Abstract
Co-evolution of mammals and their gut microbiota has profoundly effected their radiation into myriad habitats. We used shotgun sequencing of microbial community DNA and targeted sequencing of bacterial 16S rRNA genes to understand how microbial communities adapt to extremes of diets, sampling fecal DNAs from 33 mammalian species and 18 humans who kept detailed diet records. We found that microbiota adaptation to diet is reproducible across different mammalian lineages. Functional repertoires of microbiome genes, such as those encoding carbohydrate-active enzymes and proteases, can be predicted from bacterial species assemblages. These results illustrate the value of characterizing vertebrate gut microbiomes to fully understand host evolutionary histories at a supra-organismal level.
Comparative culture-independent metagenomic studies of the microbial species assemblages that comprise mammalian gut microbiota, and the functions that these communities encode in their aggregate genomes (microbiomes) can provide a complementary perspective to comparative studies of host genomes. A previous bacterial 16S rRNA-based study of 59 mammalian species revealed that their fecal microbiota clustered according to diet rather than host phylogeny (1). This finding raises several questions. What is the functional evolution of the gut microbiome in relation to diet? Is the process unique to each mammalian lineage? To what extent does microbial phylogeny predict function within microbial communities? Could analysis of inter-specific differences among mammals create a pipeline for deciphering intra-specific differences among humans in response to varied diets or other factors? Therefore, we have extended our 16S rRNA studies to a broader sampling of microbial genes in total fecal community DNA prepared from herbivores, omnivores, and carnivores.
We generated shotgun pyrosequencing datasets from 33 mammalian species, along with newly collected bacterial 16S rRNA data. These adult animals represent 10 Orders, and varied digestive physiologies (hindgut-fermenters, foregut-fermenters, simple-gut). In some cases free-living and captive representatives of a given species were sampled (Table 1, Table S1). Methods for classifying diets, as well as collecting and processing fecal samples for metagenomic analyses have been described (1). Multiplex pyrosequencing of amplicons generated from the V2 region of bacterial 16S rRNA genes yielded 149,675 high-quality, de-noised reads (average 3,838±1,080/sample; Table S2) (2). After chimera removal, 8,541 operational taxonomic units (OTUs) were identified in the combined dataset (an OTU was defined as reads sharing ≥97% nucleotide sequence identity). Shotgun sequencing of the same fecal DNA preparations produced 2,163,286 reads (mean 55,469±28,724 (S.D.)/sample; 261+83 nt/read) (Table S3)(2). Shotgun reads were functionally annotated using KEGG [KEGG Orthology (KO) groups and Enzyme Commission (E.C.) numbers], CAZy (carbohydrate-active enzymes) and MEROPS (peptidase) databases (3–5). When shotgun reads were assigned to phylogenetic bins using the program MEGAN (6), the results revealed that fecal microbiomes were dominated by members of Bacteria, had low levels of Eukarya (0.15–5.35% of identifiable reads), and archaeons were variably represented (0–1.77% of assignable reads with none detected in any carnivore microbiome). Seventeen samples had reads assigned to known viruses (Table S4)(2).
Table 1.
Overview of mammals in this study
| Foregut Fermenting Herbivores | Hindgut Fermenting Herbivores |
|---|---|
| Big Horn Sheep 1 (BigHornSD)* | African Elephant (AfElphSD3)* |
| Big Horn Sheep 2 (BigHornW)‡ | Black Rhinoceros (BlackRhino1)† |
| Colobus (Colobus)† | Capybara (Capybara)† |
| Gazelle (Gazelle3)† | Gorilla (GorillaSTL)† |
| Giraffe (Giraffe2)† | Horse (Horse1)‡ |
| Rock Hyrax 1 (HyraxSD)* | Orangutan (Orang1)† |
| Rock Hyrax 2 (HyraxSTL)† | European Rabbit (Rabbit)† |
| Kangaroo (Kroo3)† | Zebra (ZebraSTL1)† |
| Okapi 1 (Okapi1)† | |
| Okapi 2 (Okapi2)† | Omnivores |
| Springbok (SpgbkW)‡ | Baboon 1 (BaboonSTL)† |
| Transcaspian Urial Sheep (Urial2)† | Baboon 2 (BaboonW)‡ |
| Visayan Warty Pig (VWPig)* | Black Bear (BlackBr2)† |
| Black Lemur (BlackLemur)† | |
| Carnivores | Callimicos (Callimicos)† |
| Armadillo (Armadillo)† | Chimpanzee 1 (Chimp1)† |
| Bush Dog (BushDog1)† | Chimpanzee 2 (Chimp2)† |
| Echidna (Echidna)† | Ringtailed Lemur (RTLemur)† |
| Hyena (Hyena2)† | Saki (Saki)† |
| Lion 1 (Lion1)† | Spectacled Bear (SpecBr2)† |
| Lion 2 (Lion2)† | Squirrel (Squirrel)† |
| Polar Bear (PolarBr2)† |
Legend:
=San Diego Zoological Park.
