Abstract
Developmental organophosphate exposure reduces the numbers of neural cells, contributing to neurobehavioral deficits. We administered chlorpyrifos or diazinon to newborn rats on postnatal days 1–4, in doses straddling the threshold for barely-detectable cholinesterase, and evaluated gene expression in the cell cycle and apoptosis pathways on postnatal day 5. Both organophosphates evoked transcriptional changes in 20–25% of the genes in each category; chlorpyrifos and diazinon targeted the same genes, with similar magnitudes of change, as evidenced by high concordance. Furthermore, the same effects were obtained with doses above or below the threshold for cholinesterase inhibition, indicating a mechanism unrelated to anticholinesterase actions. We then evaluated the effects of chlorpyrifos in undifferentiated and differentiating PC12 cells and found even greater targeting of cell cycle and apoptosis genes, affecting up to 40% of all genes in the pathways. Notably, the genes affected in undifferentiated cells were not concordant with those in differentiating cells, pointing to dissimilar outcomes dependent on developmental stage. The in vitro model successfully identified 60–70% of the genes affected by chlorpyrifos in vivo, indicating that the effects are exerted directly on developing neural cells. Our results show that organophosphates target the genes regulating the cell cycle and apoptosis in the developing brain and in neuronotypic cells in culture, with the pattern of vulnerability dependent on the specific stage of development. Equally important, these effects do not reflect actions on cholinesterase and operate at exposures below the threshold for any detectable inhibition of this enzyme.
Keywords: Apoptosis, Cell cycle, Chlorpyrifos, Developmental neurotoxicity, Diazinon, Gene transcription, Microarray, Organophosphate insecticides, PC12 cells
INTRODUCTION
Organophosphates are among the most widely-used insecticides and exposure of the human population is virtually ubiquitous [5]. Over the past decade, a number of countries have imposed restrictions on the residential use of several members of this class, including chlorpyrifos and diazinon, largely because of their propensity to elicit developmental neurotoxicity [7,15,26]. In turn, this reflects increasing recognition that damage to the developing brain occurs at exposures below the threshold for acute signs of intoxication and even at low levels that are insufficient to cause cholinesterase inhibition [26,28]. Irreversible inhibition of acetylcholinesterase by the active oxon metabolites is the mechanism underlying the systemic effects of organophosphates and, through plasma butyrylcholinesterase, provides a surrogate biomarker of exposure [5,17]. However, work over the past two decades shows conclusively that the native compounds are themselves developmental neurotoxicants at low, nonsymptomatic exposures and that consequently, the cholinesterase biomarker is inadequate to monitor safety [7,15,26]. Indeed, three recent studies documented cognitive loss in children exposed prenatally to organophosphates in residential and agricultural settings, involving exposures far too low to have elicited either signs of intoxication or cholinesterase inhibition [3,10,21], confirming the predictions of developmental neurotoxicity made from animal models [26,28].
A number of studies on the cholinesterase-unrelated effects underlying the developmental neurotoxicity of organophosphates have pinpointed the combination of antimitotic and pro-apoptotic mechanisms operating within a critical window of neurodifferentiation, leading to deficits in the numbers of neural and/or glia [12,15,24,26,28]. However, it is unclear whether these effects represent direct actions of the organophosphates on developing neural cells or indirect consequences of interference with more complex processes of brain development, such as cell-to-cell communication, oxidative stress or other aspects of altered metabolism or architectural assembly. Antimitotic and pro-apoptotic outcomes are also seen in vitro [12,16,19,23,43], supporting a direct mechanism; however, in vitro models cannot mimic the totality of the mechanisms contributing to organophosphate-induced developmental neurotoxicity, and therefore it would be pivotal to show that the antimitotic and proapoptotic effects in vitro are indeed reflecting actions on the same cellular mechanisms that occur in vivo. In the current study, we conducted a comparative examination of the effects of organophosphates on gene transcription known to be involved in the control of cell cycle and apoptosis. First, we compared the effects of in vivo exposures of neonatal rats to chlorpyrifos and diazinon, utilizing treatment paradigms known to produce neural cell loss and neurobehavioral abnormalities, focusing on doses well below those necessary for acute symptoms of exposure and straddling the threshold for barely-detectable inhibition of cholinesterase [41,42]. We then contrasted these effects with those seen with chlorpyrifos exposure in undifferentiated and differentiating neuronotypic PC12 cells, using treatment paradigms that do not cause overt cytotoxicity but that compromise neurodifferentiation [26,28]. We found 60–70% overlap in the pattern of affected genes between the two models, providing some of the first essential evidence that organophosphates directly target neural cell mitosis and apoptosis during a critical stage of neurodifferentiation.
MATERIALS AND METHODS
Animal treatments
No additional animal studies were conducted for this work. Instead, we mined gene expression data from microarrays used in earlier studies of the effects of chlorpyrifos and diazinon in neonatal rats [32]; here, we focused on cell cycle and apoptosis, as defined by the corresponding Gene Ontology categories [2]. Details of the animal treatments have been published [32] and therefore will be described only briefly. Neonatal rats received daily subcutaneous injections of chlorpyrifos or diazinon (both obtained from Chem Service, West Chester, PA) in dimethylsulfoxide vehicle on postnatal days 1–4, whereas control animals received equivalent injections of the vehicle (1 ml/kg). For both agents, we utilized doses below the threshold for growth retardation and the first signs of systemic toxicity [4,29,45]: 1 mg/kg for chlorpyrifos and 1 or 2 mg/kg for diazinon. The chlorpyrifos treatment and the higher dose of diazinon elicit less than 20% cholinesterase inhibition, well below the 70% threshold necessary for symptoms of cholinergic hyperstimulation [6], whereas the lower dose of diazinon produces no measurable inhibition [26,27,41,42,45]. On postnatal day 5 (24 hr after the last dose), one male pup was selected from each of five litters in each treatment group and separate determinations were made for the forebrain and brainstem from each animal.
Cell cultures
Because of the clonal instability of the PC12 cell line [11], the experiments were performed on cells that had undergone fewer than five passages. As described previously [20,43], PC12 cells (American Type Culture Collection CRL-1721, obtained from the Duke Comprehensive Cancer Center, Durham, NC) were seeded onto poly-D-lysine-coated plates in RPMI-1640 medium (Invitrogen, Carlsbad, CA) supplemented with 10% inactivated horse serum (Sigma Chemical Co., St. Louis, MO), 5% inactivated fetal bovine serum (Sigma), and 50 μg/ml penicillin streptomycin (Invitrogen). Incubations were carried out with 7.5% CO2 at 37°C, standard conditions for PC12 cells. To initiate neurodifferentiation [13,31,44] twenty-four hours after seeding, the medium was changed to include 50 ng/ml of 2.5 S murine nerve growth factor (Invitrogen). Determinations were made in either undifferentiated or differentiating cells in the presence or absence of 30 μM chlorpyrifos, a concentration chosen from earlier studies that demonstrated adverse effects on mitotic activity and neurodifferentiation of PC12 cells without outright cytotoxicity [14,19,31,39]. Chlorpyrifos was dissolved in dimethylsulfoxide (final concentration 0.1%), which was also added to the control cultures; this concentration of dimethylsulfoxide has no effect on PC12 cell growth or differentiation [19,20,43]. Cultures were examined 24 and 72 hr after commencing exposure, with 5 independent cultures evaluated for each treatment at each time point. We used two time points so as to be able to evaluate changes in gene expression regardless of whether the mRNA for a given gene has a rapid turnover (and hence can rise rapidly) or a slower turnover that would require a longer period to show corresponding increases or decreases. Each culture was run on a separate array.
