Abstract
Endogenous retroviruses have the ability to become permanently integrated into the genomes of their host, and they are generally transmitted vertically from parent to progeny. With the exception of gypsy, few endogenous retroviruses have been identified in insects. In this study, we describe the tirant endogenous retrovirus in a subset of Drosophila simulans natural populations. By focusing on the envelope gene, we show that the entire retroviral cycle (transcription, translation, and retrotransposition) can be completed for tirant within one population of this species.
INTRODUCTION
Endogenous retroviruses are genomic sequences that are widely dispersed throughout the host genome. These sequences constitute 8% of the human genome and represent the remnants of ancient infections by retroviruses (1). Some of these retroviruses were domesticated and generated cellular neogenes, such as the syncytin gene (9, 14). The structure of a canonical endogenous retrovirus consists of three open reading frames (ORFs), which are bordered by long terminal repeats (LTRs). The third ORF encodes the envelope protein (Env), which possesses fusogenic properties and is responsible for the infectious behavior of exogenous retroviruses. In insects, several endogenous retroviruses have been described, and most of them possess a complete retroviral structure. Representative insect endogenous retroviruses (IERVs) or insect ErantiViruses (24) include the following: gypsy, ZAM, Idefix, tirant, 17.6, 297, and nomad in Drosophila melanogaster; tom in Drosophila ananassae; Tv1 in Drosophila virilis; TED in Trichoplusia ni; Osvaldo in Drosophila buzzatii; and Yoyo in Ceratitis capitata (see reference 19 for a review). With the exception of nomad and Tv1, each of these displays a complete env ORF. Thus far, only the well-known gypsy element of D. melanogaster has been shown to possess infectious properties (10), as Moloney murine leukemia virus pseudotypes with the gypsy Env protein were shown to infect insect cells (25).
The tirant LTR retrotransposon of D. melanogaster was previously described to share sequence similarity with the fusion proteins of certain baculoviruses (13, 16, 20) (see Fig. 1 for the tirant structure). In a phylogenetic study of numerous elements from insects, Terzian et al. (24) proposed that the tirant element from D. melanogaster (GenBank accession number X93507) belongs to the IERVs. Thus, the tirant element was placed into this clade of retroviruses, which uses a tRNA-Ser binding site to prime reverse transcription (RT). In the study described in this report, we examined this ERV family in Drosophila simulans using a collection of strains with variable numbers of genomic insertions (7), and we demonstrated that this family was able to produce Env proteins in the ovaries of the host. We found that tirant was capable of performing the first step required for infection, i.e., the production of a functional Env protein, which suggests that it could be classified as an active endogenous retrovirus of D. simulans. Furthermore, we demonstrated that the complete retrotransposition cycle of tirant occurs within a particular natural strain of D. simulans, which indicates that the endogenization of tirant in the natural populations of this species is an ongoing process. In addition, this experimental system is of particular interest because our panel of natural strains displayed three distinct states in regard to tirant dynamics and thus will be ideal for deciphering the fine regulatory mechanisms: (i) the absence of transcription in most strains, (ii) transcription but the absence of translation, and (iii) transcription and translation both occurring in a single strain.
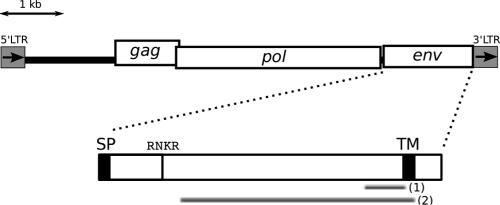
Fig 1.
Structure of tirant env. SP, putative signal peptide, as predicted previously (4); TM, transmembrane domain, as predicted previously (27); RNKR, peptide sequence of the furin cleavage site, as determined previously (16); fragment (1), RT-qPCR amplicon; fragment (2), probe for in situ hybridization of the transcripts.
MATERIALS AND METHODS
Drosophila strains.
We analyzed samples of flies collected from several geographically distinct natural populations of D. simulans. These populations were maintained in the laboratory at 25°C as isofemale lines or as small-mass cultures with approximately 50 pairs in each generation.
The D. simulans strains used were previously described by Fablet et al. (7). They differed in the number of euchromatic copies of tirant that could be detected by in situ hybridization on polytene chromosomes. These strains and their respective copy numbers of tirant were as follows: Chicharo (Portugal, 0 copies), Makindu (Kenya, 5 copies), Zimbabwe (2 copies), Mayotte (Indian Ocean, 2 copies), and Brazzaville (Congo, 0 copies). Each population is known to display specific, unique euchromatic insertion sites, although bands representing heterochromatic sites may appear to be shared between populations in transposon display experiments.
