Abstract
Fruit quality traits are major breeding targets in the Rosaceae. Several of the major Rosaceae species are current or ancient polyploids. To dissect the inheritance of fruit quality traits in polyploid fleshy fruit species, we used a cultivated strawberry segregating population comprising a 213 full-sibling F1 progeny from a cross between the variety ‘Capitola’ and the genotype ‘CF1116’. We previously developed the most comprehensive strawberry linkage map, which displays seven homoeology groups (HG), including each four homoeology linkage groups (Genetics 179:2045–2060, 2008). The map was used to identify quantitative trait loci (QTL) for 19 fruit traits related to fruit development, texture, colour, anthocyanin, sugar and organic acid contents. Analyses were carried out over two or three successive years on field-grown plants. QTL were detected for all the analysed traits. Because strawberry is an octopolyploid species, QTL controlling a given trait and located at orthologous positions on different homoeologous linkage groups within one HG are considered as homoeo-QTL. We found that, for various traits, about one-fourth of QTL were putative homoeo-QTL and were localised on two linkage groups. Several homoeo-QTL could be detected the same year, suggesting that several copies of the gene underlying the QTL are functional. The detection of some other homoeo-QTL was year-dependent. Therefore, changes in allelic expression could take place in response to environmental changes. We believe that, in strawberry as in other polyploid fruit species, the mechanisms unravelled in the present study may play a crucial role in the variations of fruit quality.
Electronic supplementary material
The online version of this article (doi:10.1007/s00122-011-1769-3) contains supplementary material, which is available to authorized users.
Introduction
Polyploidy or whole genome duplication has long been recognised as a major force in evolution (Ohno 1970). This process is especially prevalent in plants, where it is estimated that 50–70% of flowering plants are polyploids (Masterson 1994). In addition to the plethora of examples represented by polyploid complexes, all the sequenced plant genomes previously considered as “diploids” (e.g. Arabidopsis, rice, poplar or grape vine) have revealed superimposed traces of past genome duplication events (The Arabidopsis Genome Initiative 2000; Tuskan et al. 2006; Jaillon et al. 2007). Polyploidization is usually followed by a process of diploidization (Bowers et al. 2003; Paterson et al. 2004) whereby gene redundancy is reduced via gene silencing, sequence elimination and rearrangement, demethylation of retroelements and relaxation of imprinting (see reviews in Chen 2007; Doyle et al. 2008).
To date, quantitative trait loci (QTL) have been characterised in few polyploid species. These include commercial species such as tetraploid cotton (e.g. Paterson et al. 2003; Rong et al. 2007; Chen et al. 2009), tetraploid wheat (Somers et al. 2006) and wild wheat (Howard et al. 2011), the fleshy fruit species allotetraploid sour cherry (Wang et al. 2000), the autotetraploid potato (Sagredo et al. 2009; McCord et al. 2011) and the hexaploid wheat (Ma et al. 2010). In more genetic complex polyploids such as sugarcane (2n = 8 − 18x), QTL have been also identified (Ming et al. 2001; Ming and Wang 2002; Hoarau et al. 2002; Aitken et al. 2006, 2008; Aljanabi et al. 2007). More recently, QTL approaches have been undertaken for the analysis of day-neutrality (Weebadde et al. 2008) and of agronomic and fruit quality traits (Zorrilla-Fontanesi et al. 2011) in the cultivated octoploid strawberry, Fragaria × ananassa as well as for analysing the genetic architecture of sexual dimorphism in the octoploid Fragaria virginiana (Spigler et al. 2011).
QTL analysis of polyploids not only provides information on the genetic control of traits useful for breeding new varieties through marker-assisted-selection, but also on the relationships among genes or gene copies influencing the traits. Considering the polyploid nature, each trait is likely controlled by homoeologous gene series (homoeoalleles). Homoeoalleles are alleles located at orthologous positions that belong to the different genomes that compose the polyploid species. One major question is to know how the different homoeoalleles contribute to the control of a specific trait. The prerequisite for answering this question is first to construct a genetic linkage map on which the homoeologous linkage groups are ranged by homoeology groups and then to detect the QTL controlling the traits of interest. This approach allowed the detection of homoeoalleles controlling cane yield trait in sugarcane (Aitken et al. 2008) or drought resistance in durum wheat (Peleg et al. 2009). The octoploid cultivated strawberry, for which homoeology groups have been recently identified (Rousseau-Gueutin et al. 2008; Sargent et al. 2009), appears as a good model for studying the control of fruit quality in polyploid fleshy fruit species.
Fragaria, which encompasses all soft-fruited strawberry species, is comprised of a limited number of species ranging from diploids (2n = 2x = 14) to decaploid (2n = 8x = 56) (Staudt 1989, 2009; Hummer et al. 2009). It is belonging to the Rosaceae family, which comprises many economically important species such as the apple, peach, and plum and to the Rosodeae supertribe (Potter et al. 2007) which includes various edible and cultivated berries (e.g. Rubus and Fragaria) and ornamental plants (e.g. Rosa and Potentilla). In the Fragaria genus, the most economically relevant species is the octoploid cultivated strawberry, Fragaria × ananassa (2n = 8x = 56) which is a hybrid species which originated by chance following the hybridization of the two New World strawberry species, F. chiloensis and F. virginiana, in the botanical gardens of Europe in the mid-18th century (Darrow 1966). The high ploidy level of the cultivated strawberry has likely delayed the development of genomics tools such as linkage maps. Nevertheless, recent comparative mapping analysis using diploid and octoploid genetic reference maps of Fragaria (Rousseau-Gueutin et al. 2008; Sargent et al. 2009) highlighted high macrosynteny and colinearity levels between Fragaria genomes thereby supporting the hypothesis that major chromosomal rearrangements have not been frequent along the evolution of the Fragaria genus. Some rearrangements and duplications were, however, detected in specific regions (Sargent et al. 2009; Goldberg et al. 2010; Spigler et al. 2010). In addition, linkage maps in the octoploid Fragaria showed major disomic behaviour at meiosis both in the cultivated octoploid Fragaria species Fragaria × ananassa (Lerceteau-Köhler et al. 2003; Rousseau-Gueutin et al. 2008; Weebadde et al. 2008) and in the wild octoploid species, F. virginiana (Spigler et al. 2010), which facilitates the mapping and QTL detection studies.
