO.DCMV.TetO, resulting in pLKO.DCMV.TetO.CANc-HA. To produce the tetracycline-inducible cell line, we transfected HA-TetR cells already expressing the EGFPnlsTetR protein (21, 22) with the constructed pLKO.DCMV.TetO.CANc-HA plasmid and selected for positive cells by puromycin resistance. These cells are derived from HepaRG cells, which were recently shown to be susceptible and permissive for Ad5 infection and to fully support the adenoviral life cycle, although they display delayed kinetics similar to what has been observed with other normal human cells (70). The generated cell line was used throughout this work and is referred to as HA-CANc. To evaluate the expression of CANc, a clone derived from the constructed cell line was analyzed at various times after tetracycline induction. Clearly detectable levels of CANc were obtained at 3 h after tetracycline treatment (Fig. 1A). The CANc levels increased to a stable steady-state concentration between 4.5 and 6 h posttreatment and were maintained through the last time point postinduction (24 h) (Fig. 1A). Significantly, the steady-state concentrations of CRM1 were not affected upon CANc expression (Fig. 1A), suggesting that neither expression nor stability of CRM1 were affected by expression of CANc. In addition, expression of CANc did not result in reduced cell viability, since only minor decreases in viable cells were induced after 12, 24, or 48 h postinduction, an effect that was comparable to that of 12 or 24 h of LMB treatment in A549 cells (Fig. 1B). To test the effect of CANc expression on CRM1-dependent nucleocytoplasmic transport, we used two approaches. First, we transfected the HA-CANc cells with p3_eGFP-CD83complete, a plasmid expressing the mRNA of CD83 linked downstream of a GFP coding sequence. Since the CD83 mRNA depends on CRM1 for export to the cytoplasm (61), the CD83-GFP mRNA was used as a reporter for functional CRM1 transport, resulting in accumulation of GFP. As expected inhibition of CRM1 transport by CANc resulted in reduced GFP expression compared to the untreated control (Fig. 1C), indicating that CANc expression inhibited CRM1-dependent CD83 mRNA export. The constitutively expressed Tet repressor (EGFPnlsTetR) served as a loading control. As a second approach, we tested the impact of CANc on CRM1-dependent transport of a NES-containing protein by immunofluorescence analysis. As discussed in the introduction, the adenoviral E1B-55K contains an NES dependent on CRM1 (20, 46); therefore, induced expression of CANc should result in the inhibition of E1B-55K export to the cytoplasm. In this experiment the viral E4orf3 protein was used as a control, since it does not depend on CRM1 to exit the nucleus. HA-CANc cells were infected with the Ad5 wt H5pg4100 at a multiplicity of 10 FFU/cell, and tetracycline or ethanol were added 21 h p.i., followed by methanol fixation at 36 h p.i. These times postinfection correspond to a period after transition into the late phase has occurred in adenovirus-infected HepaRG cells, when the E1B-55K displays the typical nuclear and cytoplasmic distribution (Fig. 1Da) that has been described previously (32, 50, 58). As expected, induction of CANc led to a complete redistribution of the E1B-55K protein to the nuclear compartment (Fig. 1Dd), while the intracellular localization of E4orf3 remained unaltered (Fig. 1D, compare panels b and e). Taken together, these results indicate that while CANc did not affect CRM1 expression or stability and did not impair cell viability, the expression of CANc abrogated export of bona fide CRM1-dependent mRNA and protein cargo. Consequently, the role of CRM1 in both early and late phenotypes could be analyzed in detail, taking advantage of inducible CANc expression.
Fig 1.
Induction of CANc-HA by tetracycline and the effect of CRM1 inhibition on cell viability and cargo transport. (A) Tetracycline-inducible accumulation of CANc-HA. HA-CANc cells were cultivated in the presence of 1 μg of tetracycline/ml and harvested after indicated times posttreatment, and total cell extracts were prepared. Proteins (15 μg) from each time point were separated by SDS–10% PAGE and subjected to immunoblotting with anti-HA rat MAb 3F10, anti-CRM1 mouse MAb exportin-1 (CRM1), or anti-β-actin mouse MAb AC-15. (B) Effect of CRM1 inhibition on viability of the cells used in the present study. HA-CANc cells were treated with 1 μg of tetracycline/ml. A549 cells were treated with 20 nM leptomycin B (LMB). Living cells were counted at indicated times posttreatment using staining of cells with trypan blue and are presented as percentage of total cells. The results represent the average of three independent experiments. Error bars indicate the standard deviation of the mean. (C) Effect of CRM1 inhibition on mRNA cargo transport. HA-CANc cells were transfected with p3_eGFP-CD83complete and treated either with 1 μg of tetracycline/ml to induce CANc expression or absolute ethanol as a negative control 4 h posttransfection. Cells were harvested at 24 h posttransfection, and total cell extracts were prepared. Proteins (50-μg samples for GFP; 10-μg samples for HA) were separated by SDS–10% PAGE and subjected to immunoblotting with anti-GFP rabbit polyclonal antibody GFP (FL) and anti-HA rat MAb 3F10. (D) Effect of CRM1 inhibition on protein cargo transport. HA-CANc cells were infected with wt virus at a multiplicity of 10 FFU per cell, treated either with 1 μg of tetracycline/ml (+Tet) to induce CANc expression or absolute ethanol (−Tet) as a negative control 21 h p.i. and fixed 36 h p.i. Cells were labeled in situ with anti-E1B-55K mouse MAb 2A6 (α-E1B) and anti-E4orf3 rat MAb 6A11 (α-E4orf3) as primary antibodies and with Texas Red and fluorescein isothiocyanate-conjugated secondary antibodies, respectively. Representative anti-E1B (red) (a and d) and anti-E4orf3 (green) (b and e) staining patterns are shown. Nuclei were visualized using DAPI. Overlays of DAPI staining (blue) with the green and red images (merge) are also shown (c and f). Magnification, ×7,600.
CRM1 activity is required for efficient viral progeny production.