As controls, we used a strain of Drosophila pseudoobscura, which is a species devoid of any tirant copies (17), that was kindly provided by F. Lemeunier from Gif-sur-Yvette, France, and a D. melanogaster isolate of a natural population from Senegal, which was grown in the laboratory and contains 9 euchromatic insertions of tirant that can be detected by in situ hybridization on polytene chromosomes (2). It should also be noted that D. melanogaster populations display much less variability of the tirant copy number than D. simulans populations (2, 7).
Analysis of genomic copies of tirant env. (i) Screening for env full-length ORFs in natural populations of D. simulans.
To search for full-length ORFs for tirant env in natural populations of D. simulans, we used the protein truncation test (PTT) protocol, which consists of PCR amplification of the env gene followed by in vitro transcription and translation. A T7-coupled reticulocyte lysate system (Promega) was used for the PTT analysis, according to the protocol recommended by the manufacturer (the TNT quick-coupled transcription/translation system and the Transcend nonradioactive translation detection system from Promega). The primers used for env amplification were designed from the tirant sequence (GenBank accession number AC0054444, positions 50203 to 58729) and consisted of the following sequences: forward (Fw) primer 5′ GGA TCC TAA TAC GAC TCA CTA TAG GGA GCC ACC ATG GAA CAA TTC 3′ and reverse (Rv) primer 5′ CCT GAA TTT TTT TTT GGG TTA GAG GCG 3′. The upstream primer was positioned at the beginning of ORF3, which allowed transcription to begin at the underlined ATG. We also used D. pseudoobscura, a species devoid of tirant insertions, as a negative control.
(ii) Analysis of tirant env sequence polymorphism.
We isolated tirant env sequences from a subset of the pool of natural populations by PCR using the following primers: Fw primer 5′ TGT ATG GAA CAA TTC CAT CTG AC 3′ and Rv primer 5′ TTG TTT AGA GGC GTG GGG GT 3′. These PCR products were then cloned (TOPO TA cloning kit; Invitrogen) and sequenced (Sanger method; GenoScreen, France). The DNA sequences were aligned and clustered using the Seaview software (version 4) (8; http://pbil.univ-lyon1.fr/software/seaview.html), and DNA polymorphisms were measured using the DnaSP software (version 5) (21). The alignment of the sequences is provided in the supplemental material.
Analysis of tirant env transcription. (i) Isolation of tirant mRNAs and synthesis of cDNA.
Total RNA was extracted from pools of 20 to 30 ovaries from adult females using an RNeasy kit (Qiagen) and subsequently treated with DNase (DNA-free kit; Ambion). One microgram of the total RNA was then converted into cDNA using a Thermoscript Invitrogen kit primed with a mix of oligo(dT)20 and random primers.
(ii) Quantification of env transcripts of tirant by qPCR.
The cDNAs were diluted 50 times and then quantified using SYBR green quantitative PCR (qPCR) in a LightCycler apparatus (Roche Diagnostics) with primers specific to the tirant env gene (Fw primer 5′ ACA CGT TCC CTG AAC AGA CG 3′ and Rv primer 5′ GAA CGT TAC CAA TCC GAG CA 3′) (see Fig. 1 for the position of the amplicon). The transcript quantity was estimated relative to that of the rp49 gene, for which we used the following primers: Fw primer 5′ CGG ATC GAT ATG CTA AGC TGT 3′ and Rv primer 5′ GCG CTT GTT CGA TCC GTA 3′. The rp49 gene demonstrated equivalent amounts of transcripts for all of the strains analyzed, which enabled the amount of env transcripts to be compared. Measurements were obtained from two independent experiments; i.e., RNAs were extracted from two different pools of ovaries. For each experiment, the reactions were performed in duplicate, and the standard curves were calculated using serial dilutions of genomic DNA. The primers were tested for all the strains, and an efficiency value greater than 1.9 was maintained.
(iii) Detection of tirant env transcripts by in situ hybridization in ovaries.
