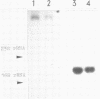
Abstract
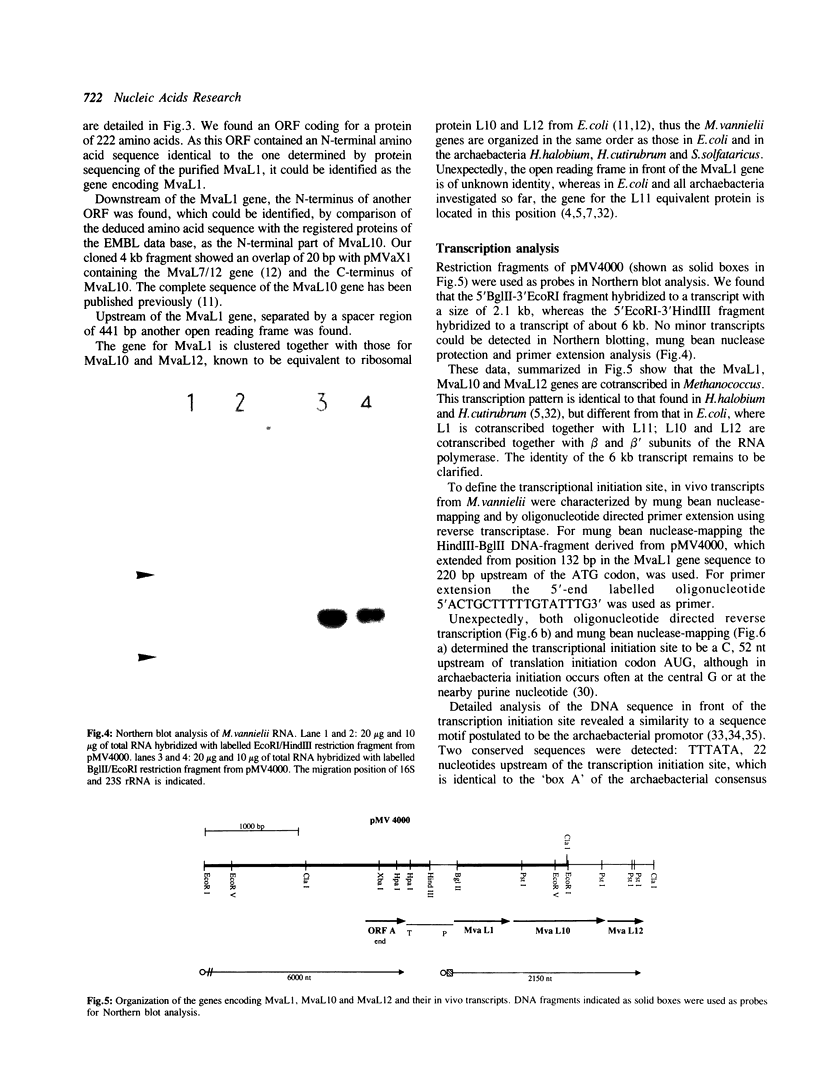
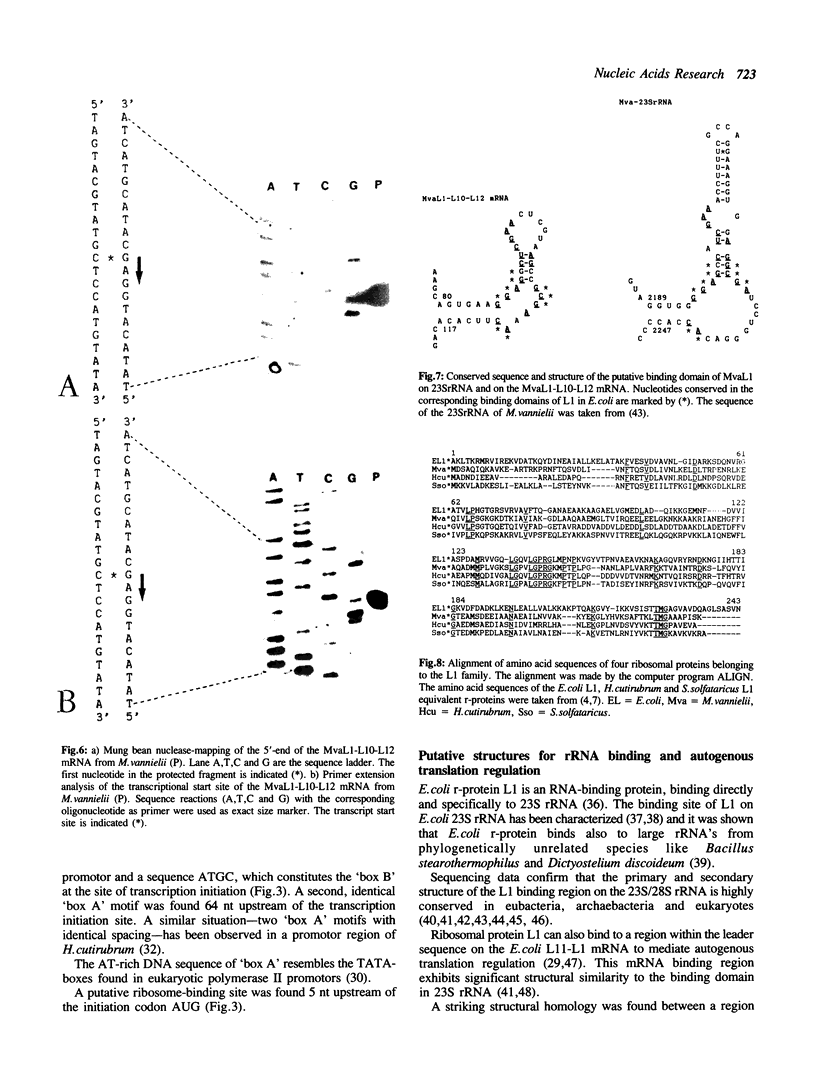
The gene for ribosomal protein MvaL1 from the arachaebacterium Methanococcus vannielii was cloned and characterized. It is clustered together with the genes for MvaL10 and MvaL12, thus is organized in the same order as in E.coli and other archaebacteria. Unexpectedly, analysis of the sequence in front of the MvaL1 gene revealed an ORF of unknown identity, whereas in E.coli, Halobacterium and Sulfolobus solfataricus the gene for the L11 equivalent protein is located in this position. Northern blot analysis revealed a single tricistronic transcript encoding proteins MvaL1, MvaL10 and MvaL12. The 5'-end of the MvaL1-L10-L12 transcript contains a region that has a sequence and structure almost identical to a region on the 23S rRNA which is the putative binding domain for MvaL1, and is highly similar to the E.coli L11-L1 mRNA leader sequence that has been implicated in autogenous translational regulation. Amino acid sequence comparison revealed that MvaL1 shares 30.5% identity with ribosomal protein L1 from E.coli and 41.5% and 33.3% identity with the L1-equivalent proteins from the archaebacteria H.cutirubrum and S.solfataricus respectively.
Full text
PDF
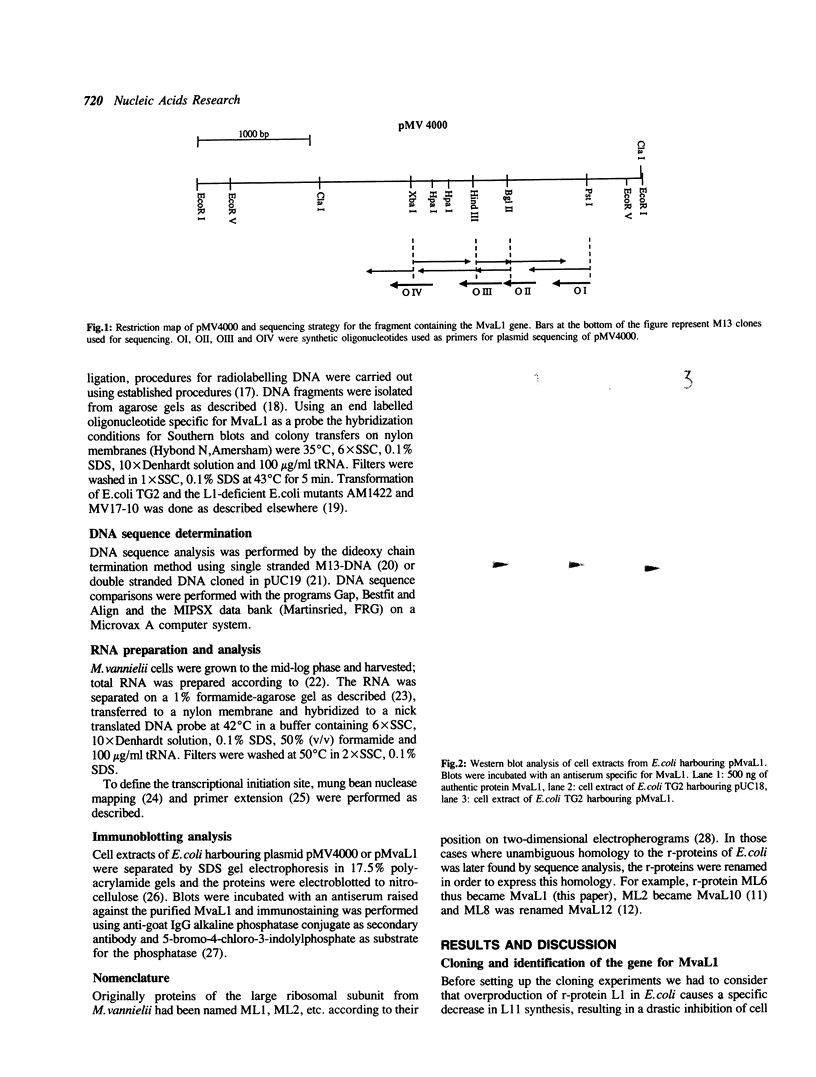


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldea M., Claverie-Martín F., Díaz-Torres M. R., Kushner S. R. Transcript mapping using [35S]DNA probes, trichloroacetate solvent and dideoxy sequencing ladders: a rapid method for identification of transcriptional start points. Gene. 1988 May 15;65(1):101–110. doi: 10.1016/0378-1119(88)90421-0. [DOI] [PubMed] [Google Scholar]
- Auer J., Lechner K., Böck A. Gene organization and structure of two transcriptional units from Methanococcus coding for ribosomal proteins and elongation factors. Can J Microbiol. 1989 Jan;35(1):200–204. doi: 10.1139/m89-031. [DOI] [PubMed] [Google Scholar]
- Auer J., Spicker G., Böck A. Organization and structure of the Methanococcus transcriptional unit homologous to the Escherichia coli "spectinomycin operon". Implications for the evolutionary relationship of 70 S and 80 S ribosomes. J Mol Biol. 1989 Sep 5;209(1):21–36. doi: 10.1016/0022-2836(89)90167-8. [DOI] [PubMed] [Google Scholar]