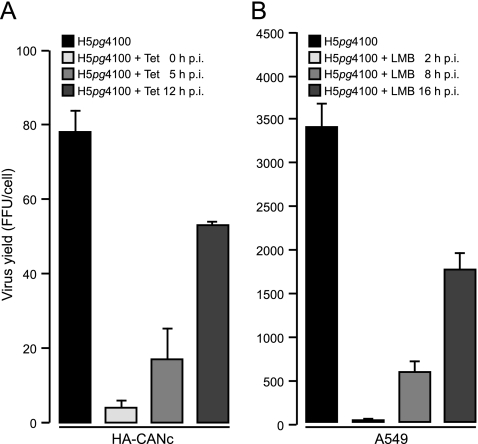
To examine the role of CRM1 at different times postinfection after viral entry into the nucleus, the impact of inhibiting CRM1 activity on virus growth, using the inducible expression of CANc in the HepaRG cells, was initially evaluated. It has been shown that pVII and viral DNA reach the nucleus within the first 60 min p.i. (54). Therefore, to avoid an inhibitory effect on viral entry into the nucleus, these experiments were designed to inhibit CRM1 activity beginning at 2 to 3 h postinfection, a time postinduction at which CANc could first be detected. After the addition of 1 μg of tetracycline/ml to the medium at 0, 5, or 12 h p.i., Ad5-infected HA-CANc cells were harvested 48 h p.i. In parallel, virus growth was also measured in A549 cells in the presence of LMB. To compensate for the time of initial CANc expression after induction (Fig. 1A), the addition of 20 nM LMB to Ad5-infected A549 cells was deferred to 2, 8, and 16 h p.i. and harvested at 26 h p.i. Virus progeny production was affected in a gradual manner in both HA-CANc (Fig. 2A) and LMB-treated A549 cells (Fig. 2B). Although the total numbers of progeny virions differed between the two cell types, the effect on virus production displayed a similar pattern. Inhibition of CRM1 either through CANc (HepaRG cells) or LMB (A549 cells) induced 20 (Fig. 2A) to >200-fold (Fig. 2B) reductions in virus yield when CRM1 was inhibited at times after infection that correspond to the early phase of viral replication (2 to 8 h p.i.). Interestingly, ca. 30% (HepaRG) to 50% (A549) reductions in virus production were also observed when CRM1 was inhibited at later time points.
Fig 2.
Effect of CRM1 inhibition on virus growth. (A) HA-CANc cells were infected with wt virus at a multiplicity of 50 FFU per cell. Tetracycline was added to the culture medium at different time points after infection as indicated. Viral particles were harvested 48 h p.i. (B) A549 cells were infected with wt virus at a multiplicity of 10 FFU per cell. Leptomycin B (LMB) was added to the culture medium at different times after infection as indicated. Viral particles were harvested 26 h p.i. Virus yield was determined by quantitative E2A-72K immunofluorescence staining on HEK293 cells. The results represent the averages from at least three independent experiments. Error bars indicate the standard deviation of the mean.
Inhibition of CRM1-mediated transport reduces synthesis of early and late adenoviral proteins.
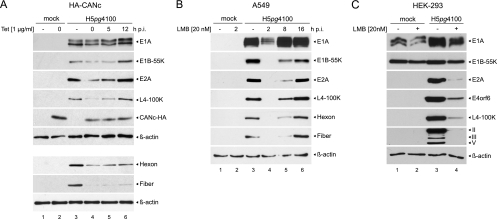
To determine the contribution of CRM1 shuttling on early and late protein synthesis the levels of the adenoviral E1A, E1B-55K, and E2A (DBP) early proteins, as well as Hexon, Fiber, and L4-100K late proteins, were determined in wt H5pg4100-infected cells in the presence or absence of CANc (Fig. 3A) or LMB (Fig. 3B), at different times postinfection. After the addition of 1 μg of tetracycline/ml to the medium at 0, 5, or 12 h p.i., Ad5-infected HA-CANc cells were harvested 26 or 48 h p.i., and early and late protein levels were compared to those of untreated and/or uninfected controls (Fig. 3A). As before, in the case of H5pg4100-infected A549 cells, 20 nM LMB was added at 2, 8, and 16 h p.i. to compensate for the delayed CANc expression, and the cells were harvested at 26 h p.i. (Fig. 3B). Consistent with previous reports on the effect of LMB in infected HeLa cells during the early phase of adenoviral replication (11, 62), the levels of the late proteins Hexon, Fiber, and L4-100K were severely reduced when CRM1 shuttling was blocked in the first hours after Ad infection, either with CANc or LMB (Fig. 3). Similar to the relative impact of CANc and LMB on virus growth, this effect was more pronounced in cells treated with LMB than in cells treated with CANc. However, since cell viability was not significantly compromised by either LMB or CANc (Fig. 1B), it is possible that differences might result from the mode of action of the inhibitors. Overexpression of CANc leads to a competitive inhibition of CRM1-dependent nuclear export by selectively binding to CRM1 and thereby preventing its association with the NPC (6, 27, 42, 84). In contrast, LMB associates covalently with the NES binding region of CRM1 and irreversibly blocks CRM1-mediated export (26, 29), thereby providing an explanation for the stronger phenotype observed upon LMB treatment (Fig. 2 and 3). CRM1 inhibition by CANc expression at times corresponding to the early phase led to clear reductions of both early and late proteins (Fig. 3A, lanes 4 and 5). In contrast, inhibition at ∼15 h p.i. displayed no effect on early proteins. However, late protein levels were still clearly reduced (Fig. 3A, lane 6). LMB induced severe reductions in both early and late proteins when cells were treated at the earliest time after infection (2 h p.i.; Fig. 3B, lane 4). Inhibition of CRM1 close to the onset of viral DNA replication (at ∼8 h p.i.) (59) also led to drastically reduced early and late protein levels (Fig. 3B, lane 5). Upon LMB treatment, this effect was less dramatic than in cells treated at 2 h p.i. (Fig. 3B, lane 4). At a later time postinfection (16 h p.i.), inhibition of CRM1 shuttling displayed no effect on further accumulation of viral proteins, when early and late proteins reached levels comparable to untreated cells (Fig. 3B, compare lanes 3 and 6).
Fig 3.
