Abstract
Despite intensive research, there are very few reagents with which to modulate and dissect the mRNA splicing pathway. Here, we describe a novel approach to identify such tools, based on detection of the exon junction complex (EJC), a unique molecular signature that splicing leaves on mRNAs. We developed a high-throughput, splicing-dependent EJC immunoprecipitation (EJIPT) assay to quantitate mRNAs spliced from biotin-tagged pre-mRNAs in cell extracts, using antibodies to EJC components Y14 and eukaryotic translation initiation factor 4aIII (eIF4AIII). Deploying EJIPT we performed high-throughput screening (HTS) in conjunction with secondary assays to identify splicing inhibitors. We describe the identification of 1,4-naphthoquinones and 1,4-heterocyclic quinones with known anticancer activity as potent and selective splicing inhibitors. Interestingly, and unlike previously described small molecules, most of which target early steps, our inhibitors represented by the benzothiazole-4,7-dione, BN82685, block the second of two trans-esterification reactions in splicing, preventing the release of intron lariat and ligation of exons. We show that BN82685 inhibits activated spliceosomes' elaborate structural rearrangements that are required for second-step catalysis, allowing definition of spliceosomes stalled in midcatalysis. EJIPT provides a platform for characterization and discovery of splicing and EJC modulators.
INTRODUCTION
Splicing of pre-mRNA is an essential step in gene expression that is mediated by a modular, multi-small nuclear RNP (multi-snRNP) complex called the spliceosome, a myriad of RNA binding proteins, and various enzymes that regulate their activity (20, 53, 60). The intricacy and importance of this process have been underscored by recent evidence indicating that 94% of genes undergo alternative splicing and that an estimated 15% of genetically inherited disorders arise from mutations in either cis- or trans-acting elements required for splicing (7, 14, 30, 58, 63). Similar to their use to study other cellular processes including transcription and translation, small molecules provide a powerful tool to dissect the splicing process and study its regulation (18, 59). Reporter constructs, including a recently developed system that rapidly responds to splicing modulation to minimize compound toxicity, have been used to identify effectors of alternative and constitutive splicing (45, 54, 65). Serendipitously, phenotypic screens of anticancer reagents in tumor cells have also identified splicing inhibitors, spliceostatin A and pladienolide-B (21, 29). Additional splicing modifiers have been identified from in vitro chemical screens based on the known dependence of splicing on the phosphorylation status of serine-arginine-rich (SR) splicing factors, for example, by measuring effects on the kinase activity of the Clk/Sty proteins, SRPK, or spliceosome-associated enzymes like topoisomerase I (16, 42, 46, 52, 57). More recently, the effect of protein acetylation/deacetylation inhibitors on in vitro splicing was examined and shown to block the splicing process at various stages (31). Despite these advances, very few potent and selective splicing inhibitors are available, largely due to the lack of suitable high-throughput in vitro assays that could be used for systematic discovery of splicing modulators.
Splicing leaves in its wake a unique molecular signature in the form of the exon junction complex (EJC) that assembles ∼20 to 24 nucleotides (nt) upstream of splice junctions (25, 27, 34). Four proteins comprise the core of the EJC, including eukaryotic translation initiation factor 4A III (eIF4AIII), which directly binds the mRNA, Y14, Magoh, and MLN51. These proteins remain bound at their original positions after export to the cytoplasm, communicating the splicing “history” of the mRNA and determining its export, localization, and translation efficiency, as well as mediating turnover of mRNAs that contain premature termination codons by nonsense-mediated decay (15, 35, 40). Based on this unique splicing-dependent assembly of EJCs, we have developed a rapid and sensitive high-throughput assay that measures the interaction of eIF4AIII or Y14 with spliced mRNA to provide a quantification of EJC formation and splicing. In addition to capturing known splicing modifiers in our screen, we describe the identification of 1,4-naphthoquinones and 1,4-heterocyclic quinones as potent and selective splicing inhibitors. We further show that this class of compounds, previously known also for their cancer-inhibitory activity, blocks splicing at a discrete step during catalysis, defining a spliceosome pathway intermediate.
MATERIALS AND METHODS
Chemical compounds.
Screening was performed on the compounds contained in the Library of Pharmacologically Active Compounds (LOPAC; Sigma/RBI), Natural Product Collection (MicroSource), and the Kinase/Phosphatase Inhibitor (BioMol) libraries. Additional compounds were purchased from commercial resources: NSC95397, NSC663284, 2-chloromethyl-3-methyl-1,4-naphthoquinone, 2,3-bis(methylthio)-1,4-naphthoquinone, and menadione were from Sigma; benzothiazole-4,7-dione (BN82685) was from Calbiochem; okadaic acid and calyculin A were from BioMol. Compound 5 (Cpd5) and PM-20 were kind gifts of Brian Carr (Thomas Jefferson University, Philadelphia, PA). Control (5′-CCTCTTACCTCAGTTACAATTTATA-3′) and U2 (5′-TGATAAGAACAGATACTACACTTGA-3′) antisense morpholinos (AMO) were synthesized by Gene Tools, LLC.
In vitro transcription of 32P- and biotin-labeled pre-mRNA.
[32P]UTP-labeled pre-mRNAs of an adenovirus type 2 construct with a deletion of intervening sequence (Ad2), CDC14-15, and IgM C3-C4 were prepared according to the Riboprobe Combination SP6/T7 system (Promega). Megascript T7 or SP6 kits (Ambion) were used to generate large amounts of biotin-labeled pre-mRNAs. The ratio of biotin-16-UTP (Roche) to cold UTP was 1:14 (final concentration, 350 μM) to incorporate approximately three biotin molecules per spliced Ad2 mRNA. Pre-mRNAs were gel purified, phenol-chloroform extracted, ethanol precipitated, and resuspended in nuclease-free water.
Preparation of splicing extracts, in vitro splicing, RNA isolation, and 32P quantification of gel bands.
Whole-cell splicing extracts from HEK 293T cells were prepared, and in vitro splicing reactions of 32P-labeled Ad2 and CDC14-15 pre-mRNAs were performed as described previously (24). HeLa nuclear extracts were purchased from Computer Cell Culture Centre (Mons, Belgium) and used for splicing of IgM C3-C4 pre-mRNA. Radiolabeled RNA for gels was purified using TRIzol reagent (Invitrogen). RNA for Northern dot blots was purified from 96-well plates on a KingFisher 96 magnetic particle processor (Thermo Lab Systems) using nucleic acid-binding magnetic beads, according to instructions of the MagMAX-96 Total RNA Isolation kit (Ambion). Radiolabeled band intensities were quantified on a Typhoon Trio phosphorimager (Amersham) using ImageQuant software.
High-throughput in vitro splicing assay in 384-well plates.
A Beckman Coulter Biomek FX robot was used to assemble 20-μl splicing reaction mixtures consisting of 1× SP buffer (0.5 mM ATP, 20 mM creatine phosphate, 1.6 mM MgCl2), 80 μg of whole-cell splicing extract, 40 nM biotin-labeled Ad2ΔIVS pre-mRNA, 1 U/μl of RNasin RNase inhibitor (Promega), and 20 μM each individual compound (final concentration, 1% dimethyl sulfoxide [DMSO]). After 1.5 h at 30°C, reaction mixtures were diluted with 40 μl of HNT buffer (20 mM HEPES-KOH, pH 7.9, 150 mM NaCl, 0.5% Triton X-100), and 50 μl was transferred to black-well NeutrAvidin-coated plates (Pierce) for a 1-h incubation at room temperature. Samples were aspirated using an ELx405 microplate washer (Bio-Tek), and 40 μl of primary anti-eIF4AIII (3F1) antibody (11) diluted 1:350 in HNT buffer with 1 μg/ml bovine serum albumin (BSA) was added to each well of the plate using a Thermo Multidrop and incubated for 1 h at room temperature. Plates were washed in six cycles with HNT buffer on the ELx405, and then a horseradish peroxidase (HRP)-conjugated anti-mouse IgG secondary antibody (Jackson Immunologicals) diluted 1:10,000 in HNT buffer was added and incubated for 1 h. After a final series of identical washes, 50 μl of Super Signal ELISA (enzyme-linked immunosorbent assay) Fempto chemiluminescent substrate (Pierce) was added, and luminescence signals were measured on a Perkin Elmer Envision reader.
Ninety-six-well magnetic bead assay.
