Abstract
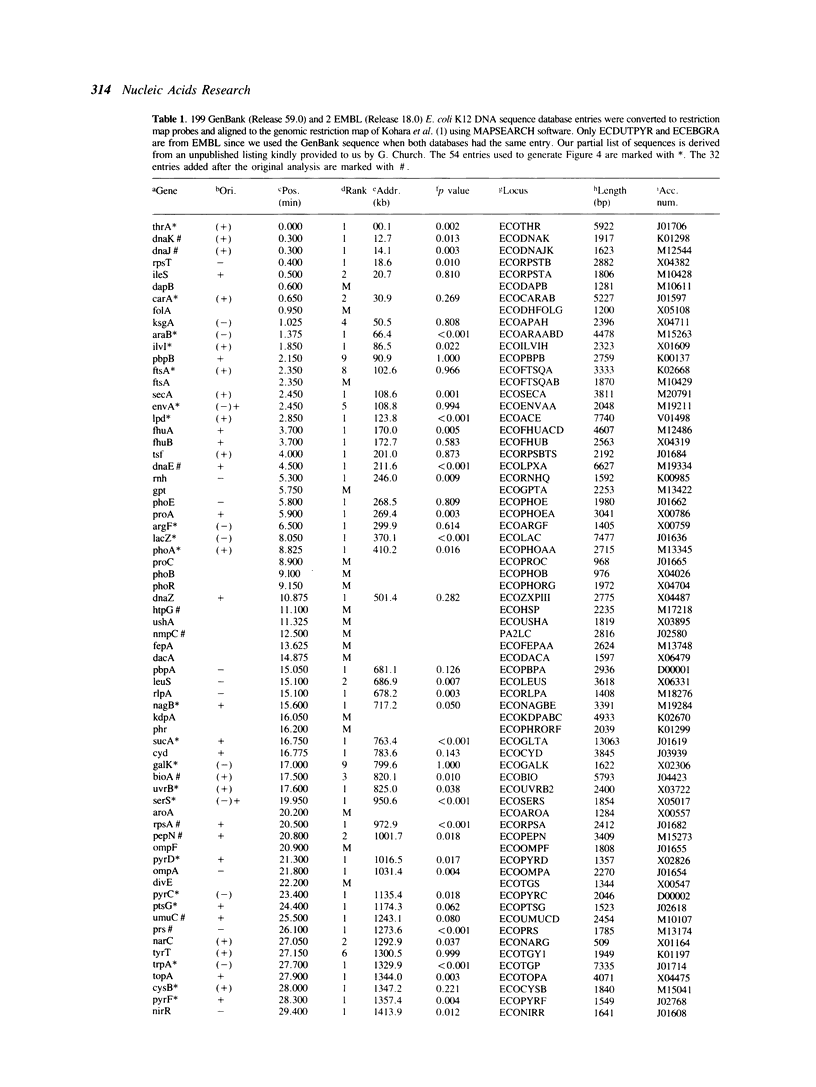
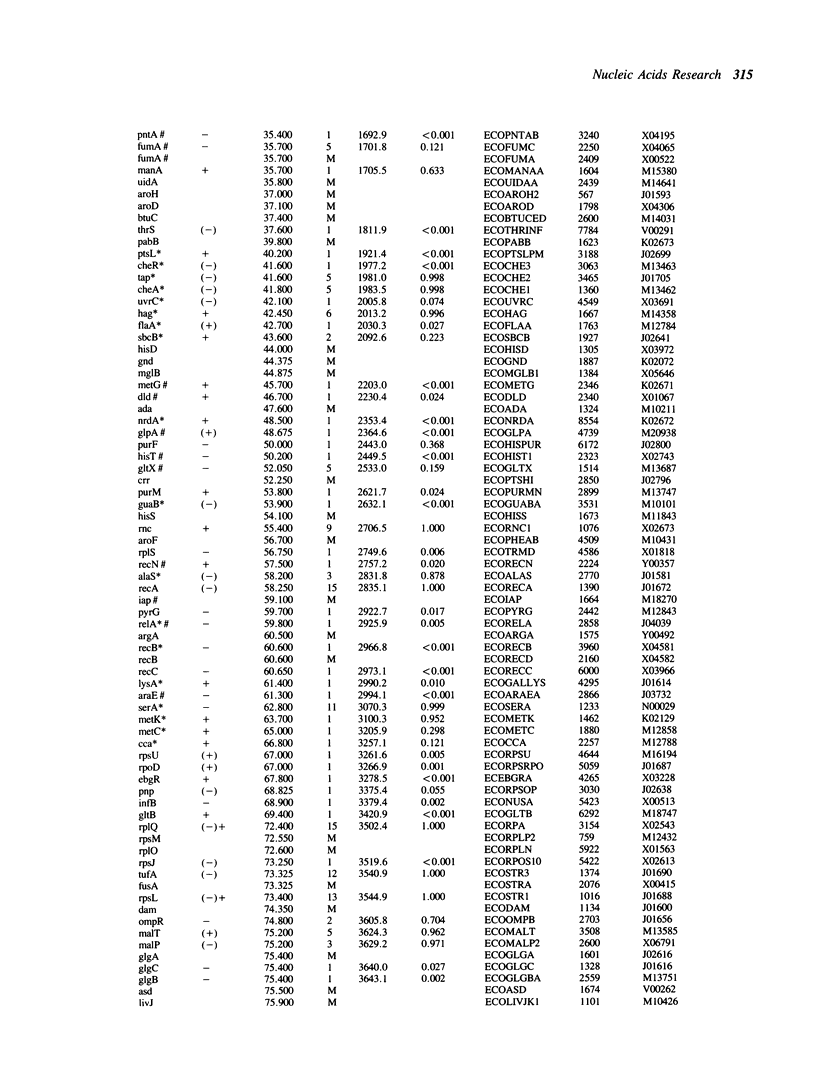
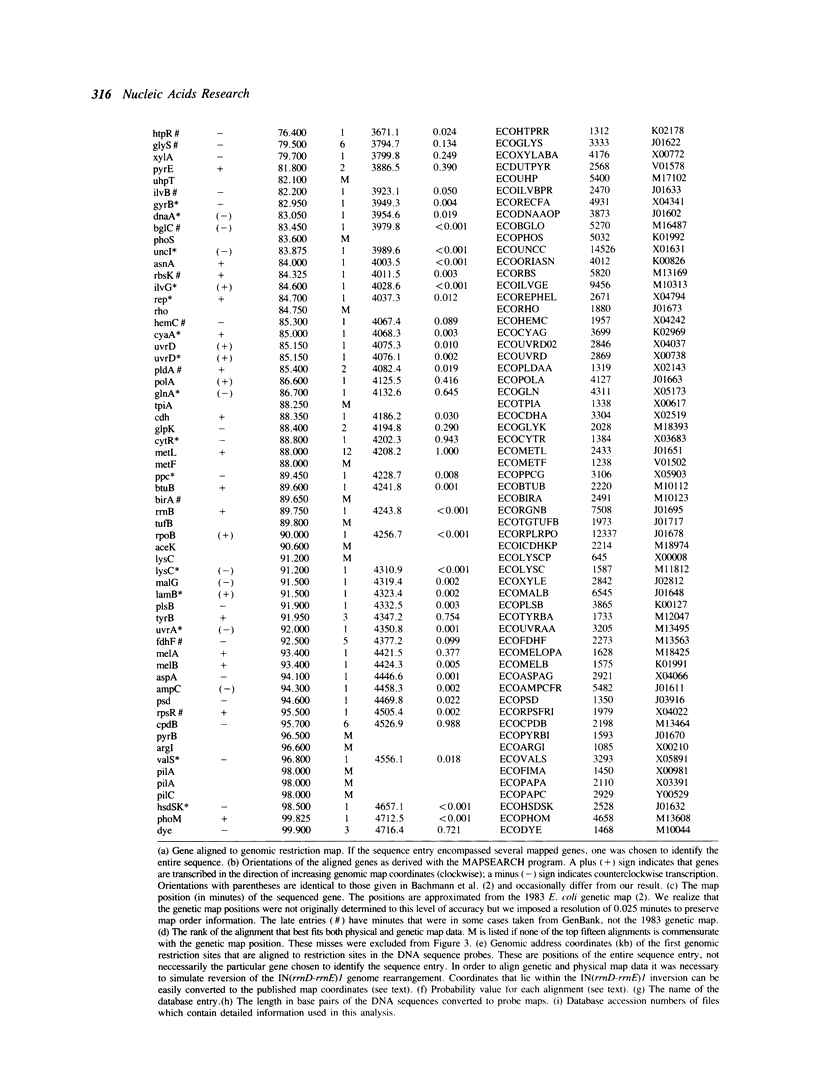
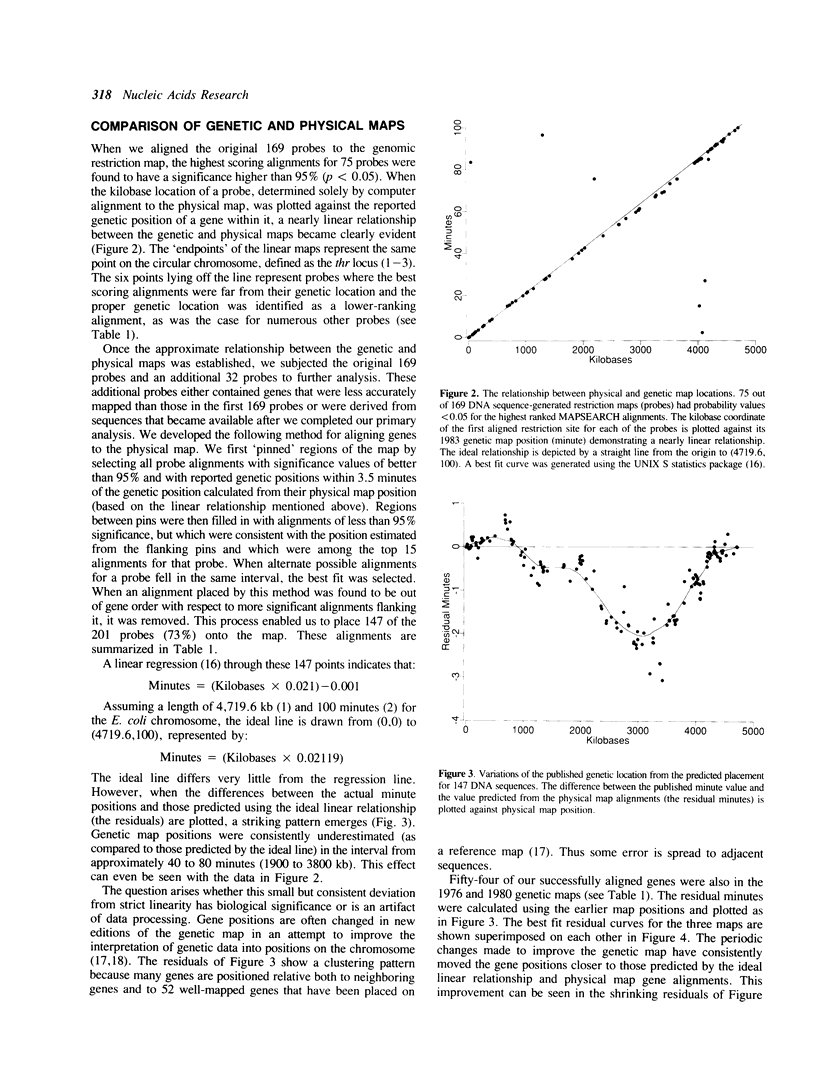
We use the extensive published information describing the genome of Escherichia coli and new restriction map alignment software to align DNA sequence, genetic, and physical maps. Restriction map alignment software is used which considers restriction maps as strings analogous to DNA or protein sequences except that two values, enzyme name and DNA base address, are associated with each position on the string. The resulting alignments reveal a nearly linear relationship between the physical and genetic maps of the E. coli chromosome. Physical map comparisons with the 1976, 1980, and 1983 genetic maps demonstrate a better fit with the more recent maps. The results of these alignments are genomic kilobase coordinates, orientation and rank of the alignment that best fits the genetic data. A statistical measure based on extreme value distribution is applied to the alignments. Additional computer analyses allow us to estimate the accuracy of the published E. coli genomic restriction map, simulate rearrangements of the bacterial chromosome, and search for repetitive DNA. The procedures we used are general enough to be applicable to other genome mapping projects.