Microarray determinations
Our earlier studies detailed the techniques for mRNA isolation, preparation of cDNA, conversion to cRNA incorporating cyanine-3 (reference RNA) or cyanine-5 (sample RNA), verification of RNA purity and quality, hybridization to the microarrays, washing and scanning [32,38,39]. These all involve commercial kits and standardized procedures, and since the current studies were done identically, the techniques will not be described here. The mRNA used for the reference standard was created by pooling aliquots from each of the samples in the study so as to ensure measurable levels of all genes expressed over the background. Array normalizations and error detection were also carried out by standard procedures described previously [32,38,39]. We used type G4131A Agilent Whole Rat Genome Arrays (Agilent Technologies, Palo Alto, CA), selecting the Gene Ontology categories for cell cycle and apoptosis within the GeneSpring™ platform.
For many of the genes, the arrays contain multiple probes and/or replicates of the same probe in different locations on the chip, and these were used to verify the reliability of values and the validity of the measures on the chip. In these cases, to avoid artificially inflating the number of genes utilized for statistical analysis, we limited each gene to a single set of values, selecting those obtained for the probe showing the smallest intragroup variance. The other values for that gene were used only to corroborate direction and magnitude of change. We also validated the readings on the arrays through the use of duplicate arrays for one sample selected from each treatment group [32,38].
Statistical procedures
Results were compiled as means and standard errors of the normalized expression ratios from the arrays, with statistical analyses performed on log-transformed values, since the data are in the form of ratios. For the in vivo studies, our design involved planned comparisons of four different treatments (control, 1 mg/kg chlorpyrifos, 1 mg/kg diazinon, 2 mg/kg diazinon) in two different brain regions (brainstem, forebrain), across hundreds of genes in each category. It was therefore important to protect against the increased probability of type 1 errors engendered by repeated testing of the same data base. Accordingly, before looking for effects on individual genes, we performed a global ANOVA incorporating all the variables in a single comparison. Lower-order ANOVAs on subdivisions of the data set were then carried out as permitted by the interactions of treatment with the other variables; however, where there were no interactions, we report only main treatment effects without further subdivision. Differences for individual treatments were evaluated with Fisher’s Protected Least Significant Difference test. Similarly, for the in vitro studies, our design involved planned comparisons of two treatments (control, chlorpyrifos) in two differentiation states (undifferentiated vs. differentiating) at two time points and for hundreds of genes in each category. Accordingly, the global ANOVA included the factors of treatment, time, and all genes involved in the planned comparisons; lower-order tests were then carried out using the same procedures.
In addition to these parametric tests of the direction and magnitude of changes in gene expression, we evaluated the incidence of significant differences as compared to the predicted false positive rate, using the χ2 test. This was necessary because the large numbers of genes in each category (252 genes for cell cycle, 217 for apoptosis) generate 11–12 false positive significant differences; accordingly, an interpretation of a significant impact on either process requires a higher incidence of significant increases and/or decreases in gene expression than the false positive rate. Unlike the parametric ANOVA, which weights the results according to the magnitude of the effect, this nonparametric approach simply determines whether a given gene shows a significant difference from the corresponding control, regardless of the absolute magnitude of the difference. The alternate approach thus addresses the important contribution of differences in variability among genes: a large difference for a gene with high variability may not be statistically significant, whereas a smaller difference for a tightly-controlled gene may be both significant and biologically relevant [32]. To classify a given gene as showing a significant increase or decrease, we tested the values using a two-factor ANOVA, with factors of treatment and region for the in vivo studies, and treatment and time for the in vitro studies. Where a main treatment effect was identified, the result for that gene was classified as an increase or a decrease. Where there was only an interaction of treatment × region (in vivo) or treatment × time (in vitro), we evaluated each region or time point for a significant treatment effect, with three possible outcomes. Where neither region or time point showed a significant treatment effect, that gene was classified as “unchanged,” despite the significant interaction term. Where the interaction reflected the fact that only one region or time point showed a significant treatment effect, that gene was classified as “increased” or “decreased” depending on the direction of change for the individual value. Finally, where there was an interaction and both regions or time points showed significant changes, but in opposite directions from each other, that value was tabulated as ½ increase and ½ decrease. The χ2 test was then run for the distribution of observed values in the three categories (increased, decreased, unchanged) as compared to the values expected from a random event: at p < 0.05, the false-positive rate corresponds to a statistically significant increase in 2.5% of the genes, a significant decrease in 2.5%, and 95% unchanged.
Finally, we conducted concordance analysis to evaluate: (a) the effects of chlorpyrifos and diazinon for each of the two gene categories in vivo, so as to evaluate the degree to which the two organophosphates target the same genes; (b) the effects of chlorpyrifos on undifferentiated vs. differentiating cells in vitro, so as to determine whether the differentiation state of the cells alters the overall pattern of which genes are targeted; and (c) the effects of chlorpyrifos in vivo vs. those seen in vitro. These were all performed by linear regression analysis of pairwise comparisons [35,36,40].
For all tests, treatment effects were considered significant at p < 0.05 (two-tailed).
RESULTS
Validation of gene groupings
For the in vivo study, transcripts of 252 of the genes designated as “cell cycle-related” were detected and passed the quality-control filters of the GeneSpring™ platform and indeed, the same transcripts were detected in both the brainstem and forebrain samples (Supplemental Table 1). Similarly, for the genes designated as “apoptosis-related,” 217 were detected and passed quality-control, with the same gene list in either brain region (Supplemental Table 2). For the in vitro samples, the same exact cell cycle (Supplemental Table 3) and apoptosis (Supplemental Table 4) transcripts were detected, with the exception of one gene in the cell cycle category (btc; failed quality-control because of excessive variability); because this gene was unaffected by the in vivo organophosphate exposures, its inclusion or exclusion did not change any of the outcomes. Twenty-eight genes appeared on both the cell cycle and apoptosis lists (aatf, ahr, akt1, appbp1, atm, bcl10, brca1, casp3, cdkn1a, dcc, ddit3, ets1, gsk3b, gspt1, il18, inha, myc, mycs, notch2, pglyrp1, plagl1, pten, siah2, tgfa, tp53, trp63, ube1c, vegf) and again, these were duplicated exactly between the in vivo and in vitro results.
