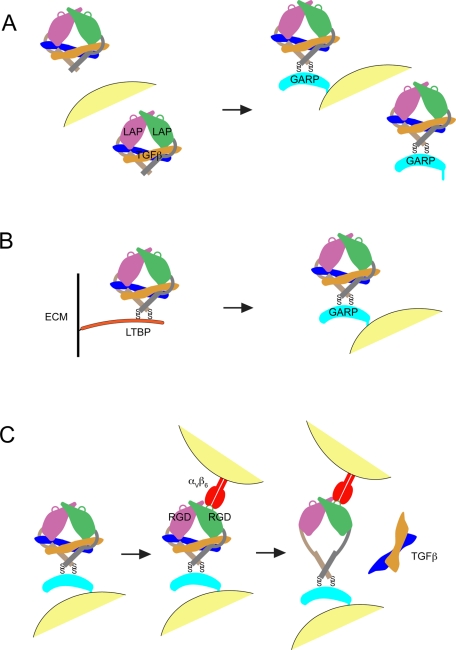
GARP disulfide links to latent TGFβ on the cell surface, which serves as a reservoir for TGFβ activation by αVβ6 and to a lesser extent αVβ8. Activation requires the RGD motif of latent TGFβ, disulfide linkage between GARP and latent TGFβ, and membrane association of GARP.
Abstract
Glycoprotein-A repetitions predominant protein (GARP) associates with latent transforming growth factor-β (proTGFβ) on the surface of T regulatory cells and platelets; however, whether GARP functions in latent TGFβ activation and the structural basis of coassociation remain unknown. We find that Cys-192 and Cys-331 of GARP disulfide link to the TGFβ1 prodomain and that GARP with C192A and C331A mutations can also noncovalently associate with proTGFβ1. Noncovalent association is sufficiently strong for GARP to outcompete latent TGFβ-binding protein for binding to proTGFβ1. Association between GARP and proTGFβ1 prevents the secretion of TGFβ1. Integrin αVβ6 and to a lesser extent αVβ8 are able to activate TGFβ from the GARP–proTGFβ1 complex. Activation requires the RGD motif of latent TGFβ, disulfide linkage between GARP and latent TGFβ, and membrane association of GARP. Our results show that GARP is a latent TGFβ-binding protein that functions in regulating the bioavailability and activation of TGFβ.
INTRODUCTION
Transforming growth factor-β (TGFβ) is a pleiotropic cytokine with potent immunoregulatory properties, which manifests in TGFβ1-knockout mice as multifocal inflammatory disorders and death within 4 wk of birth (Shull et al., 1992; Kulkarni et al., 1993). TGFβ1 is produced by every leukocyte lineage and controls the differentiation, proliferation, and other functions of immune cells (Li et al., 2006; Yoshimura et al., 2010). For example, TGFβ is involved in the generation and function of T regulatory cells (Treg) and T helper 17 cells (Th17; Nakamura et al., 2004; Veldhoen et al., 2006). TGFβ also induces the expression of αEβ7 integrin in intraepithelial lymphocytes (Kilshaw and Murant, 1991; Cepek et al., 1993). In addition, TGFβ regulates immunoglobulin A isotype expression in B cells (Coffman et al., 1989).
TGFβ1, 2, and 3 are synthesized as precursor polypeptides (pro+TGFβ), which dimerize and are proteolytically cleaved by furin prior to secretion to yield pro-TGFβ. (Here we use pro-TGFβ for furin-cleaved latent TGFβ, pro+TGFβ for uncleaved latent TGFβ, and proTGFβ to refer to the cDNA or a mixture of pro-TGFβ and pro+TGFβ protein products.) Pro-TGFβ contains a ∼250-residue prodomain known as latency-associated peptide (LAP) and a ∼110-residue mature TGFβ growth factor domain. The prodomain remains noncovalently associated with TGFβ after secretion, thereby conferring latency (Gentry et al., 1987; Wakefield et al., 1988; Khalil, 1999). Latent TGFβ does not have biological activity, and the release of TGFβ from LAP therefore is a critical regulatory step for TGFβ function and signaling. The LAPs of TGFβ1, 2, and 3 are denoted LAP1, 2, and 3, respectively.
LAP1 and LAP3 contain an RGD motif, which is recognized by some αV integrins (Rifkin, 2005). αVβ6 and αVβ8 activate TGFβ through binding to the RGD motif; mice lacking both αVβ6 and αVβ8 integrins recapitulate all major phenotypes of TGFβ1 and β3 double-deficient mice (Aluwihare et al., 2009), demonstrating the critical roles of αVβ6 and αVβ8 in TGFβ1 and β3 activation. Furthermore, knock-in mice with the RGD motif of TGFβ1 mutated to RGE phenotypically resemble mice with complete deficiency of TGFβ1 (Yang et al., 2007). Therefore, although multiple mechanisms that include thrombospondin and metalloproteases have been implicated in activation of TGFβ1 and TGFβ3, recognition by αV integrins of the RGD motif has a central role in activation in vivo.
The latent TGFβ-binding proteins (LTBPs) are important in the biosynthesis, storage, and activation of TGFβ (Rifkin, 2005). Association with and disulfide linkage to LTBP targets proTGFβ to the extracellular matrix (ECM; Miyazono et al., 1991). There are four different LTBPs, and at least three of them bind to proTGFβ. LTBPs are large proteins related to fibrillins, which have a large number of calcium-binding epidermal growth factor–like domains and a smaller number of TGFβ-binding (TB) domains. One of these TB domains specifically associates with proTGFβ, and two cysteines in the TB domain disulfide link to Cys-4 in each of the prodomains, yielding an unusual 1:2 LTBP:proTGFβ-monomer stoichiometry. Other domains in LTBPs cause them to coassemble with fibrillins in elastic fibrils in the ECM, where latent TGFβ is stored until activation (Rifkin, 2005). Association with the ECM and the β6 cytoplasmic domain is required for latent TGFβ activation by αVβ6, and it has been suggested that tensile force exerted across the complex by the actin cytoskeleton is also required for activation by αVβ6 (Annes et al., 2004; Wipff et al., 2007; Wipff and Hinz, 2008).
The structure of latent TGFβ is ring-like. The two prodomains form two arms, which are disulfide linked in a bowtie at a neck and have RGD motifs in their shoulders. The growth factor monomers locate to the forearms. They are surrounded by a prodomain straightjacket element that includes an α1-helix, a latency lasso, and a clasp between the arm domain and the α1-helix. The Cys-4 residues in the α1-helix that link to LTBP, and RGD motifs that bind to integrins, locate to opposite sides of the ring, so that tensile force exerted across them would elongate the α1-helix and latency lasso and release TGFβ. The structure is incompatible with binding of either type I or type II receptor to TGFβ in its latent form (Shi et al., 2011).
Recently glycoprotein-A repetitions predominant protein (GARP, also known as LRRC32) was shown to associate with LAP (Stockis et al., 2009; Tran et al., 2009). However, whether GARP functions analogously to LTBP in TGFβ activation is unknown, and there are numerous distinctions between these proteins. First, the four LTBP isoforms are broadly expressed in a variety of cell types (Rifkin, 2005), whereas GARP expression has only been detected in activated (FoxP3+) Tregs and platelets (Macaulay et al., 2007; Wang et al., 2008). Second, LTBP targets proTGFβ into the ECM, whereas GARP has a transmembrane domain and associates with LAP on the cell surface (Stockis et al., 2009; Tran et al., 2009). Finally, the LAP-binding motif in LTBP is a TB domain (Rifkin, 2005), whereas the extracellular domain of GARP is composed of leucine-rich repeats (LRRs) and it has no TB domain (Ollendorff et al., 1994). The role of LTBP in TGFβ assembly and activation is well established; association of proTGFβ with LTBP and incorporation of LTBP into the ECM are required for activation (Rifkin, 2005). Association between GARP and LAP has been shown by immunoprecipitation (IP) followed by Western blotting, and binding of GARP-Fc to proTGFβ was shown by fluorescence (Tran et al., 2009); however, whether they are covalently linked by disulfide bonds is unknown. Small interfering RNA to GARP has been shown to decrease surface expression of LAP and to moderately decrease Treg-mediated suppression in vitro (Tran et al., 2009). However, whether a proTGFβ complex with GARP can provide a cell-surface reservoir of latent TGFβ for αV integrin–dependent activation and how GARP coexpression affects secretion and bioavailability of TGFβ remain unknown. Here we address gaps in understanding of the role of GARP in TGFβ function. Our findings support the idea that GARP is a new latent TGFβ-binding protein that regulates the bioavailability of TGFβ and provides a cell surface platform for αV integrin–dependent TGFβ activation.