Ten-microliter splicing reaction mixtures consisting of 1× SP buffer, 40 μg of extract, 20 nM biotin pre-mRNA, and 1 U/μl of RNasin were assembled on ice in 96-well plates with or without drug and incubated for 1.5 h. All steps were performed at 30°C in an Eppendorf tabletop shaker set at 750 rpm. Prior to splicing, antibodies were immobilized on protein A magnetic beads (Invitrogen) for >1 h at 4°C in 1× phosphate-buffered saline (PBS)–0.1% NP-40, and then beads were extensively washed. Immunoprecipitation (IP) reaction mixtures (100 μl) consisting of 8.6 μl of bead suspension, primary antibody (0.7 μl of 4C4, 4F4, and 4B10; 1.7 μl for 3F1), and 0.2 U/μl of RNasin in HNT buffer were added to each well, and IPs were carried out for 1 h. Beads were washed five times with 200 μl of HNT buffer on a KingFisher apparatus. Biotinylated RNA/protein complexes bound to antibodies were then reacted with 120 μl of an avidin-HRP conjugate (1:25,000 in HNT buffer; Pierce) for another hour. Following five more automated washes (200 μl of HNT) on the KingFisher, 150 μl of Super Signal ELISA Fempto chemiluminescent substrate was added to beads. Samples were transferred to Costar black assay plates for reading of luminescence.
Northern blot analysis.
For dot blots, RNA from drug-treated (40 μM) or untreated splicing reaction mixtures was diluted with SSC buffer having a final concentration of 0.9 M NaCl plus 0.09 M sodium citrate 6 ×) and transferred along with known amounts of pre-mRNA and mRNA in separate wells to a nylon N+ Hybond membrane under vacuum using an S&S Minifold dot blot system. Membranes were UV cross-linked and baked for 1 h at 75°C. mRNA and pre-mRNA were detected with 3′ digoxigenin (DIG) end-labeled oligonucleotide probes complementary to the Ad2 exon 1-exon 2 junction (5′-CAACCGCGAGCCCAACAGCTG) or the exon 1-intron border (5′-GAGAGGGAGTACTCCTCCCAACAGCTGGCC). Membranes were prehybridized for >3 h in DIG granules and hybridized overnight with 300 pmol of probe at 42°C. Wash and detection procedures were performed as described in the DIG chemiluminescence kit (Roche). Filters were exposed to X-ray film, and scanned images were quantified in Photoshop.
For snRNAs and the Ad2 pre-mRNA/mRNA shown in Fig. 7A, RNA from streptavidin-biotin precipitates was eluted from magnetic beads (Dynabeads M-280; Invitrogen) and purified with TRIzol. RNA was resolved on 8% urea-PAGE gels, transferred to a nylon membrane, UV cross-linked, and probed with α-32P-labeled oligonucleotides overnight at 40°C with OligoHyb buffer (Ambion). Membranes were stripped by boiling in 10 mM Tris, pH 7.5, with 0.1% SDS and reprobed. Antisense probe sequences (5′ to 3′) are as follows: U1, GCACATCCGGAGTGCAATG; U2, TCGGATAGAGGACGTATCAGATATTA; U4, CCAGTGCCGACTATATTGCAAGT; U5, CTCAAAAAATTGGGTTAAGACTCAGA; U6, ACGAATTTGCGTGTCATCCTT; Ad2 exon 2, GACGGGTTTCCGATCCAAGAGT (detects both mRNA and pre-mRNA).
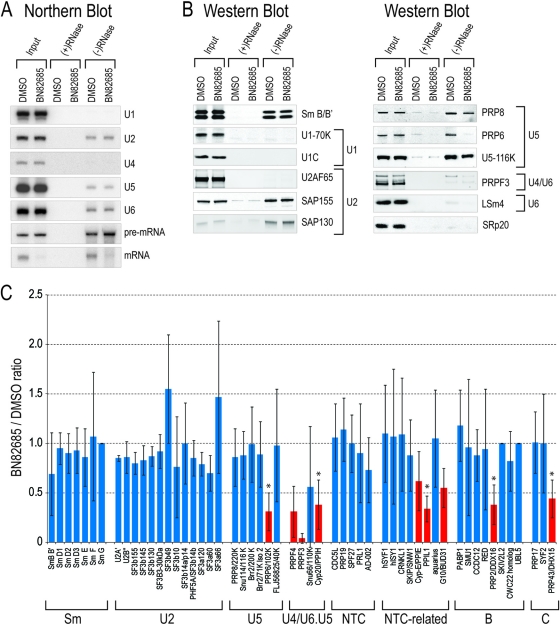
Fig 7.
BN82685 freezes spliceosomes in a postactivation state. (A) Ninety-minute in vitro splicing reactions of biotin-labeled Ad2 pre-mRNA containing either DMSO or 20 μM BN82685 were incubated with streptavidin-coated beads. Precipitates were treated with (+) or without (−) RNase A, beads were washed, and RNAs were isolated and probed by Northern blotting. (B) For the reactions in panel A, coprecipitating proteins, as indicated to the right of the gels, were probed by Western blotting. (C) Streptavidin precipitates were treated as described in the Materials and Methods and analyzed by mass spectrometry. The number of peptide spectrums captured for individual proteins in BN82685 samples for three independent experiments are expressed as an average ratio compared to DMSO controls (set as 1). Error bars, standard deviation; *, P < 0.05 by paired-end Student t test.
Spliceosomal complex analysis by native gels.
Splicing reactions and native gel electrophoresis were performed essentially as described previously (28). Briefly, 10-μl reaction mixtures (with or without 20 μM BN82685) containing 6,000 cpm of radiolabeled Ad2ΔIVS pre-mRNA were incubated at 30°C and stopped at the indicated time points with a 5-mg/ml final concentration of heparin. Samples in native gel loading buffer (10% glycerol, 0.1% xylene cyanol) were placed on ice, resolved on 3.5% (80:1, acrylamide-bisacrylamide) gels run with 0.5× Tris-borate-EDTA (TBE) buffer at 10 mA in 4°C, dried under vacuum, and then autoradiographed at −80°C with an intensifying screen. For Sephadex G-50 nick column fractionations shown in Fig. 6B, 10-μl reaction mixtures were diluted to 100 μl with 0.5× TBE buffer and eluted with 1.2 ml of 0.5× TBE buffer. One-drop (∼50-μl) fractions were collected, and Cerenkov counts were measured on a TriCarb 2900TR liquid scintillation analyzer (Perkin Elmer). RNA was isolated by phenol-chloroform extraction followed by ethanol precipitation and then resolved by urea-PAGE.
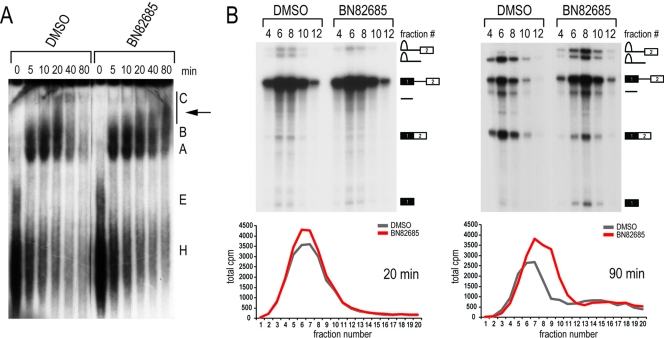
Fig 6.
BN82685 stalls the progression of complex C. (A) DMSO or 20 μM BN82685 was added to splicing reaction mixtures containing radiolabeled Ad2 pre-mRNA and stopped by the addition of heparin (5 mg/ml) at the indicated time points (0 to 80 min). Positions of splicing complexes resolved on 3.5% native acrylamide gels are listed to the right. The arrow indicates the presence of a stalled complex in BN82685-treated reactions. (B) Size exclusion elution profiles of 20-min (left) and 90-min (right) reactions were generated from radioactivity counts (bottom), and even-numbered fractions were run on denaturing gels (top).
Spliceosome association in vitro.