To further validate that the gene expression patterns reflected biologically meaningful processes, we compared the values for brainstem vs. forebrain in the control group. By postnatal day 5 in the rat brain, neurogenesis is essentially complete in the brainstem but is still ongoing to some extent in some areas of the forebrain, whereas apoptosis, a later developmental event, is greater in the brainstem and has not yet peaked in the forebrain [22]. In accord with this known temporal relationship, we found a significantly greater overall expression of cell cycle genes in the forebrain (main effect of region, p < 0.0001; Supplemental Table 1) but higher expression of apoptosis genes in the brainstem (main effect of region, p < 0.0001; Supplemental Table 2). We then carried out the same comparisons for the in vitro study. Cell cycle genes were more highly expressed in undifferentiated control PC12 cells as compared to differentiating cells (main effect of differentiation state, p < 0.005; Supplemental Table 3), whereas apoptosis genes were higher in differentiating cells (main effect of differentiation state, p < 0.0001; Supplemental Table 4), in keeping with the elevated apoptotic rate during differentiation [9,18].
In vivo effects
Early postnatal exposure to organophosphates had highly significant (p < 0.0001) effects on the expression of cell cycle-related genes (Supplemental Table 1) and apoptosis-related genes (Supplemental Table 2), with the effects showing significant interactions with region and gene, thus requiring separate analysis for each. The global test also indicated a main treatment effect (p < 0.0009) for the apoptosis-related genes, which was significant for each of the treatments individually vs. control (p < 0.05 for chlorpyrifos, p < 0.02 for the low dose of diazinon, p < 0.0001 for the higher diazinon dose); in each case, this reflected a net decrease in expression for this gene grouping. Treatment effects remained statistically robust regardless of whether lower-order subdivisions were conducted according to individual treatments or regions (Supplemental Tables 1 and 2).
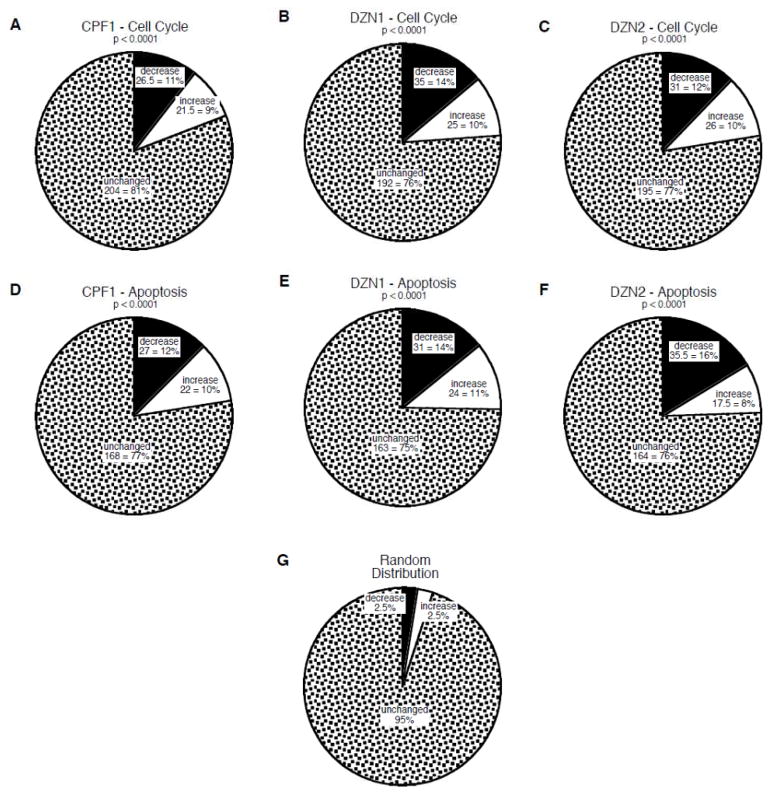
To characterize the effects on gene expression, we determined the proportion of genes showing significant alterations in expression evoked by organophosphate exposure (Figure 1). Chlorpyrifos elicited a significant decrease in expression for 11% of all the cell cycle-related genes and an increase for 9% (Figure 1A). Similarly, either dose of diazinon evoked a decrease in expression for 12–14% of the genes and an increase for about 10% (Figure 1B,C). The same pattern was seen for apoptosis-related genes, with significant decreases or increases seen for 22% of the genes for chlorpyrifos (Figure 1D), and 25% for diazinon at either the low dose (Figure 1E) or the higher dose (Figure 1F). Each of these effects was highly statistically different from what would have been at seen at random (Figure 1G), namely a 2.5% incidence of significant increases and a 2.5% incidence of significant decreases. Furthermore, the net effects were significantly skewed toward decreased gene expression (p < 0.01), in keeping with the parametric determinations (Supplemental Tables 1 and 2) described above. Of the 28 genes shared by the cell cycle and apoptosis categories, 20 were negative for organophosphate effects and the remaining 8 accounted for only 3 significant changes for chlorpyrifos, 4 for the low dose of diazinon, and 5 for the higher dose of diazinon, so these repeated values were not pivotal to the net outcome.
Figure 1.
Effects of in vivo organophosphate treatments on genes involved in cell cycle (A,B,C) and apoptosis (D,E,F): CPF1 = 1 mg/kg chlorpyrifos (A,D); DZN1 = 1 mg/kg diazinon (C,E); DZN2 = 2 mg/kg diazinon (C,F). Charts show the number and percent of genes in each category that were unchanged, or that showed significant increases or decreases from control values. P-values for each chart compare the distribution of values from those expected at random (G).
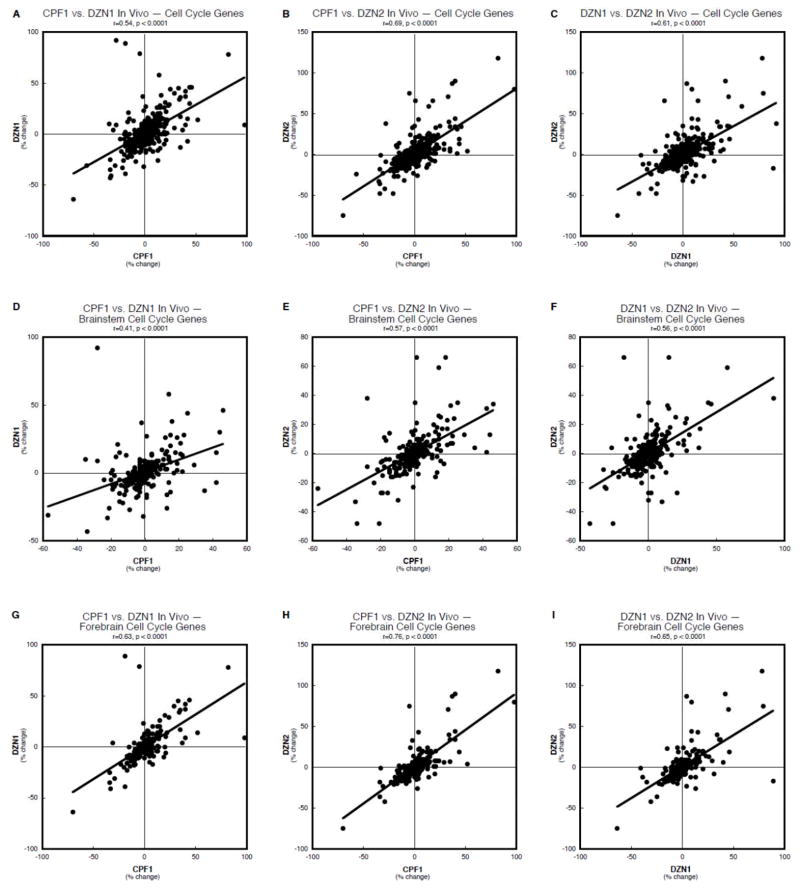
To determine whether the same genes were affected by chlorpyrifos and the two diazinon treatments, and whether these effects shared the same magnitude and regional selectivity, we performed concordance analysis to correlate the pairwise effects. First, for the cell cycle genes, considering both regions together, there was a highly significant correlation between the effects of chlorpyrifos and either the low (Figure 2A) or higher dose (Figure 2B) of diazinon; similarly, the effects of the two diazinon regimens were highly concordant (Figure 2C). Regarding only the brainstem (Figure 2D,E,F), these correlations all remained statistically robust but the coefficients were uniformly lower than for both regions taken together. In contrast, restricting the comparisons to the forebrain resulted in higher correlations than for the brainstem or for both regions taken together (Figure 2G,H,I). It should be noted that, in each case, these highly significant relationships were detected even against the background of the inclusion of 12 randomly-changed genes for each treatment, generated by the false positive rate.