In the recent years, fruit sensorial and nutritional traits have become major breeding targets in strawberry. Strawberries with intense flavour are characterised by their high titratable acidity, high soluble-solid content (Kader 1991) and high level of aromas (Aharoni et al. 2004). The main soluble sugars in the strawberry, glucose and fructose, represent more than 80% of the total sugars and 40% of the total dry weight (Wrolstad and Shallenberger 1981). The main organic acid is citric acid, which accounts for 88% of the total acid content (Green 1971). Strawberry is also, as other berries, an important source of phytochemicals. Part of them are antioxidant compounds, which are important dietary components helping to reduce risk of chronic disease (Demmig-Adams and Adams 2002; Battino et al. 2009). One of the major antioxidants in strawberry is the anthocyanin pelargonidin-3-glucoside (Wrolstad et al. 1970; Kalt et al. 1993; Tulipani et al. 2009). Genotype effect on fruit quality traits has been shown for health compounds such as phenolic compounds, ascorbic acid and ellagic acid (Atkinson et al. 2006; Tulipani et al. 2008) and for flavour (de Boishebert et al. 2004). In addition, inheritance of some strawberry quality traits has been documented for fruit size and ripening (see references in Scott and Lawrence 1975; Galletta and Maas 1990; Coman and Popescu 1997), soluble-solid content and sugars, titratable acidity and organic acids (Shaw et al. 1987; Shaw 1988; Ohtsuka et al. 2004) and ascorbic acid (Lundergan and Moore 1975; Sone et al. 2003).
In the present study, we report QTL identification for fruit quality in the octoploid cultivated strawberry using a cross between cv. Capitola and the breeding line CF1116. Fruit quality traits included fruit sensorial traits characterised by soluble sugars and organic acids, fruit colour characterised by anthocyanin content, and fruit shape characterised by fruit length and diameter. Two parental maps were developed, which comprised 28 and 26 linkage groups for the female and male parent, respectively. Most of the linkage groups have been ranged into the seven expected homoeologous groups. These two parental maps were finally merged into the most complete strawberry map developed to date (Rousseau-Gueutin et al. 2008). All the merged linkage groups of the integrated map were in coupling/repulsion phase, except two of them which were composed of two parental linkage groups that segregated in coupling. The cultivar Capitola produces large fruits with low sugar and high acidity, while the breeding line CF1116 produces small fruits with high sugar and lower acidity. QTL, including homoeo-QTL, were identified over two or three consecutive years for 19 traits among which fruit size and shape, firmness, colour, and metabolites (sugars, organic acids, anthocyanins). We further show that ca. one-fourth of the fruit quality traits may be controlled by homoeo-QTL in the cultivated strawberry. The implications of these results are most important for (1) the identification and functional analysis of candidate genes controlling major fruit traits and (2) the strategy used for breeding new strawberry varieties with improved fruit quality.
Materials and methods
Plant materials
A pseudo full-sibling F1 population of 213 individuals obtained from a cross between the variety ‘Capitola’ (‘CA75.121-101’ × ‘Parker’, University of California, Davis, USA) and the genotype ‘CF1116’ ([‘Pajaro’ × (‘Earlyglow’ × ‘Chandler’], reference from the Ciref, France) was developed. The two parents, ‘Capitola’ and ‘CF1116’, present many contrasting fruit quality traits (Moing et al. 2001).
The mapping progeny was obtained in 1999 and the field plantation was renewed each year by runner propagation. Due to propagation problems, only 193 out of the initial 213 individuals (90.6%) were maintained in the field. For each of the three consecutive study years [2001 (year 1), 2002 (year 2) and 2003 (year 3)], field plantation was conducted by planting cold stored plants under a plastic tunnel with drip irrigation.
Nineteen fruit traits or derived traits were evaluated from five randomly harvested mature fruits per genotype. The ripe fruits were harvested at maturity, i.e. when they have attained a homogenous red colouration.
Agronomic and physical traits
A total of ten agronomical and physical traits were measured for the three studied years. For each fruit, the ripening date (RD), corresponding to the harvest date, was recorded. In addition, its length (FL), diameter (FD), weight (FW), skin colour parameters (L, a, b), firmness (FIRM) were measured and its diameter/length ratio (FD/FL) was calculated. The percentage dry weight (DW) of each fruit was estimated by weighing a ca. 0.5 g portion (fresh weight) before and after drying at 60°C for 8 days. The external skin colour parameters (L, a, b) were measured using a Minolta CR2000 chromameter, where L corresponds to Luminance, and a and b to the chromaticity coordinates (on green-to-red and blue-to-yellow tones, respectively). Firmness was evaluated using a penetrometer (Setop-Giraud Technologie, Cavaillon, France). Agronomic and physical traits were measured the 3 years of observation except DW, which was measured only in years 1 and 2.