Effect of CRM1 inhibition on viral protein synthesis. (A) Steady-state levels of viral proteins upon CANc induction. HA-CANc cells were either not infected (mock) or infected with wt virus at a multiplicity of 50 FFU/cell. Cells were treated with 1 μg of tetracycline/ml (lane 2 and lanes 4 to 6) or absolute ethanol (lanes “–”; lanes 1 and 3) at the indicated times postinfection, and total cell extracts were prepared 26 h p.i. or for Hexon and Fiber proteins 48 h p.i. Proteins (15-μg samples for E1A, Hexon, Fiber, L4-100K, and β-actin; 25-μg samples for E1B-55K; 20-μg samples for E2A; 50-μg samples for CANc-HA) were separated by SDS–10% PAGE, followed by immunoblotting with anti-E1A mouse MAb M73, anti-E1B-55K mouse MAb 2A6, anti-E2A mouse MAb B6-8, anti-L4-100K rat MAb 6B10, anti-Ad5 rabbit polyclonal serum L133, anti-HA rat MAb 3F10, or anti-β-actin mouse MAb AC-15. (B) Steady-state levels of viral proteins upon LMB treatment. A549 cells were either not infected (mock) or infected with wt virus at a multiplicity of 10 FFU/cell. Cells were treated with 20 nM LMB (lane 2 and lanes 4 to 6) or DMSO (lanes “–”; lanes 1 and 3) at the indicated times p.i., and total cell extracts were prepared 26 h p.i. Proteins (10-μg samples for E1A, E2A, L4-100K, Hexon, Fiber, and β-actin; 25-μg samples for E1B-55K) were separated by SDS–10% PAGE, followed by immunoblotting with anti-E1A mouse MAb M73, anti-E1B-55K mouse MAb 2A6, anti-E2A mouse MAb B6-8, anti-L4-100K rat MAb 6B10, anti-Ad5 rabbit polyclonal serum L133, or anti-β-actin mouse MAb AC-15. (C) Steady-state levels of viral proteins upon LMB treatment of infected Ad complementing cells. HEK293 cells were either not infected (mock) or infected with wt virus at a multiplicity of 10 FFU/cell. Cells were treated with 20 nM LMB (lanes “+”; 2 and 4) or DMSO (lanes “–”; lanes 1 and 3) 2 h p.i., and total cell extracts were prepared 26 h p.i. Proteins (10-μg samples for E1A, E2A, L4-100K, capsid proteins, and β-actin; 25-μg samples for E1B-55K and E4orf6) were separated by SDS–10% PAGE, followed by immunoblotting with anti-E1A mouse MAb M73, anti-E1B-55K mouse MAb 2A6, anti-E2A mouse MAb B6-8, anti-E4orf6 mouse MAb RSA3, anti-L4-100K rat MAb 6B10, anti-Ad5 rabbit polyclonal serum L133, or anti-β-actin mouse MAb AC-15. Bands corresponding to viral late proteins Hexon (II), Penton (III), and Fiber (V) are indicated on the right.
Given that E1A activates transcription of early genes and consequently impacts activation of the major late transcription unit (MLTU) (3, 56), we decided to examine whether reduced expression of early and late proteins was due to decreased E1A protein levels induced by CRM1 inhibition. Therefore, HEK293 cells (which constitutively express the Ad5 E1 region and have been used extensively in experiments where complementation of E1A or E1B activities is required) (33) were mock infected or infected with the wild-type virus H5pg4100, treated with 20 nM LMB or dimethyl sulfoxide (DMSO) beginning at 2 h p.i. and harvested 26 h p.i. Interestingly, accumulation of early (E2A, E4orf6) and late proteins (L4-100K, II, III, and V) was significantly reduced in LMB-treated HEK-293 cells, despite expression of high levels of E1A and E1B-55K (Fig. 3C). These results suggest that expression of adenoviral early genes depends on CRM1 and that the effect of CRM1 inhibition occurs at the posttranscriptional level, leading to reduced early adenoviral protein concentrations even though early gene transcription can be induced by E1A.
Inhibition of CRM1 reduces viral early mRNA export.
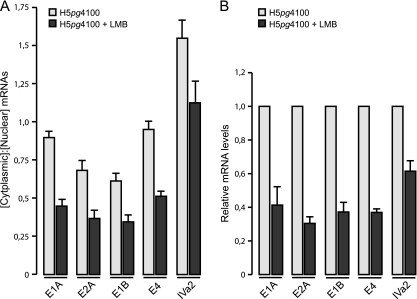
Since a detailed analysis of export of the adenoviral early transcripts has not been reported and our results indicate that CRM1 influences viral early gene expression at a posttranscriptional level, we decided to examine whether CRM1 supports export of the E1A, E2A, E1B-55K, and E4orf6 mRNA. A549 cells were infected with the wild-type virus (H5pg4100), 20 nM LMB was added to the culture medium 4 h p.i., and both nuclear and cytoplasmic mRNA were prepared after cell fractionation at 12 h p.i., a time span, in which these mRNAs are actively transcribed and exported to the cytoplasm (4). The results are shown in Fig. 4A, where in mock-treated Ad5-infected cells the ratio of cytoplasmic to nuclear E1A mRNA was approximately 1:1. This is in perfect agreement with previous studies on the subcellular distribution of newly synthesized E1A mRNA in HEK293 cells, which revealed the same nuclear/cytoplasmic ratio at both 8 and 14 h p.i. (81).
Fig 4.
Effect of CRM1 inhibition on nuclear export efficiency and accumulation of viral early and intermediate mRNAs. (A) A549 cells were infected with wt virus at a multiplicity of 10 FFU per cell. LMB was added to the culture medium 4 h p.i. (early mRNAs; E1A, E2A, E1B, and E4) or 10 h p.i. (IVa2). Steady-state concentrations of mRNAs in cytoplasm and nucleus were determined by real-time PCR at 12 h p.i. (early mRNAs) and at 18 h p.i. (IVa2). Raw numbers were corrected using GAPDH as an internal control and then used to calculate the cytoplasmic/nuclear mRNA ratios. (B) A549 cells were infected with wt virus at a multiplicity of 10 FFU per cell. LMB at 20 nM was added to the culture medium 4 h p.i. (early mRNAs) or 10 h p.i. (IVa2). Steady-state concentrations of mRNAs were determined by real-time PCR at 12 h p.i. (early mRNAs) and at 18 h p.i. (IVa2). Total RNA values were corrected using GAPDH as internal control and are expressed relative to the wt value. The results and standard deviations shown are presented for at least three independent experiments, each performed in duplicate. Error bars indicate the standard deviation of the mean.
Notably, treatment of the cells with LMB resulted in a decreased cytoplasmic accumulation of the E1A transcript of ca. 50% (Fig. 4A), providing an explanation for the lower E1A protein levels accumulated following inhibition of CRM1 transport (Fig. 3). Interestingly, blockage of CRM1-dependent shuttling also induced similar reductions in the export efficiency of the E2A, E1B-55K, and E4orf6 mRNA compared to the untreated controls (Fig. 4A). This unanticipated observation thus provides evidence for a novel function of CRM1 as export receptor of adenoviral early transcripts.