RESULTS
GARP associates with proTGFβ
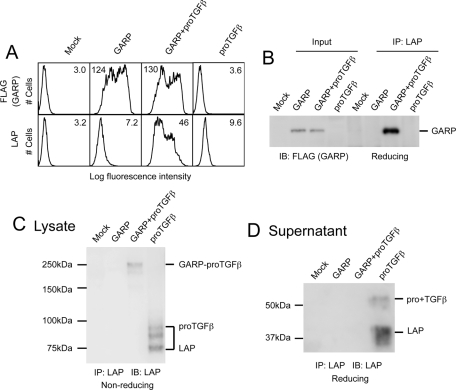
To study their interaction, we transiently expressed GARP and proTGFβ1 in 293T cells. Consistent with previous findings (Stockis et al., 2009; Tran et al., 2009), the expression level of LAP on the cell surface was greatly elevated in the presence of GARP (Figure 1A, bottom). In addition, GARP and LAP coimmunoprecipitated in cotransfected cells (Figure 1B). An ∼250-kDa species representing the GARP–proTGFβ1 complex was detected in the cotransfected cells on a 7.5% nonreduced SDS–PAGE gel (Figure 1C), indicating that GARP forms a disulfide linkage with proTGFβ1. ProTGFβ1 and LAP secretion was detected in the supernatant of cells transfected with proTGFβ1 alone but not in the supernatant of cells cotransfected with GARP and proTGFβ1 (Figure 1D), suggesting that GARP blocks direct secretion of pro+TGFβ1 and pro-TGFβ1.
FIGURE 1:
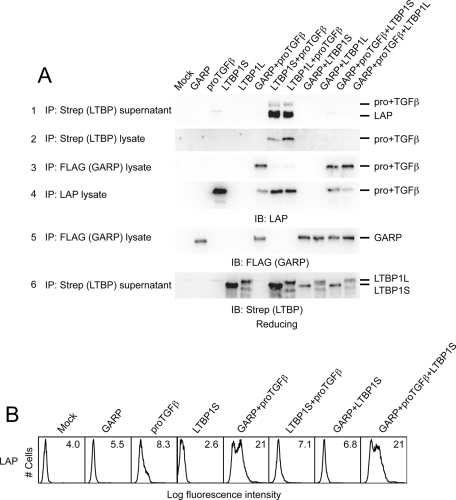
GARP regulates the secretion of TGFβ1 by forming a complex with proTGFβ1 on the cell surface. (A) LAP is coexpressed with GARP on the cell surface. 293T cells were transfected with mock, FLAG-tagged GARP, FLAG-tagged GARP + proTGFβ1, or proTGFβ1. Surface expression of FLAG-GARP and LAP1 was measured using FACS. Numbers in each histogram show the mean fluorescence intensity. GARP markedly increased LAP1 expression on the cell surface. (B) LAP is associated with GARP. 293T cells were transfected with the indicated plasmids. The clarified lysates were immunoprecipitated with anti-LAP1 antibody. The resulting samples were subjected to Western blot analysis using an anti-FLAG antibody. One-fifteenth of the cell lysates used in IP were loaded as input. (C) GARP disulfide links to proTGFβ1. 293T cells were transfected with the indicated plasmids. The cell lysates were immunoprecipitated with the anti-LAP1 antibody, analyzed by 7.5% nonreduced SDS–PAGE, and Western blotted with a different LAP1 antibody. A 250-kDa band representing the GARP–proTGFβ1 complex was detected in the lysate prepared from GARP- and proTGFβ1- cotransfected cells. (D) Association of GARP and proTGFβ1 prevents the direct secretion of proTGFβ1 into the supernatant. 293T cells were transfected with the indicated plasmids. The supernatants were immunoprecipitated with the anti-LAP1 antibody and analyzed by 10% reducing SDS–PAGE.
Cys-192 and Cys-331 of GARP disulfide link to Cys-4 of proTGFβ1
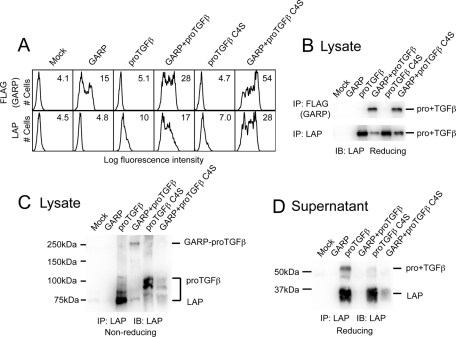
Our findings suggested that GARP disulfide links with proTGFβ1. Cys-4 in each proTGFβ1 disulfide links to LTBP, and the proTGFβ1 C4S mutant is unable to bind to LTBP (Saharinen et al., 1996). In contrast, we found that GARP was able to noncovalently associate with the proTGFβ1 C4S mutant. The C4S mutant increased LAP expression in cotransfectants similar to wild-type (WT; Figure 2A). Furthermore, both WT and C4S pro+TGFβ1 associated with GARP, as shown by coIP (Figure 2B). However, WT proTGFβ1 formed an ∼250-kDa complex with GARP in nonreducing SDS–PAGE, whereas C4S proTGFβ1 failed to do so (Figure 2C). GARP greatly attenuated the amount of secreted proTGFβ1 and LAP both for WT and the C4S mutant (Figure 2D). However, GARP essentially completely prevented secretion of WT proTGFβ1, whereas there was some leakage of C4S mutant proTGFβ1 (Figure 2D). Thus covalent linkage is important for complete association. Formation of disulfide bonds is aberrant in proTGFβ1 when Cys-4 is present in the absence of LTBP (Brunner et al., 1989), and this may account for the difference in size of products in the 100- to 75-kDa range between WT and C4S proTGFβ1 in nonreducing SDS–PAGE (Figure 2C). Taken together, our findings suggested that GARP disulfide links to the Cys-4 of proTGFβ1 and that GARP also associates relatively stably with proTGFβ1 through noncovalent interactions.
FIGURE 2:
Cys-4 of TGFβ1 disulfide links to GARP. (A) ProTGFβ1 C4S mutant is coexpressed with GARP on the cell surface. 293T cells were transfected with the indicated plasmids, and the surface FLAG-GARP and LAP1 expressions were measured by FACS. (B) ProTGFβ1 C4S mutant associates with GARP. 293T cells were transfected with the indicated plasmids. The cell lysates were immunoprecipitated with anti-FLAG or anti-LAP1 antibody, subjected to reducing SDS–10% PAGE, and blotted with a different anti-LAP1 antibody. (C) GARP disulfide links to Cys-4 of proTGFβ1. 293T cells were transfected with the indicated plasmids. The clarified lysates were immunoprecipitated with anti-LAP1 antibody, subjected to reducing SDS–7.5% PAGE, and blotted with a different anti-LAP1 antibody. (D) C4S mutation reduces the stability of the GARP–proTGFβ1 complex. 293T cells were transfected with the indicated plasmids. The supernatants were immunoprecipitated with anti-LAP1 antibody, subjected to reducing SDS–10% PAGE, and blotted with a different anti-LAP1 antibody.
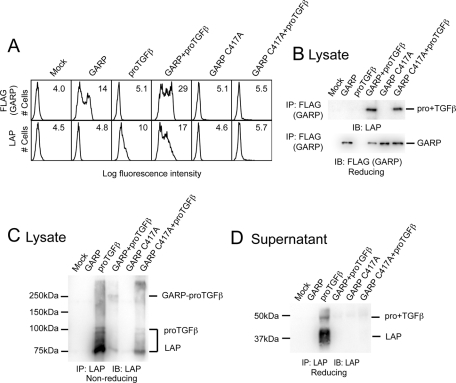
There are 15 cysteines in the extracellular domain of GARP. In choosing candidates for linkage to proTGFβ1, we excluded cysteines that are not conserved across species from fish to mammals or that aligned with cysteines in the N-cap or C-cap regions of structurally characterized LRR proteins that are known to form intra- chain disulfide bonds. This left three candidate cysteines—Cys-192, Cys-331, and Cys-417—which were tested by mutation to alanine. The C417A mutation abolished surface expression of GARP (Figure 3A). However, C417A GARP associated with proTGFβ1 inside the cell, as shown by IP of cell lysates (Figure 3B). The C417A mutant diminished amounts of free proTGFβ1 and LAP in cell lysates (Figure 3C) and completely prevented secretion of proTGFβ1 and LAP (Figure 3D). Thus these results suggest that a GARP mutant that is too aberrant to be expressed on the cell surface nonetheless can associate with proTGFβ1 and prevent its cell surface expression and secretion.