Chemical traits
A total of eight traits related to sweetness [four traits: soluble-solids content (SSC), glucose (GLU), fructose (FRU) and sucrose (SUCR) concentrations], acidity [four traits: titratable acidity (TA), pH, malate (MAL) and citrate (CIT) concentrations] were measured the first two years except SSC, TA and pH measured in fruits harvested in the three years. For each genotype, 0.5 g fresh weight per fruit was cut off from each of five fruits and extracted with 16 ml 3% metaphosphoric acid. Juices were then expressed from the remainder of the five individual fruits (0.5 ml per fruit) and were rapidly combined. Combined juices were then immediately centrifuged at 3,800×g for 15 min, and kept on ice for the evaluation of pH, TA and SSC. SSC and pH were measured according to Moing et al. (2001). Four hundred μl of the supernatant was rapidly fixed for 15 min with hot ethanol at a final concentration of 80% (v/v). Ethanolic extracts were vacuum dried and each extract corresponded to one genotype. Sucrose, glucose, fructose, malate and citrate, were analysed enzymatically (Velterop and Vos 2001) in ethanolic extracts solubilised in pure water using a microplate spectrophotometer (MR 5000, Dynatech, St. Cloud, France).
Total anthocyanin (ANTH) content was measured the two last years, 2 and 3, using a method based on the protocol of Sanz et al. (1999). The anthocyanin concentration was expressed in terms of pelargonidine-3-glucoside equivalents (eq P3G), since this is the main anthocyanin found in strawberry fruit (Wrolstad et al. 1970; Kalt et al. 1993).
Data analysis
Using measurements across years as replicates, the mean and standard deviation of each trait were calculated for the two parents and the progeny. Pairwise comparisons among parents were done using Student’s T test (P < 0.05). For each trait, differences among progeny were tested by ANOVA. Where a genotypic effect within progeny was found, broad sense heritability was evaluated from variance analysis as follows:
where σ2g is the genetic variance, σ2e is the environmental variance and n the coefficient for σ2g from the genotype expected mean square in the RANDOM statement of PROC GLM (SAS).
Trait segregation was declared transgressive when at least one progeny had a value that was higher or lower than that of the highest or lowest parent, by at least twice the standard deviation of the parents.
Pearson correlation coefficients were calculated for the different trait values, both between and within years (PROC CORR, SAS).
For traits deviating from normality, three transformations (log, square root and cubic root) were tested. The transformation providing significant results was used in the QTL analysis. Where the post transformation result was not significant, we used the transformation that gave the least-skewed results. All statistical analyses were performed with the Statistical Analysis System (SAS Institute, Inc., Cary, NC, USA).
QTL detection
QTL detection was performed by composite interval mapping (CIM) (Zeng 1993; Zeng 1994) using QTL Cartographer software (Basten et al. 1997) separately for each parent using the female and male maps developed by Rousseau-Gueutin et al. (2008). A forward–backward stepwise regression was performed to choose the five co-factors with the highest F values before performing QTL detection by CIM. A window size of 10 cM around the test interval, where the co-factors were not considered, was chosen (model 6 of QTLCartographer). The statistical significance threshold for declaring a putative QTL was determined by permutations of the data set. After 1,000 permutations, mean LOD thresholds of 2.86 and 2.90 were chosen for female and male data, respectively. This corresponded to a genome-wise significance level of α = 0.10. The proportion of phenotypic variation explained by each significant marker was estimated as the coefficient of determination (R 2) at the peak QTL position estimated by QTL Cartographer.
We examined whether clusters of unique QTL linked to related traits, e.g. a, b or L traits for fruit colour, colocalised on the same linkage groups. The probability that colocations of QTL for two related traits belonging to the same class of traits occurred by chance is given by the hypergeometric probability distribution function (Spigler et al. 2011):
where n is the total number of common intervals compared, l is the number of QTL linked to the trait exhibiting the largest number of QTL, s is the number of QTL linked to the second relative trait, and m is the number of QTL colocalised linked to the two traits. When the probability was lower than 0.05, we can conclude that the colocation of QTL underlies genetic process rather than chance. In this study, we considered the linkage group as the interval. Therefore, a total of 28 intervals were considered. The colocations of QTL linked to related traits were analysed only considering unique QTLs.
Results
Phenotypic analysis
The mean of the trait values of the two parents ‘Capitola’ and ‘CF1116’, calculated by combining the data obtained across years, differed significantly from each other for the 19 traits analysed (Table 1). The most important differences between the parents were observed for dry weight, soluble-solids, glucose, and malate (dry weight DW, soluble-solids content SSC, glucose GLU, and malate MAL, respectively). Compared to ‘CF1116’, ‘Capitola’’s fruits were more watery and acidic. ‘CF1116’ fruits had higher sugar concentrations than those of Capitola, matured earlier and were firmer. ‘Capitola’ fruits were dark red and contained a higher concentration of total anthocyanin ANTH than their ‘CF1116’ counterparts.
Table 1.