=St. Louis Zoo.
=Wild, free-living. Sample abbreviations used in figures and tables are noted in parentheses. See Table S1 for additional details.
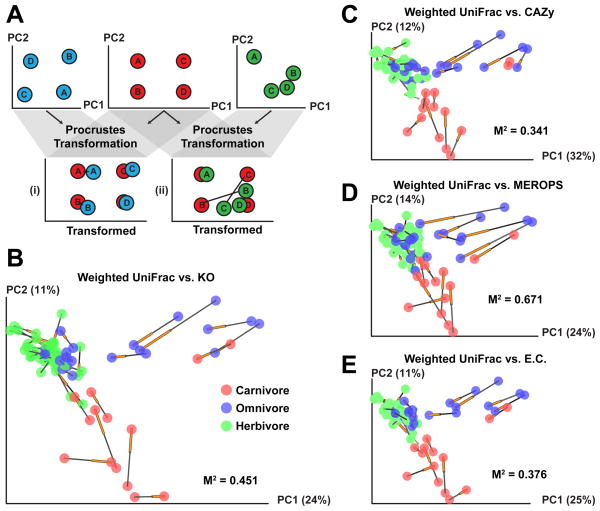
Procrustes analysis (least-squares orthogonal mapping) was used to test whether the functional properties of a microbiome can be predicted from the bacterial species that comprise it (2). Procrustes analysis attempts to stretch and rotate the points in one matrix, such as points obtained by Principal Coordinates Analysis (PCoA), to be as close as possible to points in the other matrix, thus preserving the relative distances between points within each matrix (7,8) (Fig. 1A). We first took the 16S rRNA dataset and used the UniFrac metric to compare the overlap between each pair of communities in terms of their evolutionary distance (9). The similarity in functional profiles was then determined using the Bray-Curtis distance metric applied to KO groups, E.C.s, CAZYmes, or peptidases. Principal coordinates reduction was performed separately on the 16S rRNA and annotated shotgun (microbiome) datasets, and the point clouds were aligned using Procrustes. For each comparison, the goodness of fit, or M2 value, of the transformed datasets was measured over the first three dimensions. The statistical significance of the goodness of the fit was measured by a Monte Carlo label permutation approach (2).
Figure 1. Procrustes analysis shows that mammalian bacterial lineages and metagenomic gene content give similar clustering patterns.
(A) Cartoon illustrating Procrustes analysis. The Procrustes transformation of the blue and red data types (i) results in a good fit, while the transformation of the green and red data (ii) yields a worse fit with large distances separating data from samples B and C. (B–E) Procrustes analysis of 16S rRNA sequences (weighted UniFrac) against KEGG Orthology (KO) groups, CAZymes (glycoside hydrolases), peptidases, and E.C.s. Every sphere represents a single mammalian fecal community and is colored by host diet. The black end of each line connects to the 16S data for the sample, while the orange end is connected to the functional annotation data. The fit of each Procrustes transformation over the first three dimensions, is reported as the M2 value.
