Abstract
The epidermal growth factor receptor (EGFR) pathway is one of the most dysregulated molecular pathways in human cancers. Despite its well-established importance in tumor growth, progression and drug-resistant phenotype over the past several decades, targeted therapy designed to circumvent EGFR has yielded only modest clinical success in cancer patients, except those with non-small cell lung cancer (NSCLC) carrying EGFR activation mutations. However, almost all of these NSCLC patients eventually developed resistance to small molecule EGFR kinase inhibitors. These disappointing outcomes are, in part, due to the high complexity and the interactive nature of the EGFR signaling network. More recent compelling evidence further indicates that EGFR functionality can be dependent on its subcellular location. In this regard, EGFR undergoes translocation into different organelles where it elicits distinctly different functions than its best known activity as a plasma membrane-bound receptor tyrosine kinase. EGFR can be shuttled into the cell nucleus and mitochondrion upon ligand binding, radiation, EGFR-targeted therapy and other stimuli. Nuclear EGFR behaves as transcriptional regulator, tyrosine kinase, and mediator of other physiological processes. The role of mitochondrial EGFR remains poorly understood but it appears to regulate apoptosis. While studies using patient tumors have shown nuclear EGFR to be an indicator for poor clinical outcomes in cancer patients, the impact of mitochondrial EGFR on tumor behavior and patient prognosis remains to be defined. Most recently, several lines of evidence suggest that mislocated EGFR may regulate tumor response to therapy and that plasma membrane-bound EGFR elicits survival signals independent of its kinase activity. In light of these recent progresses and discoveries, we will outline in this minireview an emerging line of research that uncovers and functionally characterizes several novel modes of EGFR signaling that take center stage in the cell nucleus, mitochondrion and other subcellular compartments. We will also discuss the clinical implications of these findings in the rationale design for therapeutic strategy that overcomes tumor drug resistance.
1. Introduction
Epidermal growth factor receptor (EGFR) was isolated approximately two decades after the discovery of its ligand EGF in 1962 [1; 2; 3]. The importance of EGFR in protein phosphorylation [1; 4; 5] and in tumorigenesis [6] was later-established. Since then, the EGF-EGFR signaling axis has taken the center stage of not only cancer research, but also developmental biology for over three decades. EGFR is best known for its classical function as a receptor tyrosine kinase localized on the plasma membrane and activated upon ligand binding (Figure 1). Activated EGFR recruits a number of downstream signaling molecules, leading to the activation of several major pathways that are important for tumor growth, progression, and survival [7; 8; 9]. The main pathways downstream of EGFR activation include those mediated by PLC-γ–PKC, Ras-Raf-MEK, PI-3K-Akt-mTOR and JAK2-STAT3. Similar to EGFR, the EGFRvIII variant is primarily localized on the cell-surface where it activates several signaling modules. However, unlike EGFR, EGFRvIII is constitutively active independent of ligand stimulation, in part, due to its loss of a portion of the ligand-binding domain [10; 11; 12; 13].
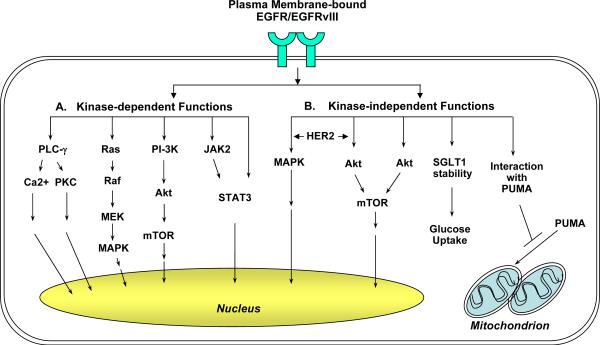
Figure 1. The plasma membrane-bound EGFR/EGFRvIII signaling is consisted of the kinase-dependent and -independent modes of actions.
A: Kinase-dependent functions. Upon ligand binding, EGFR becomes activated and phosphorylated at multiple tyrosine residues including those within its kinase domain. Phosphorylated EGFR then recruits and phosphorylates downstream signaling molecules. The major pathways downstream of EGFR include those mediated by PLC-γ–PKC, Ras-Raf-MEK, PI3-K-Akt-mTOR and JAK2-STAT3. In addition, EGFR can directly interact with and phosphorylate STAT3 transcription factor. EGFRvIII is constitutively active independent of ligand stimulation.
B: Kinase-independent functions. Co-expression of the kinase-dead EGFR K721M mutant with HER2 rescued the inability of the mutant EGFR to activate Akt and MAPK. Kinase-dead EGFR D813A mutant may activate Akt via undefined mechanisms. Independent of its kinase activity, EGFR also interacts with and stabilizes plasma membrane-bound SGLT1, leading to glucose uptake and increased intracellular glucose levels. Our laboratory recently reported that EGFR and EGFRvIII associated with and sequestered the proapoptotic protein PUMA in the cytoplasm independent on EGF stimulation or its kinase activity. The EGFR-PUMA and EGFRvIII-PUMA interactions contribute to reduced apoptosis and survival.
While EGFR overexpression is found in many types of human cancers, EGFRvIII is predominantly detected in malignant gliomas [10; 11; 12; 13]. Both EGFR and EGFRvIII play critical roles in tumorigenesis and in supporting the malignant phenotypes in human cancers. Consequently, both receptors are targets of anti-cancer therapy. Several EGFR-targeting small molecule kinase inhibitors and therapeutic antibodies have been approved by the FDA to treat patients with breast cancer, colorectal cancer, non-small cell lung cancer (NSCLC), squamous cell carcinoma of the head and neck, and pancreatic cancer. Despite extensive efforts invested in the preclinical and clinical development of EGFR-targeted therapy, the current treatments have demonstrated only modest effects on most cancer types [9; 14; 15; 16], with the exception of NSCLC that expresses gain-of-function EGFR mutants [17; 18; 19]. However, almost all of these NSCLC patients eventually developed resistance to small molecule EGFR kinase inhibitors [20; 21]. This acquired resistance has been shown to be linked to a secondary EGFR T790M mutation in about a half of patients [22; 23; 24]. The resistance can be attributed to other potential mechanisms, such as, uncontrolled activation of MET [25; 26] and subsequent MET-mediate HER3 activity [27; 28], and activated insulin-like growth factor-1 receptor [29; 30]. Since lung cancer-associated EGFR mutations are either absent or very rare in other tumor types, there is an imminent need to indentify the mechanisms underlying tumor resistance to anti-EGFR agents in order to derive sensitization strategies that can be used to overcome the resistance [31].
In addressing the need of gaining a deeper understanding of the EGFR pathway and EGFR-associated malignant biology in human cancer, compelling evidence indicates that plasma membrane-bound EGFR can mediate cellular processes independent of its kinase activity [32; 33; 34; 35; 36; 37; 38]. This atypical mode of EGFR signaling could contribute to the failure of the majority of EGFR-targeted agents that are designed to inhibit its kinase activity. Also compelling are the facts that both EGFR and EGFRvIII undergo nuclear and mitochondrial transport and that within these organelles, the receptors exert novel functions that are distinctly different from their classical role as a receptor tyrosine kinase [31; 39]. To date, EGFR nuclear accumulation has been linked to several malignant phenotypes of human cancers, including proliferation, inflammatory response, DNA repair and therapeutic resistance and poor clinical outcomes in cancer patients [40; 41; 42; 43]. Over the past few years, it has become clear that both EGFR and EGFRvIII undergo ligand- and treatment-induced mitochondrial localization. However, regulation and consequences of the mitochondrial mode of EGFR signaling are still poorly understood despite being actively investigated. These exciting discoveries and recent advances in the landscape of the EGFR signaling provide the foundation for this minireview in which we will summarize our current knowledge of the classical and atypical modes of the EGFR pathway and provide a timely outline of their impact on malignant tumor biology as well as therapeutic response of human cancers to currently available agents.
2. Cell-surface and Cytoplasmic Modes of EGFR Signaling
2.1. Kinase-dependent functions
The best known ligands of EGFR are EGF, transforming growth factor-α, and heparin-binding EGF-like growth factor. As summarized in Figure 1, upon ligand binding, activated EGFR recruits, phosphorylates and activates a number of important signaling molecules such as PLC-γ, Ras, PI-3K and JAK2 [9; 44; 45; 46; 47]. Activated EGFR also phosphorylates signal transducer and activator of transcription-3 (STAT3) at Y705 and activates its dimerization, nuclear transport and subsequent gene regulation [46; 48; 49; 50; 51]. For example, EGFR-activated STAT3 has been shown to activate the expression of an E-cadherin repressor, TWIST, and thereby-, promote epithelial-mesenchymal transition [51]. These EGFR downstream signaling cascades can also be activated via EGFR-independent mechanisms whereby regulating tumorigenesis, tumor proliferation and progression, and therapeutic resistance [6; 52; 53; 54; 55].
2.2. Kinase-independent functions
Independent of kinase activity or ligand activation, EGFR can mediate cellular processes mostly through its ability to physically interact with other proteins (Figure 1). One of the first observations suggesting this interesting phenomenon derived from the notion that loss of EGFR kinase activity did not lead to the phenotypes similar to ablation of EGFR expression. In this context, EGFR knockout animals were found to survive for up to eight days after birth and suffer from impaired epithelial development in several organs including skin, lung and gastrointestinal tract [56], whereas the animals with kinase-dead EGFR were viable despite having skin and eye abnormalities [57]. In line with these findings, Coker et al [32] showed that the kinase-dead EGFR D813A mutant retained the ability to stimulate DNA synthesis. Co-expression of the kinase-dead EGFR K721M mutant with HER2 rescued the inability of the mutant EGFR to activate Akt and MAPK, suggesting that heterodimerization with other members of the ErbB family of receptors may help support the kinase-independent function of EGFR [33].
In agreement with these reports, Ewald et al [34] showed that the kinase-dead EGFR K721R mutant retained the ability to survive serum starvation-induced death, while losing its ability to respond to EGF or to stimulate cell growth. Interestingly, the same study found another kinase-dead EGFR mutant D813A to lose both growth-stimulating and prosurvival properties, suggesting that the prosurvival activity of EGFR is independent of the kinase activity but likely dependent of its unique structural properties to associate with other cellular proteins. This notion is in line with a more recent report by Weihua et al [35] showing that loss of expression of EGFR, but not its kinase activity, resulted in autophagic cell death. The authors found that reduced intracellular glucose levels, leading to autophagy in EGFR-deficient cells, was due to the degradation of sodium/glucose cotransporter 1, SGLT1, a plasma membrane-bound protein that enables glucose uptake. Interestingly, cell-surface EGFR was found to physically interact with and stabilize SGLT1 independent of its kinase activity, thereby maintaining high glucose levels in the cells. Conversely, EGFR expression knockdown, but not kinase inhibition, led to SGLT1 degradation, reduction in intracellular glucose and subsequent autophagic cell death [35]. In support of these observations, co-expression of EGFR and SGLT1 was found to be frequent in cell lines and specimens of oral squamous cell carcinoma [58].