Recorded trait values for ‘Capitola’ and ‘CF1116’ parents and related progeny
| Traita | Unit | ‘Capitola’ | ‘CF1116’ | Progeny | h 2 | Tr | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | Mean | SD | Range | No. | Mean | SD | Range | Cap vs. CFb | No. | Mean | SD | Range | ||||
| Development | ||||||||||||||||
| FDc | mm | 21 | 36.2 | 6.7 | 24.1–48.8 | 20 | 30.8 | 5.8 | 25.4–48.1 | ** | 592d | 32.5 | 4.1 | 22.1–41.1 | 0.21 | No |
| FLc | mm | 21 | 44.1 | 5.9 | 29.4–52.6 | 20 | 40.6 | 4.3 | 35.8–54.4 | * | 594 | 42.4 | 5.5 | 30.1–53.3 | 0.51 | No |
| FD/FLc | – | 21 | 1.2 | 0.1 | 1.1–1.5 | 20 | 1.3 | 0.1 | 1.1–1.5 | ** | 592 | 1.3 | 0.2 | 1.0–1.8 | 0.72 | Yes |
| FWc | g | 21 | 24.7 | 11.4 | 6.7–48.2 | 20 | 15.8 | 9 | 9.3–48.9 | ** | 594 | 17.9 | 5.5 | 6.9–28.1 | – | No |
| DWd | g | 11 | 7.5 | 1 | 6.2–9.7 | 10 | 11 | 0.9 | 9.4–12.1 | *** | 302 | 9.3 | 1.4 | 5.5–12.4 | 0.28 | Yes |
| RDc | Julian day | 21 | 127.2 | 5.5 | 119–141 | 20 | 124.3 | 3.5 | 115–127 | * | 503 | 124.7 | 8.3 | 112–142 | – | Yes |
| Texture | ||||||||||||||||
| FIRMc | Durofel unit | 57 | 55.1 | 16.1 | 32.3–79.0 | 52 | 61.1 | 16 | 40.5–87.0 | *** | 579 | 57.7 | 14.5 | 32.1–75.7 | – | Yes |
| Colour | ||||||||||||||||
| L c | 21 | 36.2 | 2.9 | 29.3–41.7 | 20 | 38.5 | 2.5 | 30.8–41.0 | * | 594 | 37.4 | 2.9 | 31.9–46.4 | 0.61 | Yes | |
| a c | 21 | 35.5 | 3.1 | 30.8–40.5 | 20 | 38.8 | 3.1 | 31.3–43.8 | ** | 594 | 36.1 | 3.7 | 28.1–42.9 | 0.57 | Yes | |
| b c | 21 | 19.4 | 3.7 | 14.1–26.5 | 20 | 22.6 | 3.3 | 15.2–27.6 | ** | 593 | 20.4 | 4 | 13.7–30.6 | 0.52 | Yes | |
| AN-THd | eq P3G g/L juice | 17 | 0.9 | 0.1 | 0.6–1.2 | 17 | 0.4 | 0.1 | 0.3–0.8 | *** | 390 | 0.7 | 0.2 | 0.2–1.2 | 0.7 | Yes |
| Sugar-related traits | ||||||||||||||||
| SSCc | Brix | 21 | 5.5 | 0.6 | 4.4–6.7 | 20 | 8.4 | 1.2 | 6.7–10.3 | *** | 595 | 7.1 | 1.2 | 4.5–9.3 | – | No |
| GLUd | g/L juice | 11 | 13 | 2.7 | 7.9–17.1 | 10 | 24.6 | 3.5 | 18.8–28.9 | *** | 394 | 19.9 | 4.1 | 11.8–32.8 | 0.53 | Yes |
| FRUd | g/L juice | 11 | 15.9 | 2.8 | 13.6–23.5 | 10 | 26 | 4.3 | 19.6–33.6 | *** | 393 | 20.8 | 4 | 12.6–32.3 | 0.24 | No |
| SUCRd | g/L juice | 11 | 12.2 | 4.7 | 6.7–22.5 | 10 | 30.5 | 15 | 15.0–59.5 | ** | 390 | 16 | 5.9 | 2.4–29.0 | 0.55 | Yes |
| Acid-related traits | ||||||||||||||||
| PHc | 21 | 3.4 | 0.1 | 3.2–3.9 | 20 | 3.7 | 0.1 | 3.52–4.0 | *** | 595 | 3.5 | 0.1 | 3.1–3.8 | 0.53 | Yes | |
| TAc | meq/L juice | 21 | 103.9 | 13.6 | 80–130 | 20 | 88.3 | 8.4 | 70–104 | *** | 592 | 101.9 | 15.5 | 67.2–137.5 | 0.65 | Yes |
| MALd | g/L juice | 11 | 2 | 0.3 | 1.6–2.6 | 10 | 1.2 | 0.3 | 0.8–1.7 | *** | 391 | 1.6 | 0.5 | 0.6–2.8 | 0.54 | Yes |
| CITd | g/L juice | 11 | 6.4 | 0.7 | 5.8–7.6 | 10 | 5.8 | 0.7 | 4.5–6.8 | * | 393 | 5.6 | 1.1 | 3.3–9.2 | 0.53 | Yes |
The number (No.) of individuals analysed, the mean values, standard deviations (SD), range and heritability of the traits are described. The significance level of the difference between ‘Capitola’ versus ‘CF1116’ means is indicated in the Cap versus CF column. Heritability (h 2) was only calculated for traits in which ANOVA identified a significant within-progeny genotypic effect. Trait segregation was declared transgressive (Tr) when at least one progeny had a value that was higher or lower than the highest or lowest parental value by at least twice the SD of the parents
aTraits are the following: development: FD, fruit diameter; FL, fruit length; FD/FL, ratio; DW, dry weight; FW, fruit weight; RD, ripening date of the fruit. Texture: FIRM, fruit firmness. Colour: skin colour parameters, a, b and L; ANTH, total anthocyanin. Sugar-related traits: SSC, soluble-solids content; FRU, fructose; GLU, glucose; SUCR, sucrose. Acid-related traits: pH; TA, titratable acidity; CIT, citrate; MAL, malate
b*, ** and *** indicate values that are significant at 0.01 < P < 0.05, 0.001 < P < 0.01, and P < 0.001, respectively
cValues are the means of the genotype for n = 3 replicates (years)
dValues are the means of the genotype for n = 2 replicates (years)
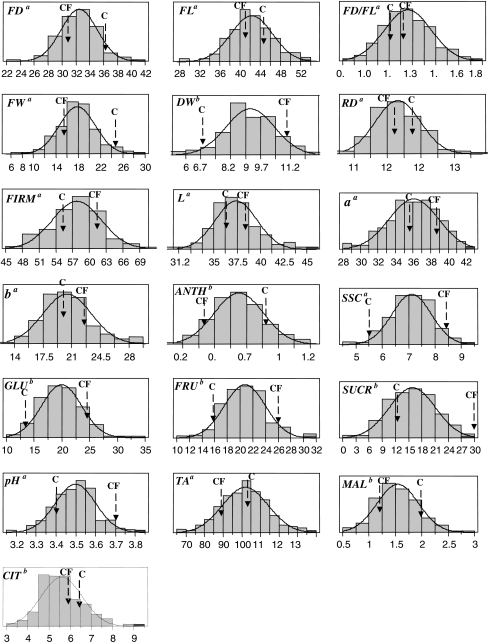