Figure 2.
Concordance analysis of the effects of in vivo organophosphate treatments on cell cycle genes, shown for brainstem and forebrain combined (A,B,C), and separately for the brainstem (D,E,F) and forebrain (G,H,I): chlorpyrifos 1 mg/kg vs. diazinon 1 mg/kg (A,D,G), chlorpyrifos 1 mg/kg vs. diazinon 2 mg/kg (B,E,H), and diazinon 1 mg/kg vs. diazinon 2 mg/kg (C,F,I). Data are shown as the percent change from control values for all genes in the cell cycle category, regardless of whether the change was statistically significant or not. The linear correlation coefficient and its statistical significance are shown at the top of each panel and the line represents the least-squares fit of the data points.
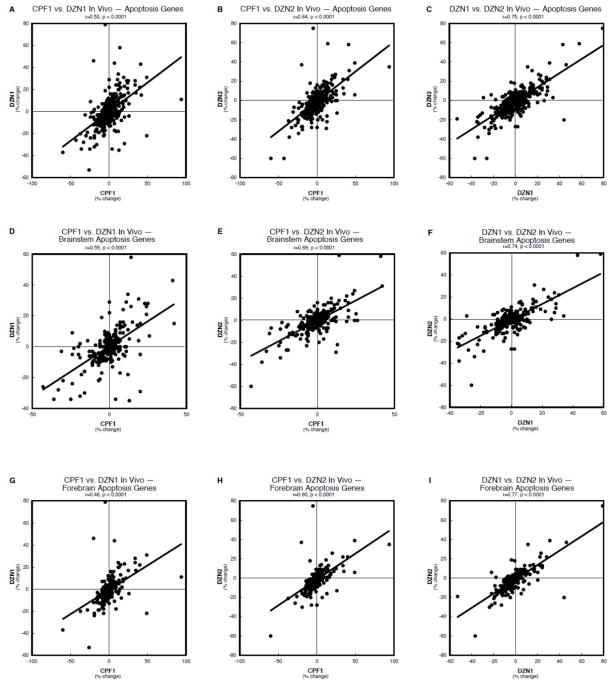
Concordance analysis of the apoptosis-related genes likewise showed highly significant correlations of the effects of chlorpyrifos with either the low (Figure 3A) or high dose (Figure 3B) of diazinon, or of the two diazinon treatments (Figure 3C). However, in this case, examining regional subdivisions did not reveal a net improvement in the outcome, with only slight changes in the correlation coefficient when the values were restricted to the brainstem (Figure 3D,E,F) or forebrain (Figure 3G,H,I).
Figure 3.
Concordance analysis of the effects of in vivo organophosphate treatments on apoptosis genes, shown for brainstem and forebrain combined (A,B,C), and separately for the brainstem (D,E,F) and forebrain (G,H,I): chlorpyrifos 1 mg/kg vs. diazinon 1 mg/kg (A,D,G), chlorpyrifos 1 mg/kg vs. diazinon 2 mg/kg (B,E,H), and diazinon 1 mg/kg vs. diazinon 2 mg/kg (C,F,I). Data are shown as the percent change from control values for all genes in the cell cycle category, regardless of whether the change was statistically significant or not. The linear correlation coefficient and its statistical significance are shown at the top of each panel and the line represents the least-squares fit of the data points.
In vitro effects
In the PC12 cell model, chlorpyrifos exposure had highly significant effects on the expression of genes involved in either cell cycle (Supplemental Table 3) or apoptosis (Supplemental Table 4). For both classes of genes, the effects of chlorpyrifos were highly dependent on the differentiation state of the cells, as indicated by significant interactions of treatment × state and/or treatment × state × other variables (time, gene). The significant effects were maintained when values were subdivided according to differentiation state. For cell cycle-related genes in the undifferentiated state, there was a main treatment effect of chlorpyrifos (p < 0.005), reflecting a net decrease in gene expression, along with gene-specific and time-specific effects (significant interactions of treatment with these other variables); the effects in differentiating cells also were highly significant but did not display a specific direction of change (interactions of treatment × gene and of treatment × gene × time, but no main treatment effect). For apoptosis-related genes, the net effects again depended highly on whether the cells were in the undifferentiated state or were differentiating (treatment × gene × state, p < 0.0001; treatment × gene × state × time, p < 0.0001), and again showed interactions with the other variables, necessitating separate consideration of the changes for each gene. However, for apoptosis, chlorpyrifos did not evoke a net direction of change in either undifferentiated or differentiating cells (no main treatment effect), indicating roughly equivalent net magnitudes up- and downregulation of the various genes.