Ovaries were dissected in PBT (1× phosphate-buffered saline [PBS], 0.2% Tween 20), fixed for 20 to 40 min, washed in PBT, and then incubated in 1× PBS containing 3.7% formaldehyde for 10 min at room temperature. Hybridization (overnight) and washing were performed at 65°C. The tirant antisense riboprobe was a 1-kb fragment corresponding to the env gene and included a T3 promoter site in the reverse primer (Fw primer 5′ ACG GGG TTT GAT TAA CG 3′ and Rv primer 5′ GGA ATT AAC CCT CAC TAA AGG GAG TGT CCA GGT GTG CTG 3′). The riboprobe was labeled by the in vitro transcription of T3 using a digoxigenin (DIG)-RNA labeling mix (Roche). DIG-labeled RNA probes were detected using anti-DIG Fab antibody fragments (1/50 dilution; Roche) and fluorescence amplification (Tyramide signal amplification kit; PerkinElmer). An RNase H control was included to prevent RNA-DNA hybridization in the experiments. Specifically, after the hybridization and washing steps, the ovaries were treated with RNase H (0.3 U/μl) for 1 h at 37°C and then evaluated by immunodetection. To stain the DNA, the ovaries were incubated in PBS containing Sytox green (Invitrogen), and visualization was performed with a Zeiss LSM510 Meta confocal microscope.
Analysis of tirant Env protein production by detection of tirant Env proteins in ovary preparations using immunochemistry. (i) Antibodies.
A polyclonal antibody against the tirant Env protein was produced via the DNA vaccination of mice. The Env cDNA sequence was synthesized and cloned into a pVax plasmid designed using the ICAntibodies technology (developed by In Cell Art for in vivo gene expression), and this plasmid was injected into a Swiss mouse.
(ii) Immunochemistry.
Ovaries were dissected in PBS on ice and fixed with 4% paraformaldehyde in PBS with 0.2% Triton X-100 at room temperature for 20 min. The tissue samples were then washed three times for 2 min each with PBS. After blocking in PBS containing 0.2% Triton X-100 and 0.1% bovine serum albumin (BSA) for 2 h at room temperature, the ovaries were incubated overnight at 4°C with the primary antibody at a 1/50 dilution in PBS containing 0.2% Triton X-100 and 0.1% BSA. After three washes in PBT, the ovaries were incubated with the secondary antibody (1/600 dilution of Alexa 488-conjugated anti-mouse IgG [H+L]; Molecular Probes) for 2 h at room temperature. For the negative controls, the ovaries were incubated with a 1/500 dilution of mouse preimmune serum for the corresponding primary antibody. The DNA was stained with propidium iodide, and confocal images were acquired using an LSM510 microscope (Carl Zeiss).
Detection of tirant retrotransposition by transposon display.
Total genomic DNA was isolated from individual flies using a standard phenol-chloroform extraction procedure after proteinase K digestion. The transposon display was performed using a modified version of the protocol employed previously (6). Briefly, genomic DNA from individual adult flies was digested with 10 units of MseI for 3 h at 37°C, and the adaptors consisted of the following sequences: 5′ AAC AGC TGG ACG ATG AGT CCT GAG A 3′ (AdaptMseI+) and 5′ TAT CTC AGG AGT GTA 3′ (AdaptMseI−). The ligation of the adaptors was performed at 37°C for 1 h with 2.5 units of T4 DNA ligase. During the first round of amplification, 0.625 units of Dream Taq polymerase (Fermentas), 0.2 μM deoxynucleoside triphosphate, 0.4 μM the adaptor-specific primer (5′ AAC AGC TGG ACG ATG AGT 3′), and 0.2 μM the tirant-specific primer (5′ GTC TTC CCG GTT GAG TGT 3′) were used. During the second amplification round, 0.2 μM the adaptor primer, 0.25 μM the nested tirant-specific primer with 5′ hexachloro-6-carboxyfluorescein (HEX) fluorescent labeling (5′ TAG AGG CGT GGG GGT TTA 3′), and 0.625 units of Dream Taq polymerase (Fermentas) were used. The PCR was performed for 35 cycles with amplification steps that lasted for 45 s. Negative controls that used the adaptor-specific primer or the element-specific primer alone were included. One microliter of the PCR products was loaded onto a 3730XL capillary DNA analyzer (Applied Biosystems) with a GS600LIZ standard size marker (Applied Biosystems). The raw data were analyzed using the GeneMapper software (Applied Biosystems).
We measured the number of novel bands that were obtained in the progeny (7 to 10 males and females) of five independent pairs for the Makindu population and four independent pairs for the Mayotte population. The bands were considered novel only when they were absent from both parental samples and were present in no more than one individual in the progeny.
Nucleotide sequence accession numbers.
The nucleotide sequences were deposited in the GenBank database with the following accession numbers: JN786085 to JN786091 for Makindu, JN786092 to JN786098 for Zimbabwe, JN786099 to JN786104 for Chicharo, and JQ219965 to JQ219971 for Mayotte.