Through physical associations but not kinase activity, EGFR can modulate protein subcellular trafficking. Our laboratory recently reported that both EGFR and EGFRvIII associate with p53-upregulated modulator of apoptosis (PUMA), a proapoptotic member of the Bcl-2 family of proteins primarily located on the mitochondria [36]. PUMA is a potent apoptosis inducer that binds to and inhibits all five anti-apoptotic proteins [59; 60], while most BH3-only proteins selectively engage anti-apoptotic proteins. PUMA also directly binds to the apoptotic executor BAX [61; 62] to induce mitochondrial outer membrane permeabilization. PUMA strongly induces apoptosis in colorectal cancer [63; 64], malignant gliomas [65], and in adult stem cells [66]. Importantly, our study [36] further demonstrated that the EGFR-PUMA and EGFRvIII-PUMA interactions are independent of EGF stimulation or kinase activity and that these interactions are constitutive and only modestly reduced following apoptotic stress. The same study also found that EGFR/EGFRvIII did not significantly interact with other proa poptotic members of Bcl-2 family of proteins, Bax and Bmf. As a consequence of the EGFR-PUMA and EGFRvIII-PUMA interactions, PUMA is sequestered in the cytoplasm and unable to translocate onto the mitochondria to initiate apoptosis. This interesting observation is in agreement with the evidence showing that PUMA is highly co-expressed with EGFR/EGFRvIII in cell lines and primary specimens of malignant gliomas and that this particular tumor type is highly resistant to apoptosis-inducing treatments [36]. We further reported in this study that a Bcl-2/Bcl-xL inhibitor that mimics PUMA's proapoptotic activity sensitized EGFR- and EGFRvIII-expressing glioblastoma cells to an EGFR kinase inhibitor Iressa, suggesting that targeting both kinase-dependent and -independent functions of EGFR/EGFRvIII could be an effective strategy to overcome tumor resistance to the agents that solely inhibit the kinase function of the receptors.
Interestingly, two recent studies indicate that EGFR localized within the lipid raft microdomain of the plasma membrane could activate Akt without EGFR kinase activity [37; 38]. Breast cancer cell lines with higher levels of EGFR in the lipid rafts appeared to be more resistant to Iressa [38]. Treating resistant cells with lovastatin to deplete cholesterol, an essential component of the lipid rafts, sensitized the cells to EGFR kinase inhibition. The mechanisms for these interesting observations remain unclear. However, it has been shown that PI-3K and c-Src co-localized and associated with EGFR in the lipid rafts [37]. These findings suggest that the lipid raft microdomain may serve as a platform for EGFR and other signaling molecules to interact with each other to transmit survival signals, independent of EGFR kinase function, and that pharmacological inhibitors for cholesterol biosynthesis may be useful in targeting some of the kinase-independent activities of cell-surface localized EGFR.
3. Nuclear Mode of EGFR Signaling
3.1. Detection of nuclear EGFR and EGFRvIII
Nuclear existence of EGFR was first observed in hepatocytes that underwent regeneration more than two decades ago [67]. EGFR ligands, EGF and pro-TGF-α, were also found to translocate into the nucleus of proliferating hepatocytes [68; 69]. Nuclear expression of EGFR was further detected in other types of normal cells and tissues, such as placenta, thyroid, immortalized epithelial cells of ovary and kidney origins, and keratinocytes [70; 71; 72; 73]. More recently, nuclear EGFR has been shown to be detected in many different types of cancer cells and specimens, including those of breast [71; 74], epidermoid [75; 76], bladder [75], ovary [77], oral cavity [74; 78], lungs [79], and pancreas [80], and also in malignant gliomas [81; 82]. Nuclear EGFR can be localized within the nucleoplasm [70; 71; 83; 84] and on the inner nuclear membrane [73; 85]. Evidence to date indicates nuclear EGFR to be the full-length receptor that originates from the cell-surface [70; 71; 74; 82]. Analysis for nuclear presence of EGFRvIII has not been extensively conducted; the presently available information from our laboratory and those of others showed that EGFRvIII can be detected in prostate cancer [86] and in malignant gliomas [81; 82].
3.2. Nuclear EGFR and EGFRvIII as transcriptional regulators
The role of EGFR in regulating gene regulation independent of its kinase activity was first suggested by a 1994 study by Eldredge et al [87]. The authors showed that a kinase-dead EGFR mutant transcriptionally activated c-fos gene expression. This interestingly finding is in line with another 1994 report by Xie et al [88] demonstrating that rat neu/p185 contained a transactivation domain in the C-terminus and that the full-length receptor underwent nuclear transport. A milestone study in 2001 by Mien-Chi Hung's group [70] defined nuclear EGFR as a transcriptional co-factor that contains a transactivation domain in its C-terminus, like rat neu/p185. This study also showed that nuclear EGFR associated with a consensus A/T-rich sequence within the human cyclin D1 promoter and that following binding, cyclin D1 gene expression was upregulated. As summarized in Figure 2, the transcriptional targets of nuclear EGFR that have been identified to date include cyclin D1 [70], inducible nitric oxide synthase (iNOS) [71], B-Myb [76], cyclooxygenase-2 (COX-2) [82], aurora A [89], c-Myc [80], and breast cancer resistance protein, BCRP [90]. Through increasing the expression of these target genes, nuclear EGFR has been linked to several malignant phenotypes of human cancers, including proliferation, inflammation and tumor drug resistance [40; 41; 42].
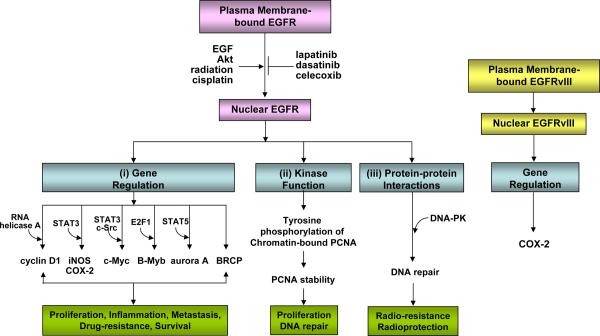
Figure 2. The nuclear mode of EGFR/EGFRvIII signaling network.
EGFR nuclear transport can be induced by EGF, Akt phosphorylation, radiation and cisplatin, and conversely, inhibited by lapatinib, dasatinib and celecoxib. Nuclear EGFR has three major functions: (i) gene regulation, (ii) kinase function, and (iii) protein-protein interactions. Via these actions, nuclear EGFR is implicated in a number of physiological and pathological processes, such as proliferation, inflammation, metastasis, DNA repair, and resistance to DNA-damaging radiation and alkylating anti-cancer agents. Nuclear EGFRvIII activates COX-2 gene expression.
Given the fact that EGFR lacks a DNA-binding domain, extensive efforts have been focused on finding its transcriptional co-regulators with DNA-binding capability. These efforts have opened up a new avenue of research. In 2005, Lo et al [71] reported that nuclear EGFR is able to associate with STAT3 oncogenic transcription factor to enhance expression of iNOS, a protein involved in inflammation, tumor progression and metastasis. The same group further reported that nuclear EGFR interacted with E2F1 to activate human B-Myb gene expression, leading to uncontrolled proliferation [76]. Nuclear EGFR has also been shown to also interact with STAT5 to enhance human aurora A gene expression, leading to chromosome instability [89].
Our laboratory recently reported, for the first time, a systemic unbiased approach to identify nuclear EGFR target genes [82]. This was accomplished using a set of three isogenic glioblastoma cell lines expressing vector control, EGFR, and nuclear entry-defective EGFR (lacking the functional nuclear localization signal within the juxtamembrane region) followed by DNA microarray for over 47,000 gene transcripts. The results indicated 19 potential target genes of nuclear EGFR of which COX-2 was subsequently validated to be a novel transcriptional target of nuclear EGFR. Our results further demonstrated that STAT3 greatly synergized with nuclear EGFR to enhance COX-2 gene expression. Importantly, we found nuclear EGFRvIII to also activate COX-2 gene expression. The impact of STAT3 on nuclear EGFRvIII-mediated COX-2 expression was found to be only modest, which is in contrast to the significant positive impact of nuclear EGFR-STAT3 complex on COX-2 gene activation [82]. Ongoing efforts are being invested on validating other potential nuclear EGFR target genes that have been identified by the gene expression profiling.
Another mechanism for nuclear EGFR-associated transcriptional regulation was suggested by Huo et al [91] that RNA helicase A serves as a DNA-binding partner for nuclear EGFR. Knockdown of RNA helicase A expression in cancer cells abolished nuclear EGFR binding to its target gene promoters and reduced EGFR-induced gene expression. Interestingly, a most recent study by Jaganathan et al [80] showed that EGFR, Src and STAT3 form a heteromeric complex in the nucleus. This nuclear complex is bound to the c-Myc gene, which may contribute to c-Myc gene overexpression in pancreatic cancer cells. Also interesting and indicative of a possible mechanism underlying the ability of nuclear EGFR to regulate gene transcription is the ability of nuclear EGFR to interact with MUC1, which may promote both the accumulation of chromatin-bound EGFR and the significant co-localization of EGFR with phosphorylated RNA polymerase II [92].
In line with the observation on rat neu/p185 [88], its human homolog HER2 can also be detected in the cell nucleus and activates COX-2 gene expression by binding to HER2-associated sequences [93]. Nuclear HER2 has been shown to associate with STAT3 to upregulate cyclin D1 gene expression [94]. This study also showed that progesterone receptor induces HER2 nuclear translocation. Interestingly, a recent report by Li et al [95] demonstrated that nuclear HER2 enhanced translation by activating transcription of ribosomal RNA genes. Taken together, these findings indicate that nuclear EGFR and EGFRvIII function as transcriptional regulators that cooperate with their transcriptional co-factors to mediate the expression of a number of important cancer-related genes and thereby, regulate many physiological and pathological processes.
3.3. EGFR as a nuclear tyrosine kinase
Evidence to date indicates that nuclear EGFR retains its tyrosine kinase activity. Wang et al [96] reported that nuclear EGFR phosphorylates proliferating cell nuclear antigen (PCNA) to promote cell proliferation and DNA repair (Figure 2). Chromatin-bound PCNA protein is phosphorylated on the Tyr-211 residue by nuclear EGFR and becomes stabilized. This important finding raised the possibility that additional nuclear proteins may be phosphorylated by both nuclear EGFR and HER2, and that the functions, stability, and/or intracellular trafficking of these target proteins may be altered as a consequence of tyrosine phosphorylation. Future efforts are needed to explore these exciting possibilities.