The agreement between phylogenetic and functional measurements was remarkable for all mammals, regardless of their diet, host lineage or gut physiology. Fig. 1B–E shows how the goodness-of-fit was robust to different functional databases. The analysis was also robust to taxon- or phylogenetic-based species classification, weighted or unweighted metrics, and whether one or more member of each mammalian species was considered (Fig. S1). For both bacterial 16S rRNA and whole community gene datasets, the PCoA plots separated carnivores and omnivores from herbivores, emphasizing the importance of diet in differentiating gut microbial communities (p<0.05, 2). Our previous study using full-length 16S rRNA sequences revealed that the fecal microbiota of conspecifics were significantly more similar than the communities of different host species (1). The V2 16S rRNA data generated in this study confirmed this result using both weighted and unweighted UniFrac distances (p<0.05 by 1,000 Monte Carlo permutations (2).
The Procrustes results prompted us to use a nearest-neighbor model to test whether the functional configuration of a microbiome could be predicted from its 16S rRNA sequences. Remarkably, using a fecal sample’s nearest neighbor, as defined by unweighted or weighted UniFrac, to predict the sample’s functional profile generated a significantly better prediction than a random neighbor; this was true for KOs, E.C.s, CAZymes, and peptidases (p <0.0001, 106 Monte Carlo permutations) (2).
The concordance of diet and microbiome structure and function raises the question of whether it is caused primarily by co-evolution between mammals and their gut microbiota/microbiome, or by the many parallel dietary shifts that have occurred over the course of mammalian evolution (10). We tested which of these hypotheses, which have traditionally been viewed as competing but need not be mutually exclusive, were supported by looking for congruence between mammalian phylogeny and subsets of bacterial species, KOs, CAZymes, peptidases, or other enzymatic activities. Briefly, the mammalian phylogenetic tree defines sets of organisms that are monophyletic, i.e. groups containing all and only the descendants of a common ancestor. We reasoned that if bacterial taxa or functions originated rarely, then these taxa or functions should be vertically transmitted during mammalian speciation. Therefore, there should be more cases in which a given taxon or function occurred in all members of a monophyletic mammalian group than chance would predict. Using this analytic approach (2), we found that the overall distribution of microbial species and microbiome functions in the gut do not mirror mammalian phylogeny. 198 different named bacterial genera were detected in our dataset – of these, only three were significantly associated with the mammalian phylogenetic tree more than would be expected by chance (Prevotella, Barnesiella, and Bacteroides). No CAZymes or peptidases and only 18 of the 3,866 KOs tested were associated with host phylogeny. We repeated the analysis using a more relaxed constraint that a taxon or function occurs in a given monophyletic group more frequently than expected by chance rather than requiring strict presence/absence agreement (2). The relaxed definition gave similar results; only three additional genera and a total of 90 KOs were detected as having a significant association with the mammalian tree. We concluded that bacterial taxa and functions are evolutionarily labile and do not explain the concordance between bacterial communities and microbiome functions.
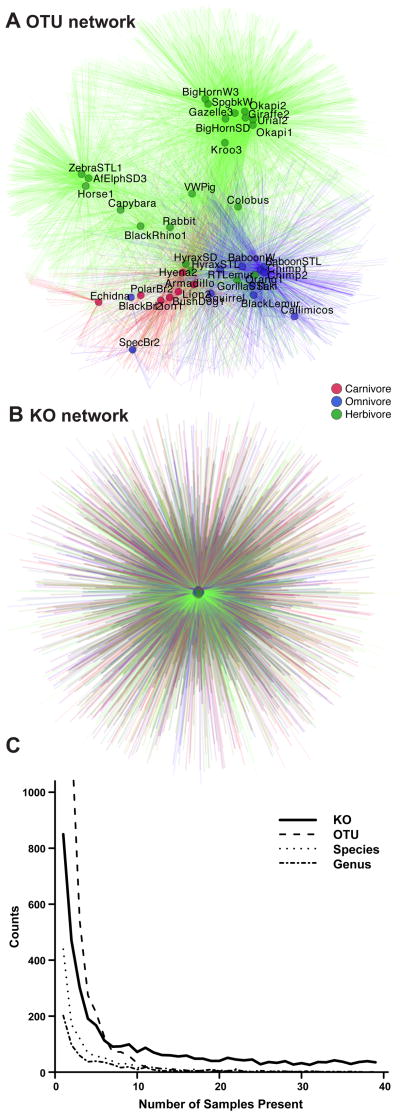
Bipartite network analysis provided an additional tool for exploring the interrelationship between host diet, host lineage, gut physiology, and shared and unique bacteria taxa (1). Mammalian hosts and bacterial OTUs were used as nodes in a bipartite graph, with edges connecting OTU nodes to the hosts in which they are found (2). Using 1,900 V2 16S rRNA sequences from each mammalian host, the network shows clear separation of fecal communities by host diet (Fig. 2A), mirroring our earlier results based on smaller numbers of full-length 16S rRNA sequences (1).