The 19 traits showed continuous variation in the progeny and normal distributions were observed for almost all traits (Fig. 1). With the exception of citrate CIT, the progeny mean for a given trait was within the value of the two parental means (Table 1). Progeny showed high variations in most trait values and a genotypic effect among progeny was observed for 15 of the 19 (79.0%) analysed traits (Table 1). For these 15 traits, broad sense heritabilities were calculated and displayed high values (h 2 > 0.5) for 12 traits (fruit length FL, fruit diameter/length ratio FD/FL, skin colour parameters L, a and b, ANTH, GLU, sucrose SUCR, pH, titratable acidity TA, MAL, and CIT) or low values (h 2 < 0.3) for three traits (fruit diameter FD, DW, fructose FRU). Transgressive segregations were detected for 14 of the 19 traits (Table 1).
Fig. 1.
Distribution of the progeny mean. aValues are the means of the genotype for n = 2 replicates (years). bValues are the means of the genotype for n = 3 replicates (years). FD, fruit diameter; FL, fruit length; FD/FL ratio; DW, dry weight; FW, fruit weight; RD, ripening date of the fruit; FIRM, fruit firmness; skin colour parameters, L, a and b; ANTH, total anthocyanin; SSC, soluble-solids content; FRU, fructose; GLU, glucose; SUCR, sucrose; pH, TA, titratable acidity; CIT, citrate; MAL, malate. Mean of the trait values of the two parents are shown, C for ‘Capitola’ and CF for ‘CF1116’
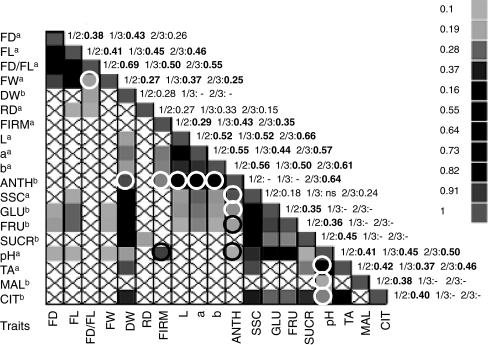
Phenotypic correlations among the 19 traits were calculated both between and within years. Due to the large amount of data collected in this study, only the data relating to year 2 are reported (Fig. 2, Online Resource 1). Even if the correlation values between traits are dependent on the year of study, the results reported for year 2 can be regarded as showing general trends. Correlation values between years 1, 2 and 3 are shown along the diagonal in Fig. 2. These between-year correlations were all highly significant.
Fig. 2.
Phenotypic correlations for the traits measured in year 2 (P < 0.05). The number of individuals per trait pair ranges from 178 to 190. The diagonal gives the three phenotypic correlations between years for each trait: 1/2, 1/3, 2/3 for the phenotypic correlation between years 1 and 2, years 1 and 3, and years 2 and 3, respectively. Correlations significant at P < 0.0001 are reported in bold. A white box with a cross represents a non significant correlation. Negative correlations are indicated with a circle (black or white). The strength of the correlation is colour coded from green-to-red, with green reflecting the lowest correlation values and the red the highest (see figure for more details on the values). FD, fruit diameter; FL, fruit length; FD/FL ratio; DW, dry weight; FW, fruit weight; RD, ripening date of the fruit; FIRM, fruit firmness; skin colour parameters, L, a and b; ANTH, total anthocyanin; SSC, soluble-solids content; FRU, fructose; GLU, glucose; SUCR, sucrose; pH, TA, titratable acidity; CIT, citrate; MAL, malate. aThree years of evaluation. bTwo years of evaluation
In each of year of study, fruit weight (FW) showed a strong positive correlation with fruit shape (FD and FL). FD and FL were positively correlated in all 3 years (P < 0.0001 to P < 0.05 depending on the year) (data not shown). Correlations between the three-colour parameters (L, a and b) were also highly significant in each of the three studied years (P < 0.0001). In addition, these parameters were highly negatively correlated with anthocyanin content (ANTH) when measured (years 2 and 3) (P < 0.0001). These colour parameters were also negatively correlated with ripening date (RD) and positively correlated with percent dry weight (DW) (years 1 and 2, different P values depending on the trait and on the year). DW was positively correlated with the different sugar-related traits, SSC, FRU, GLU and SUCR, in two (FRU, GLU, SUCR) or three (SSC) years of observation (P < 0.0001). All sugar-related traits, FRU, GLU, SUCR as SSC, were positively correlated between them for the 2 years and the correlation is highly significant between FRU and GLU. Same relationship occurred between acid related traits, CIT, pH and TA, except for MAL which is minor in strawberry. Correlations were observed between the various acid and sugar-related traits, both in years 1 and 2. pH was positively correlated with soluble-solids and sugar concentration (SSC, FRU, GLU and SUCR, different P values depending on the trait). Thus, fruits with a lower acidity tended to have a higher sugar concentration.