3.4. Nuclear EGFR as a modulator of DNA repair
Nuclear EGFR also plays an essential role in DNA repair following radiation therapy (Figure 2). The first two studies that reported radiotherapy-induced EGFR nuclear transport in cancer cells and the consequences of this process were conducted by Dittmann and colleagues [97; 98]. They showed that upon radiation therapy induced EGFR nuclear entry, EGFR localized in the nucleus interacts with DNA-dependent protein kinase (DNA-PK), leading to repair of radiation-induced DNA double-strand breaks in bronchial carcinoma cells. A non-steroid anti-inflammatory drug celecoxib has been shown to facilitate tumor cell radiosensitization by inhibiting radiation-induced nuclear EGFR transport and DNA repair [99]. This action of celecoxib appears to be independent of its COX-2 inhibitory effect since radiosensitization was correlated with neither COX-2 expression nor prostaglandin E2 levels. Another study by Hsu et al [100] further showed that nuclear EGFR is required for tumor resistance to DNA damage induced by the DNA alkylating agent, cisplatin. Collectively, these studies suggest a negative impact of nuclear EGFR on tumor sensitivity to DNA-damaging radiation therapy and anti-cancer alkylating agents. A more recent study by Liccardi [101] provided a potential mechanism for nuclear EGFR-mediated tumor resistance to cisplatin. This study showed that cisplatin induces binding of nuclear EGFR and EGFRvIII to DNA-PK, leading to DNA repair. Similar to EGFR, HER2 nuclear transport can be induced by radiation [102]. Interestingly, Herceptin appears to inhibit radiation-induced HER2 nuclear accumulation, suggesting a potential benefit of combining Herceptin with radiation in treating breast cancer patients with HER2-positive tumors.
Conversely, nuclear EGFR may protect normal cells from unwanted DNA damage caused by ultraviolet and γ irradiations. Ultraviolet irradiation has been shown to induce EGFR nuclear translocation in human keratinocytes [103]. The mechanisms for the observed protective effects of nuclear EGFR in normal skin cells are still unclear. However, it has been shown that following irradiation and treatment of the radioprotector Bowman-Birk protease inhibitor, nuclear EGFR is able to associate with p53 and MDC1 protein, both of which are essential for formation of DNA repair foci [104]. Another radioprotector O-phospho-l-tyrosine has been shown to activate PKC-epsilon and to trigger nuclear EGFR import and phosphorylation of DNA-PK, leading to repair of DNA double-strand breaks [105].
3.5. Nuclear EGFR and EGFR-targeted therapy
Presently, five EGFR-targeted agents have been approved by the FDA for treating cancer patients. Among them, three are small molecule tyrosine kinase inhibitors and two are therapeutic antibodies. (1) Gefitinib (ZD1839, Iressa), a small molecular weight EGFR kinase inhibitor, is being used for locally advanced and metastatic NSCLC. (2) Erlotinib (OSI-774, Tarceva), a sma ll molecule EGFR kinase inhibitor has been approved both to treat metastatic NSCLC as single agent and to be used in combination with gemcitabine for pancreatic cancer that cannot be removed by surgery or has metastasized. (3) Lapatinib (GW572016, Tykerb/Tyverb) is an EGFR/HER2-dual targeting small molecule inhibitor [106] that is used in combination with capecitabine in women with HER2-positive breast cancer whose disease (or condition) has not responded to other chemotherapy. Lapatinib is also approved for combined therapy with letrozole in postmenopausal women with HER2-positive and hormone receptor-positive breast cancer who need hormonal therapy. (4) Cetuximab (C225, Erbitux), a humanized monoclonal antibody that recognizes the extracellular domain of both EGFR [106] and EGFRvIII [107], has been approved for squamous cell carcinoma of the head and neck that has metastasized or recurred after chemotherapy, and as a first-line treatment with radiation therapy for advanced squamous cell carcinoma of the head and neck. Cetuximab is also used for colorectal cancer that has metastasized after chemotherapy has failed and, in combination with irinotecan, for metastatic colorectal cancer patients who have not responded to irinotecan alone. (5) Panitumumab (ABX-EGF, Vectibix), a humanized monoclonal antibody, has been approved to treat metastatic colorectal cancer that has failed other therapies and has metastasized [108].
Constitutive nuclear presence of EGFR can be constitutive, in part, attributed to ligand-activated nuclear transport and the ability of some cancer cells to secret EGF and to activate an EGF-EGFR autocrine loop [70; 71; 76; 84]. Constitutive presence of EGFR in the tumor nuclei may contribute to intrinsic therapeutic resistance. In this context, nuclear existence of EGFR may be beneficial to the tumors encountering EGFR-targeted therapeutic antibodies and small molecule inhibitors [16; 43]. Furthermore, a study by Li et al [79] demonstrated that NSCLS cells that had acquired resistance to cetuximab expressed increased levels of nuclear EGFR and that forced expression of a nuclear localization sequence-tagged EGFR rendered cetuximab-sensitive cells resistant to cetuximab, both in vitro and in mouse xenografts.
The effects of EGFR-targeted therapy and other anti-cancer treatments on the extent of EGFR nuclear translocation are still unclear, with mixed results being reported. Ionizing radiation induces EGFR nuclear transport [97; 104; 109], which can be inhibited by cetuximab [98]. Both lapatinib [110] and the Src family kinase inhibitor, dasatinib [79] block EGFR nuclear entry (Figure 2). Celecoxib has been shown to inhibit radiation-induced nuclear EGFR transport [99]. In contrast, Liao and Carpenter [111] showed that cetuximab is able to activate EGFR nuclear transport by promoting receptor endocytosis and activating receptor intracellular trafficking to the endoplasmic reticulum.
Overall, these observations have provided rationales for selecting novel combination treatments that can overcome nuclear EGFR-mediated therapeutic resistance. For example, celecoxib [99] and dasatinib [79] could be useful in blocking EGFR nuclear entry. Akt inhibition could be used to suppress EGFR nuclear transport because Akt-mediated EGFR phosphorylation at Ser-229 has been shown to be required for EGFR nuclear entry [90]. In summary, despite significant advances in our understanding of the impact of nuclear EGFR on intrinsic and acquired tumor drug resistance as well as the reciprocal effects of anti-cancer agents on EGFR nuclear transport, our current knowledge regarding both aspects remains very limited. In particular, no information is available, to date, on the relationship between nuclear EGFRvIII and tumor drug resistance.
3.6. Nuclear EGFR and EGFRvIII as indicators for poor clinical outcome
There are only a few studies that have been conducted to elucidate the prognostic value of nuclear EGFR and EGFRvIII, with the first study published by Mien-Chi Hung's group in 2005 using 130 primary breast carcinomas [74]. In this study, immunohistochem ical staining was performed to detect the levels of nuclear and non-nuclear EGFR, as well as expression of cyclin D1 and Ki-67 in the tumors. The study found for the first time that 37.7% of the cohort was immunostained positively for nuclear EGFR with 6.9% having high levels of expression. Importantly, a significant inverse correlation was observed between high nuclear EGFR expression and overall survival of breast cancer patients [74]. In contrast, expression of non-nuclear EGFR did not significantly correlate with the overall survival rate of breast cancer patients. There were also positive associations of nuclear EGFR with cyclin D1 and Ki-67. In agreement with previous findings reported by Lo et al [74], Hadzisejdic et al [112] found in a cohort of 113 breast carcinomas that nuclear EGFR was detected in 40% of cases with 12% expressing high degrees of nuclear EGFR. Nuclear EGFR was found to be positively correlated with tumor size, lymph node metastasis, Nottingham prognostic index, estrogen receptor expression, and shortened overall survival.
In oral squamous carcinomas, Lo et al [74] analyzed 37 cases and reported that 24.3% of the samples contained moderate/high levels of nuclear EGFR and that those with high nuclear EGFR had the tendency to survive poorly. In line with these observations, Psyrri et al [78] analyzed 95 oropharyngeal carcinomas and observed an inverse correlation of nuclear EGFR with disease-free survival. In ovarian cancer, Xia et al. [77] investigated 221 cases of patient tumors and observed that 28.3% of the cohort had high levels of nuclear EGFR and that there was an inverse correlation of high nuclear EGFR with overall survival. Using 74 matched hormone-sensitive and hormone-refractory prostate tumors, Edwards et al [86] reported that in patients with hormone-refractory tumors, high levels of nuclear EGFRvIII were associated with poor overall survival. These promising results not only support future uses of nuclear EGFR and nuclear EGFRvIII as prognostic indicators for poor clinical outcome, but also validate the unique ability of nuclear EGFR/EGFRvIII to support the aggressiveness of tumor cells.
3.7. Trafficking of cell-surface EGFR to the nucleus
The mechanisms underlying nuclear transport of EGFR begin with endocytosis, which occurs following ligand-induced activation, as the ligand-bound receptors are internalized through clathrin-coated pits that pinch off from the plasma membrane in a dynamin-dependent manner [113]. After the endocytic vesicle fuses with the early endosome, the internalized EGFR can be (a) recycled back to the plasma membrane, (b) sorted to late endosomes and, eventually, to lysosomes for degradation, or (c) further transported into the nucleus. Playing a crucial role in the first two possible outcomes are the various members of the Rab family GTPases [114]. GTP-bound Rab5, for example, assembles on the membrane of early endosomes and recruits Rab tethering proteins to capture the initial clathrin-coated vesicles that pinch off from the cell surface. Additionally, Rab4 and Rab11 have been implicated to play a role in mediating the budding of recycling vesicles that return EGFR back to the plasma membrane [115]. Rab7 has also been shown to mediate the flow, and subsequent degradation, of EGFR out of the late endosome [116].
Early endosomal EGFR destined for the nucleus can undergo transport via several proposed models, each of which is dependent on the interaction between a nuclear localization signal (NLS) within EGFR and importin proteins [83; 84; 117]. Importin-β, either alone or as a heterodimer with importin-α, can bind to NLS of NLS-containing proteins as well as to components of nuclear pore complexes (NPCs), thereby directing these proteins for entry into the nucleus [118]. In the case of EGFR and HER2, a putative NLS has been both identified within the juxtamembrane region [71; 93; 119] and shown to interact with importin-β [84; 117]. For HER2, one proposed model suggests that importin-β associates with the NLS of endosome-bound HER2 and directs it to the nucleus by interacting with the nuclear pore protein Nup358 [117], a constituent of NPCs.