Figure 2. Mammalian gut bacterial communities share a functional core.
(A–B) Bipartite network diagrams of evenly sampled bacterial 16S rRNA-derived OTUs (A) or KOs (B). Edges connecting mammalian nodes (circles) to species-level OTUs or KOs found in that sample are colored by host diet. Sample labels are removed from the KO diagram for legibility (high-resolution image of removed labels presented in Fig. S2). (C) Mammalian gut communities share a core suite of KOs. Using evenly subsampled OTU or KO datasets, the distribution of counts is plotted as a function of the number of mammalian host microbiomes where the KO or phylotype was detected. The results demonstrate exponential decay for the 16S rRNA data, with no OTU or bacterial species found in all samples, although a “core” set of KOs is detectable in all fecal communities sampled.
We reasoned that the bipartite graph approach could also be used to connect mammalian samples to individual microbial gene functions from shotgun reads. The power of the bipartite graph approach is to represent both genes and mammalian species explicitly as nodes, thus visualizing which genes connect with which species. The clear separation by diet disappears when we consider gene functions (Fig. 2B, Fig. S2), suggesting that rather than a diet- or physiology-specific set of genes, the relationship among mammalian gut microbiomes is that they share a large core repertoire of functions. We confirmed this result by plotting the frequency of shared taxa in the 39 mammalian fecal samples, and also species- and genus-level OTU bins (2). All of the curves demonstrate an essentially exponential decay as successive samples are added, with no OTUs found in more than 30 samples (Fig. 2C). However, the plot of KO frequency flattens out, with 35 KOs found in all samples. This effect cannot be due to differences in the number of OTUs relative to KOs: there are more OTUs than KOs, and fewer assigned species or assigned genera, yet all the taxonomic curves show the same rapid decay, unlike the KOs.
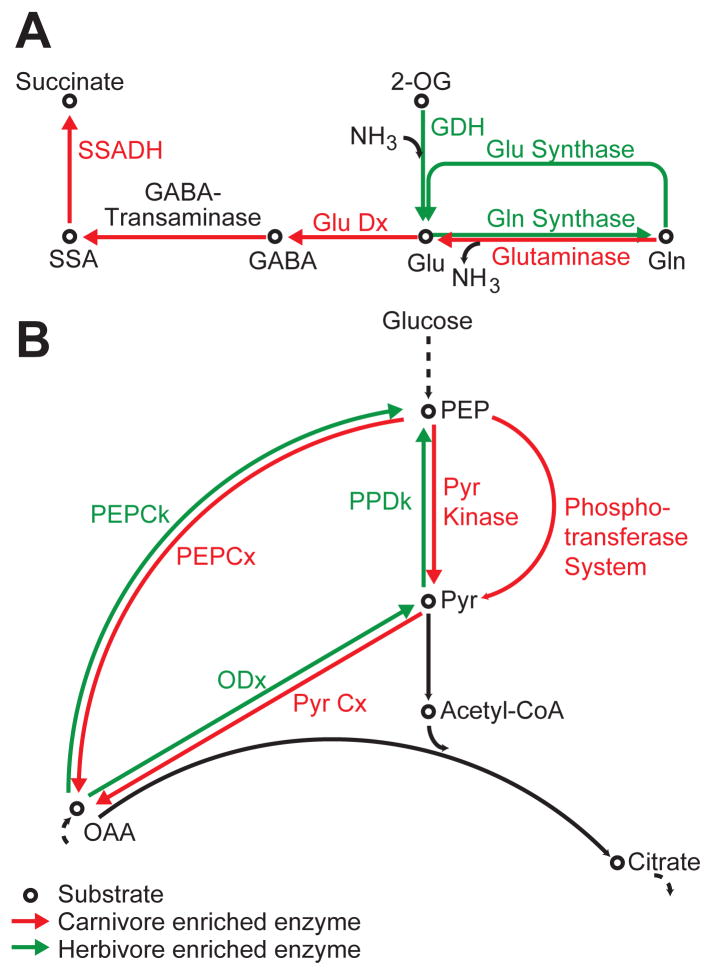
This result does not imply that there are no differences among the functional configurations of microbiomes of host species having different diets. Rather, it suggests that the differences between microbiomes likely stem from differing abundances of shared functions, such as enzymes that break down chemical substrates in host diet. We identified 495 E.C.s with significantly different proportional abundance in the 7 carnivorous and 21 