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Erickson B. W. A nonlinear measure of subalignment similarity and its significance levels. Bull Math Biol. 1986;48(5-6):617–632. doi: 10.1007/BF02462327. [DOI] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 7. Microbiol Rev. 1983 Jun;47(2):180–230. doi: 10.1128/mr.47.2.180-230.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J., Low K. B., Taylor A. L. Recalibrated linkage map of Escherichia coli K-12. Bacteriol Rev. 1976 Mar;40(1):116–167. doi: 10.1128/br.40.1.116-167.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Kieffer-Higgins S. Multiplex DNA sequencing. Science. 1988 Apr 8;240(4849):185–188. doi: 10.1126/science.3353714. [DOI] [PubMed] [Google Scholar]
- Ellwood M., Nomura M. Chromosomal locations of the genes for rRNA in Escherichia coli K-12. J Bacteriol. 1982 Feb;149(2):458–468. doi: 10.1128/jb.149.2.458-468.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C. W., Harnish B. W. Inversions between ribosomal RNA genes of Escherichia coli. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7069–7072. doi: 10.1073/pnas.78.11.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kröger M. Compilation of DNA sequences of Escherichia coli. Nucleic Acids Res. 1989;17 (Suppl):r283–r309. doi: 10.1093/nar/17.suppl.r283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N., Gielow W., Martin R., Hamilton E., Fowler A. The organization of the araBAD operon of Escherichia coli. Gene. 1986;47(2-3):231–244. doi: 10.1016/0378-1119(86)90067-3. [DOI] [PubMed] [Google Scholar]