- Beauclerk A. A., Hummel H., Holmes D. J., Böck A., Cundliffe E. Studies of the GTPase domain of archaebacterial ribosomes. Eur J Biochem. 1985 Sep 2;151(2):245–255. doi: 10.1111/j.1432-1033.1985.tb09095.x. [DOI] [PubMed] [Google Scholar]
- Blake M. S., Johnston K. H., Russell-Jones G. J., Gotschlich E. C. A rapid, sensitive method for detection of alkaline phosphatase-conjugated anti-antibody on Western blots. Anal Biochem. 1984 Jan;136(1):175–179. doi: 10.1016/0003-2697(84)90320-8. [DOI] [PubMed] [Google Scholar]
- Branlant C., Korobko V., Ebel J. P. The binding site of protein L1 on 23-S ribosomal RNA from Escherichia coli. 3. Nucleotide sequence. Eur J Biochem. 1976 Nov 15;70(2):471–482. doi: 10.1111/j.1432-1033.1976.tb11038.x. [DOI] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt A., Ebel J. P. The secondary structure of the protein L1 binding region of ribosomal 23S RNA. Homologies with putative secondary structures of the L11 mRNA and of a region of mitochondrial 16S rRNA. Nucleic Acids Res. 1981 Jan 24;9(2):293–307. doi: 10.1093/nar/9.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone F. J., Britten R. J., Davidson E. H. Mapping of gene transcripts by nuclease protection assays and cDNA primer extension. Methods Enzymol. 1987;152:611–632. doi: 10.1016/0076-6879(87)52069-9. [DOI] [PubMed] [Google Scholar]
- Cerretti D. P., Mattheakis L. C., Kearney K. R., Vu L., Nomura M. Translational regulation of the spc operon in Escherichia coli. Identification and structural analysis of the target site for S8 repressor protein. J Mol Biol. 1988 Nov 20;204(2):309–329. doi: 10.1016/0022-2836(88)90578-5. [DOI] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Dabbs E. R. The ribosomal components responsible for kasugamycin dependence, and its suppression, in a mutant of Escherichia coli. Mol Gen Genet. 1980 Jan;177(2):271–276. doi: 10.1007/BF00267438. [DOI] [PubMed] [Google Scholar]
- Dean D., Nomura M. Feedback regulation of ribosomal protein gene expression in Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3590–3594. doi: 10.1073/pnas.77.6.3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjiolov A. A., Georgiev O. I., Nosikov V. V., Yavachev L. P. Primary and secondary structure of rat 28 S ribosomal RNA. Nucleic Acids Res. 1984 Apr 25;12(8):3677–3693. doi: 10.1093/nar/12.8.3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T. Complete nucleotide sequence of the ribosomal 'A' protein operon from the archaebacterium, Halobacterium halobium. Eur J Biochem. 1988 Sep 15;176(2):297–303. doi: 10.1111/j.1432-1033.1988.tb14281.x. [DOI] [PubMed] [Google Scholar]