Another proposed model involving EGFR retrograde transport suggests that after early endosomal sorting, ErbB family of proteins destined for the nucleus are trafficked via the Golgi to the ER in COPI-coated vesicles [120]. ER-bound EGFR then interacts with Sec61 translocon [83; 121], passing through the channel in a similar manner as misfolded proteins undergoing ER-associated protein degradation (ERAD) and entering into the cytosol where it can be picked up by importin-β and transported into the nucleus. This retrotranslocation from the ER to the cytosol of full-length EGFR with its hydrophobic transmembrane domain requires the presence of cytosolic chaperone HSP70 [83], which may possibly play a role in solubilizing the receptor and preventing aggregation. Alternatively, ER-bound EGFR may also enter the nucleus via lateral diffusion from the ER membrane through the nuclear pore complex and into the inner nuclear membrane mediated by NLS-importin interaction, as suggested by evidence showing EGFR localized at the inner nuclear membrane [85] and nuclear matrix [122]. Although nuclear export signals have yet to be identified in ErbB family of receptors, the exportin CRM1 has been found to interact with EGFR and HER2 [84; 117], and inhibition of CRM1 using leptomycin B has led to increased accumulation of nuclear EGFR, HER2, and HER3 [84; 117; 123].
Other proteins reported to be involved in EGFR nuclear trafficking include Epstein-Barr virus (EBV) encoded latent membrane protein 1 [124], which was shown to regulate nuclear EGFR translocation in a dose-dependent manner, and PIKfyve kinase [75], which has been demonstrated to play a role in nuclear transport of EGFR via its interaction with cytoplasmic EGFR upon HB-EGF induced activation. Interestingly, a recent study found that Akt phosphorylation of EGFR is required for both EGFR nuclear translocation and acquisition of Iressa resistance via upregulation of BCRP by nuclear EGFR in breast cancer cells [90], indicating that advances in our understanding of nuclear EGFR trafficking can lead to further insight into the various approaches to EGFR-targeted therapy.
4. Mitochondrial Mode of EGFR Signaling
Mitochondrial detection of EGFR was first reported by Sarah Parsons' group in 2004 [125]. This important study demonstrated that EGFR translocated to the mitochondria after EGF stimulation (Figure 3). While localized on the mitochondria, EGFR interacts with cytochrome c oxidase subunit II (CoxII), a mitochondrion-encoded protein and a critical component of the oxidative phosphorylation pathway. CoxII binds to EGFR but not mutant EGFR (Y845F) that can not be phosphorylated by c-Src. EGFR Y845F mutant undergoes EGF-induced mitochondrial transport similar to EGFR, suggesting that c-Src-mediated EGFR phosphorylation is not required for EGFR mitochondrial import. This group further reported that c-Src translocated to the mitochondria with similar kinetics as EGFR after EGF stimulation, and that c-Src kinase activity/overexpression enhanced EGFR localization to the mitochondria [126]. This study also showed that CoxII can be phosphorylated by EGFR and c-Src; however, the consequences of this phosphorylation are still unknown. The authors also reported that clathrin-mediated endocytosis was shown to be essential in regulating EGFR translocation to the mitochondria, suggesting the origin of mitochondrial EGFR to be the plasma membrane-bound EGFR [126]. In contrast to this finding, Yao et al [127] reported that EGFR mitochondrial transport is independent of endocytosis.
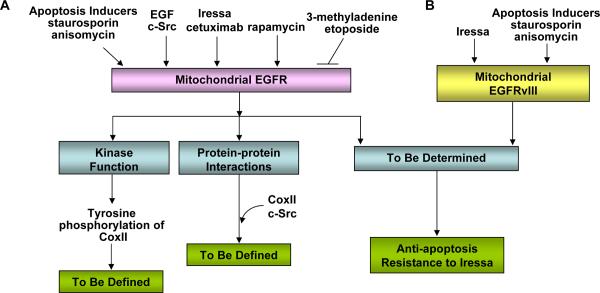
Figure 3. The mitochondrial mode of EGFR/EGFRvIII signaling pathway.
A: EGFR mitochondrial import can be constitutive and the extent can be enhanced by apoptosis inducers (staurosporine and anisomycin), EGF, c-Src, Iressa, cetuximab and rapamycin. Conversely, EGFR mitochondrial transport can be blocked by 3-methyadenine (inhibitor of autophagy and PI-3K) and etoposide. Mitochondrial EGFR retains its tyrosine kinase activity and phosphorylates CoxII; however, the consequence of the phosphorylation is not yet defined. The mitochondrial EGFR-CoxII complex can include c-Src; however, the effects of this interaction are still unknown. Furthermore, mitochondrial accumulation of EGFR led to compromised apoptotic response and resistance to Iressa treatments, while the underlying mechanisms are still undetermined.
B: EGFRvIII mitochondrial import is constitutive and can be further enhanced by apoptosis inducers (staurosporine and anisomycin) and by Iressa. Mitochondrial accumulation of EGFRvIII rendered tumor cells highly resistant to apoptotic death and to Iressa treatments, while the mechanisms are still not identified.
Demory et al [126] reported the identification of a potential mitochondrial localization signal contained in the juxtamembrane region of EGFR (residues 645–666). The authors also showed that EGFR decreased ATP biosynthesis in the cells under serum-starved conditions; expression of an EGFR deletion mutant (Δ645–666) restored ATP levels to about 80% [126]. Similarly, Yao et al [127] reported that EGFR deletion mutant (Δ646–660) was unable to enter the mitochondria. Since the nuclear localization signal of EGFR is also localized in the same juxtamembrane region (residues 645–657), [71; 82; 119] overlapping with the potential mitochondrial localization signal (Δ645–666), it remains unclear if the changes in ATP levels were attributed to mitochondrial or nuclear EGFR, or potentially to both. Also unclear is whether the region of interest (residues 645–666) is responsible for both nuclear and mitochondrial transports of EGFR. Furthermore, Yue et al [128] reported that EGFR mitochondrial translocation can be increased by a mTOR inhibitor, rapamycin. Autophagy inhibition by 3-methyladenine (an inhibitor of autophagy and a PI-3K inhibitor) and Beclin 1 expression knockdown leads to a reduction of rapamycin-induced mitochondrial import of EGFR. Etoposide also appears to decrease EGFR mitochondrial transport [128].
Most recently, our laboratory reported for the first time that EGFRvIII can be detected in tumor cell mitochondrion [129]. We showed that both EGFR and EGFRvIII are constitutively present in the mitochondria. Importantly, the degrees of EGFR and EGFRvIII mitochondrial transport were greatly enhanced following treatments with the apoptosis inducers, staurosporine and anisomycin, and with an EGFR kinase inhibitor, Iressa (Figure 3). Using mutant EGFR/EGFRvIII receptors engineered to undergo enriched intracellular trafficking into the mitochondria (but not into the cell nucleus), we showed that glioblastoma cells expressing the mitochondrially enriched EGFRvIII were more resistant to staurosporineand anisomycin-induced growth suppression and apoptosis. The tumor cells with mitochondrial EGFRvIII accumulation were highly resistant to Iressa-mediated growth inhibition. These findings indicate that apoptosis inducers and EGFR-targeted inhibitors enhance mitochondrial translocation of both EGFR and EGFRvIII, and that mitochondrial accumulation of these receptors contributes to tumor drug resistance. The findings also provide evidence for a potential link between the mitochondrial EGFR/EGFRvIII pathway and apoptosis.
Interestingly, Dreier et al [130] recently reported that cetuximab induced mitochondrial transport of EGFRvIII, but not EGFR, while we showed that Iressa increased mitochondrial transport of both receptors [129]. This could be attributed to the ability of cetuximab to increase EGFRvIII internalization. These findings together suggest that different EGFR-targeted treatments, e.g. tyrosine kinase inhibitors versus therapeutic antibodies, may differentially impact EGFR and EGFRvIII in their capability to undergo mitochondrial transport. Future investigations are clearly needed in this area to understand the mechanisms in order to provide better clinical therapeutic guidance.
HER4 intracellular domain (4ICD) has been shown by Frank Jones' group to be a BH3-only protein that undergoes ligand-induced mitochondrial transport [131]. The authors showed that 4ICD interacts with the anti-apoptotic protein Bcl-2 in a similar fashion to the BH3-only proteins. However, unlike other BH3-only proteins that depend on BAX/BAK to initiate apoptosis, 4ICD is essential for BAK-transmitted apoptosis. The same group further reported that mitochondrial 4ICD contributes to tamoxifen sensitivity in breast cancer cells [132].
In summary, several recent reports have uncovered a novel mode of EGFR signaling that takes place in the mitochondria, the central organelles that produce energy and initiate apoptosis. The mitochondrial transport of EGFR and/or EGFRvIII can be constitutive and further enhanced by EGF, rapamycin, apoptosis inducers, c-Src and EGFR inhibition (Figure 3). Conversely, the receptor mitochondrial import can be suppressed by 3-methyladenine and by etoposide. The origin of mitochondrial EGFR and EGFRvIII remains to be defined given the mixed results. While localized to the mitochondria, EGFR interacts with and phosphorylates CoxII though its impact on CoxII and CoxII-mediated ATP biosynthesis are still not known. Accumulation of EGFR and EGFRvIII in the tumor mitochondria could contribute to tumor resistance to apoptosis although the underlying mechanisms have yet to be defined. The anti-apoptotic effects of mitochondrial EGFR/EGFRvIII are in contrast to the pro-apoptotic effects of 4ICD. Overall, the nature and consequences of the mitochondrial mode of EGFR signaling are still not well understood. Consequently, there is an urgent need to further investigate this exciting new avenue of cancer research.
5. Perspectives and Future Directions
The EGFR signaling pathway is one of the most dysregulated and most extensively investigated molecular pathways in human cancers. Despite significant advances made in our understanding of the EGFR signaling network, studies in the past ten years have uncovered several atypical modes of the pathway that are intriguing and of both biological importance and therapeutic implications. These studies have helped the scientific community to reshape the subcellular landscape of the EGFR signaling network, in particular, by expanding from its classical location on the plasma membrane to the nucleus and mitochondrion, as well as, to microdomains within the plasma membrane.
Despite much skepticism, this unique unconventional field of EGFR research has attracted substantial positive attentions and tremendous interests. Consequent to this, the landscape of the EGFR pathway is now viewed as a web of signaling networks that has three major subcellular hubs. In addition to the obvious differences in the subcellular location, the evidence to date indicates that EGFR at the different subcellular locations elicits distinctly different and also overlapping signals. In this regard, mitogenic signals can be emitted from the plasma membrane-bound EGFR in both kinase-dependent and -independent fashions in which the kinase-independent activity can originate from the EGFR within the lipid rafts. Pro-survival signals can be transmitted from the EGFR/EGFRvIII on the cell-surface as well as from those on the mitochondria. The kinase-independent activity of cell-surface EGFR can modulate glucose uptake and autophagy, while EGFR mitochondrial import may be modulated by autophagy inhibition. Overall, these newly uncovered atypical modes of EGFR signaling have been established to control an array of cellular processes that post critical importance in tumor cell growth, progression, death and survival.