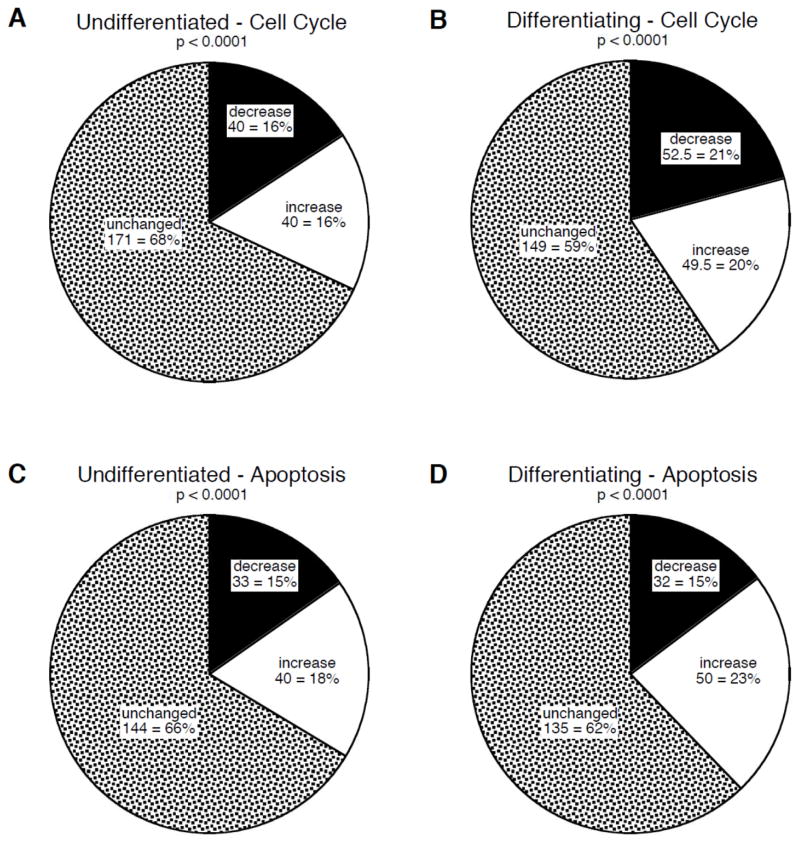
Chlorpyrifos exposure produced significant alterations in the expression of 32–41% of the cell cycle genes in either the undifferentiated state (Figure 4A) or in differentiating cells (Figure 4B), with approximately equivalent proportions of the genes showing increases or decreases; as before, this distribution was highly significantly different from the value of 5% (2.5% increased, 2.5% decreased) that would have been seen at random (Figure 1G). Chlorpyrifos evoked significant changes in a similar proportion of genes in the apoptosis category, again spanning both the undifferentiated cells (Figure 4C) and those undergoing differentiation (Figure 4D). Notably, across both gene categories, a higher proportion of genes was affected in differentiating cells as compared to undifferentiated cells (χ2=4.2, p < 0.05). Just as for the in vivo studies, 28 genes were shared by the cell cycle and apoptosis categories, and in this case, 9 showed significant effects of chlorpyrifos in undifferentiated cells and 10 in differentiating cells.
Figure 4.
Effects of in vitro chlorpyrifos exposure on genes involved in cell cycle (A,B) and apoptosis (C,D) in undifferentiated (A,C) and differentiating (B,D) PC12 cells. Charts show the number and percent of genes in each category that were unchanged, or that showed significant increases or decreases from control values. P-values for each chart compare the distribution of values from those expected at random (Figure 1G).
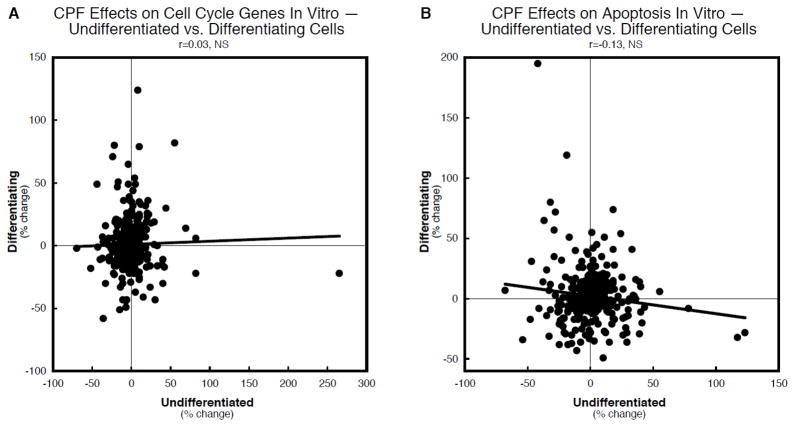
We then determined whether the effects on undifferentiated vs. differentiating cells involved the same genes, with a similar magnitude and time course of effect, correlating the values for the two differentiation states across the two time points (24h, 72h). There was a very slight, nonsignificant positive correlation for cell cycle-related genes (Figure 5A) and a small, nonsignificant negative correlation for apoptosis-related genes (Figure 5B).
Figure 5.
Concordance analysis of the effects of in vitro chlorpyrifos on genes for cell cycle (A) and apoptosis (B), comparing undifferentiated to differentiating PC12 cells. Data are shown as the percent change from control values for all genes in the appropriate category, regardless of whether the change was statistically significant or not. The linear correlation coefficient and its statistical significance are shown at the top of each panel and the line represents the least-squares fit of the data points. NS = not significant.
Finally, we examined whether the in vitro model produced changes in gene expression that paralleled those seen with chlorpyrifos treatment in vivo. Approximately 60–70% of the genes that were significantly affected in the neonatal rat brain regions were also detected as significantly changed in the cell culture model (Supplementary Tables 1–4). Nevertheless, when we subjected the data to concordance analysis, we found that the magnitude and direction of effect did not correspond closely between the two models. There were 8 possible pairwise comparisons (2 brain regions in vivo × 2 differentiation states in vitro × 2 time points in vitro), none of which showed significant concordance (data not shown).
DISCUSSION
Our findings show that organophosphates greatly target the gene families involved in the control of cell cycle and apoptosis both in vivo and in vitro, that there is a high degree of concordance in the effects of chlorpyrifos and diazinon, and that the overwhelming majority of genes affected with in vivo exposure are changed likewise in cell cultures. Taken together, these findings strongly support the conclusion that organophosphates exert a direct effect on cell replication and on programmed cell death, over and above additional effects that may be exerted at the level of cell-to-cell interactions, architectural assembly or higher levels of brain complexity. Further, the stark differences in the effects on transcriptional patterns between undifferentiated and differentiating neuronotypic PC12 cells reinforce in vivo findings of a critical window in which neural development is most vulnerable to organophosphates [26,28].
Exposure of neonatal rats to either chlorpyrifos or diazinon produced significant transcriptional changes in genes for both the cell cycle and apoptosis categories, with the number of genes affected (20–25% in each category) far exceeding the 5% incidence that would have been seen at random. Furthermore, the effects seen across all genes within each grouping (i.e. regardless of whether individual changes were statistically significant) were highly concordant between chlorpyrifos and diazinon. This means that not only were the same genes targeted by both organophosphates but the magnitude and direction of change across all the genes were decidedly similar; indeed the highly significant concordance was found despite the obligatory inclusion of up to 12 false positive changes for each treatment in each gene category, effects that are highly unlikely to be shared between the two organophosphates and that would tend to reduce concordance. This strong relationship stands in contrast to the disparities between chlorpyrifos and diazinon for gene families that almost certainly involve complex or indirect effects, such as excitotoxicity, neurotransmitter receptors or neurotransmitter transporters [30,34,35,37]. Accordingly, the current findings support the conclusion that organophosphates act more directly on cell cycle and apoptosis than they do on other features known to be part of their spectrum of developmental neurotoxic actions; as will be seen below, this is more categorically reinforced by the results in cell cultures.