RESULTS
D. simulans strains display potentially functional env genes.
We evaluated the integrity of the tirant env genes in our strains using a PTT, which consists of the amplification of genomic DNA sequences by PCR, followed by in vitro transcription and translation. In this technique, subsequent Western blotting reveals whether the env gene contains a full-length ORF. A premature stop codon or internal deletions at the genomic level appear as bands of smaller mass compared with those of the expected full-length protein. Our assay revealed the presence of complete ORFs for tirant env in the tested natural strains of D. simulans, which demonstrates that these sequences could potentially be functional in vitro (Fig. 2). Similarly, the tirant env ORFs of D. melanogaster were found to be full-length ORFs. Indeed, Western blotting after in vitro translation indicated two specific bands that corresponded to approximately 56 kDa and 52 kDa. As the expected product would give a band corresponding to 56.2 kDa, we assumed that the observed signal represented a complete and potentially functional Env protein. The shorter products may correspond to the use of a weaker downstream AUG signal (located 177 bp downstream) or to the presence of a premature stop codon or deletions in certain tirant copies that conserve the reading frame.
Fig 2.
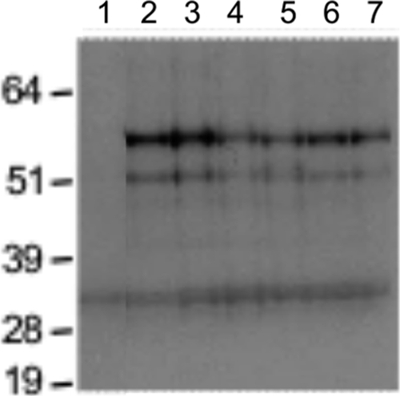
Protein truncation test for tirant env in D. simulans natural strains. Western blotting of the products from in vitro transcription and translation. Lanes: 1, D. pseudoobscura negative control; 2, D. melanogaster strain from Senegal; 3 to 7, D. simulans strains from Mayotte, Brazzaville, Zimbabwe, Makindu, and Chicharo, respectively. The molecular mass ladder (in kDa) is included on the left side of the image. All of the tested D. simulans natural populations displayed identical profiles that corresponded to a potentially functional env ORF. The fainter, lower 33-kDa band is not tirant specific because we also found this band in the D. pseudoobscura negative control (a species devoid of tirant insertions).
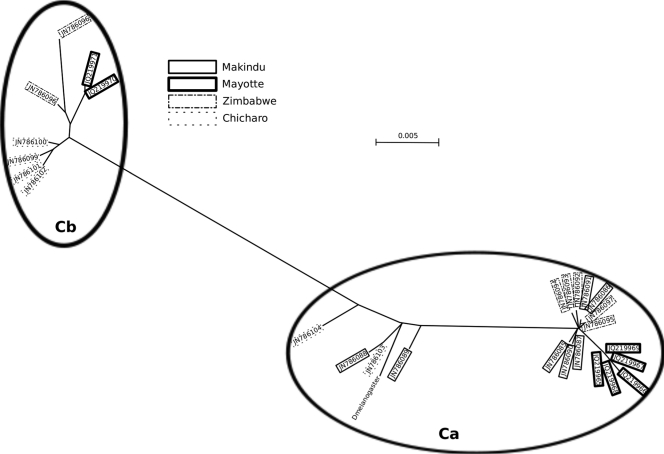
We isolated and analyzed in detail the sequences of tirant env from a subset of these populations (Makindu, Mayotte, Zimbabwe, and Chicharo) (see the supplemental material for sequence alignment). We did not observe length polymorphism in the PCR products, although the sequencing of a few clones revealed the existence of two sequence variants (Fig. 3). A previous study of tirant sequences identified two subfamilies in natural populations of D. simulans, based on differences in the sequences of the regulatory regions of the element: the C subfamily is transcriptionally active and found in both euchromatin and heterochromatin, and the S subfamily is silenced and exclusively heterochromatic (7). Both of the variants identified here belonged to the transcriptionally active C subfamily of the tirant copies. One of the sequence variants (referred to here as variant Ca) was found in all four tested strains (GenBank accession numbers JN786085 to JN786091 for Makindu, JQ219965 to JQ219969 for Mayotte, JN786092 to JN786095 and JN786097 for Zimbabwe, and JN786103 and JN786104 for Chicharo), and the sequences were highly similar (nucleotide diversity [π] = 0.0079). This variant presumably corresponds to the translated sequence from the PTT assay and is thus a putatively functional sequence. The other variant (referred to here as variant Cb) was found in all the samples except that of the Makindu strain (GenBank accession numbers JQ219970 and JQ219971 for Mayotte, JN786096 and JN786098 for Zimbabwe, and JN786099 to JN786102 for Chicharo), and this variant also displayed high levels of sequence similarity among the strains (π = 0.0059). However, this variant presented a premature stop codon. The calculation of π for the consensus of each variant resulted in a value of 0.0429. The sequencing of the env cDNAs that were obtained from the total mRNA in the Mayotte sample revealed that both variants are transcribed.