Several major knowledge gaps still exist in our understanding towards the nuclear and mitochondrial modes of the EGFR and EGFRvIII signaling pathways. (1) It remains unknown the relationship between the atypical EGFR/EGFRvIII pathways and tumorigenesis in any type of cancers. (2) The biological and pathological consequences of nuclear EGFRvIII in human cancers are still largely unknown. (3) The exact modes of actions of mitochondrial EGFR and EGFRvIII have remained undefined. (4) The impacts of nuclear and mitochondrial EGFR/EGFRvIII on tumor response to therapy and on intrinsic and acquired resistance to treatments are still poorly understood. Gaining a deeper insight into these processes could help us derive rationales to overcome the drug resistant phenotype frequently observed in tumors with dysregulated EGFR and EGFRvIII signaling. (5) Are there common factors beside ligands that activate EGFR nuclear and mitochondrial transport? Targeting these factors could potentially overcome atypical EGFR signaling mediated resistance to various therapies. (6) Are the atypical EGFR and EGFRvIII pathways involved in embryonal development or cancer stem cell biology? (7) It remains uninvestigated whether the classical and atypical EGFR/EGFRvIII pathways cooperate to regulate the malignant phenotypes of human cancers. Likely, tumor cells work to mobilize and/or switch off different modes of EGFR signaling in order to maximize its growth and survival. (8) How do we derive therapeutic strategy that will simultaneously and fully inhibit a diverse array of EGFR/EGFRvIII activities that are elicited from different subcellular locations in tumor cells?
Indeed, addressing the above mentioned knowledge gaps and unanswered questions will not only better our biological understanding of the EGFR signaling network in human cancers, but also help us to generate novel insights into the nature of other plasma membrane-bound receptors that also undergo nuclear transport. These include receptor tyrosine kinases, such as, rat p185neu, HER2, HER3, HER4, FGFR, TrkA/B, FGFR, VEGFR-2, c-Met and orphan receptor 1, as well as cytokine receptors for IL-1, IL-5, interferon-γ, type I TGF-β and prolactin [69; 88; 123; 133; 134; 135; 136; 137; 138; 139; 140; 141; 142; 143; 144]. Undoubtedly, the newly gained knowledge will facilitate future development of more effective treatments for tumors with hyperactive EGFR and EGFRvIII pathways.
Acknowledgements
The author's work was supported by grants 5K01-CA118423 from the National Cancer Institute, USA, W81XWH-07-1-0390 and W81XWH-11-1-0600 from the U.S. Department of Defense, the Beez Foundation of Childhood Cancer and the Dani P. Bolognesi, Ph.D. Award (Department of Surgery at Duke University).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Statement None Declared
References
- [1].Cohen S, Carpenter G, King L., Jr. Epidermal growth factor-receptor-protein kinase interactions. Co-purification of receptor and epidermal growth factor-enhanced phosphorylation activity. J Biol Chem. 1980;255:4834–4842. [PubMed] [Google Scholar]
- [2].Cohen S, Ushiro H, Stoscheck C, Chinkers M. A native 170,000 epidermal growth factor receptor-kinase complex from shed plasma membrane vesicles. J Biol Chem. 1982;257:1523–1531. [PubMed] [Google Scholar]
- [3].Cohen S. Isolation of a mouse submaxillary gland protein accelerating incisor eruption and eyelid opening in the new-born animal. J Biol Chem. 1962;237:1555–1562. [PubMed] [Google Scholar]
- [4].Cohen S, Carpenter G, King L., Jr. Epidermal growth factor-receptor-protein kinase interactions. Prog Clin Biol Res. 1981;66(Pt A):557–567. [PubMed] [Google Scholar]
- [5].Cohen S, Fava RA, Sawyer ST. Purification and characterization of epidermal growth factor receptor/protein kinase from normal mouse liver. Proc Natl Acad Sci U S A. 1982;79:6237–6241. doi: 10.1073/pnas.79.20.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Sporn MB, Todaro GJ. Autocrine secretion and malignant transformation of cells. N Engl J Med. 1980;303:878–880. doi: 10.1056/NEJM198010093031511. [DOI] [PubMed] [Google Scholar]
- [7].Lo HW, Hung MC. Nuclear EGFR signalling network in cancers: linking EGFR pathway to cell cycle progression, nitric oxide pathway and patient survival. Br J Cancer. 2006;94:184–188. doi: 10.1038/sj.bjc.6602941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Yarden Y, Shilo BZ. SnapShot: EGFR signaling pathway. Cell. 2007;131:1018. doi: 10.1016/j.cell.2007.11.013. [DOI] [PubMed] [Google Scholar]
- [9].Lo HW. EGFR-targeted therapy in malignant glioma: novel aspects and mechanisms of drug resistance. Current molecular pharmacology. 2010;3:37–52. doi: 10.2174/1874467211003010037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yamazaki H, Ohba Y, Tamaoki N, Shibuya M. A deletion mutation within the ligand binding domain is responsible for activation of epidermal growth factor receptor gene in human brain tumors. Jpn J Cancer Res. 1990;81:773–779. doi: 10.1111/j.1349-7006.1990.tb02644.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ekstrand AJ, Sugawa N, James CD, Collins VP. Amplified and rearranged epidermal growth factor receptor genes in human glioblastomas reveal deletions of sequences encoding portions of the N- and/or C-terminal tails. Proc Natl Acad Sci U S A. 1992;89:4309–4313. doi: 10.1073/pnas.89.10.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wong AJ, Ruppert JM, Bigner SH, Grzeschik CH, Humphrey PA, Bigner DS, Vogelstein B. Structural alterations of the epidermal growth factor receptor gene in human gliomas. Proc Natl Acad Sci U S A. 1992;89:2965–2969. doi: 10.1073/pnas.89.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Sugawa N, Ekstrand AJ, James CD, Collins VP. Identical splicing of aberrant epidermal growth factor receptor transcripts from amplified rearranged genes in human glioblastomas. Proc Natl Acad Sci U S A. 1990;87:8602–8606. doi: 10.1073/pnas.87.21.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].van den Bent MJ, Brandes AA, Rampling R, Kouwenhoven MC, Kros JM, Carpentier AF, Clement PM, Frenay M, Campone M, Baurain JF, Armand JP, Taphoorn MJ, Tosoni A, Kletzl H, Klughammer B, Lacombe D, Gorlia T. Randomized phase II trial of erlotinib versus temozolomide or carmustine in recurrent glioblastoma: EORTC brain tumor group study 26034. J Clin Oncol. 2009;27:1268–1274. doi: 10.1200/JCO.2008.17.5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Reardon DA, Desjardins A, Vredenburgh JJ, Gururangan S, Friedman AH, Herndon JE, 2nd, Marcello J, Norfleet JA, McLendon RE, Sampson JH, Friedman HS. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J Neurooncol. 2009 doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Brand TM, Iida M, Wheeler DL. Molecular mechanisms of resistance to the EGFR monoclonal antibody cetuximab. Cancer Biol. Ther. 2011;11:777–792. doi: 10.4161/cbt.11.9.15050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, Naoki K, Sasaki H, Fujii Y, Eck MJ, Sellers WR, Johnson BE, Meyerson M. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- [18].Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG, Louis DN, Christiani DC, Settleman J, Haber DA. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- [19].Mitsudomi T, Morita S, Yatabe Y, Negoro S, Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, Asami K, Katakami N, Takada M, Yoshioka H, Shibata K, Kudoh S, Shimizu E, Saito H, Toyooka S, Nakagawa K, Fukuoka M. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): an open label, randomised phase 3 trial. The lancet oncology. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- [20].Bonanno L, Jirillo A, Favaretto A. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors and new therapeutic perspectives in non small cell lung cancer. Curr Drug Targets. 2011;12:922–933. doi: 10.2174/138945011795528958. [DOI] [PubMed] [Google Scholar]
- [21].Nguyen KS, Kobayashi S, Costa DB. Acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancers dependent on the epidermal growth factor receptor pathway. Clin Lung Cancer. 2009;10:281–289. doi: 10.3816/CLC.2009.n.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ogino A, Kitao H, Hirano S, Uchida A, Ishiai M, Kozuki T, Takigawa N, Takata M, Kiura K, Tanimoto M. Emergence of epidermal growth factor receptor T790M mutation during chronic exposure to gefitinib in a non small cell lung cancer cell line. Cancer Res. 2007;67:7807–7814. doi: 10.1158/0008-5472.CAN-07-0681. [DOI] [PubMed] [Google Scholar]
- [24].Bell DW, Gore I, Okimoto RA, Godin-Heymann N, Sordella R, Mulloy R, Sharma SV, Brannigan BW, Mohapatra G, Settleman J, Haber DA. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nat Genet. 2005;37:1315–1316. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- [25].Engelman JA, Janne PA. Mechanisms of acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2008;14:2895–2899. doi: 10.1158/1078-0432.CCR-07-2248. [DOI] [PubMed] [Google Scholar]
- [26].Beau-Faller M, Ruppert AM, Voegeli AC, Neuville A, Meyer N, Guerin E, Legrain M, Mennecier B, Wihlm JM, Massard G, Quoix E, Oudet P, Gaub MP. MET gene copy number in non-small cell lung cancer: molecular analysis in a targeted tyrosine kinase inhibitor naive cohort. J Thorac Oncol. 2008;3:331–339. doi: 10.1097/JTO.0b013e318168d9d4. [DOI] [PubMed] [Google Scholar]
- [27].Arteaga CL. HER3 and mutant EGFR meet MET. Nat Med. 2007;13:675–677. doi: 10.1038/nm0607-675. [DOI] [PubMed] [Google Scholar]