- Köpke A. K., Baier G., Wittmann-Liebold B. An archaebacterial gene from Methanococcus vannielii encoding a protein homologous to the ribosomal protein L10 family. FEBS Lett. 1989 Apr 24;247(2):167–172. doi: 10.1016/0014-5793(89)81326-2. [DOI] [PubMed] [Google Scholar]
- Lechner K., Heller G., Böck A. Organization and nucleotide sequence of a transcriptional unit of Methanococcus vannielii comprising genes for protein synthesis elongation factors and ribosomal proteins. J Mol Evol. 1989 Jul;29(1):20–27. doi: 10.1007/BF02106178. [DOI] [PubMed] [Google Scholar]
- Lehrach H., Diamond D., Wozney J. M., Boedtker H. RNA molecular weight determinations by gel electrophoresis under denaturing conditions, a critical reexamination. Biochemistry. 1977 Oct 18;16(21):4743–4751. doi: 10.1021/bi00640a033. [DOI] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post L. E., Strycharz G. D., Nomura M., Lewis H., Dennis P. P. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit beta in Escherichia coli. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez C., Shimmin L. C., Newton C. H., Matheson A. T., Dennis P. P. Structure and evolution of the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can J Microbiol. 1989 Jan;35(1):234–244. doi: 10.1139/m89-036. [DOI] [PubMed] [Google Scholar]
- Reiter W. D., Palm P., Zillig W. Analysis of transcription in the archaebacterium Sulfolobus indicates that archaebacterial promoters are homologous to eukaryotic pol II promoters. Nucleic Acids Res. 1988 Jan 11;16(1):1–19. doi: 10.1093/nar/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Dennis P. P. Characterization of the L11, L1, L10 and L12 equivalent ribosomal protein gene cluster of the halophilic archaebacterium Halobacterium cutirubrum. EMBO J. 1989 Apr;8(4):1225–1235. doi: 10.1002/j.1460-2075.1989.tb03496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin L. C., Newton C. H., Ramirez C., Yee J., Downing W. L., Louie A., Matheson A. T., Dennis P. P. Organization of genes encoding the L11, L1, L10, and L12 equivalent ribosomal proteins in eubacteria, archaebacteria, and eucaryotes. Can J Microbiol. 1989 Jan;35(1):164–170. doi: 10.1139/m89-025. [DOI] [PubMed] [Google Scholar]