There were three additional features of the results of in vivo exposure worth noting. First, there was a preponderance of decreased gene expression as compared to increases, somewhat more notable for diazinon than for chlorpyrifos, and particularly so for apoptosis. Indeed, the parametric ANOVA results (which consider magnitude of change rather than numbers of genes affected) revealed a main effect of the organophosphate treatments, representing a significant net decrease in expression. The larger impact of diazinon on apoptosis-related genes is in accord with previous results showing a dichotomy between mechanisms of neural cell injury and death between the two organophosphates, with diazinon having a greater impact on oxidative stress whereas chlorpyrifos targets excitotoxicity [25,30,35].
The second notable feature was the similarity of the results from the two different doses of diazinon, whether toward cell cycle genes or genes involved in apoptosis, regardless of whether the criterion was the number of genes affected in each category or overall concordance. The two doses chosen for this study straddle the threshold for inhibition of cholinesterase [41], so that the basic similarity of the findings for low and high doses demonstrates that the underlying mechanisms are independent of effects on cholinesterase. Our findings thus reinforce the growing evidence that the developmental neurotoxicity of organophosphates involves mechanisms unrelated to their shared property as cholinesterase inhibitors and, equally important, remains prominent at exposures that are below the detection threshold for this biomarker. Such direct effects could readily explain the adverse outcomes seen in children exposed to organophosphates prenatally in standard residential or agricultural settings [3,10,21].
The third feature of the in vivo results was our finding of higher concordance between the organophosphates for effects on cell cycle in the forebrain as compared to the brainstem. Neural cell replication proceeds much earlier in the brainstem than in the forebrain [22], so when measurements are made in the early postnatal period, the forebrain represents a far more effective target for toxicants that affect the cell cycle. Indeed, even in the control group, we found greater overall expression of cell cycle genes in the forebrain, contrasted with higher expression of apoptosis genes in the brainstem, as would be expected from the underlying differences in developmental stage. These findings thus buttress the concept of a window of heightened vulnerability to adverse effects of neurotoxicants on mitotic and apoptotic activity, a conclusion that is further reinforced by the effects seen with the cell culture model.
Our in vitro studies were designed to test the two specific interpretations raised by the in vivo findings, namely that the organophosphate effects on cell cycle and apoptosis are mediated directly on developing cells, and that vulnerability depends highly on the developmental stage. Just as was found for the in vivo results, chlorpyrifos exposure in PC12 cells in either the undifferentiated or differentiating state, produced a change in a large proportion of the genes involved in cell cycle or apoptosis, much greater than would have been expected at random and indeed, exceeding the number of genes that were changed in vivo. The in vitro model successfully identified 60–70% of the cell cycle and apoptosis genes that were targeted in vivo, as well as pinpointing additional genes that were not found to be significantly changed in the intact brain. The disparity likely represents the fact that adverse effects directed at specific cell populations in the intact brain can be diluted by the inclusion of mRNA from larger amounts of unaffected tissue; organophosphate-induced developmental neurotoxicity targets specific brain regions and neuronal populations rather than producing global injury, highly dependent on the developmental stage of particular brain nuclei during the exposure period [26,28]. With the cell culture model, we are dealing with a single type of cell and a uniform timing of differentiation, thus increasing the sensitivity to detect effects on gene expression. Indeed, the in vitro findings could readily be used to guide future in vivo studies to evaluate heretofore unsuspected gene groupings that may be targeted by neurotoxicants. The main point, though, is that the culture model, which does not share the complex cell-to-cell interactions or architectural features of the developing brain, nevertheless shows targeting of gene expression involved in cell cycle and apoptosis closely related to the effects seen in vivo, supporting the conclusion that the organophosphates exert direct effects on these two developmental processes.
At first blush, it might seem that, since chlorpyrifos affected a similar number of genes in both undifferentiated and differentiating PC12 cells, there is no distinct period of vulnerability. However, more detailed analysis of the gene changes shows that this is not the case. First, there was no concordance between the effects of chlorpyrifos in undifferentiated vs. differentiating cells, meaning that the genes that were changed were not the same in the two states and/or that the direction and magnitude of effects were entirely distinct; indeed, the same stage-selective expression found in the in vivo control samples was also apparent in the in vitro model, with cell cycle genes more highly expressed in the undifferentiated cells (undergoing active replication), whereas apoptosis-related genes were higher in the differentiating cells. Second, when considering not only the number of genes changed but also the magnitude of change, we found a main effect of chlorpyrifos (ANOVA) to decrease cell cycle gene expression only in replicating (undifferentiated) cells; thus, although the numbers of genes increased or decreased were the same, the magnitude of the decreases was much larger than that of the increases, as might be expected from an agent that exerts antimitotic actions. Third, considering all the genes that were measured, differentiating cells showed a higher proportion of changes than did undifferentiated cells, consistent with earlier findings that showed that the most sensitive period for changing phenotypic outcomes centers around the transition from cell replication to neurodifferentiation [1,13,14,34,37,39]. Our results thus indicate that, although chlorpyrifos targets these gene families in both the undifferentiating and differentiating states, the pattern of gene changes, and the eventual outcomes, are distinctly different, supporting the existence of discrete windows of vulnerability to organophosphates [26,28].
Although the in vitro model successfully identified the majority of the genes that were changed with in vivo treatment, when we applied more stringent criteria of direction and magnitude of change, we did not find significant concordance between the two models. This is not surprising, given the inclusion of a heterogeneous population of cells in the intact brain, each with different phenotypes and in a different state of differentiation, as contrasted to a single cell type and uniform state of differentiation in the PC12 model. Furthermore, the treatment paradigms differ in that there is continuous exposure in the cell culture model, whereas the in vivo treatments, of necessity, involve episodic exposure via daily administration. As found in earlier work with gene expression profiling, this leads to a situation where the same genes may be identified as targets in both models, but the direction and magnitude do not correspond [1,8,33,39]; for example, direct suppression of gene expression would likely lead to a rebound elevation when sampled 24 hr later in vivo, whereas the same gene would remain suppressed with continuous exposure in vitro. Although these considerations make it difficult to draw an exact parallel between results obtained in vivo and in vitro, the main conclusion remains that the same genes are targeted by chlorpyrifos in either model, and both models reinforce the concept of stage-specific effects that depend on cell differentiation state.
The microarray approach used in this study has both advantages and limitations when compared to more traditional uses of microarrays. Here, we relied on a statistical comparison of the numbers of genes affected in biological pathways that were chosen because they are known or suspected targets for the organophosphates; this information was available from both in vivo and in vitro studies of the specified antimitotic and proapoptotic outcomes. Accordingly, we used the overall patterns of gene effects to compare neurotoxicants from the same or different classes of compounds in a global manner; likewise, we contrasted the patterns of effects seen in vivo with those obtained from a cell culture model. The similarities and differences identified by this approach were clear-cut and statistically robust. In contrast, using microarrays to determine which specific genes are affected and the precise magnitude of effect, would require RT-PCR validation to ensure that those particular genes were not among the false positives; further interpretation of whether a given gene was, for example, antiapoptotic or proapoptotic could then be undertaken if one also identified the time course of effects so as to determine direct vs. reactive changes in expression. These more detailed analyses are clearly beyond the scope of the current study, which instead uses global expression patterns to characterize the impact of neurotoxicant exposures on entire pathways rather than a specific gene or subset of genes. Indeed, one of the points of this study is to illustrate the value of this alternative use of microarray data.