herbivorous mammalian microbiomes using the program Shotgun FunctionalizeR (adjusted p<0.001 after multiple hypothesis correction, Table S5) (11). Many of the enzymes distinguishing carnivorous and herbivorous fecal microbiomes are involved in amino acid metabolism. Enzymes enriched in samples from herbivores mapped to biosynthetic reactions for 12 amino acids, while no enzymes for amino acid biosynthesis were enriched in sampled carnivores (Table S6). In contrast, nine amino acid degradation pathways contained enzymes enriched in carnivores, while the only degradative enzymes enriched in herbivores were for the breakdown of branched-chain amino acids (Val, Leu, Ile). Glutamate metabolism is particularly illustrative of these trends. Both the ATP-dependent and ATP-independent pathways for glutamate biosynthesis are significantly enriched in herbivore microbiomes, while the catabolic reactions to break down glutamate and glutamine are enriched in carnivores (Fig. 3A). These results suggest carnivorous microbiomes have specialized to degrade proteins as an energy source, while herbivorous communities have specialized to synthesize amino acid building blocks.
Figure 3. Differences in metabolic features encoded in fecal microbiomes among herbivores versus carnivores.
(A) Carnivorous and herbivorous microbiomes indicate opposing directionality for amino acid metabolism. Colored arrows denote enzyme functions significantly (p<0.001) enriched in the fecal microbiomes of herbivores (green) or carnivores (red). (B) Carnivorous and herbivorous microbiomes suggest opposing directionality at the central PEP-Pyruvate-Oxaloacetate node. Coloring scheme as in panel A. Abbreviations: 2-OG, alpha-ketoglutarate; GABA, γ–aminobutyrate; DH, Dehydrogenase; OAA, Oxaloacetate; Dx, Decarboxylase; Ck, Carboxykinase; Cx, Carboxykinase; PEP, Phosphoenolpyruvate; Pyr, Pyruvate; SSA, Succinate-Semialdehyde.
The distinctiveness of carnivorous and herbivorous microbiomes was also revealed at a central anaplerotic node (Fig 3B). When gluconeogenesis is required, oxaloacetate (OAA) can be converted to phosphoenolpyruvate (PEP) and pyruvate. When TCA cycle intermediates are withdrawn for biosynthesis, they are replenished by converting PEP and pyruvate directly to OAA (12). All of the genes encoding enzymes catalyzing OAA production from pyruvate or PEP are significantly increased in the carnivore microbiomes, while the reverse reactions are catalyzed by enzymes whose representation is increased in herbivore microbiomes.