- Lin H. C., Lei S. P., Studnicka G., Wilcox G. The araBAD operon of Salmonella typhimurium LT2. III. Nucleotide sequence of araD and its flanking regions, and primary structure of its product, L-ribulose-5-phosphate 4-epimerase. Gene. 1985;34(1):129–134. doi: 10.1016/0378-1119(85)90303-8. [DOI] [PubMed] [Google Scholar]
- Lin H. C., Lei S. P., Wilcox G. The araBAD operon of Salmonella typhimurium LT2. I. Nucleotide sequence of araB and primary structure of its product, ribulokinase. Gene. 1985;34(1):111–122. doi: 10.1016/0378-1119(85)90301-4. [DOI] [PubMed] [Google Scholar]
- Lin H. C., Lei S. P., Wilcox G. The araBAD operon of Salmonella typhimurium LT2. II. Nucleotide sequence of araA and primary structure of its product, L-arabinose isomerase. Gene. 1985;34(1):123–128. doi: 10.1016/0378-1119(85)90302-6. [DOI] [PubMed] [Google Scholar]
- Lin R. J., Capage M., Hill C. W. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol. 1984 Jul 25;177(1):1–18. doi: 10.1016/0022-2836(84)90054-8. [DOI] [PubMed] [Google Scholar]
- Muramatsu S., Kato M., Kohara Y., Mizuno T. Insertion sequence IS5 contains a sharply curved DNA structure at its terminus. Mol Gen Genet. 1988 Nov;214(3):433–438. doi: 10.1007/BF00330477. [DOI] [PubMed] [Google Scholar]