In conclusion, our results show that organophosphates directly target the genes regulating the cell cycle and apoptosis in the developing brain and in neuronotypic cells in culture. The same genes are affected in vivo and in vitro and both models indicate distinct patterns of vulnerability corresponding to the specific stage of development. Equally important, these effects do not reflect actions on cholinesterase and operate at exposures below the threshold for any detectable inhibition of this enzyme. Although there are clear limitations to the in vitro approach, the greater sensitivity of the culture model can help guide future in vivo work by identifying additional genes and pathways targeted by the organophosphates, that can then be related to adverse neurodevelopmental outcomes.
Supplementary Material
Highlights.
Chlorpyrifos or diazinon exposure in neonatal rats targeted genes involved in cell cycle & apoptosis
Effects were highly significantly concordant between the two organophosphates
The same effects were seen at doses above or below the threshold for cholinesterase inhibition
The same pathways were affected in vitro in PC12 cells
Organophosphates directly affect cell cycle and apoptosis by mechanisms unrelated to cholinesterase inhibition
Acknowledgments
Research was supported by NIH ES010356. TAS has provided expert witness testimony in the past three years at the behest of the following law firms: The Calwell Practice (Charleston WV), Weltchek Mallahan & Weltchek (Lutherville MD), Finnegan Henderson Farabow Garrett & Dunner (Washington DC), Carter Law (Peoria IL), Gutglass Erickson Bonville & Larson (Madison WI), The Killino Firm (Philadelphia PA), Alexander Hawes (San Jose, CA), Pardieck Law (Seymour, IN), Tummel & Casso (Edinburg, TX) and the Shanahan Law Group (Raleigh NC).
Abbreviations
- ANOVA
analysis of variance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Adigun AA, Seidler FJ, Slotkin TA. Disparate developmental neurotoxicants converge on the cyclic AMP signaling cascade, revealed by transcriptional profiles in vitro and in vivo. Brain Res. 2010;1316:1–16. doi: 10.1016/j.brainres.2009.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouchard MF, Chevrier J, Harley KG, Kogut K, Vedar M, Calderon N, Trujillo C, Johnson C, Bradman A, Barr DB, et al. Prenatal exposure to organophosphate pesticides and IQ in 7-year old children. Environ Health Perspect. 2011 doi: 10.1289/ehp.1003185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell CG, Seidler FJ, Slotkin TA. Chlorpyrifos interferes with cell development in rat brain regions. Brain Res Bull. 1997;43:179–189. doi: 10.1016/s0361-9230(96)00436-4. [DOI] [PubMed] [Google Scholar]
- 5.Casida JE, Quistad GB. Organophosphate toxicology: safety aspects of nonacetylcholinesterase secondary targets. Chem Res Toxicol. 2004;17:983–998. doi: 10.1021/tx0499259. [DOI] [PubMed] [Google Scholar]
- 6.Clegg DJ, van Gemert M. Determination of the reference dose for chlorpyrifos: proceedings of an expert panel. J Toxicol Environ Health. 1999;2:211–255. doi: 10.1080/109374099281179. [DOI] [PubMed] [Google Scholar]
- 7.Colborn T. A case for revisiting the safety of pesticides: a closer look at neurodevelopment. Environ Health Perspect. 2006;114:10–17. doi: 10.1289/ehp.7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crumpton TL, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos in vivo and in vitro: effects on nuclear transcription factor involved in cell replication and differentiation. Brain Res. 2000;857:87–98. doi: 10.1016/s0006-8993(99)02357-4. [DOI] [PubMed] [Google Scholar]
- 9.Ekshyyan O, Aw TY. Decreased susceptiblity of differentiated PC12 cells to oxidative challenge: relationship to cellular redox and expression of apoptotic protease activator factor-1. Cell Death Differ. 2005;12:1066–1077. doi: 10.1038/sj.cdd.4401650. [DOI] [PubMed] [Google Scholar]
- 10.Engel SM, Wetmur J, Chen J, Zhu C, Barr DB, Canfield RL, Wolff MS. Prenatal exposure to organophosphates, paraoxonase 1, and cognitive development in childhood. Environ Health Perspect. 2011 doi: 10.1289/ehp.1003183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujita K, Lazarovici P, Guroff G. Regulation of the differentiation of PC12 pheochromocytoma cells. Environ Health Perspect. 1989;80:127–142. doi: 10.1289/ehp.8980127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia SJ, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos: targeting glial cells. Environ Toxicol Pharmacol. 2005;19:455–461. doi: 10.1016/j.etap.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 13.Jameson RR, Seidler FJ, Qiao D, Slotkin TA. Chlorpyrifos affects phenotypic outcomes in a model of mammalian neurodevelopment: critical stages targeting differentiation in PC12 cells. Environ Health Perspect. 2006;114:667–672. doi: 10.1289/ehp.8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jameson RR, Seidler FJ, Slotkin TA. Nonenzymatic functions of acetylcholinesterase splice variants in the developmental neurotoxicity of organophosphates: chlorpyrifos, chlorpyrifos oxon and diazinon. Environ Health Perspect. 2007;115:65–70. doi: 10.1289/ehp.9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mauro RE, Zhang L. Unique insights into the actions of CNS agents: lessons from studies of chlorpyrifos and other common pesticides. CNS Agents Med Chem. 2007;7:183–199. [Google Scholar]
- 16.Monnet-Tschudi F, Zurich MG, Schilter B, Costa LG, Honegger P. Maturation-dependent effects of chlorpyrifos and parathion and their oxygen analogs on acetylcholinesterase and neuronal and glial markers in aggregating brain cell cultures. Toxicol Appl Pharmacol. 2000;165:175–183. doi: 10.1006/taap.2000.8934. [DOI] [PubMed] [Google Scholar]
- 17.Pope CN. Organophosphorus pesticides: do they all have the same mechanism of toxicity? J Toxicol Environ Health. 1999;2:161–181. doi: 10.1080/109374099281205. [DOI] [PubMed] [Google Scholar]
- 18.Powers CM, Wrench N, Ryde IT, Smith AM, Seidler FJ, Slotkin TA. Silver impairs neurodevelopment: studies in PC12 cells. Environ Health Perspect. 2010;118:73–79. doi: 10.1289/ehp.0901149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiao D, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos modeled in vitro: comparative effects of metabolites and other cholinesterase inhibitors on DNA synthesis in PC12 and C6 cells. Environ Health Perspect. 2001;109:909–913. doi: 10.1289/ehp.01109909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiao D, Seidler FJ, Violin JD, Slotkin TA. Nicotine is a developmental neurotoxicant and neuroprotectant: stage-selective inhibition of DNA synthesis coincident with shielding from effects of chlorpyrifos. Dev Brain Res. 2003;147:183–190. doi: 10.1016/s0165-3806(03)00222-0. [DOI] [PubMed] [Google Scholar]