FIGURE 3:
C417A mutation abolishes the surface expression of GARP. (A) 293T cells were transfected with the indicated plasmids. Surface FLAG-GARP and LAP1 expression were measured by FACS. (B, C) The GARP C417A mutant associates with proTGFβ1. 293T cells were transiently transfected with the indicated plasmids. The clarified lysates were immunoprecipitated with the indicated antibodies, subjected to reducing SDS–10% PAGE (B) or nonreducing SDS–7.5% PAGE (C), and blotted with an anti-LAP1 antibody. (D) The GARP C417A mutant prevents secretion of proTGFβ1. 293T cells were transfected with the indicated plasmids. The supernatants were immunoprecipitated with an anti-LAP1 antibody, subjected to reducing SDS–10% PAGE, and blotted with a different anti-LAP1 antibody.
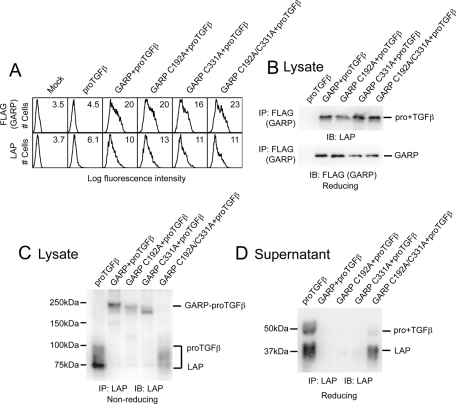
Cys-192 and Cys-331 were found to be responsible for the disulfide linkage with proTGFβ1. The GARP C192A, C331A, and C192A/C331A double mutants were expressed at similar levels on the cell surface, and the mutants were able to support surface LAP expression (Figure 4A). In addition, all the GARP mutants were able to noncovalently associate with proTGFβ1 (Figure 4B). However, the C192A/C331A double mutant was unable to form the disulfide-linked complex with proTGFβ1 seen in nonreducing SDS–PAGE (Figure 4C). The complex formed by proTGFβ1 and C192A or C331A single mutants migrated slightly differently than the complex formed by WT GARP (Figure 4C). These differences in migration are expected based on the difference in topology between LAP dimers linked through two disulfides to GARP or through one disulfide at different positions on the GARP polypeptide. Either one of the two GARP cysteines, Cys-192 or Cys-331, was sufficient to prevent secretion of proTGFβ1 and LAP (Figure 4D). Lack of both GARP cysteines resulted in proTGFβ1 and LAP secretion; however, secretion was less than in absence of GARP (Figure 4D), consistent with noncovalent association between C192A/C331A GARP and proTGFβ1. These results were similar to those seen with GARP and the proTGFβ1 C4S mutant (Figure 2D). We conclude that GARP uses Cys-192 and Cys-331 to disulfide link to the two Cys-4's of proTGFβ1.
FIGURE 4:
Cys-192 and Cys-331 of GARP disulfide link to proTGFβ1. (A) 293T cells were transfected with the indicated plasmids, and the surface FLAG-GARP and LAP1 expressions were measured by FACS. (B) The mutated GARPs associate with proTGFβ1. The cell lysates were immunoprecipitated with anti-FLAG antibody, subjected to reducing SDS–10% PAGE, and blotted with the indicated antibodies. (C) Cys-192 and Cys-331 of GARP disulfide link to proTGFβ1. 293T cells were transfected with the indicated plasmids. The clarified lysates were immunoprecipitated with anti-LAP1 antibody, subjected to reducing SDS–7.5% PAGE, and blotted with a different anti-LAP1 antibody. (D) C192A/C331A double mutation in GARP reduces the stability of the GARP–proTGFβ1 complex. 293T cells were transfected with the indicated plasmids. The supernatants were immunoprecipitated with an anti-LAP1 antibody, subjected to reducing SDS–10% PAGE, and blotted with a different anti-LAP1 antibody.
A point mutation in GARP, R395W, has been associated through genetic linkage with Usher syndrome type 1, an autosomal recessive disease characterized by profound congenital sensorineural deafness, vestibular dysfunction, and progressive visual loss (Bonne-Tamir et al., 1997). However, the R395W mutation does not affect noncovalent association with proTGFβ1, as shown with proTGFβ1 C4S mutant (Supplemental Figure S1).
GARP outcompetes LTBP1 for proTGFβ binding
Both GARP and LTBP disulfide link to Cys-4 of proTGFβ1. To investigate whether GARP and LTBP compete for proTGFβ1 binding, we performed IP experiments using cell lysates or supernatants from cells transfected with proTGFβ1, GARP, and/or short or long alternatively spliced isoforms of LTBP1 (LTBP1S and LTBP1L, respectively; Figure 5A). LTBP1S and LTBP1L complexed with proTGFβ1 were found in both the cell supernatant (Figure 5A, row 1) and lysate (Figure 5A, row 2), in contrast to the GARP complex, which was present only in lysates (Figure 5A, row 3) and not in supernatant, as shown earlier (Figure 1D).
FIGURE 5:
GARP outcompetes LTBP for proTGFβ binding. (A) 293T cells were transfected with the indicated plasmids. The clarified lysates and supernatants were immunoprecipitated with the indicated antibodies and blotted with the indicated antibodies. (B) 293T cells were transfected with the indicated plasmids. Surface LAP1 expression was measured by FACS.
Of interest, GARP outcompeted both LTBP1S and LTBP1L for proTGFβ1. When cells were cotransfected with GARP and either LTBP1S or LTBP1L, proTGFβ1 was found only in association with GARP (Figure 5A, row 3) and not with LTBP (Figure 5A, rows 1 and 2). Moreover, LAP was found on the cell surface only when GARP was present but not when LTBP1S was present; LTBP1S did not diminish GARP-dependent LAP surface expression (Figure 5B). Furthermore, the GARP C192A/C331A double mutant also outcompeted LTBP1 for proTGFβ1 binding (unpublished data), suggesting that the noncovalent association between GARP and proTGFβ1 is sufficient for GARP to outcompete LTBP.
TGFβ can be activated from the GARP–proTGFβ complex by integrins
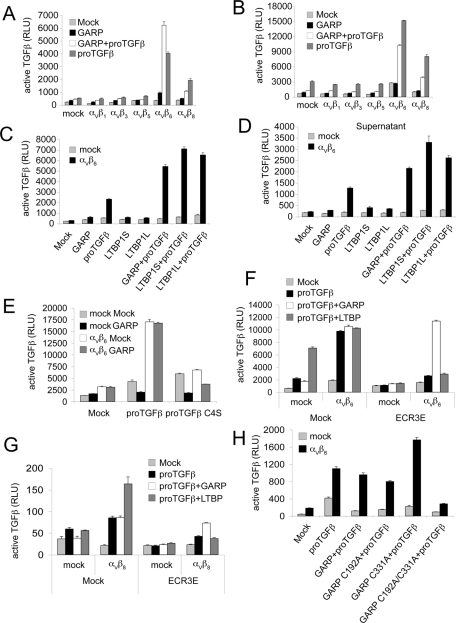
We next studied whether the GARP–proTGFβ complex could serve as a source of activated TGFβ. Several αV integrins were shown to activate TGFβ in different settings (Munger et al., 1999; Mu et al., 2002; Ludbrook et al., 2003; Wipff et al., 2007). Stable transfectants of 293 cells expressing αV and each of the five β subunits known to associate with αV (Supplemental Figure S2A) were further transfected with GARP and proTGFβ1 and cocultured with the transformed mink lung TGFβ-reporter cell line (TMLC; Abe et al., 1994). αVβ6 strongly activated TGFβ from GARP- and proTGFβ1-cotransfected cells (Figure 6A). αVβ8 also activated TGFβ, but to a lesser extent. In contrast, αVβ1, αVβ3, and αVβ5 transfectants showed no more TGFβ activation than did mock transfectants (Figure 6A). Similar results were obtained when αV integrins and the GARP–proTGFβ1 complex were expressed on different cells, demonstrating transactivation (Figure 6B).