Our studies comparing mammalian species revealed a relationship between host diet and gut microbial community structure and function. We next asked if similar trends could be detected using diet variation within a single free-living host species, namely humans. Quantitative studies of diet in most human populations are complicated by the known inaccuracy of self-reported data (13), so we turned to a group of adults known to keep meticulous records about their daily food composition and consumption. The selected cohort consisted of 18 lean members of the Calorie Restriction Society who typically measure and record all components of their diets on a daily basis with computer software to insure optimal nutrition despite reduced energy intake (14,15). We collected their dietary records for a four-day period (conservatively encompassing at least one complete intestinal transit time) prior to obtaining a single fecal sample, and analyzed macro- and micro-nutrient consumption using a validated protocol (2,17). An average of 3,642±3,826 bacterial V2 16S rRNA reads and 54,295 ±28,086 shotgun reads were obtained per sample (Tables S7–S10).
Procrustes analysis revealed a significant association between the bacterial phylogenetic structure of their fecal communities (16S rRNA) and the functions encoded in their microbiomes (p<0.05 for KOs, E.C.s, and CAZymes (glycoside hydrolases); not significant for peptidases [p=0.061]; Fig. S3). These results suggest that the processes that drive functional differentiation of microbiomes within an individual host species may be fundamentally similar to those that drive their differentiation across mammalian evolution.
Documentation of the weight of each ingredient in each meal consumed by these individuals (Table S7) allowed us to perform a follow-up analysis examining the impact on fecal bacterial community configuration of three dietary components (total protein, carbohydrate, and insoluble fiber intake). We chose these diet categories because protein intake is markedly different between carnivores and herbivores, and because an extensive literature exists about the impact of ingested polysaccharides and fiber on the gut microbiota (17). Linear regression of the three dietary categories against the position of each individual’s microbiome along Principal Coordinate 1 of the PCoA plots revealed that total protein intake was significantly associated with KO data (adjusted R2 value=0.307, adjusted p-value=0.030)(2). In contrast, insoluble dietary fiber was significantly associated with bacterial OTU content (Bray-Curtis metric; adjusted R2 value=0.371; adjusted p-value=0.013) (Table S11). These results confirm that within a single free-living species, both the structure and function of the gut microbiome are significantly associated with dietary intake.
Taken together with our prior work (1), these results teach us that even fecal samples from mammals living in zoos and human samples from a single self-selected population can provide insights into the factors driving the evolution of the gut microbiome. They also compel us, at a time when complete genomes are to be sequenced for 10,000 vertebrates (18), to take the next step and perform systematic studies that rigorously test specific mechanisms that drive the co-evolution of hosts and their (gut) microbial symbionts. These studies should be guided by experts who can choose taxa that radiated at different points in their evolutionary history, with parallel shifts in their diet, morphology, biogeography, or other key factors known or hypothesized to influence evolution. The results should help address questions such as what functional features in host intestinal environments (e.g., the biochemical characteristics of mucosal surfaces) are related to the representation of specific bacterial taxa and microbiome functions, and how readily microbial populations have been acquired and re-acquired during the course of vertebrate evolution. Additionally, our findings emphasize the need to sample humans across the globe with a variety of extreme diets and lifestyles, including relatively ancestral hunter-gatherer lifestyles, in order to provide new insights into the limits of variation within a host species and the possibility that our microbes, in coevolving with our bodies and our cultures, have help shaped our physiological differences and environmental adaptations.
Supplementary Material
Acknowledgments
We thank Jill Manchester and Sabrina Wagoner for technical assistance, Brandi Cantarel, Vincent Lombard, Corinne Rancurel, and Pedro Coutinho for CAZyme annotation, Ruth Ley and members of the Gordon lab for their suggestions; and Stephen Bircher Rob Ramey, Michael Schlegel, Mark Schrenzel, Tammy Tucker, and Peter Turnbaugh for past help in procuring mammalian fecal samples. This work was supported by grants from the NIH (DK30292, DK70977, DK078669, UL1 RR024992), the Crohn’s and Colitis Foundation of America, plus NIH Institutional Training Grant T32-A1007172 (to B.D.M.). 16S rRNA and fecal microbiome datasets have been deposited in MG-RAST.
Footnotes
Materials and Methods
References and Notes
- 1.Ley RE, et al. Evolution of mammals and their gut microbes. Science. 2008;320:1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.See supporting material on Science Online.