It has been established that export of adenoviral late transcripts occurs independently of CRM1 (25, 44, 69). Different transport mechanisms are therefore used for the export of early and late mRNA, raising the question of how intermediate adenoviral transcripts, such as IVa2, are exported to the cytoplasm. To test this, we decided to also measure IVa2 mRNA export efficiency. Therefore, 20 nM LMB was added to H5pg4100-infected A549 cells at 10 h p.i., and nuclear and cytoplasmic mRNAs were prepared 18 h p.i. Interestingly, the cytoplasmic accumulation of the IVa2 transcript (Fig. 4A) was reduced ca. 30% in LMB-treated cells compared to the control, indicating that CRM1-dependent shuttling is required for transport of IVa2 transcripts to the cytoplasm, as observed for viral early mRNAs (Fig. 4A).
Taken together, these data indicate that the defects observed on virus replication upon CRM1 inhibition during the early phase of the viral life cycle (Fig. 2 and 3) are a direct consequence of the reduced cytoplasmic accumulation of viral early and/or intermediate mRNAs (Fig. 4A). Thus, in addition to its role in the disassembly of the adenoviral virion and the nuclear export of various adenoviral proteins, these experiments provide evidence for a novel function of CRM1 as an export receptor of adenoviral early and intermediate mRNA.
Inhibition of CRM1 reduces viral early mRNA accumulation.
It is well established that specific species of mRNAs that are exported via the CRM1 transport pathway are stabilized during posttranscriptional gene regulation by this mechanism (9). Therefore, we next decided to examine the accumulation of viral early and intermediate mRNA in the absence or presence of LMB. For this purpose, A549 cells were infected with H5pg4100, 20 nM LMB was added starting at 4 (early mRNA) or 10 h p.i. (IVa2 mRNA), and total viral mRNA was prepared 12 h (early mRNA) or 18 h (IVa2 mRNA) after infection. Figure 4B shows the levels of early E1A, E2A, E1B-55K, and E4orf6, as well as intermediate IVa2 adenoviral transcripts, upon LMB treatment, each compared to an untreated control. Interestingly, CRM1 inhibition led to a 60 to 70% decrease of the early mRNA levels compared to the untreated control, as well as a 30% decrease in IVa2 mRNA levels (Fig. 4B). Such reduced amounts could be due to an effect of LMB on these transcripts at a transcriptional or posttranscriptional level. It is likely that the observed block in mRNA export (Fig. 4A) leads to reduced mRNA stability and thereby to decreased total mRNA levels.
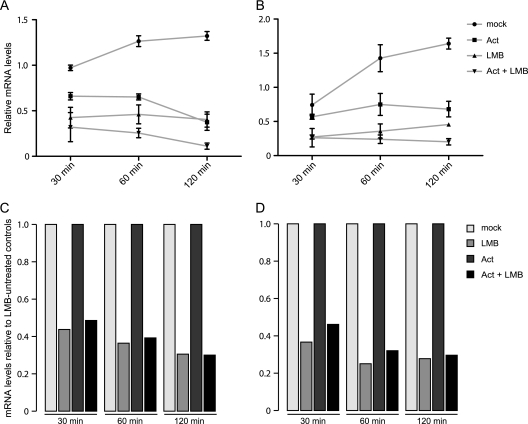
However, to distinguish between this possibility and a possible role of CRM1 in transcription of early adenoviral genes and/or a direct effect of LMB on this, we used actinomycin D, a potent transcriptional inhibitor (73), in parallel with LMB treatment, to determine total E1A and E1B mRNA levels at various times posttreatment. Consistent with the previous mRNA measurements, A549 cells were infected with the wild-type virus H5pg4100, and 20 nM LMB was added 4 h p.i. In addition, 5 μg of actinomycin D/ml was added to the medium 10 h p.i., and total mRNA was prepared 10.5, 11, and 12 h p.i. These times correspond to 30, 60, and 120 min after actinomycin D treatment (Fig. 5). The values of E1A and E1B mRNA are presented relative to an untreated control, harvested at 10 h p.i. The data indicate that both E1A (Fig. 5A) and E1B (Fig. 5B) mRNA levels were clearly compromised upon actinomycin D and/or LMB treatment, in contrast to the continuous accumulation observed for the mRNA in untreated cells. Furthermore, simultaneous treatment of the cells with both inhibitors resulted in significantly lower E1A and E1B mRNA levels (Fig. 5A and B) compared to separate treatment.
Fig 5.
Effect of CRM1 inhibition on mRNA stability. (A and B) Viral early mRNA levels upon parallel treatment with actinomycin D (Act) and LMB. A549 cells were infected with wt virus at a multiplicity of 10 FFU per cell. 20 nM LMB was added to the cell culture medium 4 h p.i. and/or 5 μg of actinomycin D/ml at 10 h p.i. Steady-state concentrations of E1A (A) and E1B (B) mRNA were determined by real-time PCR 30, 60, and 120 min after actinomycin D treatment. Total RNA values were corrected using GAPDH as internal control and are expressed relative to the untreated control, which was harvested at 10 h p.i. The results and standard deviations shown are presented for three independent experiments, each performed in duplicate. Error bars indicate the standard deviation of the mean. (C and D) Viral early mRNA levels relative to LMB-untreated samples. To visualize the direct effect of LMB treatment on steady-state concentrations of E1A (C) and E1B (D), the LMB-treated samples (LMB; Act + LMB) were calculated relative to the untreated control samples (mock; Act), which were set to 1.
To discern whether LMB treatment impacts mRNA biogenesis at a transcriptional or posttranscriptional level, the values for LMB-treated cells shown in Fig. 5A and B were calculated relative to mock- and relative to actinomycin D-treated samples, which were arbitrarily set to 1 (Fig. 5C and D). The data clearly demonstrate that LMB inhibited early mRNA accumulation to the same extent, independently of whether cells were treated with actinomycin D or not. These results therefore exclude the possibility that LMB may affect the transcription of early adenoviral mRNA and support our hypothesis that inhibition of CRM1 reduces the stability of early mRNAs by blocking their export. Altogether, these observations indicate for the first time that adenoviral early and intermediate mRNA are transported to the cytoplasm by a CRM1-dependent mechanism and that export of these mRNAs through the CRM1 pathway leads to their stabilization.
Inhibition of CRM1-dependent transport indirectly affects adenoviral genome replication.