Fig 3.
Neighbor-joining reconstruction of the relationships between tirant env sequences. Included is a reference from a tirant sequence from the D. melanogaster genome, which was extracted from contig AC005444 at positions 50203 to 58729. Variant Ca, which was found in all four of the tested strains, is the canonical sequence for the tirant env gene, whereas variant Cb contains a premature stop codon.
tirant produces env transcripts in the ovaries of certain D. simulans strains.
We tested four D. simulans strains for the presence of tirant env transcripts. Our RT-qPCR experiments showed that only two strains, Makindu and Mayotte, had significant levels of tirant env transcripts in the ovaries (Fig. 4A). Even the D. melanogaster strain, which harbors significantly more insertions within the euchromatin, did not contain tirant transcripts in the ovaries.
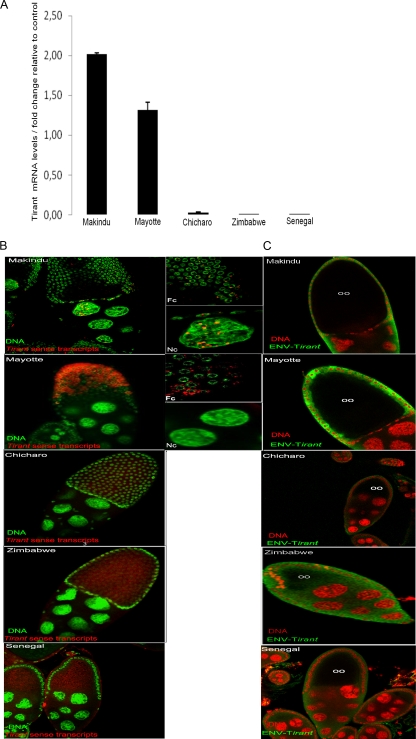
Fig 4.
Expression of the tirant env gene in the ovaries of natural strains of D. simulans. (A) RT-qPCR of the ovaries. The transcript levels were estimated relative to those of the rp49 gene. Significant levels of tirant env transcripts were detected in the Makindu and Mayotte populations. D. melanogaster Senegal was used as a control strain. (B) In situ hybridization of tirant transcripts (red). DNA is labeled in green (Sytox Green). Fc, follicle cells; Nc, nurse cells. (Left) View of ovary chambers (magnification, ×40); (right) details of the left panel. tirant transcripts were found in the nurse cells and follicle cells of the Makindu strain and in the follicle cells of the Mayotte strain. No staining was observed in the other strains. (C) Detection of tirant Env protein by immunochemistry (green) in the Mayotte strain. DNA is stained in red (propidium iodide). oo, oocyte. The green halos in the other strains do not represent Env staining but rather correspond to autofluorescence, as they were also observed following staining with preimmune serum.
To visualize the localization of the tirant transcripts in the germ line, we performed RNA fluorescent in situ hybridization for the ovaries. As expected, no transcripts were detected in the strains that showed no RT-qPCR signal, namely, Zimbabwe and Chicharo, or in the D. melanogaster strain (Fig. 4B). We observed that tirant transcripts accumulated in the mature egg chambers of the ovaries of the Makindu and Mayotte strains (a mature egg chamber consists of the oocyte and nurse cells, which both constitute the germ line, surrounded by the somatically derived follicle cells) (Fig. 4B). However, the patterns of transcript localization were different between these two strains; tirant transcripts were detected in both the follicle and nurse cells for the Makindu strain, whereas these transcripts accumulated substantially in the follicle cells and were absent from the nurse cells for the Mayotte strain. This observation may also explain why the transcript levels, as determined by RT-qPCR, were higher in the Makindu strain than the Mayotte strain.
tirant env transcription results in protein production in one D. simulans strain.
We evaluated the production of tirant Env proteins using immunochemistry, which was performed on whole ovaries using the tirant Env antibody. We observed staining for tirant Env in the ovaries of the Mayotte strain, whereas no clear signal for Env was detected in the other samples (Fig. 4C). Our immunofluorescence analysis revealed the presence of the Env protein at a late stage (stage 9) of oogenesis in the follicle cells of the Mayotte strain.
tirant is active in the Mayotte strain.