- Neumaier P. S. A program package applicable to the detection of overlaps between restriction maps. Nucleic Acids Res. 1986 Jan 10;14(1):351–362. doi: 10.1093/nar/14.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadosky A. B., Davidson A., Lin R. J., Hill C. W. rhs gene family of Escherichia coli K-12. J Bacteriol. 1989 Feb;171(2):636–642. doi: 10.1128/jb.171.2.636-642.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston J., Mallett F., Staden R., Durbin R., Horsnell T., Coulson A. Software for genome mapping by fingerprinting techniques. Comput Appl Biosci. 1988 Mar;4(1):125–132. doi: 10.1093/bioinformatics/4.1.125. [DOI] [PubMed] [Google Scholar]
- Tabata S., Higashitani A., Takanami M., Akiyama K., Kohara Y., Nishimura Y., Nishimura A., Yasuda S., Hirota Y. Construction of an ordered cosmid collection of the Escherichia coli K-12 W3110 chromosome. J Bacteriol. 1989 Feb;171(2):1214–1218. doi: 10.1128/jb.171.2.1214-1218.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman M. S., Smith T. F., Katcher H. L. Algorithms for restriction map comparisons. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):237–242. doi: 10.1093/nar/12.1part1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams K. M. Version 5 of the Mount-Conrad-Myers Sequence Analysis Software Package now available. Comput Appl Biosci. 1988 Mar;4(1):211–211. doi: 10.1093/bioinformatics/4.1.211. [DOI] [PubMed] [Google Scholar]
- Zehetner G., Lehrach H. A computer program package for restriction map analysis and manipulation. Nucleic Acids Res. 1986 Jan 10;14(1):335–349. doi: 10.1093/nar/14.1.335. [DOI] [PMC free article] [PubMed] [Google Scholar]