The CRM1-dependent export of adenoviral early and intermediate mRNAs could explain the defects observed in adenoviral replication upon CRM1 inhibition in the early phase of infection. Nevertheless, induction of CANc and LMB treatment of cells also resulted in reduced virus growth (Fig. 2) and a decreased accumulation of viral early and late proteins (Fig. 3) at later times of infection, indicating that CRM1 may support additional steps in the replication cycle of the virus. Viral DNA replication is initiated at ∼8 h p.i (59) and is known to be required for transcriptional activation of viral late genes (56). Therefore, we decided to examine whether CRM1 inhibition affects the efficiency of viral DNA replication.
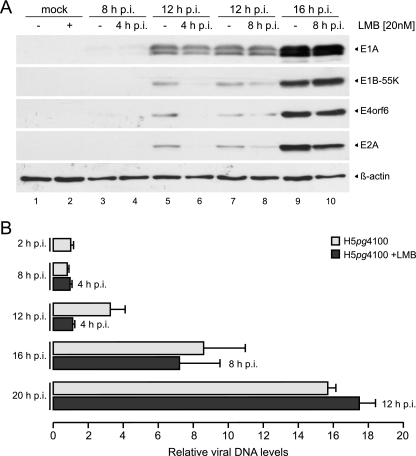
These experiments were designed to inhibit CRM1-dependent transport during both the early phase of infection and at the onset of viral DNA replication 8 h p.i (59), i.e., after the appearance of early mRNAs in the cytoplasm (4). To test whether possible effects on viral DNA synthesis are independent of or correlate with reduced levels of early proteins, we decided to simultaneously examine the accumulation of early proteins. Therefore, A549 cells were mock infected or infected with the wild-type virus, 20 nM LMB (Fig. 6A, lanes 2, 4, 6, 8, and 10) or DMSO (“–” lanes [Fig. 6A, lanes 1, 3, 5, 7, and 9]) were added to the medium at 4 or 8 h p.i., and cell lysates were prepared at 8, 12, and 16 h p.i. Consistent with previous experiments showing an effect on adenoviral early gene expression (Fig. 3 and 4), LMB treatment inhibited accumulation of E1A, E1B-55K, E4orf6, and E2A proteins, particularly after treatment from 4 to 12 h p.i. (Fig. 6A, compare lanes 5 and 6). This time span corresponds to a time when adenoviral early mRNAs are exported to the cytoplasm (4). Significantly, after the establishment of efficient accumulation of early mRNAs in the cytoplasm at 8 h p.i. (4), no reduction of early protein levels could be observed upon LMB treatment (Fig. 6A, compare lanes 7 and 9 with lanes 8 and 10).
Fig 6.
Effect of CRM1 inhibition on early protein expression and viral DNA accumulation. (A) Viral early protein accumulation upon LMB treatment. A549 cells were either not infected (mock) or infected with wt virus at a multiplicity of 40 FFU/cell. Cells were treated with 20 nM LMB (lanes 2, 4, 6, 8, and 10) or DMSO (lanes “–”; lanes 1, 3, 5, 7, and 9) at the indicated times p.i. Total cell extracts were prepared at indicated times p.i. (top row). Proteins (10-μg samples for β-actin; 50-μg samples for E1A, E1B-55K, E4orf6, and E2A) were separated by SDS–10% PAGE, followed by immunoblotting with anti-E1A mouse MAb M73, anti-E1B-55K mouse MAb 2A6, anti-E4orf6 rabbit polyclonal antibody 1807, anti-E2A mouse MAb B6-8, or anti-β-actin mouse MAb AC-15. (B) Viral DNA synthesis upon LMB treatment. A549 cells were infected with wt virus at a multiplicity of 20 FFU per cell. Cells were treated with 20 nM LMB at the indicated times postinfection (right site of bars), or untreated, total nuclear DNA was isolated at indicated times postinfection (left site of bars) and subjected to quantitative PCR. PCR products were analyzed and quantified by using the ChemiDoc system and GeneTools software (Syngene). DNA values are expressed relative to the input value (2 h p.i.). The results and standard deviations shown represent the averages from three independent experiments. Error bars indicate the standard error of the mean.
To measure viral genome replication, A549 cells were untreated or treated with 20 nM LMB starting at 4, 8, and 12 h p.i., and viral DNA was isolated at 8, 12, 16, and 20 h p.i. and subjected to quantitative PCR. To determine the input of viral genomes by infecting viral particles, A549 cells were harvested at 2 h p.i. Viral DNA levels are presented relative to this input value, which was set to 1 (Fig. 6B). An increase in viral DNA levels compared to the input and hence active DNA replication could first be detected at 12 h p.i. Interestingly, inhibition of CRM1 from 4 to 12 h p.i. led to a 65% reduction in viral DNA levels. However, blocking CRM1 transport at later times, from 8 to 16 h p.i. or from 12 to 20 h p.i., did not abrogate efficient viral genome replication, despite the fact that viral DNA levels increased between 12 and 20 h p.i. (Fig. 6B).
Taken together, these data indicate that LMB treatment has no effect on viral DNA synthesis after efficient early protein expression is established. The close correlation between efficient early protein synthesis and viral DNA production in response to CRM1 inhibition (Fig. 6) implies that, as expected, synthesis of adenoviral genomes requires efficient viral early protein expression and, furthermore, that viral genome replication is not directly dependent on active CRM1 transport. Rather, the reduced DNA replication observed upon CRM1 inhibition is an indirect consequence of lower early gene expression.
DISCUSSION
Several steps of the adenoviral replication cycle require active transport of macromolecules from the nuclear compartment to the cytoplasm. The accumulated evidence indicates that the activity of mRNA export receptors, such as Nxf1/TAP, and that of karyopherins, such as CRM1, may participate in a coordinated transport of viral proteins and mRNA at different stages of viral replication. It was recently shown that Nxf1/TAP is required for viral late mRNA export (82). In contrast, several studies have addressed the role of NES-dependent CRM1 export of adenoviral proteins (11, 16, 19, 25, 41, 44, 62, 69, 78). Although the majority of these studies have examined the participation of this export pathway in viral late mRNA transport (11, 25, 44, 62, 69), the role of CRM1 during different stages of viral replication is only partially understood.
However, now we have found that active CRM1-dependent transport is required during the early phase of viral replication. Significantly, our data indicate that the defects observed upon CRM1 inhibition are a direct consequence of reduced viral early mRNA export.