The insertion of a retrotranscribed copy into a new genomic site would represent the final step of the endogenous retroviral cycle of tirant. To assess whether this step occurs in the Mayotte population, we performed a transposon display. This technique consists of digesting genomic DNA, ligating adaptors to the obtained fragments, and performing a nested PCR with one primer specific to the adaptor and the other primer specific to tirant. The observation of the amplified fragment profile allows the detection of insertions of tirant in a given genome, and the comparison of the transposon display profiles among the mother, the father, and the progeny subsequently allows the identification of new bands, which are present in only one unique descendant and are absent from both parents. The analysis of four pairs of individuals from the Mayotte strain revealed the presence of novel tirant bands in the progeny, which were numbered between 2 and 22, depending on the pair analyzed (Table 1). This finding indicates that tirant does retrotranspose in this population.
Table 1.
Transposon display results
| Strain | Pair no. | No. of male/no. of female progeny | No. of male/no. of female novel bands | No. of male/no. of female progeny without novel bands |
|---|---|---|---|---|
| Makindu | 1 | 10/9 | 0/0 | 10/9 |
| 2 | 10/9 | 0/0 | 10/9 | |
| 3 | 9/10 | 1/1 | 9/9 | |
| 4 | 9/10 | 3/1 | 8/9 | |
| 5 | 10/10 | 4/1 | 8/9 | |
| Mayotte | 1 | 8/10 | 3/4 | 6/8 |
| 2 | 10/10 | 0/2 | 10/8 | |
| 3 | 10/7 | 8/10 | 7/4 | |
| 4 | 10/10 | 7/15 | 8/3 |
Furthermore, we repeated the in situ hybridization on polytene chromosomes, which we had performed approximately 10 years ago (2). Whereas tirant had previously been estimated to display two insertions in the Mayotte strain, the current study was able to identify seven of these insertions in the same strain. It should be noted that transposon display and in situ hybridization on polytene chromosomes are not expected to provide identical results. Indeed, heterochromatic copies were revealed by the transposon display technique only, which explains the greater number of bands observed in the transposon display experiments.
Interestingly, in the Makindu population, we identified between zero and five novel bands by the transposon display, which suggests the existence of either a tirant provirus amplification process that is independent of Env production or chromosomal rearrangement events.
DISCUSSION
tirant, a newly discovered active endogenous retrovirus of Drosophila simulans.
Similar to other IERVs, tirant was previously shown to contain copies with a complete third ORF (4, 15, 27). Short transcripts that potentially correspond to the subgenomic spliced form of env were detected by Northern blotting in young D. melanogaster embryos (15), but no substantial evidence was found to indicate the potential activity of this gene. In the present study, we have demonstrated at least two natural strains of D. simulans in which the tirant env gene is transcribed, and we found that the corresponding Env protein is produced in one of these strains (Table 2). Therefore, tirant appears to be a newly identified active endogenous retrovirus of D. simulans, a species in which no other active endogenous retrovirus has been reported thus far. Furthermore, we found that this IERV was not active in D. melanogaster, which suggests that the same endogenous retrovirus can display different behaviors in different but closely related species. This is not the first report to describe such differences between D. melanogaster and D. simulans. For example, one recent study revealed that the respective contents of the transposable elements in these species are quite different; D. melanogaster harbors mainly complete copies, whereas D. simulans displays many deleted and degenerated copies of elements within the same family (12).
Table 2.
Results obtained with the different strains
| Population | No. of euchromatic copiesa | mRNA levelb | In situ localization of mRNAs | PTT result | Env protein |
|---|---|---|---|---|---|
| Makindu | 5 | 1.53 | Nurse cells +, follicle cells + | Complete ORF | NDc |
| Mayotte | 2 | 1 | Follicle cells +++ | Complete ORF | Follicle cells |
| Zimbabwe | 2 | 0.02 | ND | Complete ORF | ND |
| Chicharo | 0 | 4.10−4 | ND | Complete ORF | ND |
| Senegal (D. melanogaster) | 9 | 3.10−5 | ND | Complete ORF | ND |
As previously determined by in situ hybridization on polytene chromosomes (2).
Relative to the Mayotte level.
ND, not detected.
These findings indicate that each strain of D. simulans studied possesses at least one potentially complete functional tirant env gene in its genome. A detailed sequence analysis revealed that each of these populations had highly similar sequences for tirant env. Moreover, another variant of env that contains a premature stop codon was also found in most of the studied populations (in all except Makindu).