QTL analysis
QTL analyses were performed and detected for all the 19 quantitative traits using composite interval mapping for either ‘Capitola’ or ‘CF1116’ (Table 2, Online Resource 2). When considering both parents and the 2 (2001 and 2002) or 3 years (2001, 2002, 2003) of observation, a total of 115 QTL were detected. They were summarised in 87 unique QTL, which distribution per HG and LG is given in Table 3. It is important to note that the letters naming one homoeologous linkage group were arbitrarily allocated (e.g. Linkage Group a, b, c or d) (Rousseau-Gueutin et al. 2008), meaning that a LG with the same letter does not necessarily belong to the same subgenome. QTL were considered unique when the same regions overlapped with one LOD support confidence interval in the parental map (male or female) where the QTL were detected (e.g. FD_IIIa_m1-m2 for the QTL of fruit diameter detected on the linkage group IIIa, Fig. 3a-LGIIIa, Online Resource 2) or when QTL were detected on the same LG in the male and female linkage maps (e.g. FIRM_Vc_m3/f3 for the QTL of firmness detected on the linkage group Vc on male and female linkage maps) (Online Resource 2). Per trait, a mean of 4.6 QTL was identified. FL and FIRM are controlled by the largest number of loci (7), followed by RD, a and CIT (6). FRU and SUCR are controlled by the fewest number of loci (2). The percentage of variance explained by a QTL ranged from 5% (FD/FL-IIId-m1) to 17% (ANTH-VIa-m3), and about a quarter of the QTL (23.5%) had a value higher than 10%.
Table 2.
Significant QTL detected for all the traits based on CIM analysis with LOD > LOD threshold (2.86 for the female map and 2.90 for the male map)
| Traitsa | No. QTLb | Location of QTLc (nb of year of detection)d |
|---|---|---|
| Development | ||
| FD | 4 | IIb-m (2), IIIa-m (2), IIIc1-m (1), IVa-m (1) |
| FL | 7 | IIa-f (1), IIc-m (3), IIIa-m (1), IIId-m (1), IVd2-f (1), Vb-f (1), VIb-m (1) |
| FD/FL | 5 | IIIa-m (3)/f (2), IIId-m (2), Va-m (2), Vb-m (1), VIa-m (2) |
| DW | 3 | IIIa-f (1), VIa-m (1), VIIb-m (1) |
| FW | 3 | Ia-f (1), IIb-m (2), IIIc1-m (1) |
| RD | 6 | IIb-m (1), IIId-f (1), IVc-m (1), Vc-m (1), VIb-m (1), VIIa-m (2) |
| Firmness | ||
| FIRM | 7 | IIb-m (1), IIIa-m (1), IIIa-f (1), IIIb-m (1), IIIc-f (1), IVa-f (1), Vc-m/f (1) |
| Colour | ||
| a | 6 | Ia-f (1), IIa-f (3), IVd-m (1), Vb-m (1), VIa-m/f (2), VIb-f (1) |
| b | 4 | Ia-m (1), IIIa-f (1), VIa-m/f (2), VIb-f (1) |
| L | 4 | IIa-f (1), IIIa-m/f (2), IVd-m (1), VIa-m (2) |
| ANTH | 5 | Ia-f (1), IIa-f (2), IIIa-f (1), VIa-m/f (2), VIb-f (1) |
| Sugar-related traits | ||
| SSC | 3 | IIIa-m/f (1), Va-f (1), VIa-m/f (2) |
| FRU | 2 | IVb-m (1), Vc-f (1) |
| GLU | 5 | Ib-f (1), IIIa-f (1), IIIc-f (1), IVa-f (1), VIa-f (1) |
| SUCR | 2 | VIa-m (1), VIIa-f (2) |
| Acid-related traits | ||
| pH | 6 | Ia-m (1), IIIb-m (1), IIIc-f (1), IVc-f (1), Vb-f (1), VIa-f (1) |
| TA | 5 | Ia-m (1), IIc-m (1), IIId-m (1), IVa-f (1), Vb-m/f (2) |
| CIT | 6 | Ia-f (1), IIc-m (1), IIIa-f (1), Vb-f (1), VIa-f (1), VIIb-f (1) |
| MAL | 4 | Vd-f (1), VIa-m (1), VIb-m (1), VIId-m (1) |
aTraits are the following: development: FD, fruit diameter; FL, fruit length; FD/FL, ratio; DW, dry weight; FW, fruit weight; RD, ripening date of the fruit. Texture: FIRM, fruit firmness. Colour: skin colour parameters, a, b and L; ANTH, total anthocyanin. Sugar-related traits: SSC, soluble-solids content; FRU, fructose; GLU, glucose; SUCR, sucrose. Acid-related traits: pH; TA, titratable acidity; CIT, citrate; MAL, malate
bNumber of unique QTL. QTL were considered unique when, the same regions overlapped with one LOD support confidence interval
cName of the linkage group according to Rousseau-Gueutin et al. (2008). The Roman number indicates the homoeologous group number. The letter, a, b c, or d, indicates one of the four homoeologous linkage groups and the final letter, m or f, indicates if the linkage group belongs to the male or the female linkage maps
dIn brackets, the number of detection years of the individual QTL
Table 3.