FIGURE 6:
Integrins αVβ6 and αVβ8 can activate TGFβ from the GARP–pro-TGFβ1 complex. (A) Mock or different αV integrin-expressing cells were transfected with the indicated plasmids and cocultured with TMLC to measure active TGFβ production. Data represent mean + SEM of triplicate samples. (B) 293T cells were transfected with indicated plasmids and cocultured with mock or αV integrin-expressing 293 cells, as well as the TMLC reporter cell line. (C, D) GARP and LTBP1 support αVβ6-mediated TGFβ activation at comparable levels. Mock or αVβ6-expressing cells were transfected with indicated plasmids. Cells (C) or the supernatants 24 h posttransfection (D) were cocultured with TMLC to assess active TGFβ production. (E, H) αVβ6 is unable to activate TGFβ from either the GARP–pro-TGFβ1 C4S complex (E) or the GARP C192A/C331A–pro-TGFβ1 complex (H). Mock or αVβ6-expressing cells were transfected with indicated plasmids and were cocultured with TMLC to assess active TGFβ production. (F, G). The ECR3E fragment does not interfere with αVβ6- or αVβ8-mediated TGFβ activation from the GARP–pro-TGFβ1 complex. Mock or αVβ6- or αVβ8-expressing cells were transfected with the indicated plasmids. The transfected cells were cocultured with TMLC to measure active TGFβ production.
GARP and LTBP1 supported αVβ6-mediated TGFβ activation at comparable levels (Figures 6, C and D). αVβ6 also activated TGFβ from cells transfected only with proTGFβ1 (Figures 6, A–C). This may be due to endogenous LTBP expression in 293 cells, since this activation was greatly reduced in proTGFβ1 C4S-transfected cells or in the presence of LTBP1 ECR3E fragment, as previously reported (Annes et al., 2004; Figure 6, F and G).
An αVβ6-dependent release of TGFβ into culture supernatants was also seen. Activation of latent TGFβ associated with endogenous LTBP is consistent with the presence of TGFβ activity in supernatants of cells transfected with proTGFβ1 (Figure 6D). TGFβ activity in supernatants was also seen with cells cotransfected with GARP and proTGFβ1 (Figure 6D). In all cases, release of TGFβ into supernatants was αVβ6 dependent (Figure 6D).
The ECR3E fragment contains the LAP-binding TB domain of LTBP, and the ECR3E fragment has been shown to compete with LTBP1 for proTGFβ1, thereby inhibiting TGFβ activation by αVβ6 (Annes et al., 2004; Figure 6F). However, the ECR3E fragment had little effect on αVβ6-mediated activation of the GARP–pro-TGFβ1 complex (Figure 6F). Similar results were obtained with αVβ8-mediated TGFβ activation (Figure 6G). This finding is consistent with our IP experiments showing that GARP interacted with proTGFβ1 in the presence of the ECR3E fragment (Supplemental Figure S2B). These results further confirmed our conclusion that GARP outcompetes LTBP for proTGFβ1 binding.
The αVβ6–mediated TGFβ activation from the GARP–pro-TGFβ complex requires the disulfide linkage between GARP and proTGFβ, the RGD motif in LAP, and membrane association of GARP
The C4S mutation in proTGFβ1 greatly reduced TGFβ activation from the GARP–pro-TGFβ1 complex (Figure 6E). The GARP C192A or GARP C331A single mutants, which supported disulfide linkage to proTGFβ1, each enabled αVβ6-mediated TGFβ activation (Figure 6H). In contrast, the C192A/C331A double mutant, which did not support disulfide linkage to GARP, failed to activate TGFβ (Figure 6H). These results demonstrated that the disulfide linkage between GARP and proTGFβ1 is critical for αVβ6-mediated TGFβ activation.
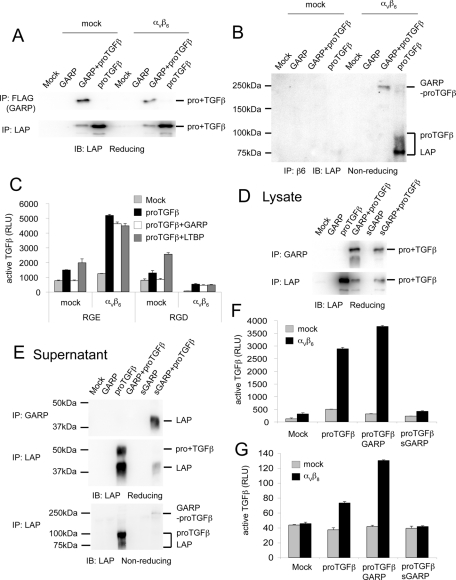
To exclude the possibility that αVβ6 interferes with the interaction between GARP and proTGFβ1, we performed IP experiments to examine the association between GARP and proTGFβ1 in the presence of αVβ6. GARP interacted with proTGFβ1 in αVβ6-expressing cells (Figure 7A). Furthermore, αVβ6, GARP, and proTGFβ1 formed a complex in cotransfected cells (Figure 7B). Therefore αVβ6 did not interfere with the interaction between GARP and proTGFβ1.
FIGURE 7:
The αVβ6-mediated TGFβ activation from the GARP–proTGFβ complex requires the RGD motif in LAP and membrane association of GARP. (A, B) αVβ6 does not interfere with the interaction between GARP and proTGFβ1. Mock or αVβ6-expressing cells were transfected with the indicated plasmids. The clarified lysates were immunoprecipitated with the indicated antibodies, subjected to reducing SDS–10% PAGE (A) or nonreducing SDS–7.5% PAGE (B), and blotted with an anti-LAP1 antibody. (C) An RGD peptide interferes with αVβ6-mediated TGFβ activation. Mock or αVβ6-expressing cells were transfected with the indicated plasmids and cocultured with TMLC in the presence of 0.5 mM RGE (GRGESP; control peptide) or RGD peptide (GRGDSP). (D, E) sGARP interacts with proTGFβ1. The extracellular domain of GARP was fused to a His-SBP tag to generate the soluble GARP construct. 293T cells were transfected with the indicated plasmids. The clarified lysates (D) and supernatants (E) were immunoprecipitated with our in-house GARP antibody (GARP2) or anti-LAP1 antibody and blotted with a different anti-LAP1 antibody. (F, G) Membrane association is required for GARP to support αVβ6- or αVβ8-mediated TGFβ activation. Cells were transfected with the indicated plasmids and cocultured with TMLC to assess active TGFβ production.
The αVβ6 binding to and activation of latent TGFβ depends on the RGD motif in the prodomain (Munger et al., 1999). Inhibition by RGD peptide, and not RGE peptide, demonstrated RGD dependence of activation of the GARP–pro-TGFβ1 complex and confirmed RGD dependence of activation of the LTBP1–pro-TGFβ1 complex (Figure 7C).
To test requirement of membrane anchoring for activation, the transmembrane and cytoplasmic domains of GARP were deleted. Soluble GARP (sGARP) associated with proTGFβ1 and was secreted as a complex (Figure 7, D and E); however, it was unable to support αVβ6- or αVβ8-mediated TGFβ activation (Figure 7, F and G).
Electron microscopy of complexes with GARP, proTGFβ, and integrin αVβ6
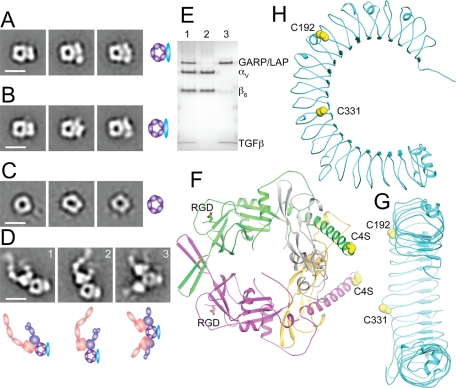
The noncovalently associated proTGFβ C4S mutant complex with sGARP was stable to gel filtration and was subjected to negative-stain electron microscopy (EM) with particle alignment and class averaging (Figure 8A). The covalent proTGFβ complex with sGARP was similarly subjected to EM (Figure 8B). ProTGFβ is ring-like, as previously described (Shi et al., 2011; Figure 8C). The noncovalent and covalent proTGFβ complexes with GARP are very similar and show an elongated and more or less linear or slightly curved density for GARP that is associated with the periphery of the proTGFβ ring (Figure 8, A and B).