In this study, we found one D. simulans strain in which tirant env was both transcribed and translated. In this strain, the staining for Env proteins was observed in the follicle cells of the ovaries during the later stages of development. Other IERVs have also been shown to display activity in the ovaries of certain strains that are referred to as “permissive” or “unstable.” In these strains, for example, Tanda et al. (22) found that tom RNAs accumulate in the germarium, in nurse cells (germ line cells), and in the oocyte nucleus. idefix was shown to be expressed in very specific early follicle cells (somatic cells) within the ovaries of unstable strains (23). gyspy, which is the only infectious IERV identified to date, was shown to produce Env proteins that accumulated in the follicle cells of permissive females at stage 10 of oogenesis (18). Leblanc et al. (11) studied ZAM, which is the IERV most closely related to tirant, and found ZAM Env proteins in the follicle cells surrounding the posterior portion of the oocytes in D. melanogaster ovaries at stages 9 and 10.
For gypsy and ZAM, the somatic expression of the env genes was thus demonstrated. However, only a transfer from the soma to the oocyte (germ line cell) can result in an increase in the copy number of these IERVs in the following generation. This mechanism was demonstrated for gypsy (5) and ZAM (3), and it likely involves the yolk protein traffic machinery that operates between the follicle cells and the oocyte (3). In the Mayotte strain, tirant transcripts are located in the follicle cells, which is also the case for gypsy and ZAM. Our transposon display results, together with the more recent in situ hybridization experiments on polytene chromosomes, show that tirant does transpose in this strain. Therefore, we may assume that the infection of an oocyte by tirant viral particles occurs and that these particles are produced in the surrounding follicle cells as they are for gypsy and ZAM.
The tirant element: a model for the study of ERV proliferation in natural populations.
As described for other endogenous retroviruses of Drosophila, such as ZAM and gypsy, most strains are classified as “restrictive” or “stable,” which means that the activity and copy number of the endogenous retroviruses are limited. In addition, there are certain “permissive” or “unstable” strains for which this inhibition is no longer detected. Our study assessed the original features of tirant in a collection of natural populations, and we detected the following three categories of strains that differed according to their tirant dynamics: (i) the Chicharo category had zero (or very few, as in the case of Zimbabwe) euchromatic copies of tirant and no detectable production of transcripts; (ii) the Mayotte category, which displays the qualities of a permissive strain, possessed tirant copies that were able to undergo the entire retrotransposition cycle; and (iii) the Makindu category had an intermediate state, where tirant was transcribed but not translated. Most natural populations worldwide behave similarly to those within the Chicharo category and present no detectable tirant activity. Our results indicate that these strains have potentially functional tirant env genes, which are, however, silenced.
Therefore, at least in the Mayotte strain, tirant is able to complete the retroviral cycle and can therefore be considered an active endogenous retrovirus of D. simulans. However, because we do not have the sequences of all the tirant entire copies, we cannot exclude the possibility that the expression of the other ORFs may originate from different copies and that the retroviral cycle is achieved through complementation. It remains unclear why tirant could not be translated in the Makindu strain. However, we can postulate that a posttranscriptional control of tirant occurs in the Makindu strain, which is not the case in the Mayotte strain. Our collection of natural populations, which provides examples of different tirant dynamics, is an ideal tool for deciphering such mechanisms.
Furthermore, it is tempting to propose that these different natural strains illustrate different time periods of the history of the tirant endogenous retrovirus in the D. simulans genome (26). When an active copy of tirant is inserted into a favorable genomic site, it can be transcribed and translated and can retrotranspose to other sites, thereby increasing its copy number. This was likely the situation for the Mayotte strain. In the context of the “arms race” hypothesis, the D. simulans genome likely develops strategies to combat this retroviral invasion, such as posttranscriptional locking, which appears to be the case in the Makindu strain. Such locking may also occur in D. melanogaster populations, where the number of euchromatic insertions is relatively high compared with D. simulans, although no significant transcript levels can be detected in these populations. However, because both species display very different dynamics related to transposable elements (12), we may assume that tirant regulation is under the control of many distinct mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the ANR grant GENEMOBILE from the CNRS, FINOVI, IUF, and CIBLE 2008 (Région Rhône Alpes).
We thank Christian Biémont for his useful discussions and Geoffrey Hutinet for his technical help. The English quality was certified by American journal experts.