Distribution of unique QTL locations per homoeologous groups (HG) and linkage group (LG)
| HGa | No. of QTL by LGb | Total (%) | |||
|---|---|---|---|---|---|
| a | b | c | d | ||
| I | 7 | 1 | 0 | 0 | 8 (9) |
| II | 4 | 4 | 3 | 0 | 11 (13) |
| III | 12 | 2 | 5 | 4 | 23 (27) |
| IV | 4 | 1 | 2 | 3 | 10 (11) |
| V | 2 | 6 | 3 | 1 | 12 (14) |
| VI | 12 | 6 | 0 | 0 | 18 (21) |
| VII | 2 | 2 | 0 | 1 | 5 (5) |
Percentage of QTL on total QTL is given in brackets
aThe Roman number indicates the homoeologous group (HG) number according to Rousseau-Gueutin et al. (2008)
bThe letter, a, b c, or d, indicates one of the four homoeologous linkage groups. The number of QTL is considered unique, i.e. the same regions overlapped with one LOD support confidence interval
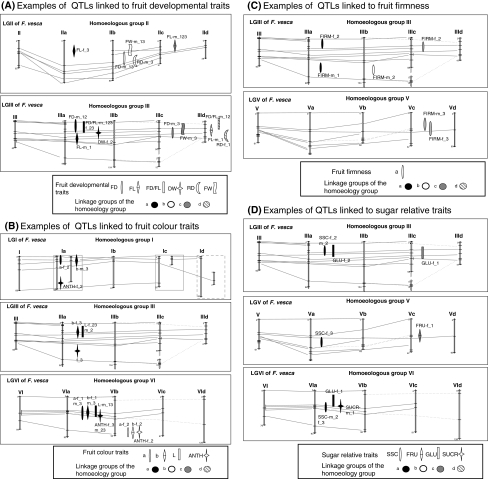
Fig. 3.
Examples of QTL locations of fruit traits (development, texture, colour, sugar-related traits, acid-related trait) on linkage groups ranged by homoeologous groups. The linkage map is the consensus map obtained using JoinMap (Rousseau-Gueutin et al. 2008). Linkage group names for the octoploid consist of the number of the HG to which they belong followed by a letter arbitrarily allocated (a, b, c or d). The position of the QTL on the consensus map is the one of the closest marker to the QTL on the parental maps. The first linkage group belongs to the diploid reference linkage map and allow to anchor common markers with and between Fragaria × ananassa linkage groups. Fruit traits are the following: development: FD, fruit diameter; FL, fruit length; FD/FL ratio; DW, dry weight; FW, fruit weight; RD, ripening date of the fruit. Texture: FIRM, fruit firmness. Colour: skin colour parameters, L, a and b; ANTH, total anthocyanin. Sugar-related traits: SSC, soluble-solids content; FRU, fructose; GLU, glucose; SUCR, sucrose. Acid-related traits: pH, TA, titratable acidity; CIT, citrate; MAL, malate
Among the 12 traits that were analysed three consecutive years, 16 (27%) of the 60 unique QTL were detected at least in 2 of the 3 years (Online Resource 2). Among these three QTL linked to FL, FD/FL and a were detected in the 3 years. For those analysed two consecutive years, three (11%) QTL linked to ANTH and SUCR out of the 27 unique QTL were detected for the 2 years. In total, 47.0% of the QTL had an effect in the expected direction based on parental mean comparison (49.1% of the QTL detected for ‘Capitola’ and 45.2% for ‘CF1116’) (Online Resource 2). Nine QTL were observed on both maps, either the same year (e.g. FD/FL-IIIa-m2 and -f2 or -m3 and -f3, L-IIIa-m2 and -f2), or different years (e.g. a-VIa-f1 and -m3, SSC-IIIa-m2 and f2) (Online Resource 2). This suggests presence of potential allelic forms of the same gene responsible for the variation of quantitative characters.