FIGURE 8:
GARP–proTGFβ and GARP–proTGFβ–αVβ6 protein complexes. (A–D) Negative-stain EM of sGARP–proTGFβ1 C4S (A), sGARP–proTGFβ1 (B), proTGFβ1 for comparison (Shi et al., 2011; C), and αVβ6 integrin complex with sGARP–proTGFβ1 (D). Scale bars, 10 nm. Schematic representations are shown to the right (A–C) or below (D). (E) Nonreducing SDS–4–15% gradient PAGE of the αVβ6 complex with sGARP–proTGFβ1 from gel filtration (lane 1), αVβ6 alone (lane 2), and sGARP–proTGFβ1 alone (lane 3) stained with Coomassie blue. (F) Architecture of proTGFβ1 (Shi et al., 2011). (G, H) Architecture of the GARP homology model, shown at the same scale as proTGFβ and in an appropriate orientation for disulfide linkage to proTGFβ (G), and so the horseshoe is in the plane of the page (H). Cartoon representations, with relevant Cys side chains shown in orange spheres in (F–H), were made with PyMOL.
To better appreciate the mode of association shown by EM, we made a homology model of GARP (Figure 8, G and H). LRR are horseshoe-shaped proteins, as shown for GARP using cryo-EM (Probst-Kepper et al., 2009). Each LRR makes one complete turn around the horseshoe. The cysteines forming the intermolecular disulfides, Cys-192 and Cys-331, locate to one side of the horseshoe, between the concave and convex faces, and near the middle of the horseshoe (Figure 8, G and H). Placing Cys-192 and Cys-331 on the flat side of the GARP model in close opposition to Cys-4 on the outer edge of the proTGFβ ring (Figure 8, F and G) recreates the orientation seen in EM (Figure 8, A and B). Furthermore, the two Cys-4 residues in proTGFβ1 are 40 Å apart (Shi et al., 2011), an appropriate spacing for binding to Cys-192 and Cys-331, which are 35 Å apart from each other in the GARP homology model (Figure 8, G and H).
Complexes between the ectodomain of integrin αVβ6 and sGARP–proTGFβ were isolated by gel filtration and subjected to EM (Figure 8D). The two RGD motifs to which integrins bind reside on the shoulders of proTGFβ1, on the opposite side of the ring from Cys-4 (Figure 8F). Representative class averages showed either one (Figure 8C, 1 and 2) or two (Figure 8C, 3) αVβ6 ectodomains bound per proTGFβ1; αVβ6 bound with its lower legs extended and its headpiece open, that is, in the high-affinity conformation. The proTGFβ1-binding site in αVβ6 was at the interface between large and small densities, corresponding to the αV β-propeller domain and β6 βI domain, respectively. This is the crystallographically determined binding site for RGD ligands in αVβ3 and αIIbβ3 (Xiong et al., 2002; Xiao et al., 2004). The negative-stain EM class averages clearly demonstrated the relationship between the GARP- and αVβ6-binding sites on the proTGFβ1 ring in ternary complexes (Figure 8D). sGARP and αVβ6 bound to opposite sides of the ring of proTGFβ1. The spatial relationships on the periphery of the proTGFβ1 ring for integrin binding and GARP binding are as predicted from the positions of the RGD motifs and Cys-4 in the proTGFβ1 crystal structure (Figure 8F). The ring-like structure of proTGFβ was similar in the absence and presence of αVβ6 (Figure 8, A, B, and D). Furthermore, SDS–PAGE of the same gel filtration fraction as subjected to EM of the αVβ6 complex with sGARP–proTGFβ showed the presence of TGFβ in the complex (Figure 8E, lane 1), suggesting that binding of αVβ6 was not sufficient to induce release of TGFβ.
DISCUSSION
The pivotal role of TGFβ in immune regulation emphasizes the need for a better understanding of the mechanisms for TGFβ storage and activation. In the present study, we characterized the structural basis and functional significance of the interaction between GARP and TGFβ and defined a critical role for GARP in regulating bioavailability of TGFβ.
Previous studies demonstrated coassociation of GARP and proTGFβ (Stockis et al., 2009; Tran et al., 2009), and yet the structural basis for this interaction was not clear. Here we present the first demonstration that GARP disulfide links with proTGFβ1 and that noncovalent bonds are also sufficient for association. The disulfide interaction was mediated by Cys-192 and Cys-331 of GARP and Cys-4 of proTGFβ1, suggesting that one GARP protein associates with one proTGFβ1 dimer. Such a complex has an estimated polypeptide molecular mass of 153,200 Da; with 11 N-linked sites at 2500 Da each, the estimated mass is 180,700 Da, close to the mass measured by multiangle light scattering of 176,000 ± 3500 Da. Although the disulfide linkage is not required for GARP–proTGFβ association, the noncovalent interaction between GARP and proTGFβ1 alone could not stably present proTGFβ1 on the cell surface because in the absence of the disulfide linkage, GARP was unable to prevent proTGFβ1 from leaking into the supernatant.
We defined by EM and confirmed with a homology model the structure of the complex between GARP and proTGFβ. Cys-192 and Cy-s331 are located in the 7th and 12th LRR of GARP, respectively. The distance between the two Cα atoms of Cys-192 and Cys-331 in our GARP homology model is ∼35 Å, whereas the distance between the two Cα atoms of the two C4S mutant residues of our proTGFβ1 homodimer crystal structure is ∼40 Å (Shi et al., 2011; Protein Data Bank [PDB] code, 3RJR). Thus disulfide linkage of Cys-192 and Cys-331 in GARP with the two Cys-4 residues in proTGFβ1 is structurally feasible. Negative-stain class averages showed overall similarity between noncovalent sGARP–proTGFβ1 C4S and covalent sGARP–proTGFβ1 complexes, although the appearance of the sGARP moiety was variable. The class averages and the positions of disulfide-linked cysteines in GARP are consistent with the disulfide linkage of the ring of proTGFβ1 to the side of GARP, with the planes of the proTGFβ1 ring and the GARP horseshoe more normal to one another than coplanar. Thus, with the proTGFβ1 ring lying flat on the EM carbon substrate, the large horseshoe of GARP may collapse at variable orientations onto the substrate. Although GARP may have some flexibility, flexibility was not evident in previous EM studies of GARP alone (Probst-Kepper et al., 2009).
Two integrin αVβ6 molecules could bind simultaneously to the proTGFβ1–GARP complex. The orientations around the proTGFβ1 ring were as predicted based on locations of RGD motifs and Cys-4 residues in the crystal structure of latent TGFβ. As previously described for latent TGFβ, αVβ6 bound in the extended-open, high-affinity conformation, and the affinity for the proTGFβ1–GARP complex is unusually high for an integrin, allowing isolation by gel filtration under nonactivating conditions, that is, in buffer with Ca2+ and Mg2+ (Shi et al., 2011). Furthermore, there was no evidence for disruption of the ring-like structure of proTGFβ upon αVβ6 integrin binding, and TGFβ remained present in the complex, as shown by SDS–PAGE. This suggests that binding of αVβ6 is not sufficient to release TGFβ from the GARP–proTGFβ complex, as previously reported for proTGFβ (Shi et al., 2011).
Both GARP and LTBP disulfide link to the same cysteine, Cys-4, in proTGFβ1. We found that GARP strongly outcompetes LTBP1 for associating with proTGFβ1. Several lines of evidence support this conclusion. First, in cells transfected with equal cDNA amounts of GARP, proTGFβ1, and LTBP1, GARP but not LTBP1 became associated with proTGFβ1. Second, whereas GARP presents proTGFβ1 on the cell surface and LTBP localizes proTGFβ1 to the ECM, LAP was present on the cell surface in GARP-, proTGFβ1-, and LTBP1-cotransfected cells; furthermore, GARP abolished coassociation of LTBP and LAP in cell supernatants. Third, ECR3E, the LAP-binding motif in LTBP1 that was previously shown to interfere with the interaction between LTBP and proTGFβ1 and block activation by αVβ6 (Annes et al., 2004), failed to block αVβ6-mediated TGFβ activation from the GARP–pro-TGFβ1 complex.