Ninety-minute in vitro splicing reactions were performed with biotin-labeled Ad2 pre-mRNA with either DMSO or 20 μM BN82685; reactions were stopped with heparin (5 mg/ml), and reaction mixtures were then incubated with M-280 streptavidin-coated magnetic beads for 1 h at 4°C in IP buffer (50 mM Tris-HCl, pH 7.6, 2 mM MgCl2, 0.5 mM dithiothreitol [DTT], and 0.05% NP-40) with 100 mM NaCl. Reaction mixtures were then not treated or treated with RNase A for 15 min at 30°C, followed by five washes of 500 μl of IP buffer with 150 mM NaCl. Beads were resuspended in 1× sample buffer, and proteins were resolved by SDS-PAGE, followed by Western blotting using LiCor detection and quantification with the indicated antisera: mouse anti-Sm (Y12; 1:200), goat anti-U1-70K (where U1-70K represents a U1-associated protein with a molecular weight of 70,000) (dilution, 1:200; Santa Cruz), rat anti-U1C (1:1,000; Cosmo Bio Co.), mouse anti-SAP155 (1:1,500; Millipore), goat anti-SAP130 (1:1,000; Abcam), rabbit anti-PRP8 (1:200; Santa Cruz), goat anti-PRP6 (1:200; Santa Cruz), rabbit anti-U5-116K (1:1500; Proteintech), rabbit anti-LSm4 (1:1,000; Proteintech), rabbit anti-PRPF3 (1:1,000; Proteintech), mouse anti-SRp20 (1:500; Zymed), and mouse anti-U2AF65 (1:1,500).
Mass spectrometry.
Proteins isolated from streptavidin bead precipitates shown in Fig. 7B (90-min splicing reactions of 40 nM biotin-labeled Ad2 pre-mRNA) were separated by a short run on 12% SDS-PAGE. Each sample lane was cut into four gel slices, and proteins were eluted and trypsinized for analysis by nano-liquid chromatography (nanoLC)/nanospray/linear trap quadrupole (LTQ) mass spectrometry at the Proteomics Core facility of the University of Pennsylvania School of Medicine. The results were analyzed by Scaffold, version 3.2, software. The threshold for protein identification was set at >95% and, for peptides, at >99%. Three independent streptavidin precipitation and mass spectrometry experiments were performed, along with a technical replicate of the third run. The average and standard deviations of the spectrum count ratio for each protein identified were calculated.
RESULTS
A high-throughput splicing-dependent assay measuring EJC binding to mRNA.
A hallmark of pre-mRNA splicing is the assembly of EJCs upstream of the splice junction. As shown previously (11, 25, 27) and illustrated in Fig. S1 in the supplemental material using radiolabeled pre-mRNA substrate and denaturing RNA gels, monoclonal antibodies to the EJC components, such as eIF4AIII and Y14, specifically immunoprecipitate mRNAs generated by splicing, in contrast to an antibody against hnRNP C1/C2 proteins, which predominantly immunoprecipitates intron-containing RNA species. We used this unique splicing-dependent association of eIF4AIII and Y14 with spliced mRNA to establish a high-throughput assay in a scheme similar to that previously devised for measuring Sm core assembly on snRNAs (61). The assay, termed EJIPT, for EJC immunoprecipitation, uses tagged pre-mRNAs and EJC antibodies to readily quantify mRNAs produced by splicing and/or EJC formation in vitro (Fig. 1). Splicing reaction mixtures containing extract, biotin-labeled pre-mRNA substrate, ATP, and various compounds were incubated in 384-well plates for 90 min and then transferred to a NeutrAvidin-coated plate to capture the RNAs. Anti-eIF4AIII or -Y14 antibody was then added to detect the EJC-containing mRNP formed as a result of splicing. The EJC antibody was then reacted with an HRP-conjugated secondary antibody. Automated washing was performed after each antibody incubation step, followed by the addition of an HRP substrate for enzymatic amplification and detection of chemiluminescence signals with a plate reader.
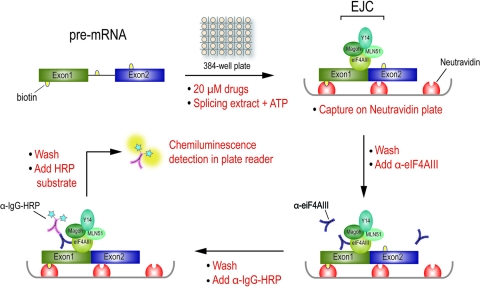
Fig 1.
A high-throughput assay (EJIPT) utilizing EJC binding to spliced mRNAs to measure splicing in vitro. Splicing reaction mixtures containing biotin-labeled pre-mRNA, 20 μM each compound, splicing extract, and 0.5 mM ATP are assembled by liquid handler in 384-well plates and incubated for 1.5 h at 30°C. Reaction mixtures are then transferred to a NeutrAvidin-coated plate, which captures biotin-labeled RNAs/RNPs postsplicing. Anti-eIF4AIII (3F1) antibody is added to detect splicing-dependent EJC formation on spliced mRNAs. Following a washing step, incubation with an HRP-conjugated anti-mouse IgG secondary antibody, and then addition of chemiluminescent substrate, signals representing eIF4AIII/mRNA complexes are obtained by an automatic plate reader.
In contrast to the time-consuming and labor-intensive 32P-based method, the EJIPT assay rapidly and accurately recapitulated the results of splicing gels, as quantifications of the effect of known conditions that inhibit splicing (U2 snRNA antisense morpholino oligonucleotide and okadaic acid) (41) closely mirrored the levels of inhibition seen on the gels (compare Fig. 2A and B). The assay has a signal-to-background ratio between 8 (for no RNA) (in Fig. S2C in the supplemental material, compare 40 nM to 0 nM pre-mRNA) and 15 (for no extract) (Fig. 2B, compare Control to no extract), a coefficient of variation (CV) of 10%, and a screening window coefficient (Z′ factor) value of 0.655. Only background levels of nonspecific binding were obtained without ATP, with AMP-PNP, with mRNA of the same sequence not generated by splicing, or with a control antibody (SP2/0) used for immunoprecipitation instead of anti-EJC antibodies (see Fig. S2A in the supplemental material). These findings demonstrate that EJIPT is a stable and robust assay suitable for high-throughput screening (HTS). We further established several secondary assays to validate and characterize hits from the primary screen as described below.
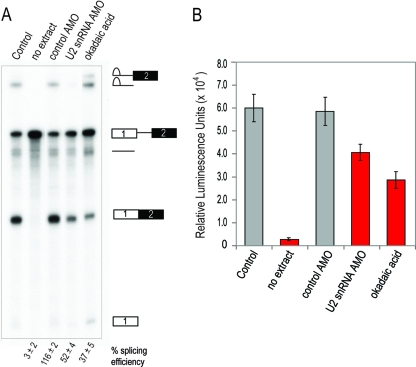
Fig 2.
The analysis of in vitro splicing by the splicing gel method and by the EJIPT assay. (A) 32P-labeled Ad2 pre-mRNA was spliced in the presence of no drug (Control), no extract, 10 μM control AMO, 10 μM U2 snRNA AMO, or 2.5 μM okadaic acid. RNAs after splicing were isolated and resolved on denaturing PAGE gels. Identities of splicing intermediates are depicted to the right of the gels, and quantifications of the splicing efficiency (mRNA/pre-mRNA ratio) by phosphorimaging from two independent experiments are shown below each lane. (B) The EJIPT assay was performed in 384-well microplates using biotin-labeled Ad2 pre-mRNA spliced under the same conditions as described for panel A.
Identification of a novel splicing inhibitor.
We used HEK 293T whole-cell splicing extracts and the commonly used Ad2 pre-mRNA substrate (23, 24) to screen a collection of 2,100 annotated bioactive compounds at 20 μM concentrations. Other biotin-labeled pre-mRNA minigenes as well as HeLa nuclear extracts could be readily substituted in the EJIPT assay (see below). The EJIPT HTS was performed with eIF4AIII antibody (3F1) (11) because, among core EJC proteins, eIF4AIII directly binds RNA (4, 51), and 3F1 produced more consistent signals in the 384-well assay format than the Y14 antibody (4C4) did. Anti-Y14 immunoprecipitates less pre-mRNA by comparison (see Fig. S1 in the supplemental material) and was used for subsequent confirmation experiments. The concentrations of biotin-labeled pre-mRNA and extract were optimized to maximize signal intensity while also ensuring that the signals were being detected in the linear range (see Fig. S2B and C). Compounds showing a statistically significant effect of more than three times the standard deviation (>3SD) were cherry picked for confirmation in a 96-well magnetic bead assay, using anti-eIF4AIII or -Y14 antibody immobilized on magnetic protein A beads to capture biotinylated mRNA, followed by detection with avidin-HRP (see Materials and Methods).