- Stanley J., Sloof P., Ebel J. P. The binding site of ribosomal protein L1 from Escherichia coli on the 23-S ribosomal RNA from Bacillus stearothermophilus. A possible base-pairing scheme differing from that proposed for Escherichia coli. Eur J Biochem. 1978 Apr;85(1):309–316. doi: 10.1111/j.1432-1033.1978.tb12240.x. [DOI] [PubMed] [Google Scholar]
- Strobel O., Köpke A. K., Kamp R. M., Böck A., Wittmann-Liebold B. Primary structure of the archaebacterial Methanococcus vannielii ribosomal protein L12. Amino acid sequence determination, oligonucleotide hybridization, and sequencing of the gene. J Biol Chem. 1988 May 15;263(14):6538–6546. [PubMed] [Google Scholar]
- Subramanian A. R., Dabbs E. R. Functional studies on ribosomes lacking protein L1 from mutant Escherichia coli. Eur J Biochem. 1980 Nov;112(2):425–430. doi: 10.1111/j.1432-1033.1980.tb07222.x. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- Thomas M. S., Nomura M. Translational regulation of the L11 ribosomal protein operon of Escherichia coli: mutations that define the target site for repression by L1. Nucleic Acids Res. 1987 Apr 10;15(7):3085–3096. doi: 10.1093/nar/15.7.3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomm M., Wich G. An archaebacterial promoter element for stable RNA genes with homology to the TATA box of higher eukaryotes. Nucleic Acids Res. 1988 Jan 11;16(1):151–163. doi: 10.1093/nar/16.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wich G., Hummel H., Jarsch M., Bär U., Böck A. Transcription signals for stable RNA genes in Methanococcus. Nucleic Acids Res. 1986 Mar 25;14(6):2459–2479. doi: 10.1093/nar/14.6.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Fox G. E. Phylogenetic structure of the prokaryotic domain: the primary kingdoms. Proc Natl Acad Sci U S A. 1977 Nov;74(11):5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Olsen G. J. Archaebacterial phylogeny: perspectives on the urkingdoms. Syst Appl Microbiol. 1986;7:161–177. doi: 10.1016/s0723-2020(86)80001-7. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yates J. L., Nomura M. Feedback regulation of ribosomal protein synthesis in E. coli: localization of the mRNA target sites for repressor action of ribosomal protein L1. Cell. 1981 Apr;24(1):243–249. doi: 10.1016/0092-8674(81)90520-1. [DOI] [PubMed] [Google Scholar]
- Zillig W., Palm P., Reiter W. D., Gropp F., Pühler G., Klenk H. P. Comparative evaluation of gene expression in archaebacteria. Eur J Biochem. 1988 May 2;173(3):473–482. doi: 10.1111/j.1432-1033.1988.tb14023.x. [DOI] [PubMed] [Google Scholar]