TGFβ regulates immune responses primarily by inducing tolerance and controlling inflammatory responses. For instance, TGFβ induces Treg generation and mediates Treg infectious tolerance through cell–cell contact (Andersson et al., 2008). On the other hand, integrins are important in TGFβ activation and immune regulation. Notably, loss of αVβ8 in dendritic cells (DCs) causes severe inflammatory bowel disease and age-related autoimmunity in mice, due to their inability to induce and maintain tissue Tregs (Travis et al., 2007). Yet it was unclear how the integrin mediates Treg generation. Recently GARP was found to be substantially and specifically upregulated in Tregs among lymphocytes (Wang et al., 2008; Oida and Weiner, 2010). GARP expression was shown to correlate with Treg suppressive activity (Wang et al., 2009); silencing GARP in Tregs significantly impaired the suppressive activity of these cells (Probst-Kepper et al., 2009; Wang et al., 2009). Of importance, a GARP-Fc fusion protein rescued the suppressive function of TGFβ-induced Tregs in NOD mice (D'Alise et al., 2011), suggesting that proTGFβ presentation by GARP plays a pivotal role in Treg function.
In the present study, we found that αVβ6 and αVβ8 integrins could activate TGFβ from the GARP–proTGFβ1 complex. This activation was highly specific because it was not given by integrins αVβ1, αVβ3, and αVβ5 expressed at comparable levels on the cell surface. Specificity was further demonstrated by ability of RGD peptide, but not RGE peptide, to completely abolish αVβ6-mediated TGFβ activation. We further showed that a secreted form of GARP was unable to support αVβ6- and αVβ8-mediated TGFβ activation, demonstrating that cell-surface GARP contributes to this activation. To the best of our knowledge, this is the first time that a molecularly defined form of TGFβ has been shown to be activated on cell surfaces and the first time that a molecularly defined mechanism, through αV integrins, has been demonstrated for activation of cell-surface TGFβ. TGFβ activated from the GARP–proTGFβ complex in Tregs may convert nearby naive cells to Tregs. Our study therefore supports a possibility that αVβ8 expressed by DCs releases TGFβ from the GARP–proTGFβ complex in Tregs via cell–cell contact, which in turn induces a larger Treg pool through the infectious tolerance mechanism. TGFβ also contributes to Th17 generation (Veldhoen et al., 2006). Recently αVβ8 expressed by DCs was implicated to regulate Th17 differentiation (Melton et al., 2010). Our results may also suggest a role of GARP in Th17 generation.
Previous studies suggested that tensile force exerted by integrin is required for activation of the proTGFβ–LTBP complex (Annes et al., 2004; Wipff et al., 2007). The crystal structure of the proTGFβ1 homodimer shows that the TGFβ growth factor dimer is sequestered by LAP straightjacket elements (Shi et al., 2011). The α1-helix, latency lasso, and clasp of the straightjacket lock the TGFβ against the prodomain arm domain. These prodomain elements shield TGFβ from recognition by both its type I and type II receptors and also change its conformation. Tensile forces exerted across the proTGFβ ring on the straightjacket would break the noncovalent structural restraints and release mature TGFβ dimer into the extracellular milieu. The conditions required for this activation include the binding of αVβ6 to the RGD motif of LAP; the incorporation of proTGFβ into the ECM by LTBP; the C-terminal portion of the β6 cytoplasmic domain; an intact cytoskeleton to generate cell traction forces and/or to provide mechanical resistance; and a mechanically resistant ECM (Wipff and Hinz, 2008).
Here we show that TGFβ can also be activated from the GARP–proTGFβ1 complex by αVβ6 and αVβ8 integrins. The αVβ6-mediated activation also requires the interaction of integrin to the RGD motif of LAP, suggesting that TGFβ is activated via similar mechanisms, whether presented by LTBP in the ECM or GARP on cell surface. Membrane anchoring of GARP is required, as a soluble form of GARP is unable to support αVβ6-mediated TGFβ activation despite forming an sGARP–proTGFβ1 complex. Furthermore, the disulfide linkage between GARP and proTGFβ1 is required, as TGFβ could not be activated in the absence of disulfide linkage and presence only of noncovalent association between proTGFβ and GARP. In addition, complex formation between purified αVβ6 and GARP–proTGFβ1 did not release TGFβ. These results suggest that αVβ6-dependent activation of TGFβ from the GARP–proTGFβ1 complex also requires tensile force. Negative-stain EM class averages showed that in the sGARP–proTGFβ1–αVβ6 ternary complex, GARP and αVβ6 bind to opposite sides of the proTGFβ1 ring. This arrangement is important for exerting tensile force through this ternary complex for releasing mature TGFβ to bind its receptors.
Although most of our experiments were conducted using cells cotransfected with GARP, proTGFβ1, and αV integrins, we have no evidence that αV integrins can activate the GARP–proTGFβ complexes in-cis on the same cell, since activation could have occurred in-trans in cell culture. We only have evidence for activation in-trans, from experiments in which the proTGFβ/GARP and αV integrins were expressed on different cells. It is known that some integrin–ligand pairs cannot interact with one another when expressed on the same cell, such as LFA-1 and ICAM-1 (Wang and Springer, 1998).
We propose three mechanisms by which GARP regulates TGFβ bioavailability at cell surfaces (Figure 9). First, GARP prevents release of free and possibly misassembled proTGFβ into the extracellular environment and thereby helps maintain its latency (Figure 9A). Second, GARP inhibits secretion of proTGFβ in association with LTBP and hence its assembly into fibrils in the ECM (Figure 9B). Third, GARP provides a cell-surface platform for presentation of latent TGFβ to αV integrins, including αV integrins on the surface of other cells, for activation of TGFβ in the context of cell–cell adhesive interactions (Figure 9C).
FIGURE 9:
Models of how GARP helps regulate TGFβ activation. (A) GARP prevents secretion of proTGFβ and displays it on the cell surface. (B) GARP outcompetes LTBP for assembly into complexes with proTGFβ during biosynthesis. (C) αVβ6 integrin on the surface of one cell binds to the RGD motif in LAP and collaborates with GARP on another cell to generate tensile force across the complex and thereby induce the conformational changes in LAP, which lead to the release and hence activation of TGFβ.
MATERIALS AND METHODS
Subcloning
Transfection-ready, untagged human GARP cDNA was purchased from Origene (Rockville, MD). Human LTBP1 cDNA was provided by Vesna Todorovic (New York University, New York, NY). TGFβ1 cDNA was provided by Katri Koli (University of Helsinki, Helsinki, Finland). GARP was subcloned into a modified pLEXm vector (Aricescu et al., 2006) with a FLAG tag at the N-terminus. LTBP1S, LTBP1L, and the ECR3E domain of LTBP1 were subcloned into a modified pIRES2-EGFP vector (BD Biosciences, San Diego, CA), which contains a streptavidin-binding peptide (SBP) tag at the C-terminus. sGARP was constructed by fusing the extracellular domain of GARP to a histidine (His)–SBP tag, followed by a 3C protease site (Shi et al., 2011) at the N-terminus. GARP and TGFβ1 point mutations were generated using the QuikChange II XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), following the manufacturer's instructions. αV was cloned into a modified pEF1 vector (Invitrogen, Carlsbad, CA) with puromycin resistance. β subunits of αV integrins were cloned into pcDNA3.1 with neomycin resistance (Invitrogen).
Antibodies and other reagents
The following antibodies were used in the present study: anti-FLAG antibody (Sigma-Aldrich, St. Louis, MO), mouse anti-LAP1 antibody for IP and fluorescence-activated cell sorting (FACS; 37232; R&D Systems, Minneapolis, MN), biotinylated goat anti-LAP1 antibody for Western blot (BAF246; R&D Systems), anti-LTBP1 antibody (R&D Systems), anti-αV antibody (Santa Cruz Biotechnology, Santa Cruz, CA), anti-β5 antibody (Millipore, Billerica, MA), anti-β6 antibody (a kind gift of Dean Sheppard, University of California, San Francisco), anti-β8 antibody (a kind gift of Stephen Nishimura, University of California, San Francisco), phycoerythrin (PE)-labeled goat anti–mouse immunoglobulin G (IgG; BD Biosciences), and horseradish peroxidase (HRP)-conjugated sheep anti–mouse IgG and streptavidin-HRP (GE Healthcare, Piscataway, NJ).