Twelve confirmed compounds were further validated by Northern dot blot assay, an EJC-independent modality that directly measures the levels of splicing products using specific oligonucleotide probes for pre-mRNA and mRNA (Fig. 3A). A decreased ratio of mRNA to pre-mRNA indicated that the compound inhibited splicing, while those that did not likely affected EJC formation or its retention on the mRNA (Fig. 3B); it is also possible that a compound's effect may be technical in nature, interfering with mRNA immobilization or antibody detection. A control reaction mixture without extract or with known splicing inhibitory conditions produced the expected decrease in this ratio (Fig. 3A, row 5, and B, red bars). Further testing of candidates that decreased the mRNA/pre-mRNA ratio by >3SD (Fig. 3B, blue bars below red dotted line) on splicing gels demonstrated that the dot blot assay reliably identified eight out of nine inhibitors (Fig. 3B and data not shown). Among the validated effectors were the following: 5-iodotubericidin (Fig. 3A and B, compound D1), a broad-spectrum kinase inhibitor recently shown to decrease spliceosome assembly; cantharidin/cantharidic acid (compounds B4 and D4), both protein phosphatase 1 (PP1)/PP2A inhibitors that are expected to interfere with splicing; and cisplatin (compound B1) (36, 48, 50, 54). The identification of these known splicing inhibitors from the screen, despite their relatively modest effects, demonstrated that the EJIPT assay is sensitive and reliable.
Fig 3.
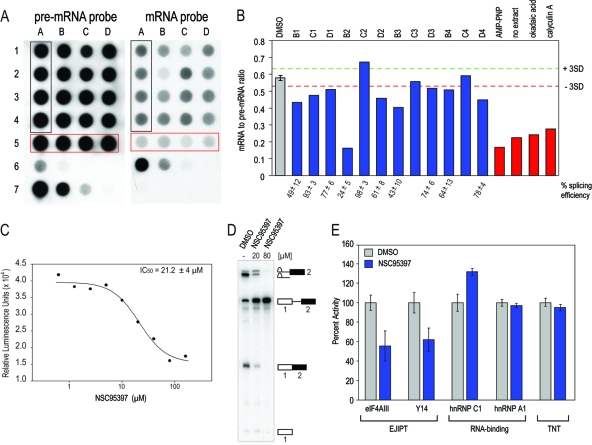
Identification, validation, and selectivity of the hits from the EJIPT HTS. (A) RNA from splicing reactions was probed for pre-mRNA and mRNA by Northern dot blot analysis. Column A samples (black box; rows 1 to 4) consist of DMSO controls. Samples in row 5 (red box, columns A to D) represent splicing-inhibitory conditions, as follows: A, 5 mM AMP-PNP; B, no extract; C, 2.5 μM okadaic acid; D, 2.5 μM calyculin A. Row 6 samples represent a standard curve of mRNA from 12.5 to 0.2 ng in 1:4 serial dilutions. Row 7 samples represent a standard curve of pre-mRNA from 29 to 0.4 ng in 1:4 serial dilutions. Samples in columns B to D (rows 1 to 4) represent splicing reactions with candidate compounds at a 40 μM final concentration. (B) Ratios of mRNA to pre-mRNA from the Northern dot blot assay in panel A were plotted for DMSO samples (gray), compounds (blue), and inhibitory controls (red). Compounds outside the range of 3SD (red and green dotted lines) of the DMSO samples were tested on splicing gels, and percent splicing efficiencies are reported. (C) Dose-response curve of NSC95397 (compound B2 in panels A and B) determined by the EJIPT assay using anti-Y14 (4C4) antibody. The IC50 is calculated from fitting the dose-response data using Sigma Plot software. (D) 32P-labeled CDC14-15 pre-mRNA was spliced in the presence of DMSO or 20 or 80 μM NSC95397 and resolved on denaturing PAGE gels. Identities of splicing intermediates are depicted to the right of gels. (E) Biotin-labeled Ad2 pre-mRNA was spliced in the presence of DMSO or 20 μM NSC95397 and measured by the EJIPT assay with eIF4AIII (3F1) or Y14 (4C4) antibodies and the RNA-binding assays with hnRNP C1/C2 (4F4) or hnRNP A1 (4B10) antibodies. The effect of the compound in the TNT assay is also reported. Results are expressed as percentages relative to DMSO (100%) and are averages of three or more independent experiments. The error bars represent standard deviations of the repeats.
The most potent splicing inhibitor we discovered was NSC95397 [2,3-bis (hydroxyethylthio)-1,4-naphthoquinone] (Fig. 3A and B, compound B2). Its calculated in vitro 50% inhibitory concentration (IC50) of 21 μM from the EJIPT dose-response assay is consistent with the estimation from the splicing gel (Fig. 3C and D). At 20 μM, while NSC95397 inhibited the association of both EJC components (eIF4AIII and Y14) with mRNA by ∼50% in the EJIPT assay, it increased the level of RNAs precipitated with hnRNP C1/C2 protein, which was measured by employing an RNA-binding assay analogous to the EJIPT assay using anti-hnRNP C1/C2 antibody instead of anti-EJC (Fig. 3E). Since hnRNP C1/C2 preferentially binds to intronic sequences (55), this indicates an accumulation of unspliced pre-mRNAs and further confirms that NSC95397 inhibits splicing.
Two additional experiments were carried out to examine the selectivity of NSC95397 (Fig. 3E). In vitro transcription and translation (TNT) reactions of a luciferase plasmid in rabbit reticulocyte lysates, followed by measurement of luciferase activity, were used to identify general inhibitors of ATP synthesis/hydrolysis. Replacing an anti-EJC antibody with anti-hnRNP A1 in the EJIPT assay was used to determine if a compound is a nonspecific inhibitor of RNA-protein interactions. At 20 μM, which is close to its IC50 for splicing, NSC95397 had no effect in either of these assays, suggesting that it is a selective splicing inhibitor in vitro (Fig. 3E).
Structure and activity probing of splicing inhibition by 1,4-naphthoquinones and related compounds.