Footnotes
Published ahead of print 25 January 2012
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1. Belshaw R, et al. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc. Natl. Acad. Sci. U. S. A. 101:4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Biémont C, et al. 2003. Worldwide distribution of transposable element copy number in natural populations of Drosophila simulans. Evolution 57:159–167 [DOI] [PubMed] [Google Scholar]
- 3. Brasset E, et al. 2006. Viral particles of the endogenous retrovirus ZAM from Drosophila melanogaster use a pre-existing endosome/exosome pathway for transfer to the oocyte. Retrovirology 3:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cañizares J, Grau M, Paricio N, Moltó MD. 2000. Tirant is a new member of the gypsy family of retrotransposons in Drosophila melanogaster. Genome 43:9–14 [DOI] [PubMed] [Google Scholar]
- 5. Chalvet F, et al. 1999. Proviral amplification of the gypsy endogenous retrovirus of Drosophila melanogaster involves env-independent invasion of the female germline. EMBO J. 18:2659–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Esnault C, et al. 2008. High genetic differentiation between the M and S molecular forms of Anopheles gambiae in Africa. PLoS One 3:e1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fablet M, McDonald JF, Biémont C, Vieira C. 2006. Ongoing loss of the tirant transposable element in natural populations of Drosophila simulans. Gene 375:54–62 [DOI] [PubMed] [Google Scholar]
- 8. Galtier N, Gouy M, Gautier C. 1996. SEAVIEW and PHYLO_WIN: two graphic tools for sequence alignment and molecular phylogeny. Comp. Appl. Biosci. 12:543–548 [DOI] [PubMed] [Google Scholar]
- 9. Heidmann O, Vernochet C, Dupressoir A, Heidmann T. 2009. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 6:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim A, et al. 1994. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. U. S. A. 91:1285–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leblanc P, et al. 2000. Life cycle of an endogenous retrovirus, ZAM, in Drosophila melanogaster. J. Virol. 74:10658–10669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lerat E, Burlet N, Biémont C, Vieira C. 2011. Comparative analysis of transposable elements in the melanogaster subgroup sequenced genomes. Gene 473:100–109 [DOI] [PubMed] [Google Scholar]
- 13. Malik HS, Henikoff S, Eickbush TH. 2000. Poised for contagion: evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 10:1307–1318 [DOI] [PubMed] [Google Scholar]
- 14. Mallet F, et al. 2004. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl. Acad. Sci. U. S. A. 101:1731–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Marsano RM, et al. 2000. The complete tirant transposable element in Drosophila melanogaster shows a structural relationship with retrovirus-like retrotransposons. Gene 247:87–95 [DOI] [PubMed] [Google Scholar]
- 16. Misseri Y, Labesse G, Bucheton A, Terzian C. 2003. Comparative sequence analysis and predictions for the envelope glycoproteins of insect endogenous retroviruses. Trends Microbiol. 11:253–256 [DOI] [PubMed] [Google Scholar]
- 17. Moltó MD, Paricio N, López-Preciado MA, Semeshin VF, Martínez-Sebastián MJ. 1996. Tirant: a new retrotransposon-like element in Drosophila melanogaster. J. Mol. Evol. 42:369–375 [PubMed] [Google Scholar]
- 18. Pélisson A, et al. 1994. Gypsy transposition correlates with the production of a retroviral envelope-like protein under the tissue-specific control of the Drosophila flamenco gene. EMBO J. 13:4401–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pelisson A, Mejlumian L, Robert V, Terzian C, Bucheton A. 2002. Drosophila germline invasion by the endogenous retrovirus gypsy: involvement of the viral env gene. Insect Biochem. Mol. Biol. 32:1249–1256 [DOI] [PubMed] [Google Scholar]
- 20. Rohrmann GF, Karplus PA. 2001. Relatedness of baculovirus and gypsy retrotransposon envelope proteins. BMC Evol. Biol. 1:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rozas J, Rozas R. 1995. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Comput. Appl. Biosci. 11:621–625 [DOI] [PubMed] [Google Scholar]
- 22. Tanda S, Mullor JL, Corces VG. 1994. The Drosophila tom retrotransposon encodes an envelope protein. Mol. Cell. Biol. 14:5392–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tcheressiz S, et al. 2002. Expression of the Idefix retrotransposon in early follicle cells in the germarium of Drosophila melanogaster is determined by its LTR sequences and a specific genomic context. Mol. Genet. Genomics 267:133–141 [DOI] [PubMed] [Google Scholar]
- 24. Terzian C, Pélisson A, Bucheton A. 2001. Evolution and phylogeny of insect endogenous retroviruses. BMC Evol. Biol. 1:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Teysset L, et al. 1998. A Moloney murine leukemia virus-based retroviral vector pseudotyped by the insect retroviral gypsy envelope can infect Drosophila cells. J. Virol. 72:853–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vieira C, Fablet M, Lerat E. 2009. Infra- and transspecific clues to understanding the dynamics of transposable elements, p 115–123 In Transposons and the dynamic genome, genome dynamics and stability. Springer-Verlag, Berlin, Germany [Google Scholar]
- 27. Viggiano L, Caggese C, Barsanti P, Caizzi R. 1997. Cloning and characterization of a copy of tirant transposable element in Drosophila melanogaster. Gene 197:29–35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.