To generate monoclonal anti-human GARP antibodies, a stable 293S cell line expressing sGARP was generated. After affinity purification of sGARP, the His-SBP tag was removed from sGARP by 3C protease digestion. sGARP was then further purified and used for immunizing mice. Several in-house anti-GARP antibodies (mouse IgG1; GARP2, GARP5 and GARP6) were confirmed to bind GARP in assays, including enzyme-linked immunosorbent assay, flow cytometry, IP, and Western blot analysis (unpublished data). The RGE (GRGESP) and RGD (GRGDSP) peptides were purchased from Bachem Americas (Torrance, CA). All other chemicals and reagents were obtained from Sigma-Aldrich, unless otherwise indicated.
Cell culture and transfection
HEK293 and 293T cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), 4 mM l-glutamine, 1% nonessential amino acids, and penicillin/streptomycin. All cells were cultured at 37°C in a humidified 5% CO2 atmosphere. For transient transfection, cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. To make stable cell lines expressing αV integrins, HEK293 cells were cotransfected with constructs encoding αV and β subunit. At 40 h posttransfection, cells were selected for the ability to proliferate in medium containing puromycin (1 μg/ml) and G418 (400 μg/ml). Live cells were FACS sorted 1 wk later into single clones based on surface integrin expression. Integrin expression was confirmed via FACS analysis 2 wk postsorting.
FACS
Cells were stained and analyzed as described previously (Wang et al., 2009). In brief, cells were incubated with primary antibody in FACS buffer (phosphate-buffered saline [PBS] with 2% FCS and 0.02% NaN3) on ice for 30 min. After washing, the cells were incubated with anti-mouse PE for 30 min and analyzed by FACScan (BD Biosciences).
IP
Cells were collected, washed once with PBS, and lysed in lysis buffer (1% Triton in Tris-buffered saline with proteinase inhibitor cocktail [Roche; Mannheim, Germany]) at 4°C for 30 min. The lysate was clarified by centrifugation at 12,000 × g for 10 min at 4°C, and the clarified lysate was incubated with antibodies overnight at 4°C on a rocking platform. Protein G–Sepharose (GE Healthcare) was then added and incubated at 4°C for another 1 h. The Sepharose was sedimented and washed three times with lysis buffer. Bound proteins were eluted by heating in SDS sample buffer, separated by SDS–PAGE, and immunoblotted with the indicated antibodies. To immunoprecipitate SBP-tagged proteins and their binding partners, streptavidin-conjugated Sepharose (GE Healthcare) was used. Data shown are representative of at least two independent experiments.
TGFβ bioassay
The TGFβ reporter cell line TMLC was a kind gift of Daniel Rifkin (New York University). The TGFβ bioassay was performed as previously described (Abe et al., 1994; Annes et al., 2003). In brief, in each well of a 96-well white plate, 15,000 TMLC cells were cocultured with 15,000 293 cells transfected with indicated plasmids for 16–24 h. In some experiments, 10,000 293 cells stably expressing integrins and 10,000 transfected 293T cells were cocultured with 15,000 TMLC cells. For the supernatant experiments, 100 μl of supernatants from transfected cells was cocultured with 15,000 TMLC cells. The cells were then processed using the Luciferase Assay System (Promega, Madison, WI) and analyzed by Synergy 2 Multi-Mode Microplate Reader (BioTek, Winooski, VT). Data are presented as the mean + SEM of triplicate samples.
Negative-stain electron microscopy
Affinity-tagged sGARP was purified from supernatant of 293S cells as described previously for proTGFβ1 (Shi et al., 2011). To purify the sGARP–proTGFβ1 complex, sGARP-stable cells were transiently transfected with proTGFβ1-encoding plasmid. To obtain the sGARP–proTGFβ–αVβ6 ternary complex, the purified sGARP-proTGFβ1 complex was mixed with purified αVβ6 in the presence of 1 mM CaCl2 and 1 mM MgCl2. Peak fractions of the purified proteins or complexes from S200 chromatography were subjected to negative-stain electron microscopy. Data processing was performed as previously described (Shi et al., 2011).
Model for GARP
We found no LRR structure with the same number of LRRs as GARP (23 LRRs). Therefore the template was constructed from multiple portions of different LRR proteins, and these were superimposed on TLR3 (PDB code 12IW), which has 24 LRRs. For some LRRs, multiple templates were used. The segments used were the N-cap and LRR1-4 of variable lymphocyte receptor Vlra.R5.1 (PDB code 3M19) for the N-cap and LRR1-4 of GARP; the LRR2-6 of mouse toll-like receptor 3 (PDB code 3CIY) for LRR4-7 of GARP; the LRR8-11 of Lrim1 leucine-rich repeat domain (PDB code 3O53) for LRR7-10 of GARP; the LRR2-6 of the hagfish variable lymphocyte receptors (PDB code 2O6S) for LRR10-14 of GARP; the LRR5-8 of glycoprotein Ib (PDB code 3PMH) for LRR14-17 of GARP and the LRR2-8 and C-cap of neuronal leucine-rich repeat protein Amigo-1 (PDB code 2XOT) for the LRR17-23 and C-cap of GARP. The model was built using MODELLER (Eswar et al., 2003).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant HL103526 and a fellowship from GlaxoSmithKline. We thank D. Rifkin (New York University) for providing the TMLC cell line, Dean Sheppard (University of California, San Francisco) for the anti-β6 antibody, Stephen Nishimura (University of California, San Francisco) for the anti-β8 antibody, Vesna Todorovic (New York University) for the LTBP1 cDNA, and Katri Koli (University of Helsinki) for the TGFβ1 cDNA.
Abbreviations used:
- ECM
extracellular matrix
- EM
electron microscopy
- GARP
Glycoprotein-A repetitions predominant protein
- LAP
latency-associated peptide
- LTBP
latent TGFβ-binding protein
- TGFβ
transforming growth factor-β
- TMLC
transformed mink lung TGFβ-reporter cell line
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-12-1018) on January 25, 2012.