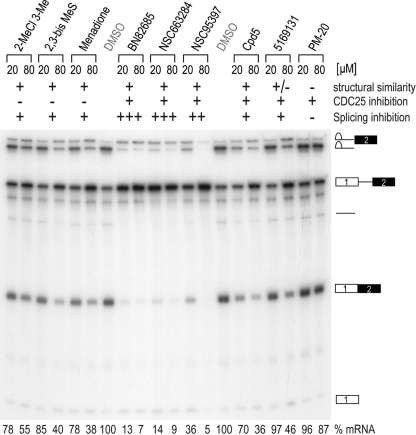
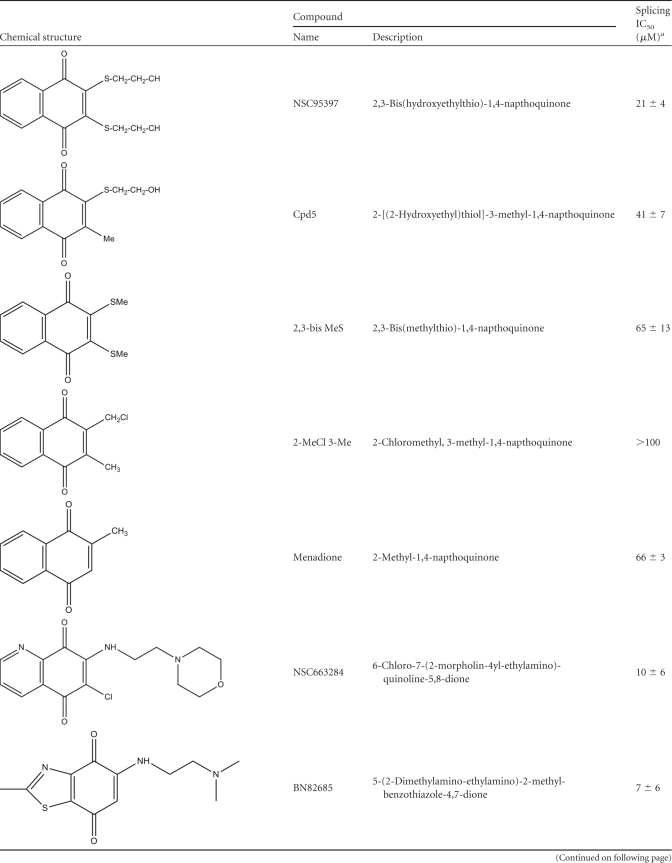
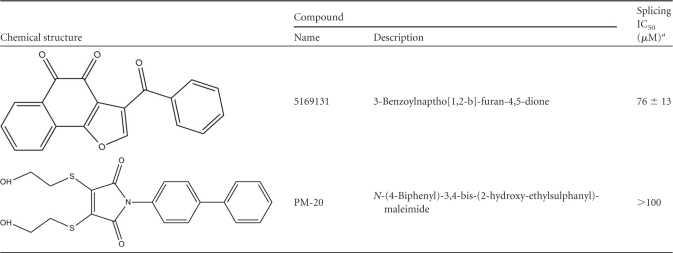
NSC95397 is a 1,4-naphthoquinone previously described as a CDC25 dual-specificity phosphatase inhibitor (33). Several commercially available compounds related to NSC95397 by structure or known to inhibit CDC25 were also tested. As shown in Fig. 4 and Table 1, structurally similar compounds, including the vitamin K analog, menadione, other 1,4-naphthoquinones including Cpd5, and an orthoquinone (5169131) (10, 56), also inhibited splicing but less potently than NSC95397, with IC50s in the 40 to 80 μM range. On the other hand, BN82865 [5-(2-dimethylamino-ethylamino)-2-methyl-benzothiazole-4,7-dione] and NSC663284 [6-chloro-7(−2 morpholin-4yl ethylamino quinoline-5,8-dione], two heteroaryl 1,4-quinones with known CDC25 inhibitory activity (9, 32), were more potent than NSC95397, with IC50s of 7 and 10 μM, respectively (Fig. 4 and Table 1). We also tested the effect of compounds in the snRNP assembly assay, which measures the formation of Sm cores on U snRNAs, a key step in the biogenesis of the major components of the spliceosome (61). This showed that unlike NSC95397, which showed a dose-dependent inhibition on snRNP assembly, BN82865 and NSC663284 had a negligible effect (see Fig. S3 in the supplemental material), demonstrating that they are more selective splicing inhibitors. Another CDC25 inhibitor with a maleimide moiety instead of 1,4-quinone, PM-20, (22), had little or no effect on splicing (Fig. 4 and Table 1). These structure-activity relationship (SAR) studies suggest that the 1,4-naptho- or 1,4-heteroaryl-quinone scaffold plays an important role in splicing inhibition and that CDC25 inhibition does not account for the mechanism by which these compounds inhibit splicing.
Fig 4.
NSC95397 and related compounds contain 1,4-naphthoquinone and 1,4-heterocyclic quinone scaffolds that confer splicing inhibition in vitro. Compounds were added to splicing reaction mixtures with CDC14-15 pre-mRNA at a 20 or 80 μM final concentration. Radiolabeled RNA was purified and resolved by denaturing PAGE, and gels were autoradiographed. Structural similarity to NSC95397 and reported inhibition of CDC25 are listed above. Phosphorimaging quantification of mRNA is expressed as a percentage compared to DMSO after values were normalized to total counts in the lane (below).
Table 1.
Structure-activity relationship summary
Mean values represent the combination of at least two dose-response experiments performed by the EJIPT assay with anti-eIF4AIII or anti-Y14 antibodies and by phosphorimaging analysis of gels.
BN82685 arrests the spliceosome during catalysis.
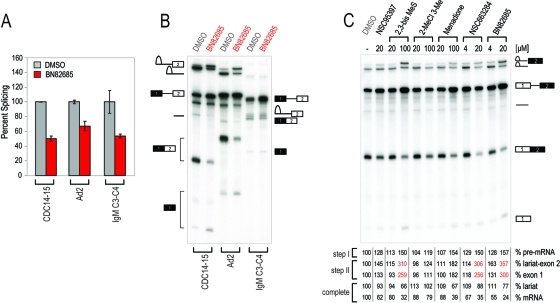
EJIPT was used to investigate the effects of the most potent drug, BN82685, on other pre-mRNAs and to probe for its mode of action. At 10 μM, BN82685 inhibited splicing of each of the pre-mRNAs tested (Ad2, CDC14-15, or IgM C3-C4) by ∼50%, which closely reflected the levels of inhibition seen for this compound by gel electrophoresis (Fig. 5A and 5B). In addition to decreasing mRNA levels, BN82685 caused accumulation of lariat-exon 2 and exon 1 intermediates and decreased the intron lariat and debranched lariat products, particularly evident for both Ad2 and CDC14-15 pre-mRNAs (Fig. 5B). Importantly, all of the 1,4-quinones that inhibited splicing showed similar profiles when tested at concentrations close to their IC50s, whereby a modest accumulation (∼120 to 150%) of pre-mRNA indicated a block preceding catalytic step I, but a much more pronounced effect (∼250 to 350%) was observed for the accumulation of lariat-exon 2 and exon 1 (Fig. 5C and Fig. 4). This suggests that this scaffold preferentially inhibits splicing after the first and prior to the second catalytic trans-esterification steps.
Fig 5.
BN82685 is a general splicing inhibitor that acts after the first catalytic step. (A) In vitro splicing reaction mixtures containing biotin-labeled Ad2, CDC14-15, or IgM C3-C4 pre-mRNA substrates in the presence of DMSO or 10 μM BN82685 were measured by the EJIPT assay using anti-Y14 antibody. (B) Mixtures described in panel A with 32P-labeled pre-mRNAs spliced in vitro were analyzed on denaturing PAGE gels. The identities of splicing intermediates for Ad2 and CDC14-15 are shown on the left, and intermediates for IgM C3-C4 are shown on the right. (C) Compounds were added to splicing reaction mixtures with CDC14-15 pre-mRNA at the final indicated concentrations. Phosphorimaging quantification of each intermediate on the splicing gel is expressed as a percentage compared to DMSO after normalizing to total counts in the lane (below).
Analysis of splicing complex formation on Ad2 pre-mRNA at different time points by native gel electrophoresis showed that complexes H, E, A, and B (corresponding to early RNPs containing no snRNPs, U1 snRNP alone, U1 and U2 snRNPs, and all five snRNPs, respectively) were unaffected; however, a slower-migrating complex above complex B and below what appears to be C (U2, U5, and U6 snRNPs) accumulated with BN82685 (Fig. 6A). This transitory intermediate (indicated by an arrow) was detected even in DMSO samples at 20 min but failed to resolve over time in BN82685. Using size exclusion columns to fractionate splicing reactions and confirm the presence of this complex, we observed similar elution profiles at 20 min for DMSO and BN82685 (Fig. 6B). However, denaturing gels revealed that while all splicing intermediates were already evident in DMSO at this time point, only step I intermediates (lariat-exon 2 and exon 1) were produced in BN82685 and in amounts similar to that of the control (compare fractions 6 and 8 between samples). This indicated that step I occurred with normal kinetics and that this intermediate complex found in both samples is a form of complex C in which catalysis is taking place. At 90 min, the elution profiles differ between BN82685 and DMSO, likely reflecting that the spliceosomes have begun to disassemble in DMSO reactions while stalling in BN82685 (Fig. 6B, fractions 8 and 10 form shoulder peaks not seen in DMSO). While second-step products (lariat and mRNA) were now present in BN82685 reactions at 90 min, they were at greatly reduced levels, the amount of which did not increase upon longer incubation (up to 3 h) (data not shown). These results place BN82685 inhibition after assembly of spliceosomal B complex, during the transition of step I to II in catalytic C complex, and additionally indicate that BN82685 caused a decreased efficiency of the second step and not simply an overall delay in kinetics.
BN82685 stalls spliceosomes in a postcatalytic activation state.