- 21.Rauh V, Arunajadai S, Horton M, Perera F, Hoepner L, Barr DB, Whyatt R. 7-Year neurodevelopmental scores and prenatal exposure to chlorpyrifos, a common agricultural pesticide. Environ Health Perspect. 2011 doi: 10.1289/ehp.1003160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodier PM. Structural-functional relationships in experimentally induced brain damage. Prog Brain Res. 1988;73:335–348. doi: 10.1016/S0079-6123(08)60514-2. [DOI] [PubMed] [Google Scholar]
- 23.Roy TS, Andrews JE, Seidler FJ, Slotkin TA. Chlorpyrifos elicits mitotic abnormalities and apoptosis in neuroepithelium of cultured rat embryos. Teratology. 1998;58:62–68. doi: 10.1002/(SICI)1096-9926(199808)58:2<62::AID-TERA7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Roy TS, Sharma V, Seidler FJ, Slotkin TA. Quantitative morphological assessment reveals neuronal and glial deficits in hippocampus after a brief subtoxic exposure to chlorpyrifos in neonatal rats. Dev Brain Res. 2005;155:71–80. doi: 10.1016/j.devbrainres.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Rush T, Liu XQ, Hjelmhaug J, Lobner D. Mechanisms of chlorpyrifos and diazinon induced neurotoxicity in cortical culture. Neuroscience. 2010;166:899–906. doi: 10.1016/j.neuroscience.2010.01.025. [DOI] [PubMed] [Google Scholar]
- 26.Slotkin TA. Cholinergic systems in brain development and disruption by neurotoxicants: nicotine, environmental tobacco smoke, organophosphates. Toxicol Appl Pharmacol. 2004;198:132–151. doi: 10.1016/j.taap.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 27.Slotkin TA. Developmental cholinotoxicants: nicotine and chlorpyrifos. Environ Health Perspect. 1999;107(suppl 1):71–80. doi: 10.1289/ehp.99107s171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Slotkin TA. Developmental neurotoxicity of organophosphates: a case study of chlorpyrifos. In: Gupta RC, editor. Toxicity of Organophosphate and Carbamate Pesticides. Elsevier Academic Press; San Diego: 2005. pp. 293–314. [Google Scholar]
- 29.Slotkin TA, Levin ED, Seidler FJ. Comparative developmental neurotoxicity of organophosphate insecticides: effects on brain development are separable from systemic toxicity. Environ Health Perspect. 2006;114:746–751. doi: 10.1289/ehp.8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slotkin TA, Lobner D, Seidler FJ. Transcriptional profiles for glutamate transporters reveal differences between organophosphates but similarities with unrelated neurotoxicants. Brain Res Bull. 2010;83:76–83. doi: 10.1016/j.brainresbull.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slotkin TA, MacKillop EA, Ryde IT, Tate CA, Seidler FJ. Screening for developmental neurotoxicity using PC12 cells: comparisons of organophosphates with a carbamate, an organochlorine and divalent nickel. Environ Health Perspect. 2007;115:93–101. doi: 10.1289/ehp.9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Slotkin TA, Seidler FJ. Comparative developmental neurotoxicity of organophosphates in vivo: transcriptional responses of pathways for brain cell development, cell signaling, cytotoxicity and neurotransmitter systems. Brain Res Bull. 2007;72:232–274. doi: 10.1016/j.brainresbull.2007.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Slotkin TA, Seidler FJ. Developmental exposure to organophosphates triggers transcriptional changes in genes associated with Parkinson’s Disease in vitro and in vivo. Brain Res Bull. 2011;876:340–347. doi: 10.1016/j.brainresbull.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slotkin TA, Seidler FJ. Developmental neurotoxicants target neurodifferentiation into the serotonin phenotype: chlorpyrifos, diazinon, dieldrin and divalent nickel. Toxicol Appl Pharmacol. 2008;233:211–219. doi: 10.1016/j.taap.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slotkin TA, Seidler FJ. Oxidative and excitatory mechanisms of developmental neurotoxicity: transcriptional profiles for chlorpyrifos, diazinon, dieldrin and divalent nickel in PC12 cells. Environ Health Perspect. 2009;117:587–596. doi: 10.1289/ehp.0800251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slotkin TA, Seidler FJ. Protein kinase C is a target for diverse developmental neurotoxicants: transcriptional responses to chlorpyrifos, diazinon, dieldrin and divalent nickel in PC12 cells. Brain Res. 2009;1263:23–32. doi: 10.1016/j.brainres.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slotkin TA, Seidler FJ. Transcriptional profiles reveal similarities and differences in the effects of developmental neurotoxicants on differentiation into neurotransmitter phenotypes in PC12 cells. Brain Res Bull. 2009;78:211–225. doi: 10.1016/j.brainresbull.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slotkin TA, Seidler FJ, Fumagalli F. Exposure to organophosphates reduces the expression of neurotrophic factors in neonatal rat brain regions: similarities and differences in the effects of chlorpyrifos and diazinon on the fibroblast growth factor superfamily. Environ Health Perspect. 2007;115:909–916. doi: 10.1289/ehp.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slotkin TA, Seidler FJ, Fumagalli F. Targeting of neurotrophic factors, their receptors, and signaling pathways in the developmental neurotoxicity of organophosphates in vivo and in vitro. Brain Res Bull. 2008;76:424–438. doi: 10.1016/j.brainresbull.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Slotkin TA, Seidler FJ, Fumagalli F. Unrelated developmental neurotoxicants elicit similar transcriptional profiles for effects on neurotrophic factors and their receptors in an in vitro model. Neurotoxicol Teratol. 2010;32:42–51. doi: 10.1016/j.ntt.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slotkin TA, Tate CA, Ryde IT, Levin ED, Seidler FJ. Organophosphate insecticides target the serotonergic system in developing rat brain regions: disparate effects of diazinon and parathion at doses spanning the threshold for cholinesterase inhibition. Environ Health Perspect. 2006;114:1542–1546. doi: 10.1289/ehp.9337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Song X, Seidler FJ, Saleh JL, Zhang J, Padilla S, Slotkin TA. Cellular mechanisms for developmental toxicity of chlorpyrifos: targeting the adenylyl cyclase signaling cascade. Toxicol Appl Pharmacol. 1997;145:158–174. doi: 10.1006/taap.1997.8171. [DOI] [PubMed] [Google Scholar]
- 43.Song X, Violin JD, Seidler FJ, Slotkin TA. Modeling the developmental neurotoxicity of chlorpyrifos in vitro: macromolecule synthesis in PC12 cells. Toxicol Appl Pharmacol. 1998;151:182–191. doi: 10.1006/taap.1998.8424. [DOI] [PubMed] [Google Scholar]
- 44.Teng KK, Greene LA. Cultured PC12 cells: a model for neuronal function and differentiation. In: Celis JE, editor. Cell Biology: A Laboratory Handbook. Academic Press; San Diego: 1994. pp. 218–224. [Google Scholar]
- 45.Whitney KD, Seidler FJ, Slotkin TA. Developmental neurotoxicity of chlorpyrifos: cellular mechanisms. Toxicol Appl Pharmacol. 1995;134:53–62. doi: 10.1006/taap.1995.1168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.