- [28].Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J, Kosaka T, Holmes AJ, Rogers AM, Cappuzzo F, Mok T, Lee C, Johnson BE, Cantley LC, Janne PA. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- [29].Morgillo F, Kim WY, Kim ES, Ciardiello F, Hong WK, Lee HY. Implication of the insulin-like growth factor-IR pathway in the resistance of non-small cell lung cancer cells to treatment with gefitinib. Clin Cancer Res. 2007;13:2795–2803. doi: 10.1158/1078-0432.CCR-06-2077. [DOI] [PubMed] [Google Scholar]
- [30].Ramalingam SS, Spigel DR, Chen D, Steins MB, Engelman JA, Schneider CP, Novello S, Eberhardt WE, Crino L, Habben K, Liu L, Janne PA, Brownstein CM, Reck M. Randomized Phase II Study of Erlotinib in Combination With Placebo or R1507, a Monoclonal Antibody to Insulin-Like Growth Factor-1 Receptor, for Advanced-Stage Non-Small-Cell Lung Cancer. J Clin Oncol. 2011;29:4574–4580. doi: 10.1200/JCO.2011.36.6799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lo HW. EGFR-targeted Cancer Therapy: Promise, Problems and Potential Solutions Translational Medicine. 2011;1 doi:10.4172/2161-1025.1000105e. [Google Scholar]
- [32].Coker KJ, Staros JV, Guyer CA. A kinase-negative epidermal growth factor receptor that retains the capacity to stimulate DNA synthesis. Proc Natl Acad Sci U S A. 1994;91:6967–6971. doi: 10.1073/pnas.91.15.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Deb TB, Su L, Wong L, Bonvini E, Wells A, David M, Johnson GR. Epidermal growth factor (EGF) receptor kinase-independent signaling by EGF. J Biol Chem. 2001;276:15554–15560. doi: 10.1074/jbc.M100928200. [DOI] [PubMed] [Google Scholar]
- [34].Ewald JA, Wilkinson JC, Guyer CA, Staros JV. Ligand- and kinase activity-independent cell survival mediated by the epidermal growth factor receptor expressed in 32D cells. Exp Cell Res. 2003;282:121–131. doi: 10.1016/s0014-4827(02)00014-9. [DOI] [PubMed] [Google Scholar]
- [35].Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer Cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhu H, Cao X, Ali-Osman F, Keir S, Lo HW. EGFR and EGFRvIII interact with PUMA to inhibit mitochondrial translocalization of PUMA and PUMA-mediated apoptosis independent of EGFR kinase activity. Cancer Lett. 2010;294:101–110. doi: 10.1016/j.canlet.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Irwin ME, Bohin N, Boerner JL. Src family kinases mediate epidermal growth factor receptor signaling from lipid rafts in breast cancer cells. Cancer Biol Ther. 2011;12:718–726. doi: 10.4161/cbt.12.8.16907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Irwin ME, Mueller KL, Bohin N, Ge Y, Boerner JL. Lipid raft localization of EGFR alters the response of cancer cells to the EGFR tyrosine kinase inhibitor gefitinib. J Cell Physiol. 2011;226:2316–2328. doi: 10.1002/jcp.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hung MC, Link W. Protein localization in disease and therapy. J. Cell Sci. 2011;124:3381–3392. doi: 10.1242/jcs.089110. [DOI] [PubMed] [Google Scholar]
- [40].Wang YN, Yamaguchi H, Hsu JM, Hung MC. Nuclear trafficking of the epidermal growth factor receptor family membrane proteins. Oncogene. 2010;29:3997–4006. doi: 10.1038/onc.2010.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lo HW. Nuclear mode of the EGFR signaling network: biology, prognostic value, and therapeutic implications. Discov Med. 2010;10:44–51. [PMC free article] [PubMed] [Google Scholar]
- [42].Carpenter G, Liao HJ. Trafficking of receptor tyrosine kinases to the nucleus. Exp Cell Res. 2009;315:1556–1566. doi: 10.1016/j.yexcr.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7:493–507. doi: 10.1038/nrclinonc.2010.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Anderson D, Koch CA, Grey L, Ellis C, Moran MF, Pawson T. Binding of SH2 domains of phospholipase C gamma 1, GAP, and Src to activated growth factor receptors. Science. 1990;250:979–982. doi: 10.1126/science.2173144. [DOI] [PubMed] [Google Scholar]
- [45].Navolanic PM, Steelman LS, McCubrey JA. EGFR family signaling and its association with breast cancer development and resistance to chemotherapy (Review) Int J Oncol. 2003;22:237–252. [PubMed] [Google Scholar]
- [46].Park OK, Schaefer TS, Nathans D. In vitro activation of Stat3 by epidermal growth factor receptor kinase. Proc Natl Acad Sci U S A. 1996;93:13704–13708. doi: 10.1073/pnas.93.24.13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Bowman T, Garcia R, Turkson J, Jove R. STATs in oncogenesis. Oncogene. 2000;19:2474–2488. doi: 10.1038/sj.onc.1203527. [DOI] [PubMed] [Google Scholar]
- [48].Bowman T, Broome MA, Sinibaldi D, Wharton W, Pledger WJ, Sedivy JM, Irby R, Yeatman T, Courtneidge SA, Jove R. Stat3-mediated Myc expression is required for Src transformation and PDGF-induced mitogenesis. Proc Natl Acad Sci U S A. 2001;98:7319–7324. doi: 10.1073/pnas.131568898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Barre B, Avril S, Coqueret O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J Biol Chem. 2003;278:2990–2996. doi: 10.1074/jbc.M210422200. [DOI] [PubMed] [Google Scholar]
- [50].Lo HW, Cao X, Zhu H, Ali-Osman F. Constitutively activated STAT3 frequently coexpresses with epidermal growth factor receptor in high-grade gliomas and targeting STAT3 sensitizes them to Iressa and alkylators. Clin Cancer Res. 2008;14:6042–6054. doi: 10.1158/1078-0432.CCR-07-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei Y, Abbruzzese JL, Hortobagyi GN, Hung MC. Epidermal growth factor receptor cooperates with signal transducer and activator of transcription 3 to induce epithelial-mesenchymal transition in cancer cells via up-regulation of TWIST gene expression. Cancer Res. 2007;67:9066–9076. doi: 10.1158/0008-5472.CAN-07-0575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Bacus SS, Chin D, Yarden Y, Zelnick CR, Stern DF. Type 1 recept or tyrosine kinases are differentially phosphorylated in mammary carcinoma and differentially associated with steroid receptors. Am J Pathol. 1996;148:549–558. [PMC free article] [PubMed] [Google Scholar]
- [53].Craven RJ, Lightfoot H, Cance WG. A decade of tyrosine kinases: from gene discovery to therapeutics. Surg Oncol. 2003;12:39–49. doi: 10.1016/s0960-7404(03)00004-5. [DOI] [PubMed] [Google Scholar]
- [54].Baselga J, Norton L. Focus on breast cancer. Cancer Cell. 2002;1:319–322. doi: 10.1016/s1535-6108(02)00066-1. [DOI] [PubMed] [Google Scholar]
- [55].Bromberg JF, Wrzeszczynska MH, Devgan G, Zhao Y, Pestell RG, Albanese C, Darnell JE., Jr. Stat3 as an oncogene. Cell. 1999;98:295–303. doi: 10.1016/s0092-8674(00)81959-5. [DOI] [PubMed] [Google Scholar]
- [56].Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- [57].Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- [58].Hanabata Y, Nakajima Y, Morita KI, Kayamori K, Omura K. Coexpression of SGLT1 and EGFR is associated with tumor differentiation in oral squamous cell carcinoma. Odontology. 2011 doi: 10.1007/s10266-011-0033-2. [DOI] [PubMed] [Google Scholar]
- [59].Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DC. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- [60].Chipuk JE, Fisher JC, Dillon CP, Kriwacki RW, Kuwana T, Green DR. Mechanism of apoptosis induction by inhibition of the anti-apoptotic BCL-2 proteins. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:20327–20332. doi: 10.1073/pnas.0808036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gallenne T, Gautier F, Oliver L, Hervouet E, Noel B, Hickman JA, Geneste O, Cartron P-F, Vallette FM, Manon S, Juin P. Bax activation by the BH3-only protein Puma promotes cell dependence on antiapoptotic Bcl-2 family members. J. Cell Biol. 2009;185:279–290. doi: 10.1083/jcb.200809153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kim H, Tu HC, Ren D, Takeuchi O, Jeffers JR, Zambetti GP, Hsieh JJ, Cheng EH. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol Cell. 2009;36:487–499. doi: 10.1016/j.molcel.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Yu J, Zhang L, Hwang PM, Kinzler KW, Vogelstein B. PUMA induces the rapid apoptosis of colorectal cancer cells. Mol Cell. 2001;7:673–682. doi: 10.1016/s1097-2765(01)00213-1. [DOI] [PubMed] [Google Scholar]
- [64].Yu J, Wang Z, Kinzler KW, Vogelstein B, Zhang L. PUMA mediates the apoptotic response to p53 in colorectal cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:1931–1936. doi: 10.1073/pnas.2627984100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Ito H, Kanzawa T, Miyoshi T, Hirohata S, Kyo S, Iwamaru A, Aoki H, Kondo Y, Kondo S. Therapeutic efficacy of PUMA for malignant glioma cells regardless of p53 status. Human gene therapy. 2005;16:685–698. doi: 10.1089/hum.2005.16.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liu D, Ou L, Clemenson GD, Jr., Chao C, Lutske ME, Zambetti GP, Gage FH, Xu Y. Puma is required for p53-induced depletion of adult stem cells. Nature cell biology. 2010;12:993–998. doi: 10.1038/ncb2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Marti U, Burwen SJ, Wells A, Barker ME, Huling S, Feren AM, Jones AL. Localization of epidermal growth factor receptor in hepatocyte nuclei. Hepatology. 1991;13:15–20. [PubMed] [Google Scholar]
- [68].Raper SE, Burwen SJ, Barker ME, Jones AL. Translocation of epidermal growth factor to the hepatocyte nucleus during rat liver regeneration. Gastroenterology. 1987;92:1243–1250. doi: 10.1016/s0016-5085(87)91084-5. [DOI] [PubMed] [Google Scholar]
- [69].Schausberger E, Eferl R, Parzefall W, Chabicovsky M, Breit P, Wagner EF, Schulte-Hermann R, Grasl-Kraupp B, Chabikovsky M. Induction of DNA synthesis in primary mouse hepatocytes is associated with nuclear pro-transforming growth factor alpha and erbb-1 and is independent of c-jun. Carcinogenesis. 2003;24:835–841. doi: 10.1093/carcin/bgg027. [DOI] [PubMed] [Google Scholar]