To determine which factor(s) may be involved and more precisely define the stage of inhibition, we purified complexes using streptavidin beads after 90 min of splicing and probed for known splicing components bound to biotin-labeled Ad2 pre-mRNA and its intermediates. Material captured on streptavidin beads (using RNase-digested extract as control for substrate-dependent binding specificity) was washed extensively, eluted with TRIzol, and probed by Northern blotting (Fig. 7A) or eluted with protein sample buffer and probed by Western blotting (Fig. 7B). Since the amount of pre-mRNA (40 nM; i.e., the same as in the EJIPT assay) used here is in large excess compared to that used on 32P-labeled gels to allow for sufficient recovery of associated spliceosomal components, splicing reactions are not expected to proceed to completion. In the absence of RNase, Sm protein levels were equivalent, and nonspliceosomal proteins such as SRp20 were mostly absent, reflecting equal and specific immunoprecipitations (Fig. 7B). The detection of only U2, U5, and U6 snRNAs (and pre-mRNA/mRNA) in both samples further indicated that BN82685 inhibition occurred after U1 and U4 had already exited spliceosomes, at a point after activation (Fig. 7A). Indeed, U1 snRNP-associated proteins (U1-70K and U1C) were not detected in IPs of either sample, demonstrating that U1 snRNP displacement from the 5′ splice site by U6 (e.g., after B complex formation) is largely unaffected. Likewise, the signal for LSm4 in U6 snRNP was considerably lower than that of input, which is consistent with its absence in complex C and suggests that the unwinding of the U4-U6 di-snRNP and subsequent removal of associated components, allowing U6 base pairing to the 5′ splice site, is not impaired (5, 6, 12). In both samples the absence of U2AF65, indicative of its dissociation from the polypyrimidine tract, provides additional support that the drug-treated reaction mixtures were able to progress to complex C.
With U2/U5/U6 snRNA levels essentially unchanged (Fig. 7A), we asked if differences in individual U2- and U5-associated proteins could explain the BN82685-mediated block after catalytic step I. Quantitation from three Western blots showed that the levels of SAP155 and SAP130 of the U2 snRNP were unaffected and that the U5 components PRP8 and U5-116K were slightly reduced by ∼20%. However, the most striking effect was that U5-associated PRP6 and di-snRNP factor PRPF3 were greatly reduced, by 86% and 72%, respectively. To obtain a comprehensive survey of what other proteins might also be affected, the same purified complexes used for Western blotting were analyzed by mass spectrometry. Three independent experiments were performed for which the proteins identified and the corresponding number of spectra detected for each are listed in Table S1 in the supplemental material. Proteins were grouped into each spliceosomal subcomplex according to Bessonov et al. (5, 6), with histograms reflecting protein levels in BN82685-treated samples compared to samples treated with DMSO (Fig. 7C). The overall abundance of peptides for individual proteins showed that U1 snRNP and LSm proteins were underrepresented, while most of the U2, U5, and PRP19/CDC5L complex components were highly represented, agreeing with previous work and our assertion that the splicing reactions for both DMSO and BN82685 have proceeded past the activation step (5, 6). The presence of reported B, Bact, and C complex-specific proteins also underscored the heterogeneous nature of these samples and further suggested that splicing was still ongoing in both samples (5) (see Table S1). Consistent with the Western blot results, Sm proteins and U2 components were largely unchanged, as were most of the U5 components, whereas PRPF3, PRPF4, and PRP6 were consistently lower in BN82685 samples (5, 6). Additionally, there were statistically significant decreases in proteins associated with the human PRP19 (hPRP19)/CDC5L (NTC, for nineteen complex) complex, such as PPIE, PPIL1, and the BUD31 homolog. Also of note are decreases in PRP2/DDX16, required for catalytic activation of complex B before step I (26), and PRP43/DDX15, required for disassembly of spliceosomes after exon ligation (3). Together, BN82685-treated samples contain an inventory of proteins expected of spliceosomes stalled at the postactivation step, which blocks progression through complex C.
DISCUSSION
We show here that EJIPT is an effective assay for measuring in vitro splicing, based on the specific assembly of EJC components on spliced mRNA. EJIPT has significant advantages over the conventional splicing assay using gel electrophoresis, including speed and ease of use and the ability to provide quantitative readout. EJIPT's favorable attributes of low-percent CV and high signal-to-noise ratio make it an excellent HTS tool. Combining this with the secondary assays we implemented provides a highly effective platform to identify modifiers of splicing and EJC formation or stability.
Our HTS identified 1,4-naphthoquinones and 1,4-heterocyclic quinones as splicing inhibitors. In addition to inhibiting splicing, NSC95397 also decreased snRNP assembly in a dose-dependent manner (see Fig. S3 in the supplemental material), consistent with results from a screen identifying inhibitors of Sm core assembly (62). This activity is attributed to the compound's hydroxyethylthio side chains which generate reactive oxygen species (13), causing intermolecular cross-linking of SMN (survival of motor neurons) and thus inactivation of the SMN complex activity in snRNP assembly (62). It is likely that these moieties also contribute to its splicing inhibition activity, since 1,4 naphthoquinones without it (Table 1) have higher IC50s. The most potent compounds we found through SAR studies were the heterocyclic quinones, NSC663284 and BN82685, which inhibited in vitro splicing with low IC50s while having only a negligible effect on snRNP assembly, indicating selectivity for splicing (Fig. 3E and 4 and Table 1; see also Fig. S3). NSC95397 and several of the structurally related compounds have been described as CDC25 inhibitors (13). However, the 50-fold discrepancy between in vitro IC50s for CDC25 phosphatase inhibition (∼200 nM) versus splicing inhibition (∼7 to 20 μM) and the lack of an effect by structurally unrelated CDC25 inhibitors (e.g., PM-20) (Fig. 4 and Table 1) suggested that CDC25 is not the target of these drugs vis-a-vis splicing inhibition in vitro. Therefore, albeit at higher concentrations, these anticancer drugs also target the spliceosome.
Of the relatively few small-molecule splicing inhibitors that have been recently described, all appear to target early stages of splicing. A natural product bioflavonoid, isoginkgetin, and histone acetyltransferase inhibitors (e.g., garcinol) have been shown to arrest splicing at prespliceosomal A complex stage, while histone deacetylase inhibitors (e.g., suberoylanilide hydroxamide [SAHA] and splitomicin) inhibit splicing at complex B (31, 45). The FR901464 derivatives, pladienolide-B and spliceostatin A, are antitumor agents which target U2 snRNP-associated SF3b components SAP130 and SAP155, respectively, and interfere with the A-to-B complex transition (21, 29, 44). Meayamycin, another FR901464 analog, blocks complex A formation in vitro with increased potency (2). SR-dependent splicing inhibitors like TG003 (42), which targets the Clk1/Sty kinase, or NB-506 (46), which targets topoisomerase I, both block the phosphorylation of ASF/SF2 and prevent complex A formation. In contrast, the 1,4-naphthoquinone and 1,4-heterocyclic quinone scaffolds we identified here inhibit splicing at a later stage than these inhibitors, predominantly accumulating lariat-exon 2 and exon 1 intermediates and thus arresting splicing at a point between the first and second steps of catalysis. The persistence of complexes A and B at late times (Fig. 6A) and the accumulation of unspliced pre-mRNA (Fig. 4 and 5C), more prevalent at higher drug concentrations, suggest that these drugs may also target an earlier step. Although we did not detect convincing evidence of an effect on ASF/SF2 phosphorylation, it is possible that the benzothiazole scaffold of BN82685, which is also found in TG003, may additionally inhibit the Clk kinases and contribute to this early block to splicing (42). Nevertheless, when these compounds are used at their IC50s, a combination of denaturing gels, native gels, and size exclusion column fractionation demonstrated that spliceosome assembly and the first catalytic step typically occur with normal kinetics (Fig. 4 to 6). Splicing reaction mixtures incubated longer (e.g., 3 h) continue to be blocked at this stage and do not produce more mRNA (data not shown). Thus, it is the efficiency and the rate at which the second step proceeds that are dramatically reduced, rather than an overall delay in splicing, causing lariat-exon 2 and exon 1 intermediates to accumulate (Fig. 6B).