- 3.Kanehisa M, Goto S. KEGG: Kyoto encylocpedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cantarel BL, et al. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res. 2007;17:377–386. doi: 10.1101/gr.5969107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hurley JR, Cattell RB. The Procrustes program: Producing direct rotation to test a hypothesized factor structure. Behav Sci. 1962;7:258–262. [Google Scholar]
- 8.Gower JC. Generalized Procrustes analysis. Psychometrika. 1975;40:33–51. [Google Scholar]
- 9.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cerling TE, Ehleringer JR, Harris JM. Carbon dioxide starvation, the development of C4 ecosystems, and mammalian evolution. Philos Trans R Soc London Ser B. 1998;353:159–171. doi: 10.1098/rstb.1998.0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristiansson E, Hugenholtz P, Dalevi D. ShotgunFunctionalizeR: an R-package for functional comparison of metagenomes. Bioinformatics. 2009;25:2737–2738. doi: 10.1093/bioinformatics/btp508. [DOI] [PubMed] [Google Scholar]
- 12.Owen OE, Kalhan SC, Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem. 2002;277:30409–30412. doi: 10.1074/jbc.R200006200. [DOI] [PubMed] [Google Scholar]
- 13.Poslusna K, Ruprich J, de Vries JHM, Jakubikova M, van’t Veer P. Misreporting of energy and micronutrient intake estimated by food records and 24 hour recalls, control and adjustment methods in practice. Br J Nutr. 2009;101:S73–S85. doi: 10.1017/S0007114509990602. [DOI] [PubMed] [Google Scholar]
- 14.Fontana L, Klein S. Aging, adiposity, and calorie restriction. JAMA. 2007;297:986–994. doi: 10.1001/jama.297.9.986. [DOI] [PubMed] [Google Scholar]
- 15.Heilbronn LK, et al. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295:1539–1548. doi: 10.1001/jama.295.13.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schakel SF, Sievert YA, Buzzard IM. Sources of data for developing and maintaining a nutrient database. J Am Diet Assoc. 1988;88:1268–1271. [PubMed] [Google Scholar]
- 17.Flint HJ. Polysaccharide breakdown by anaerobic microorganisms inhabiting the Mammalian gut. Adv Appl Microbiol. 2004;56:89–120. doi: 10.1016/S0065-2164(04)56003-3. [DOI] [PubMed] [Google Scholar]
- 18.Genome 10K Community of Scientists. Genome 10K: a proposal to obtain whole-genome sequences for 10,000 vertebrate species. J Hered. 2009;100:659–674. doi: 10.1093/jhered/esp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.http://gordonlab.wustl.edu/Mammals_2011/
- 20.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeder J, Knight R. Rapidly denoising pyrosequencing reads by exploiting rank- abundance distributions. Nat Meth. 2010;7:668–669. doi: 10.1038/nmeth0910-668b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haas BJ, et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shannon P, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Youssef N, et al. Comparison of species richness estimates obtained using nearly complete fragments and simulated pyrosequencing-generated fragments in 16S rRNA gene-based environmental surveys. Appl Environ Microbiol. 2009;75:5227–5236. doi: 10.1128/AEM.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Alvarez V, Teal TK, Schmidt TM. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 26.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caspi R, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathways/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.http://www.metacyc.org/.
- 29.Dray S, Chessel D, Thioulouse J. Co-inertia analysis and the linking of ecological data tables. Ecology. 2003;84:3078–3089. [Google Scholar]
- 30.Francl KE, Schnell GD. Relationships of human disturbance, bird communities, and plant communities along the land-water interface of a large reservoir. Environ Monit Assess. 2002;73:67–93. doi: 10.1023/a:1012615314061. [DOI] [PubMed] [Google Scholar]
- 31.Caporaso JG, et al. Microbes and Health Sackler Colloquium: Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci USA. 2010 doi: 10.1073/pnas.1000080107. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight R, et al. PyCogent: a toolkit for making sense from sequence. Genome Biol. 2007;8:R171. doi: 10.1186/gb-2007-8-8-r171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Foundation for Statistical Computing. http://www.R-project.org.
- 34.Ochman H, et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8:e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.