- [70].Lin SY, Makino K, Xia W, Matin A, Wen Y, Kwong KY, Bourguignon L, Hung MC. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat Cell Biol. 2001;3:802–808. doi: 10.1038/ncb0901-802. [DOI] [PubMed] [Google Scholar]
- [71].Lo H-W, Hsu S-C, Ali-Seyed M, Gunduz M, Xia W, Wei Y, Bartholomeusz G, Shih J-Y, Hung M-C. Nuclear Interaction of EGFR and STAT3 in the Activation of iNOS/NO Pathway. Cancer Cell. 2005;7:575–589. doi: 10.1016/j.ccr.2005.05.007. [DOI] [PubMed] [Google Scholar]
- [72].Marti U, Ruchti C, Kampf J, Thomas GA, Williams ED, Peter HJ, Gerber H, Burgi U. Nuclear localization of epidermal growth factor and epidermal growth factor receptors in human thyroid tissues. Thyroid. 2001;11:137–145. doi: 10.1089/105072501300042785. [DOI] [PubMed] [Google Scholar]
- [73].Cao H, Lei ZM, Bian L, Rao CV. Functional nuclear epidermal growth factor receptors in human choriocarcinoma JEG-3 cells and normal human placenta. Endocrinology. 1995;136:3163–3172. doi: 10.1210/endo.136.7.7540549. [DOI] [PubMed] [Google Scholar]
- [74].Lo HW, Xia W, Wei Y, Ali-Seyed M, Huang SF, Hung MC. Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 2005;65:338–348. [PubMed] [Google Scholar]
- [75].Kim J, Jahng WJ, Di Vizio D, Lee JS, Jhaveri R, Rubin MA, Shisheva A, Freeman MR. The Phosphoinositide Kinase PIKfyve Mediates Epidermal Growth Factor Receptor Trafficking to the Nucleus. Cancer Res. 2007;67:9229–9237. doi: 10.1158/0008-5472.CAN-07-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Hanada N, Lo HW, Day CP, Pan Y, Nakajima Y, Hung MC. Co-regulation of B-Myb expression by E2F1 and EGF receptor. Mol Carcinog. 2006;45:10–17. doi: 10.1002/mc.20147. [DOI] [PubMed] [Google Scholar]
- [77].Xia W, Wei Y, Du Y, Liu J, Chang B, Yu YL, Huo LF, Miller S, Hung MC. Nuclear expression of epidermal growth factor receptor is a novel prognostic value in patients with ovarian cancer. Mol Carcinog. 2009;48:610–617. doi: 10.1002/mc.20504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Psyrri A, Yu Z, Weinberger PM, Sasaki C, Haffty B, Camp R, Rimm D, Burtness BA. Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin Cancer Res. 2005;11:5856–5862. doi: 10.1158/1078-0432.CCR-05-0420. [DOI] [PubMed] [Google Scholar]
- [79].Li C, Iida M, Dunn EF, Ghia AJ, Wheeler DL. Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene. 2009;28:3801–3813. doi: 10.1038/onc.2009.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Jaganathan S, Yue PB, Paladino DC, Bogdanovic J, Huo Q, Turkson J. A Functional Nuclear Epidermal Growth Factor Receptor, Src and Stat3 Heteromeric Complex in Pancreatic Cancer Cells. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019605. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [81].de la Iglesia N, Konopka G, Puram SV, Chan JA, Bachoo RM, You MJ, Levy DE, Depinho RA, Bonni A. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–462. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lo HW, Cao X, Zhu H, Ali-Osman F. Cyclooxygenase-2 is a novel transcriptional target of the nuclear EGFR-STAT3 and EGFRvIII-STAT3 signaling axes. Mol Cancer Res. 2010;8:232–245. doi: 10.1158/1541-7786.MCR-09-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Liao HJ, Carpenter G. Role of the Sec61 translocon in EGF receptor trafficking to the nucleus and gene expression. Mol Biol Cell. 2007;18:1064–1072. doi: 10.1091/mbc.E06-09-0802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 2006;98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- [85].Klein C, Gensburger C, Freyermuth S, Nair BC, Labourdette G, Malviya AN. A 120 kDa nuclear phospholipase Cgamma1 protein fragment is stimulated in vivo by EGF signal phosphorylating nuclear membrane EGFR. Biochemistry. 2004;43:15873–15883. doi: 10.1021/bi048604t. [DOI] [PubMed] [Google Scholar]
- [86].Edwards J, Traynor P, Munro AF, Pirret CF, Dunne B, Bartlett JM. The role of HER1-HER4 and EGFRvIII in hormone-refractory prostate cancer. Clin Cancer Res. 2006;12:123–130. doi: 10.1158/1078-0432.CCR-05-1445. [DOI] [PubMed] [Google Scholar]
- [87].Eldredge ER, Korf GM, Christensen TA, Connolly DC, Getz MJ, Maihle NJ. Activation of c-fos gene expression by a kinase-deficient epidermal growth factor receptor. Mol Cell Biol. 1994;14:7527–7534. doi: 10.1128/mcb.14.11.7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Xie Y, Hung MC. Nuclear localization of p185neu tyrosine kinase and its association with transcriptional transactivation. Biochem Biophys Res Commun. 1994;203:1589–1598. doi: 10.1006/bbrc.1994.2368. [DOI] [PubMed] [Google Scholar]
- [89].Hung LY, Tseng JT, Lee YC, Xia W, Wang YN, Wu ML, Chuang YH, Lai CH, Chang WC. Nuclear epidermal growth factor receptor (EGFR) interacts with signal transducer and activator of transcription 5 (STAT5) in activating Aurora-A gene expression. Nucleic Acids Res. 2008;36:4337–4351. doi: 10.1093/nar/gkn417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Huang WC, Chen YJ, Li LY, Wei YL, Hsu SC, Tsai SL, Chiu PC, Huang WP, Wang YN, Chen CH, Chang WC, Chen AJE, Tsai CH, Hung MC. Nuclear Translocation of Epidermal Growth Factor Receptor by Akt-dependent Phosphorylation Enhances Breast Cancer-resistant Protein Expression in Gefitinib-resistant Cells. Journal of Biological Chemistry. 2011;286:20558–20568. doi: 10.1074/jbc.M111.240796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Huo L, Wang YN, Xia W, Hsu SC, Lai CC, Li LY, Chang WC, Wang Y, Hsu MC, Yu YL, Huang TH, Ding Q, Chen CH, Tsai CH, Hung MC. RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc Natl Acad Sci U S A. 2010;107:16125–16130. doi: 10.1073/pnas.1000743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Bitler BG, Goverdhan A, Schroeder JA. MUC1 regulates nuclear localization and function of the epidermal growth factor receptor. J. Cell Sci. 2010;123:1716–1723. doi: 10.1242/jcs.062661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang SC, Lien HC, Xia W, Chen IF, Lo HW, Wang Z, Ali-Seyed M, Lee DF, Bartholomeusz G, Ou-Yang F, Giri DK, Hung MC. Binding at and transactivation of the COX-2 promoter by nuclear tyrosine kinase receptor ErbB-2. Cancer Cell. 2004;6:251–261. doi: 10.1016/j.ccr.2004.07.012. [DOI] [PubMed] [Google Scholar]
- [94].Beguelin W, Flaque MCD, Proietti CJ, Cayrol F, Rivas MA, Tkach M, Rosemblit C, Tocci JM, Charreau EH, Schillaci R, Elizalde PV. Progesterone Receptor Induces ErbB-2 Nuclear Translocation To Promote Breast Cancer Growth via a Novel Transcriptional Effect: ErbB-2 Function as a Coactivator of Stat3. Molecular and Cellular Biology. 2010;30:5456–5472. doi: 10.1128/MCB.00012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Li LY, Chen HY, Hsieh YH, Wang YN, Chu HJ, Chen YH, Chien PJ, Ma HT, Tsai HC, Lai CC, Sher YP, Lien HC, Tsai CH, Hung MC. Nuclear ErbB2 Enhances Translation and Cell Growth by Activating Transcription of Ribosomal RNA Genes. Cancer Research. 2011;71:4269–4279. doi: 10.1158/0008-5472.CAN-10-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Wang SC, Nakajima Y, Yu YL, Xia W, Chen CT, Yang CC, McIntush EW, Li LY, Hawke DH, Kobayashi R, Hung MC. Tyrosine phosphorylation controls PCNA function through protein stability. Nat Cell Biol. 2006;8:1359–1368. doi: 10.1038/ncb1501. [DOI] [PubMed] [Google Scholar]
- [97].Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Raju U, Milas L, Chen DJ, Kehlbach R, Rodemann HP. Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J Biol Chem. 2005;280:31182–31189. doi: 10.1074/jbc.M506591200. [DOI] [PubMed] [Google Scholar]
- [98].Dittmann K, Mayer C, Rodemann HP. Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 2005;76:157–161. doi: 10.1016/j.radonc.2005.06.022. [DOI] [PubMed] [Google Scholar]
- [99].Klaus HD, Mayer C, Petra AO, Raju U, Nickolaus HA, Luka M, Rodemann HP. Celecoxib induced tumor cell radiosensitization by inhibiting radiation induced nuclear EGFR transport and DNA-repair: A COX-2 independent mechanism. Int. J. Radiat. Oncol. Biol. Phys. 2008;70:203–212. doi: 10.1016/j.ijrobp.2007.08.065. [DOI] [PubMed] [Google Scholar]
- [100].Hsu SC, Miller SA, Wang Y, Hung MC. Nuclear EGFR is required for cisplatin resistance and DNA repair. American journal of translational research. 2009;1:249–258. [PMC free article] [PubMed] [Google Scholar]
- [101].Liccardi G, Hartley JA, Hochhauser D. EGFR Nuclear Translocation Modulates DNA Repair following Cisplatin and Ionizing Radiation Treatment. Cancer Research. 2011;71:1103–1114. doi: 10.1158/0008-5472.CAN-10-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Luo B, Yu SY, Zhuang L, Xia S, Zhao Z, Rong L. Induction of ERBB2 Nuclear Transport after Radiation in Breast Cancer Cells. J. Huazhong Univ. Sci. Tech.-Med. 2009;29:350–353. doi: 10.1007/s11596-009-0317-8. [DOI] [PubMed] [Google Scholar]
- [103].Xu YR, Shao Y, Zhou J, Voorhees JJ, Fisher GJ. Ultraviolet Irradiation-Induces Epidermal Growth Factor Receptor (EGFR) Nuclear Translocation in Human Keratinocytes. J. Cell. Biochem. 2009;107:873–880. doi: 10.1002/jcb.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Dittmann K, Mayer C, Kehlbach R, Rodemann HP. The radioprotector Bowman-Birk proteinase inhibitor stimulates DNA repair via epidermal growth factor receptor phosphorylation and nuclear transport. Radiotherapy and Oncology. 2008;86:375–382. doi: 10.1016/j.radonc.2008.01.007. [DOI] [PubMed] [Google Scholar]
- [105].Wanner G, Mayer C, Kehlbach R, Rodemann HP, Dittmann K. Activation of protein kinase Cepsilon stimulates DNA-repair via epidermal growth factor receptor nuclear accumulation. Radiother Oncol. 2008;86:383–390. doi: 10.1016/j.radonc.2007.10.041. [DOI] [PubMed] [Google Scholar]
- [106].Moy B, Kirkpatrick P, Kar S, Goss P. Lapatinib. Nature reviews. 2007;6:431–432. doi: 10.1038/nrd2332. [DOI] [PubMed] [Google Scholar]