The splicing pathway requires extensive rearrangement of proteins and RNA-RNA interactions of spliceosomal complexes, chief of which involve the remodeling of the U5/U4-U6 tri-snRNP (for a review, see reference 60). The presence of a complex comprised only of the U2/U5/U6 snRNAs in equivalent amounts at late reaction times (Fig. 7A, 90 min) and the absence of U1 and U4 indicated that BN82685-stalled splicing reactions proceed beyond activation of complex B. A decrease in PRP2, the helicase which drives spliceosomes from B* to catalysis, suggests that even the activation step may also be impaired. Biochemical characterization and mass spectrometry of streptavidin-pulled down splicing complexes (Fig. 7B and C) were oriented by an inventory of spliceosomal proteins found in each complex as systematically determined by the Lührmann group (for a review, see references 60 and 64). Importantly, BN82685 nonuniformly disrupted association of individual protein components, pointing to interference with specific rearrangements, rather than a block in the recruitment of the entire tri-snRNP. Indeed, levels of components in the stable core of U5 snRNP (PRP8, Brr2, and U5-116K) (1) were largely unaffected, as were the Sm proteins and the core PRP19/CDC5L complex, which stably interacts with U5 during both catalytic steps (12, 38). Despite the egress of U4 snRNA concomitant with activation, U4/U6-specific proteins (e.g., PRP3 and PRP4) are still variably detected in complex C (5, 6, 38). Reduced levels of PRP6 are also expected at late stages since this protein also dissociates from the U5 snRNP upon activation (60). Levels of these proteins were all dramatically decreased in BN82685 reactions compared to reactions with DMSO. Interestingly, peptidyl-prolyl cis/trans isomerases of the tri-snRNP (PPIH) and those associated with the PRP19/CDC5L complex (PPIE and PPIL1) were also consistently decreased. These enzymes are members of the cyclophilin family which associate with spliceosomes at discrete stages and are believed to play a role in both catalytic steps, possibly by determining the rate of protein folding (19, 43). Finally, given the block to step II and the failure to resolve C complexes, it is perhaps not surprising that the U2-associated DEXH/D-box helicase, PRP43, required for spliceosome disassembly, is also deficient.
Currently, it is unclear what the actual target(s) of BN82685 is and whether these changes we observe represent the cause or result of the inhibition. Considering the role currently described for PRP6, which is to bridge U5 to the U4-U6 di-snRNP in the precatalytic spliceosome, reactions would not be expected to proceed beyond complex B in its absence (17, 39, 47). Indeed, mutations in the conserved phosphorylation sites at the N terminus of PRP6 prevent spliceosome activation and cause an accumulation of complex B containing all five snRNPs (8, 37, 49). In contrast, BN82685 reactions clearly produced step I intermediates and formed complex C, with purified splicing complexes from this reaction containing only U2, U5, and U6 snRNAs. Therefore, a deficiency in PRP6, or inhibition of PRPF4 kinase, for example, which phosphorylates PRP6 and PRP31 as they are incorporated into complex B, is not likely the cause of splicing inhibition by BN82685 (49). Upon catalytic activation, while PRP8, U5-116K, and U5-200K remain tightly bound to U5 snRNA, there is an exodus of several proteins from the spliceosome, involving PRP6, PRP31, PRP3, and PRP4, all of whose associations are decreased by BN82685. Thus, whereas DMSO-treated profiles represent a mixture of completed and ongoing reactions, reactions with BN82685 are “frozen” after catalytic step I, and proteins that exit upon activation immediately prior to this are decreased by comparison (e.g., PRP6). Furthermore, as a result of not completing step II, BN82685-treated spliceosomes fail to disassemble properly (Fig. 6A) and recycle the tri-snRNP, which in turn would affect the next round of splicing. This could explain why the predominant effect of BN82685 is in the accumulation of step I intermediates (∼250 to 350%) and why the accumulation of pre-mRNA (∼120 to 150%) appears secondary. As a final note, we considered PP1/PP2A phosphatases as candidate targets. These are required to dephosphorylate SAP155 and U5-116K prior to the second step of splicing; however, our compounds had little to no effect on the phosphorylation status of these substrates, nor was the addition of purified phosphatase able to rescue drug-treated reaction mixtures (data not shown) (50).
Rearrangements during the early stages of splicing are fairly well understood, but the later steps are not as well characterized. For example, what the U2/U6 base pairing interactions immediately preceding catalysis are, whether the U4/U6 associated proteins exit catalytically activated spliceosomes en masse or in distinct steps, and a detailed description of which events occur during spliceosome dissociation are just some of the open issues remaining (60). BN82685 and the related compounds we identified can provide important tools for dissecting these steps, identifying additional rearrangements, intermediates, or players involved late in the splicing process. With these additional anticancer drugs now also shown to be splicing inhibitors, it has becoming increasingly appreciated that pre-mRNA splicing is another cellular process that can be targeted for cancer treatment (18, 59). The small but growing number of splicing inhibitors known so far and the ability of EJIPT and other HTS assays to discover additional ones should help dissect this important cellular pathway and have important biomedical applications.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hyunil Jo and the members of our laboratory, especially Byung Ran So, for stimulating discussions and comments on the manuscript. We give special thanks to Yoon Cho for technical assistance, Jeongsik Yong for technical advice, and Brian Carr for compounds (Cpd5 and PM-20). We thank Chao-Xing Yuan and staff at the University of Pennsylvania Proteomics Core facility for assistance with mass spectrometry and data analysis.
This work was supported by the Association Française Contre les Myopathies. G.D. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print 17 January 2012
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1. Achsel T, Ahrens K, Brahms H, Teigelkamp S, Luhrmann R. 1998. The human U5-220kD protein (hPrp8) forms a stable RNA-free complex with several U5-specific proteins, including an RNA unwindase, a homologue of ribosomal elongation factor EF-2, and a novel WD-40 protein. Mol. Cell. Biol. 18:6756–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Albert BJ, et al. 2009. Meayamycin inhibits pre-messenger RNA splicing and exhibits picomolar activity against multidrug-resistant cells. Mol. Cancer Ther. 8:2308–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Arenas JE, Abelson JN. 1997. Prp43: an RNA helicase-like factor involved in spliceosome disassembly. Proc. Natl. Acad. Sci. U. S. A. 94:11798–11802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ballut L, et al. 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat. Struct. Mol. Biol. 12:861–869 [DOI] [PubMed] [Google Scholar]
- 5. Bessonov S, et al. 2010. Characterization of purified human Bact spliceosomal complexes reveals compositional and morphological changes during spliceosome activation and first step catalysis. RNA 16:2384–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bessonov S, Anokhina M, Will CL, Urlaub H, Luhrmann R. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452:846–850 [DOI] [PubMed] [Google Scholar]
- 7. Blencowe BJ. 2006. Alternative splicing: new insights from global analyses. Cell 126:37–47 [DOI] [PubMed] [Google Scholar]
- 8. Bottner CA, Schmidt H, Vogel S, Michele M, Kaufer NF. 2005. Multiple genetic and biochemical interactions of Brr2, Prp8, Prp31, Prp1 and Prp4 kinase suggest a function in the control of the activation of spliceosomes in Schizosaccharomyces pombe. Curr. Genet. 48:151–161 [DOI] [PubMed] [Google Scholar]
- 9. Brezak MC, et al. 2005. Inhibition of human tumor cell growth in vivo by an orally bioavailable inhibitor of CDC25 phosphatases. Mol. Cancer Ther. 4:1378–1387 [DOI] [PubMed] [Google Scholar]
- 10. Brisson M, et al. 2004. Discovery and characterization of novel small molecule inhibitors of human Cdc25B dual specificity phosphatase. Mol. Pharmacol. 66:824–833 [DOI] [PubMed] [Google Scholar]
- 11. Chan CC, et al. 2004. eIF4A3 is a novel component of the exon junction complex. RNA 10:200–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chan SP, Kao DI, Tsai WY, Cheng SC. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279–282 [DOI] [PubMed] [Google Scholar]
- 13. Contour-Galcera MO, Sidhu A, Prevost G, Bigg D, Ducommun B. 2007. What's new on CDC25 phosphatase inhibitors. Pharmacol. Ther. 115:1–12 [DOI] [PubMed] [Google Scholar]
- 14. Cooper TA, Wan L, Dreyfuss G. 2009. RNA and disease. Cell 136:777–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dreyfuss G, Kim VN, Kataoka N. 2002. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3:195–205 [DOI] [PubMed] [Google Scholar]