Expression of the dominant-negative form of CANc in primary hepatocytes efficiently inhibited export of well-characterized CRM1-dependent mRNA and protein cargoes (CD83 mRNA and E1B-55K protein), without compromising CRM1 expression or stability or cell viability, thus making it possible to evaluate the role of CRM1 at defined times postinfection. The observed diminished production of viral progeny could result from the additive and pleiotropic effects of CRM1 inhibition on early and late gene expression, viral DNA replication, and viral late protein assembly. Therefore, it was necessary to examine the effect of CRM1 inhibition at defined times postinfection. Both CANc and LMB (which under the conditions used in these studies displayed similar phenotypes and the absence of significant toxicity) exhibited a stronger effect during the earlier stages, when viral early proteins and hence viral DNA replication were clearly diminished. However, CRM1 inhibition by LMB in A549 cells reduced virus progeny significantly more than CANc expressed in the hepatocytes. The reason(s) for this difference are not clear, but it is possible that the covalent modification of CRM1 by LMB may have a more stable effect than the dynamic/transient interaction that could be established between CANc and CRM1. The effect of LMB treatment could therefore result in stronger phenotypes, as was indeed observed for viral protein synthesis and progeny production.
The reduction of viral DNA synthesis induced by restricted CRM1 shuttling seems to be a direct consequence of lower early gene expression. Inhibition of CRM1 at 4 to 12 h p.i. resulted in lower E2A mRNA and protein levels and could thus account for a similar reduction in DNA during this time span. However, after efficient synthesis of early proteins was established (at 8 h p.i.), inhibition of CRM1 transport did not abrogate the de novo synthesis of adenoviral genomes. Although these results indicate that CRM1 transport is not directly required for viral DNA replication, they underscore the close correlation between CRM1-dependent early protein synthesis and efficient viral genome replication.
The primary effect of CRM1 inhibition can be linked directly to reduced early mRNA export: E2A (and E4orf6) protein levels were reduced upon LMB treatment in HEK293 cells, that is, in the presence of high levels of E1A, indicating that at early times postinfection a posttranscriptional block is induced by CRM1 inhibition. Such an effect was indeed demonstrated by our data showing that CRM1 mediates adenoviral early mRNA export. This novel finding raises the question of whether this pathway provides a selective advantage over cellular mRNA.
It is well established that viral late mRNAs have a competitive advantage over cellular mRNA, both at the level of translation and at the level of export (24, 79, 80). Significantly, while efficient viral late mRNA export is accompanied by a concomitant inhibition of export of most cellular mRNA, viral early mRNA do not seem to benefit from a selective export process. However, since adenovirus infection does not perturb export of cellular snRNAs or rRNAs (12, 72), which are exported by the CRM1 pathway, it is possible that export of the early mRNA through this pathway may be advantageous since they would avoid competition with bulk cellular mRNA, which exits the nucleus through the Nxf1/TAP pathway.
Since CRM1 does not bind RNA directly, it requires adaptor proteins, such as HuR and eukaryotic initiation factor 4E (eIF4E) for mRNA export (8, 17, 61). The subset of cellular mRNA that is exported through CRM1 includes certain short-lived early response genes, like proto-oncogene and cytokine mRNAs, that contain an AU-rich element in their 3′-untranslated region. HuR binds to these ARE-mRNAs to protect them from rapid degradation (9) and subsequently mediates the CRM1-dependent export of these mRNAs from the nucleus to the cytoplasm (30, 31). In contrast, the export of the IFN-α1 mRNA through CRM1 is not mediated by ARE sequences (43), suggesting that other features, unrelated to ARE sequences, can be recognized by adaptor proteins that associate with CRM1.
It is noteworthy that inhibiting CRM1-dependent transport of adenoviral early transcripts also decreased their total levels, seemingly by affecting these mRNAs' stability. Therefore, it is possible that a similar mechanism to the export of ARE-containing mRNA could explain the link between CRM1-dependent export and mRNA levels. However, as with the viral late mRNA, it would be interesting to identify the responsible adaptor proteins and the underlying molecular mechanisms. Notably, although the features of the early mRNA directing their CRM1-dependent export remain to be determined, our data indicate for the first time that adenoviral early mRNAs are both exported and stabilized by the CRM1 pathway.
ACKNOWLEDGMENTS
We thank Roger Everett for providing HA-TetR cells and plasmid pLKO.DCMV.TetO.cICPO and Dorothea Pieper and Joachim Hauber for providing plasmids pcDNA3-CANc and p3_eGFP-CD83complete.
This study was supported by the Erich and Gertrud Roggenbuck-Foundation, Hamburg, Germany. R.A.G. received a grant from CONACyT-Mexico (SEP-84582) and support from the Alexander von Humboldt-Foundation. The Heinrich-Pette-Institut is supported by the Freie und Hansestadt Hamburg and the Bundesministerium für Gesundheit.
Footnotes
Published ahead of print 14 December 2011
REFERENCES
- 1. Babiss LE, Ginsberg HS, Darnell JE., Jr 1985. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol. Cell. Biol. 5:2552–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beltz GA, Flint SJ. 1979. Inhibition of HeLa cell protein synthesis during adenovirus infection: restriction of cellular messenger RNA sequences to the nucleus. J. Mol. Biol. 131:353–373 [DOI] [PubMed] [Google Scholar]
- 3. Berk AJ, Lee F, Harrison T, Williams J, Sharp PA. 1979. Pre-early adenovirus 5 gene product regulates synthesis of early viral messenger RNAs. Cell 17:935–944 [DOI] [PubMed] [Google Scholar]
- 4. Binger MH, Flint SJ. 1984. Accumulation of early and intermediate mRNA species during subgroup C adenovirus productive infections. Virology 136:387–403 [DOI] [PubMed] [Google Scholar]
- 5. Blanchette P, et al. 2008. Control of mRNA export by adenovirus E4orf6 and E1B55K proteins during productive infection requires E4orf6 ubiquitin ligase activity. J. Virol. 82:2642–2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bogerd HP, Echarri A, Ross TM, Cullen BR. 1998. Inhibition of human immunodeficiency virus Rev and human T-cell leukemia virus Rex function, but not Mason-Pfizer monkey virus constitutive transport element activity, by a mutant human nucleoporin targeted to Crm1. J. Virol. 72:8627–8635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boivin D, Morrison MR, Marcellus RC, Querido E, Branton PE. 1999. Analysis of synthesis, stability, phosphorylation, and interacting polypeptides of the 34-kilodalton product of open reading frame 6 of the early region 4 protein of human adenovirus type 5. J. Virol. 73:1245–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan CM, Gallouzi IE, Steitz JA. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brennan CM, Steitz JA. 2001. HuR and mRNA stability. Cell. Mol. Life Sci. 58:266–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bridge E, Ketner G. 1990. Interaction of adenoviral E4 and E1b products in late gene expression. Virology 174:345–353 [DOI] [PubMed] [Google Scholar]