REFERENCES
- Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter-luciferase construct. Anal Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- Aluwihare P, Mu Z, Zhao Z, Yu D, Weinreb PH, Horan GS, Violette SM, Munger JS. Mice that lack activity of αVβ6- and αVβ8-integrins reproduce the abnormalities of TGFβ1- and TGFβ3-null mice. J Cell Sci. 2009;122:227–232. doi: 10.1242/jcs.035246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson J, Tran DQ, Pesu M, Davidson TS, Ramsey H, O'shea JJ, Shevach EM. CD4+ FoxP3 +regulatory T cells confer infectious tolerance in a TGF-β-dependent manner. J Exp Med. 2008;205:1975–1981. doi: 10.1084/jem.20080308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS, Rifkin DB. Integrin αVβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J Cell Biol. 2004;165:723–734. doi: 10.1083/jcb.200312172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annes JP, Munger JS, Rifkin DB. Making sense of latent TGFβ activation. J Cell Sci. 2003;116:217–224. doi: 10.1242/jcs.00229. [DOI] [PubMed] [Google Scholar]
- Aricescu AR, Lu W, Jones EY. A time- and cost-efficient system for high-level protein production in mammalian cells. Acta Crystallogr D Biol Crystallogr. 2006;62:1243–1250. doi: 10.1107/S0907444906029799. [DOI] [PubMed] [Google Scholar]
- Bonne-Tamir B, Nystuen A, Seroussi E, Kalinsky H, Kwitek-Black AE, Korostishevsky M, Adato A, Sheffield VC. Usher syndrome in the Samaritans: strengths and limitations of using inbred isolated populations to identify genes causing recessive disorders. Am J Phys Anthropol. 1997;104:193–200. doi: 10.1002/(SICI)1096-8644(199710)104:2<193::AID-AJPA5>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Brunner AM, Marquardt H, Malacko AR, Lioubin MN, Purchio AF. Site-directed mutagenesis of cysteine residues in the pro region of the transforming growth factor β1 precursor. Expression and characterization of mutant proteins. J Biol Chem. 1989;264:13660–13664. [PubMed] [Google Scholar]
- Cepek KL, Parker CM, Madara JL, Brenner MB. Integrin αEβ7 mediates adhesion of T lymphocytes to epithelial cells. J Immunol. 1993;150:3459–3470. [PubMed] [Google Scholar]
- Coffman RL, Lebman DA, Shrader B. Transforming growth factor β specifically enhances IgA production by lipopolysaccharide-stimulated murine B lymphocytes. J Exp Med. 1989;170:1039–1044. doi: 10.1084/jem.170.3.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alise AM, Ergun A, Hill JA, Mathis D, Benoist C. A cluster of coregulated genes determines TGF-β-induced regulatory T-cell (Treg) dysfunction in NOD mice. Proc Natl Acad Sci USA. 2011;108:8737–8742. doi: 10.1073/pnas.1105364108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eswar N, et al. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentry LE, Webb NR, Lim GJ, Brunner AM, Ranchalis JE, Twardzik DR, Lioubin MN, Marquardt H, Purchio AF. Type 1 transforming growth factor β: amplified expression and secretion of mature and precursor polypeptides in Chinese hamster ovary cells. Mol Cell Biol. 1987;7:3418–3427. doi: 10.1128/mcb.7.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil N. TGF-β: from latent to active. Microbes Infect. 1999;1:1255–1263. doi: 10.1016/s1286-4579(99)00259-2. [DOI] [PubMed] [Google Scholar]
- Kilshaw PJ, Murant SJ. Expression and regulation of β7(bp) integrins on mouse lymphocytes: relevance to the mucosal immune system. Eur J Immunol. 1991;21:2591–2597. doi: 10.1002/eji.1830211041. [DOI] [PubMed] [Google Scholar]
- Kulkarni AB, Huh C, Becker D, Geiser A, Lyght M, Flanders KC, Roberts AB, Sporn MB, Ward JM, Karlsson S. Transforming growth factor β1 null mutation in mice causes excessive inflammatory response and early death. Proc Natl Acad Sci USA. 1993;90:770–774. doi: 10.1073/pnas.90.2.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-β regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- Ludbrook SB, Barry ST, Delves CJ, Horgan CM. The integrin αVβ3 is a receptor for the latency-associated peptides of transforming growth factors β1 and β3. Biochem J. 2003;369:311–318. doi: 10.1042/BJ20020809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay IC, et al. Comparative gene expression profiling of in vitro differentiated megakaryocytes and erythroblasts identifies novel activatory and inhibitory platelet membrane proteins. Blood. 2007;109:3260–3269. doi: 10.1182/blood-2006-07-036269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melton AC, Bailey-Bucktrout SL, Travis MA, Fife BT, Bluestone JA, Sheppard D. Expression of αVβ8 integrin on dendritic cells regulates Th17 cell development and experimental autoimmune encephalomyelitis in mice. J Clin Invest. 2010;120:4436–4444. doi: 10.1172/JCI43786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K, Olofsson A, Colosetti P, Heldin CH. A role of the latent TGF-β 1-binding protein in the assembly and secretion of TGF-β1. EMBO J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu D, Cambier S, Fjellbirkeland L, Baron JL, Munger JS, Kawakatsu H, Sheppard D, Broaddus VC, Nishimura SL. The integrin αVβ8 mediates epithelial homeostasis through MT1-MMP-dependent activation of TGF-β1. J Cell Biol. 2002;157:493–507. doi: 10.1083/jcb.200109100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, et al. The integrin αVβ6 binds and activates latent TGFβ1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Kitani A, Fuss I, Pedersen A, Harada N, Nawata H, Strober W. TGF-β1 plays an important role in the mechanism of CD4+CD25+ regulatory T cell activity in both humans and mice. J Immunol. 2004;172:834–842. doi: 10.4049/jimmunol.172.2.834. [DOI] [PubMed] [Google Scholar]
- Oida T, Weiner HL. TGF-β induces surface LAP expression on murine CD4 T cells independent of Foxp3 induction. PLoS One. 2010;5:e15523. doi: 10.1371/journal.pone.0015523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollendorff V, Noguchi T, deLapeyriere O, Birnbaum D. The GARP gene encodes a new member of the family of leucine-rich repeat-containing proteins. Cell Growth Differ. 1994;5:213–219. [PubMed] [Google Scholar]
- Probst-Kepper M, et al. GARP: a key receptor controlling FOXP3 in human regulatory T cells. J Cell Mol Med. 2009;13:3343–3357. doi: 10.1111/j.1582-4934.2009.00782.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifkin DB. Latent transforming growth factor-beta (TGF-β) binding proteins: orchestrators of TGF-β availability. J Biol Chem. 2005;280:7409–7412. doi: 10.1074/jbc.R400029200. [DOI] [PubMed] [Google Scholar]
- Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-β with an eight cysteine repeat of its binding protein LTBP-1. EMBO J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. Latent TGF-β structure and activation. Nature. 2011;474:343–349. doi: 10.1038/nature10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, et al. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature. 1992;359:693–699. doi: 10.1038/359693a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockis J, Colau D, Coulie PG, Lucas S. Membrane protein GARP is a receptor for latent TGF-β on the surface of activated human Treg. Eur J Immunol. 2009;39:3315–3322. doi: 10.1002/eji.200939684. [DOI] [PubMed] [Google Scholar]
- Tran DQ, Andersson J, Wang R, Ramsey H, Unutmaz D, Shevach EM. GARP (LRRC32) is essential for the surface expression of latent TGF-β on platelets and activated FOXP3 +regulatory T cells. Proc Natl Acad Sci USA. 2009;106:13445–13450. doi: 10.1073/pnas.0901944106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis MA, et al. Loss of integrin αVβ8 on dendritic cells causes autoimmunity and colitis in mice. Nature. 2007;449:361–365. doi: 10.1038/nature06110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Smith DM, Flanders KC, Sporn MB. Latent transforming growth factor-β from human platelets. A high molecular weight complex containing precursor sequences. J Biol Chem. 1988;263:7646–7654. [PubMed] [Google Scholar]
- Wang J-H, Springer TA. Structural specializations of immunoglobulin superfamily members for adhesion to integrins and viruses. Immunol Rev. 1998;163:197–215. doi: 10.1111/j.1600-065x.1998.tb01198.x. [DOI] [PubMed] [Google Scholar]
- Wang R, Kozhaya L, Mercer F, Khaitan A, Fujii H, Unutmaz D. Expression of GARP selectively identifies activated human FOXP3+ regulatory T cells. Proc Natl Acad Sci USA. 2009;106:13439–13444. doi: 10.1073/pnas.0901965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Wan Q, Kozhaya L, Fujii H, Unutmaz D. Identification of a regulatory T cell specific cell surface molecule that mediates suppressive signals and induces Foxp3 expression. PLoS One. 2008;3:e2705. doi: 10.1371/journal.pone.0002705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wipff PJ, Hinz B. Integrins and the activation of latent transforming growth factor β1 - an intimate relationship. J Eur Cell Biol. 2008;87:601–615. doi: 10.1016/j.ejcb.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Wipff PJ, Rifkin DB, Meister JJ, Hinz B. Myofibroblast contraction activates latent TGF-β1 from the extracellular matrix. J Cell Biol. 2007;179:1311–1323. doi: 10.1083/jcb.200704042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao T, Takagi J, Wang J-H, Coller BS, Springer TA. Structural basis for allostery in integrins and binding of fibrinogen-mimetic therapeutics. Nature. 2004;432:59–67. doi: 10.1038/nature02976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong JP, Stehle T, Zhang R, Joachimiak A, Frech M, Goodman SL, Arnaout MA. Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science. 2002;296:151–155. doi: 10.1126/science.1069040. [DOI] [PubMed] [Google Scholar]
- Yang Z, Mu Z, Dabovic B, Jurukovski V, Yu D, Sung J, Xiong X, Munger JS. Absence of integrin-mediated TGFβ1 activation in vivo recapitulates the phenotype of TGFβ1-null mice. J Cell Biol. 2007;176:787–793. doi: 10.1083/jcb.200611044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura A, Wakabayashi Y, Mori T. Cellular and molecular basis for the regulation of inflammation by TGF-β. J Biochem. 2010;147:781–792. doi: 10.1093/jb/mvq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.