- [107].Patel D, Lahiji A, Patel S, Franklin M, Jimenez X, Hicklin DJ, Kang X. Monoclonal antibody cetuximab binds to and down-regulates constitutively activated epidermal growth factor receptor vIII on the cell surface. Anticancer Res. 2007;27:3355–3366. [PubMed] [Google Scholar]
- [108].Rivera F, Vega-Villegas ME, Lopez-Brea MF, Marquez R. Current situation of Panitumumab, Matuzumab, Nimotuzumab and Zalutumumab. Acta oncologica (Stockholm, Sweden) 2008;47:9–19. doi: 10.1080/02841860701704724. [DOI] [PubMed] [Google Scholar]
- [109].Dittmann K, Mayer C, Fehrenbacher B, Schaller M, Kehlbach R, Rodemann HP. Nuclear EGFR shuttling induced by ionizing radiation is regulated by phosphorylation at residue Thr654. FEBS Lett. 2010;584:3878–3884. doi: 10.1016/j.febslet.2010.08.005. [DOI] [PubMed] [Google Scholar]
- [110].Kim HP, Yoon YK, Kim JW, Han SW, Hur HS, Park J, Lee JH, Oh DY, Im SA, Bang YJ, Kim TY. Lapatinib, a Dual EGFR and HER2 Tyrosine Kinase Inhibitor, Downregulates Thymidylate Synthase by Inhibiting the Nuclear Translocation of EGFR and HER2. PLoS One. 2009;4:9. doi: 10.1371/journal.pone.0005933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Liao HJ, Carpenter G. Cetuximab/C225-induced intracellular trafficking of epidermal growth factor receptor. Cancer Res. 2009;69:6179–6183. doi: 10.1158/0008-5472.CAN-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Hadzisejdic I, Mustac E, Jonjic N, Petkovic M, Grahovac B. Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod. Pathol. 2010;23:392–403. doi: 10.1038/modpathol.2009.166. [DOI] [PubMed] [Google Scholar]
- [113].Campos ACD, Rodrigues MA, de Andrade C, de Goes AM, Nathanson MH, Gomes DA. Epidermal growth factor receptors destined for the nucleus are internalized via a clathrin-dependent pathway. Biochemical and Biophysical Research Communications. 2011;412:341–346. doi: 10.1016/j.bbrc.2011.07.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ceresa BP. Regulation of EGFR endocytic trafficking by rab proteins. Histol Histopathol. 2006;21:987–993. doi: 10.14670/HH-21.987. [DOI] [PubMed] [Google Scholar]
- [116].Ceresa BP, Bahr SJ. rab7 activity affects epidermal growth factor:epidermal growth factor receptor degradation by regulating endocytic trafficking from the late endosome. J Biol Chem. 2006;281:1099–1106. doi: 10.1074/jbc.M504175200. [DOI] [PubMed] [Google Scholar]
- [117].Giri DK, Ali-Seyed M, Li L-Y, Lee D-F, Ling P, Bartholomeusz G, Wang S-C, Hung M-C. Endosomal Transport of ErbB-2: Mechanism for Nuclear Entry of the Cell Surface Receptor. Mol. Cell. Biol. 2005;25:11005–11018. doi: 10.1128/MCB.25.24.11005-11018.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Weis K. Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell. 2003;112:441–451. doi: 10.1016/s0092-8674(03)00082-5. [DOI] [PubMed] [Google Scholar]
- [119].Hsu SC, Hung MC. Characterization of a novel tripartite nuclear localization sequence in the EGFR family. J Biol Chem. 2007;282:10432–10440. doi: 10.1074/jbc.M610014200. [DOI] [PubMed] [Google Scholar]
- [120].Wang YN, Wang HM, Yamaguchi H, Lee HJ, Lee HH, Hung MC. COPI-mediated retrograde trafficking from the Golgi to the ER regulates EGFR nuclear transport. Biochemical and Biophysical Research Communications. 2010;399:498–504. doi: 10.1016/j.bbrc.2010.07.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang YN, Yamaguchi H, Huo LF, Du Y, Lee HJ, Lee HH, Wang HM, Hsu JM, Hung MC. The Translocon Sec61 beta Localized in the Inner Nuclear Membrane Transports Membrane-embedded EGF Receptor to the Nucleus. Journal of Biological Chemistry. 2010;285:38720–38729. doi: 10.1074/jbc.M110.158659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Wang ZH, Tian XX, Cheng Y, Yam GH, Ng HK, Ding MX, Chew-Cheng SB, Chew EC. Association of EGFR gene fragments with nuclear matrices in glioblastoma cell lines. Anticancer Res. 1998;18:4329–4332. [PubMed] [Google Scholar]
- [123].Offterdinger M, Schofer C, Weipoltshammer K, Grunt TW. c-erbB-3: a nuclear protein in mammary epithelial cells. J Cell Biol. 2002;157:929–939. doi: 10.1083/jcb.200109033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Tao Y, Song X, Tan Y, Lin X, Zhao Y, Zeng L, Tang M, Li W, Wu Q, Cao Y. Nuclear translocation of EGF receptor regulated by Epstein-Barr virus encoded latent membrane protein 1. Sci China C Life Sci. 2004;47:258–267. doi: 10.1007/BF03182771. [DOI] [PubMed] [Google Scholar]
- [125].Boerner JL, Demory ML, Silva C, Parsons SJ. Phosphorylation of Y845 on the epidermal growth factor receptor mediates binding to the mitochondrial protein cytochrome c oxidase subunit II. Mol Cell Biol. 2004;24:7059–7071. doi: 10.1128/MCB.24.16.7059-7071.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Demory ML, Boerner JL, Davidson R, Faust W, Miyake T, Lee I, Hüttemann M, Douglas R, Haddad G, Parsons SJ. Epidermal Growth Factor Receptor Translocation to the Mitochondria. Journal of Biological Chemistry. 2009;284:36592–36604. doi: 10.1074/jbc.M109.000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Yao Y, Wang G, Li Z, Yan B, Guo Y, Jiang X, Xi Z. Mitochondrially localized EGFR is independent of its endocytosis and associates with cell viability. Acta Biochim Biophys Sin (Shanghai) 2010;42:763–770. doi: 10.1093/abbs/gmq090. [DOI] [PubMed] [Google Scholar]
- [128].Yue X, Song W, Zhang W, Chen L, Xi Z, Xin Z, Jiang X. Mitochondrially localized EGFR is subjected to autophagic regulation and implicated in cell survival. Autophagy. 2008;4:641–649. doi: 10.4161/auto.5971. [DOI] [PubMed] [Google Scholar]
- [129].Cao X, Zhu H, Ali-Osman F, Lo HW. EGFR and EGFRvIII undergo stress- and EGFR kinase inhibitor-induced mitochondrial translocalization: a potential mechanism of EGFR-driven antagonism of apoptosis. Mol Cancer. 2011;10:26. doi: 10.1186/1476-4598-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Dreier A, Barth S, Goswami A, Weis J. Cetuximab induces mitochondrial translocalization of EGFRvIII, but not EGFR: involvement of mitochondria in tumor drug resistance? Tumour Biol. 2011 doi: 10.1007/s13277-011-0248-4. [DOI] [PubMed] [Google Scholar]
- [131].Naresh A, Long W, Vidal GA, Wimley WC, Marrero L, Sartor CI, Tovey S, Cooke TG, Bartlett JMS, Jones FE. The ERBB4/HER4 Intracellular Domain 4ICD Is a BH3-Only Protein Promoting Apoptosis of Breast Cancer Cells. Cancer Res. 2006;66:6412–6420. doi: 10.1158/0008-5472.CAN-05-2368. [DOI] [PubMed] [Google Scholar]
- [132].Naresh A, Thor AD, Edgerton SM, Torkko KC, Kumar R, Jones FE. The HER4/4ICD Estrogen Receptor Coactivator and BH3-Only Protein Is an Effector of Tamoxifen-Induced Apoptosis. Cancer Res. 2008;68:6387–6395. doi: 10.1158/0008-5472.CAN-08-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Maher PA. Nuclear Translocation of fibroblast growth factor (FGF) receptors in response to FGF-2. J Cell Biol. 1996;134:529–536. doi: 10.1083/jcb.134.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Ni CY, Murphy MP, Golde TE, Carpenter G. gamma -Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science. 2001;294:2179–2181. doi: 10.1126/science.1065412. [DOI] [PubMed] [Google Scholar]
- [135].Ni CY, Yuan H, Carpenter G. Role of the ErbB-4 carboxyl terminus in gamma-secretase cleavage. J Biol Chem. 2003;278:4561–4565. doi: 10.1074/jbc.M210504200. [DOI] [PubMed] [Google Scholar]
- [136].Raabe TD, Deadwyler G, Varga JW, Devries GH. Localization of neuregulin isoforms and erbB receptors in myelinating glial cells. Glia. 2004;45:197–207. doi: 10.1002/glia.10311. [DOI] [PubMed] [Google Scholar]
- [137].Rakowicz-Szulczynska EM, Herlyn M, Koprowski H. Nerve growth factor receptors in chromatin of melanoma cells, proliferating melanocytes, and colorectal carcinoma cells in vitro. Cancer Res. 1988;48:7200–7206. [PubMed] [Google Scholar]
- [138].Wells A, Marti U. Signalling shortcuts: cell-surface receptors in the nucleus? Nat Rev Mol Cell Biol. 2002;3:697–702. doi: 10.1038/nrm905. [DOI] [PubMed] [Google Scholar]
- [139].Pillai G, Cook N, Turley H, Leek RD, Blasquez C, Pezzella F, Harris AL, Gatter KC. The expression and cellular localization of phosphorylated VEGFR2 in lymphoma and non-neoplastic lymphadenopathy: an immunohistochemical study. Histopathology. 2005;46:209–216. doi: 10.1111/j.1365-2559.2005.02081.x. [DOI] [PubMed] [Google Scholar]
- [140].Zwaagstra JC, Guimond A, O'Connor-McCourt MD. Predominant intracellular localization of the type I transforming growth factor-beta receptor and increased nuclear accumulation after growth arrest. Exp Cell Res. 2000;258:121–134. doi: 10.1006/excr.2000.4905. [DOI] [PubMed] [Google Scholar]
- [141].Jans DA, Hassan G. Nuclear targeting by growth factors, cytokines, and their receptors: a role in signaling? Bioessays. 1998;20:400–411. doi: 10.1002/(SICI)1521-1878(199805)20:5<400::AID-BIES7>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- [142].Tseng HC, Lyu PC, Lin WC. Nuclear localization of orphan receptor protein kinase (Ror1) is mediated through the juxtamembrane domain. BMC Cell Biol. 2010;11:48. doi: 10.1186/1471-2121-11-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Fiorillo AA, Medler TR, Feeney YB, Liu Y, Tommerdahl KL, Clevenger CV. HMGN2 Inducibly Binds a Novel Transactivation Domain in Nuclear PRLr to Coordinate Stat5a-Mediated Transcription. Mol. Endocrinol. 2011;25:1550–1564. doi: 10.1210/me.2011-0106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Gomes DA, Rodrigues MA, Leite MF, Gomez MV, Varnai P, Balla T, Bennett AM, Nathanson MH. C-met must translocate to the nucleus to initiate calcium signals. Journal of Biological Chemistry. 2008;283:4344–4351. doi: 10.1074/jbc.M706550200. [DOI] [PMC free article] [PubMed] [Google Scholar]