- 11. Carter CC, Izadpanah R, Bridge E. 2003. Evaluating the role of CRM1-mediated export for adenovirus gene expression. Virology 315:224–233 [DOI] [PubMed] [Google Scholar]
- 12. Castiglia CL, Flint SJ. 1983. Effects of adenovirus infection on rRNA synthesis and maturation in HeLa cells. Mol. Cell. Biol. 3:662–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Catalucci D, et al. 2005. An adenovirus type 5 (Ad5) amplicon-based packaging cell line for production of high-capacity helper-independent deltaE1-E2-E3-E4 Ad5 vectors. J. Virol. 79:6400–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cepko CL, Sharp PA. 1983. Analysis of Ad5 hexon and 100K ts mutants using conformation-specific monoclonal antibodies. Virology 129:137–154 [DOI] [PubMed] [Google Scholar]
- 15. Cepko CL, Sharp PA. 1982. Assembly of adenovirus major capsid protein is mediated by a nonvirion protein. Cell 31:407–415 [DOI] [PubMed] [Google Scholar]
- 16. Cuesta R, Xi Q, Schneider RJ. 2004. Structural basis for competitive inhibition of eIF4G-Mnk1 interaction by the adenovirus 100-kilodalton protein. J. Virol. 78:7707–7716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KL. 2006. eIF4E is a central node of an RNA regulon that governs cellular proliferation. J. Cell Biol. 175:415–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dallaire F, Blanchette P, Groitl P, Dobner T, Branton PE. 2009. Identification of integrin α3 as a new substrate of the adenovirus E4orf6/E1B 55-kilodalton E3 ubiquitin ligase complex. J. Virol. 83:5329–5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Dobbelstein M, Roth J, Kimberly WT, Levine AJ, Shenk T. 1997. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 16:4276–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dosch T, et al. 2001. The adenovirus type 5 E1B-55K oncoprotein actively shuttles in virus-infected cells, whereas transport of E4orf6 is mediated by a CRM1-independent mechanism. J. Virol. 75:5677–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Everett RD, Boutell C, McNair C, Grant L, Orr A. 2010. Comparison of the biological and biochemical activities of several members of the alphaherpesvirus ICP0 family of proteins. J. Virol. 84:3476–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Everett RD, Parsy ML, Orr A. 2009. Analysis of the functions of herpes simplex virus type 1 regulatory protein ICP0 that are critical for lytic infection and derepression of quiescent viral genomes. J. Virol. 83:4963–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Flint J, Shenk T. 1989. Adenovirus E1A protein paradigm viral transactivator. Annu. Rev. Genet. 23:141–161 [DOI] [PubMed] [Google Scholar]
- 24. Flint SJ, Gonzalez RA. 2003. Regulation of mRNA production by the adenoviral E1B 55-kDa and E4 Orf6 proteins. Curr. Top. Microbiol. Immunol. 272:287–330 [DOI] [PubMed] [Google Scholar]
- 25. Flint SJ, Huang W, Goodhouse J, Kyin S. 2005. A peptide inhibitor of exportin1 blocks shuttling of the adenoviral E1B 55-kDa protein but not export of viral late mRNAs. Virology 337:7–17 [DOI] [PubMed] [Google Scholar]
- 26. Fornerod M, Ohno M, Yoshida M, Mattaj IW. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051–1060 [DOI] [PubMed] [Google Scholar]
- 27. Fornerod M, et al. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16:807–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Frisch SM, Mymryk JS. 2002. Adenovirus-5 E1A: paradox and paradigm. Nat. Rev. Mol. Cell. Biol. 3:441–452 [DOI] [PubMed] [Google Scholar]
- 29. Fukuda M, et al. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308–311 [DOI] [PubMed] [Google Scholar]
- 30. Gallouzi IE, Brennan CM, Steitz JA. 2001. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7:1348–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gallouzi IE, Steitz JA. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. RNA 294:1895–1901 [DOI] [PubMed] [Google Scholar]
- 32. Gonzalez RA, Flint SJ. 2002. Effects of mutations in the adenoviral E1B 55-kilodalton protein coding sequence on viral late mRNA metabolism. J. Virol. 76:4507–4519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Graham FL, Smiley J, Russell WC, Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 34. Greber UF, Webster P, Weber J, Helenius A. 1996. The role of the adenovirus protease on virus entry into cells. EMBO J. 15:1766–1777 [PMC free article] [PubMed] [Google Scholar]
- 35. Gripon P, et al. 2002. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. U. S. A. 99:15655–15660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Groitl P, Dobner T. 2007. Construction of adenovirus type 5 early region 1 and 4 virus mutants. Methods Mol. Med. 130:29–39 [DOI] [PubMed] [Google Scholar]
- 37. Halbert DN, Cutt JR, Shenk T. 1985. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J. Virol. 56:250–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harlow E, Franza BR, Jr., Schley C. 1985. Monoclonal antibodies specific for adenovirus early region 1A proteins: extensive heterogeneity in early region 1A products. J. Virol. 55:533–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hong SS, et al. 2005. The 100K-chaperone protein from adenovirus serotype 2 (subgroup C) assists in trimerization and nuclear localization of hexons from subgroups C and B adenoviruses. J. Mol. Biol. 352:125–138 [DOI] [PubMed] [Google Scholar]
- 40. Hutten S, Kehlenbach RH. 2007. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 17:193–201 [DOI] [PubMed] [Google Scholar]
- 41. Jiang H, et al. 2006. A novel CRM1-dependent nuclear export signal in adenoviral E1A protein regulated by phosphorylation. FASEB J. 20:2603–2605 [DOI] [PubMed] [Google Scholar]
- 42. Kang Y, Cullen BR. 1999. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 13:1126–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kimura T, Hashimoto I, Nagase T, Fujisawa J. 2004. CRM1-dependent, but not ARE-mediated, nuclear export of IFN-α1 mRNA. J. Cell Sci. 117:2259–2270 [DOI] [PubMed] [Google Scholar]
- 44. Kindsmuller K, et al. 2007. Intranuclear targeting and nuclear export of the adenovirus E1B-55K protein are regulated by SUMO1 conjugation. Proc. Natl. Acad. Sci. U. S. A. 104:6684–6689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kindsmuller K, et al. 2009. A 49-kilodalton isoform of the adenovirus type 5 early region 1B 55-kilodalton protein is sufficient to support virus replication. J. Virol. 83:9045–9056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kratzer F, et al. 2000. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53, and Mdm2. Oncogene 19:850–857 [DOI] [PubMed] [Google Scholar]