- 16. Fukuhara T, et al. 2006. Utilization of host SR protein kinases and RNA-splicing machinery during viral replication. Proc. Natl. Acad. Sci. U. S. A. 103:11329–11333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galisson F, Legrain P. 1993. The biochemical defects of prp4-1 and prp6-1 yeast splicing mutants reveal that the PRP6 protein is required for the accumulation of the [U4/U6.U5] tri-snRNP. Nucleic Acids Res. 21:1555–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hagiwara M. 2005. Alternative splicing: a new drug target of the post-genome era. Biochim. Biophys. Acta 1754:324–331 [DOI] [PubMed] [Google Scholar]
- 19. Horowitz DS, Lee EJ, Mabon SA, Misteli T. 2002. A cyclophilin functions in pre-mRNA splicing. EMBO J. 21:470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jurica MS, Moore MJ. 2003. Pre-mRNA splicing: awash in a sea of proteins. Mol. Cell 12:5–14 [DOI] [PubMed] [Google Scholar]
- 21. Kaida D, et al. 2007. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3:576–583 [DOI] [PubMed] [Google Scholar]
- 22. Kar S, Wang M, Yao W, Michejda CJ, Carr BI. 2006. PM-20, a novel inhibitor of Cdc25A, induces extracellular signal-regulated kinase 1/2 phosphorylation and inhibits hepatocellular carcinoma growth in vitro and in vivo. Mol. Cancer Ther. 5:1511–1519 [DOI] [PubMed] [Google Scholar]
- 23. Kataoka N, Dreyfuss G. 2008. Preparation of efficient splicing extracts from whole cells, nuclei, and cytoplasmic fractions. Methods Mol. Biol. 488:357–365 [DOI] [PubMed] [Google Scholar]
- 24. Kataoka N, Dreyfuss G. 2004. A simple whole cell lysate system for in vitro splicing reveals a stepwise assembly of the exon-exon junction complex. J. Biol. Chem. 279:7009–7013 [DOI] [PubMed] [Google Scholar]
- 25. Kataoka N, et al. 2000. Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol. Cell 6:673–682 [DOI] [PubMed] [Google Scholar]
- 26. Kim SH, Lin RJ. 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol. 16:6810–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim VN, et al. 2001. The Y14 protein communicates to the cytoplasm the position of exon-exon junctions. EMBO J. 20:2062–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konarska MM, Sharp PA. 1986. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell 46:845–855 [DOI] [PubMed] [Google Scholar]
- 29. Kotake Y, et al. 2007. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3:570–575 [DOI] [PubMed] [Google Scholar]
- 30. Krawczak M, Reiss J, Cooper DN. 1992. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: causes and consequences. Hum. Genet. 90:41–54 [DOI] [PubMed] [Google Scholar]
- 31. Kuhn AN, van Santen MA, Schwienhorst A, Urlaub H, Luhrmann R. 2009. Stalling of spliceosome assembly at distinct stages by small-molecule inhibitors of protein acetylation and deacetylation. RNA 15:153–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lazo JS, et al. 2001. Discovery and biological evaluation of a new family of potent inhibitors of the dual specificity protein phosphatase Cdc25. J. Med. Chem. 44:4042–4049 [DOI] [PubMed] [Google Scholar]
- 33. Lazo JS, et al. 2002. Identification of a potent and selective pharmacophore for Cdc25 dual specificity phosphatase inhibitors. Mol. Pharmacol. 61:720–728 [DOI] [PubMed] [Google Scholar]
- 34. LE Hir H, Izaurralde E, Maquat LE, Moore MJ. 2000. The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J. 19:6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Le Hir H, Nott A, Moore MJ. 2003. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 28:215–220 [DOI] [PubMed] [Google Scholar]
- 36. Li YM, Casida JE. 1992. Cantharidin-binding protein: identification as protein phosphatase 2A. Proc. Natl. Acad. Sci. U. S. A. 89:11867–11870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lutzelberger M, et al. 2010. The N terminus of Prp1 (Prp6/U5-102 K) is essential for spliceosome activation in vivo. Nucleic Acids Res. 38:1610–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Makarov EM, et al. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205–2208 [DOI] [PubMed] [Google Scholar]
- 39. Makarova OV, Makarov EM, Liu S, Vornlocher HP, Luhrmann R. 2002. Protein 61K, encoded by a gene (PRPF31) linked to autosomal dominant retinitis pigmentosa, is required for U4/U6*U5 tri-snRNP formation and pre-mRNA splicing. EMBO J. 21:1148–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Maquat LE. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5:89–99 [DOI] [PubMed] [Google Scholar]
- 41. Mermoud JE, Cohen P, Lamond AI. 1992. Ser/Thr-specific protein phosphatases are required for both catalytic steps of pre-mRNA splicing. Nucleic Acids Res. 20:5263–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Muraki M, et al. 2004. Manipulation of alternative splicing by a newly developed inhibitor of Clks. J. Biol. Chem. 279:24246–24254 [DOI] [PubMed] [Google Scholar]
- 43. Nagradova N. 2007. Enzymes catalyzing protein folding and their cellular functions. Curr. Protein Pept. Sci. 8:273–282 [DOI] [PubMed] [Google Scholar]
- 44. Nakajima H, et al. 1996. New antitumor substances, FR901463, FR901464 and FR901465. II. Activities against experimental tumors in mice and mechanism of action. J. Antibiot. (Tokyo) 49:1204–1211 [DOI] [PubMed] [Google Scholar]
- 45. O'Brien K, Matlin AJ, Lowell AM, Moore MJ. 2008. The biflavonoid isoginkgetin is a general inhibitor of Pre-mRNA splicing. J. Biol. Chem. 283:33147–33154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pilch B, et al. 2001. Specific inhibition of serine- and arginine-rich splicing factors phosphorylation, spliceosome assembly, and splicing by the antitumor drug NB-506. Cancer Res. 61:6876–6884 [PubMed] [Google Scholar]
- 47. Schaffert N, Hossbach M, Heintzmann R, Achsel T, Luhrmann R. 2004. RNAi knockdown of hPrp31 leads to an accumulation of U4/U6 di-snRNPs in Cajal bodies. EMBO J. 23:3000–3009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schmittgen TD, Ju JF, Danenberg KD, Danenberg PV. 2003. Inhibition of pre-mRNA splicing by cisplatin and platinum analogs. Int. J. Oncol. 23:785–789 [PubMed] [Google Scholar]
- 49. Schneider M, et al. 2010. Human PRP4 kinase is required for stable tri-snRNP association during spliceosomal B complex formation. Nat. Struct. Mol. Biol. 17:216–221 [DOI] [PubMed] [Google Scholar]
- 50. Shi Y, Reddy B, Manley JL. 2006. PP1/PP2A phosphatases are required for the second step of Pre-mRNA splicing and target specific snRNP proteins. Mol. Cell 23:819–829 [DOI] [PubMed] [Google Scholar]
- 51. Shibuya T, Tange TO, Sonenberg N, Moore MJ. 2004. eIF4AIII binds spliced mRNA in the exon junction complex and is essential for nonsense-mediated decay. Nat. Struct. Mol. Biol. 11:346–351 [DOI] [PubMed] [Google Scholar]
- 52. Soret J, et al. 2005. Selective modification of alternative splicing by indole derivatives that target serine-arginine-rich protein splicing factors. Proc. Natl. Acad. Sci. U. S. A. 102:8764–8769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Staley JP, Guthrie C. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315–326 [DOI] [PubMed] [Google Scholar]
- 54. Stoilov P, Lin CH, Damoiseaux R, Nikolic J, Black DL. 2008. A high-throughput screening strategy identifies cardiotonic steroids as alternative splicing modulators. Proc. Natl. Acad. Sci. U. S. A. 105:11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Swanson MS, Dreyfuss G. 1988. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 7:3519–3529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tamura K, et al. 2000. Cdc25 inhibition and cell cycle arrest by a synthetic thioalkyl vitamin K analogue. Cancer Res. 60:1317–1325 [PubMed] [Google Scholar]
- 57. Tazi J, et al. 2005. Selective inhibition of topoisomerase I and various steps of spliceosome assembly by diospyrin derivatives. Mol. Pharmacol. 67:1186–1194 [DOI] [PubMed] [Google Scholar]
- 58. Tazi J, Bakkour N, Stamm S. 2009. Alternative splicing and disease. Biochim. Biophys. Acta 1792:14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tazi J, Durand S, Jeanteur P. 2005. The spliceosome: a novel multi-faceted target for therapy. Trends Biochem. Sci. 30:469–478 [DOI] [PubMed] [Google Scholar]
- 60. Wahl MC, Will CL, Luhrmann R. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701–718 [DOI] [PubMed] [Google Scholar]
- 61. Wan L, et al. 2005. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25:5543–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wan L, Ottinger E, Cho S, Dreyfuss G. 2008. Inactivation of the SMN complex by oxidative stress. Mol. Cell 31:244–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang ET, et al. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456:470–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Will CL, Luhrmann R. 2010. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3:a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Younis I, et al. 2010. Rapid-response splicing reporter screens identify differential regulators of constitutive and alternative splicing. Mol. Cell. Biol. 30:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.