- 47. Kudo N, et al. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. U. S. A. 96:9112–9117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kzhyshkowska J, Kremmer E, Hofmann M, Wolf H, Dobner T. 2004. Protein arginine methylation during lytic adenovirus infection. Biochem. J. 383:259–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Leppard KN, Shenk T. 1989. The adenovirus E1B 55 kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 8:2329–2336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lethbridge KJ, Scott GE, Leppard KN. 2003. Nuclear matrix localization and SUMO-1 modification of adenovirus type 5 E1b 55K protein are controlled by E4 Orf6 protein. J. Gen. Virol. 84:259–268 [DOI] [PubMed] [Google Scholar]
- 51. Martin-Fernandez M, et al. 2004. Adenovirus type-5 entry and disassembly followed in living cells by FRET, fluorescence anisotropy, and FLIM. Biophys. J. 87:1316–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Marton MJ, Baim SB, Ornelles DA, Shenk T. 1990. The adenovirus E4 17-kilodalton protein complexes with the cellular transcription factor E2F, altering its DNA-binding properties and stimulating E1A-independent accumulation of E2 mRNA. J. Virol. 64:2345–2359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mathews MB, Shenk T. 1991. Adenovirus virus-associated RNA and translation control. J. Virol. 65:5657–5662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mirza MA, Weber J. 1979. Uncoating of adenovirus type 2. J. Virol. 30:462–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morin N, Boulanger P. 1986. Hexon trimerization occurring in an assembly defective, 100K temperature-sensitive mutant of adenovirus 2. Virology 152:11–31 [DOI] [PubMed] [Google Scholar]
- 56. Morris SJ, Scott GE, Leppard KN. Adenovirus late-phase infection is controlled by a novel L4 promoter. J. Virol. 84:7096–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nevels M, et al. 1999. Transforming potential of the adenovirus type 5 E4orf3 protein. J. Virol. 73:1591–1600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ornelles DA, Shenk T. 1991. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J. Virol. 65:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Osborne TF, Berk AJ. 1983. Far upstream initiation sites for adenovirus early region 1A transcription are utilized after the onset of viral DNA replication. J. Virol. 45:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pilder S, Moore M, Logan J, Shenk T. 1986. The adenovirus E1B-55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol. Cell. Biol. 6:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prechtel AT, et al. 2006. Expression of CD83 is regulated by HuR via a novel cis-active coding region RNA element. J. Biol. Chem. 281:10912–10925 [DOI] [PubMed] [Google Scholar]
- 62. Rabino C, Aspegren A, Corbin-Lickfett K, Bridge E. 2000. Adenovirus late gene expression does not require a Rev-like nuclear RNA export pathway. J. Virol. 74:6684–6688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Reich NC, Sarnow P, Duprey E, Levine AJ. 1983. Monoclonal antibodies which recognize native and denatured forms of the adenovirus DNA-binding protein. Virology 128:480–484 [DOI] [PubMed] [Google Scholar]
- 64. Riley D, Flint SJ. 1993. RNA-binding properties of a translational activator, the adenovirus L4 100-kilodalton protein. J. Virol. 67:3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rosorius O, et al. 1999. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: evidence for direct interaction with the export receptor CRM1. J. Cell Sci. 112(Pt 14):2369–2380 [DOI] [PubMed] [Google Scholar]
- 66. Sang N, Caro J, Giordano A. 2002. Adenoviral E1A: everlasting tool, versatile applications, continuous contributions and new hypotheses. Front. Biosci. 7:d407–d413 [DOI] [PubMed] [Google Scholar]
- 67. Sarnow P, Sullivan CA, Levine AJ. 1982. A monoclonal antibody detecting the adenovirus type 5-E1b-58Kd tumor antigen: characterization of the E1b-58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology 120:510–517 [DOI] [PubMed] [Google Scholar]
- 68. Reference deleted.
- 69. Schmid M, et al. 2011. The E3 ubiquitin ligase activity associated with the adenoviral E1B-55K-E4orf6 complex does not require CRM1-dependent export. J. Virol. 85:7081–7094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Schreiner S, et al. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schutz S, et al. 2006. Stimulated expression of mRNAs in activated T cells depends on a functional CRM1 nuclear export pathway. J. Mol. Biol. 358:997–1009 [DOI] [PubMed] [Google Scholar]
- 72. Smiley JK, Young MA, Bansbach CC, Flint SJ. 1995. The metabolism of small cellular RNA species during productive subgroup C adenovirus infection. Virology 206:100–107 [DOI] [PubMed] [Google Scholar]
- 73. Sobell HM. 1985. Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. U. S. A. 82:5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Strunze S, Trotman LC, Boucke K, Greber UF. 2005. Nuclear targeting of adenovirus type 2 requires CRM1-mediated nuclear export. Mol. Biol. Cell 16:2999–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Svensson C, Akusjärvi G. 1984. Adenovirus VA RNAI: a positive regulator of mRNA translation. Mol. Cell. Biol. 4:736–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092–1100 [DOI] [PubMed] [Google Scholar]
- 77. Wiegand HL, et al. 2002. Formation of Tap/NXT1 heterodimers activates Tap-dependent nuclear mRNA export by enhancing recruitment to nuclear pore complexes. Mol. Cell. Biol. 22:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wodrich H, et al. 2003. Switch from capsid protein import to adenovirus assembly by cleavage of nuclear transport signals. EMBO J. 22:6245–6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Xi Q, Cuesta R, Schneider RJ. 2005. Regulation of translation by ribosome shunting through phosphotyrosine-dependent coupling of adenovirus protein 100k to viral mRNAs. J. Virol. 79:5676–5683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Xi Q, Cuesta R, Schneider RJ. 2004. Tethering of eIF4G to adenoviral mRNAs by viral 100k protein drives ribosome shunting. Genes Dev. 18:1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yang UC, Huang W, Flint SJ. 1996. mRNA export correlates with activation of transcription in human subgroup C adenovirus-infected cells. J. Virol. 70:4071–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Yatherajam G, Huang W, Flint SJ. 2011. Export of adenoviral late mRNA from the nucleus requires the Nxf1/Tap export receptor. J. Virol. 85:1429–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhang Y, Schneider RJ. 1993. Adenovirus inhibition of cellular protein synthesis and the specific translation of late viral mRNAs. Semin. Virol. 4:229–236 [Google Scholar]
- 84. Zolotukhin AS, Felber BK. 1999. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]