QTL were preferentially located in the homoeology group (HG) III and VI (26 and 21%, respectively, for unique QTL) (Table 3) and on one and two linkage groups within these two homoeology groups, namely LGIIIa (14%) for the HGIII and LGVIa (14%) and LGVIb (7%) for the HGVI (Table 3). No significant correlation between linkage groups length and QTL number was observed either when male and female maps were considered separately or when linkage groups of the integrated linkage map were compared. Many QTL co-locations were found both on these LG and on others, forming QTL clusters that represented the same class of traits. In the 28 LGs of the consensus map, a total of 15 putative clusters of unique QTL linked to the same class of traits, i.e. fruit development, colour, acidity, or sweetness, were analysed. For seven out of the 15 clusters colocations were found more often than expected by chance (P < 0.05). For example, a cluster of development-related QTL was observed on LGIIb and LGIIIc (FD-IIb-m1 and -m3 and FW_IIb-m1-m3; FD_IIIc1_m3 and FW_IIIc1_m3) (Fig. 3a-LGIIb and LGIIIc). A cluster of colour-related QTL (a-VIb-f2, b-VIb-f2, and ANTH-VIb-f2) was detected on LGVIb on the female map (Fig. 3b-LGVIb). In addition, some chromosomal regions with QTL for several types of traits were also observed. As an example, sugar- and acid-related traits clusters were observed on the LGVIa belonging to the homoeologous group VI (for sugar-related traits: SSC-VIa-m2, GLU-VIa-f1, SUCR-VIa-m1; for acid-related traits: CIT-VIa-f1, MAL-VIa-m1, pH-VIa-f3) (Fig. 3d, e-LGVIa).
Complexity of QTL and contribution of homoeo-QTL to the control of fruit quality traits
Analysis of fruit quality traits QTL further indicated that, among the 87 unique QTL locations identified, 20 (23%) were located on homoeologous groups (HGs) and most probably at orthologous regions of DNA, defined based on the presence of homoeoalleles amplified using the same SSR locus (Table 4). These QTL are thus considered as putative homoeo-QTL. Interestingly, they were exclusively detected on four of the seven HGs, namely on HGII, HGIII, HGV and HGVI. In addition, these homoeo-QTL were exclusively localised on two homoeologous linkage groups out of the four homoeologous linkage groups (Table 4).
Table 4.
Number, location and year of detection of the homoeo-QTL for fruit quality traits
| Traitsa | No. of homoeo-QTL (%)b | Location and year of detected homoeo-QTLc, d |
|---|---|---|
| Development | ||
| FD | 2 (50) | IIIa-m (1-2) and IIIc-m (3) |
| FL | 4 (57) | IIa-f (3) and IIc-m (1-2-3) |
| IIIa-m (1) and IIId-m (1) | ||
| FD/FL | 4 (80) | IIIa-m (1-2-3)/f (1-2) and IIId-m (1-2) |
| Va-m (1-2) and Vb-m (2) | ||
| Firmness | ||
| FIRM | 4 (57) | IIIa-f (2) and IIIc-f (2) |
| IIIa-m (1) and IIIb-m (2) | ||
| Sugar-related traits | ||
| GLU | 2 (40) | IIIa-f (2) and IIIc-f (1) |
| Acid-related traits | ||
| pH | 2 (33) | IIIb-m (2) and IIIc-f (2) |
| MAL | 2 (50) | VIa-m (1) and VIb-m (1) |
aTraits are the following: development: FD, fruit diameter; FL, fruit length; FD/FL ratio. Texture: FIRM, fruit firmness. Sugar-related traits: GLU, glucose. Acid-related traits: pH; MAL, malate
bThe number of unique QTL involved in homoeo-QTL are indicated. The percentage of homoeo-QTL in relation to the total number of QTL detected for each trait is indicated in bracket
cThe Roman number indicates the homoeologous group number according to Rousseau-Gueutin et al. (2008) and the letter, a, b c, or d, indicates one of the four homoeologous linkage groups
dThe year of detection of the QTL is indicated in bracket after its location on the linkage group of the homoeology group
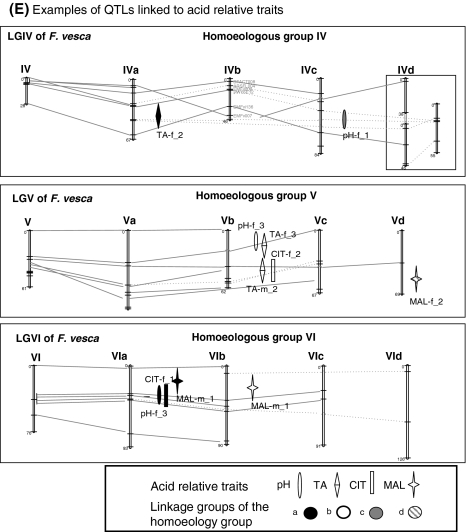
We next investigated whether the contributions of these putative homoeo-QTL were comparable for all fruit quality traits analysed (Table 4). For each class of traits, selected results are shown in Fig. 3. Detailed map locations of all the QTL are presented in extenso in supplemental material (Online Resource 3-A-to-E). It appears that numerous homoeo-QTL were observed for traits related to fruit shape and size (Fig. 3a). Other traits including firmness (Fig. 3c), glucose content (Fig. 3d), pH and malate content (Fig. 3e) were also controlled by homoeo-QTL. At the opposite, no homoeo-QTL were detected for several major fruit quality traits including fruit colour (L, a, b and ANTH; Fig. 3b), fructose content (FRU, Fig. 3d), citrate content and titratable acidity (CIT and TA, Fig. 3e).
The complexity of the genetic control of fruit quality traits in strawberry can be exemplified by the analysis of the fruit development-related traits (FL, FD, FD/FL, FW, RD, and DW) (Table 4; Fig. 3a). Several cases can be highlighted: (1) one QTL can be located on only one of the four linkage groups, as is the case with the QTL for FD observed on years 1 and 3, FD_IIb_m13, which is located on linkage group IIb of the homoeologous group II. That situation can be considered as the same as that of a similar fruit diameter QTL detected in a diploid fleshy fruit species such as tomato (Tanksley 2004); (2) one trait can be controlled by two homoeo-QTL located at orthologous positions on different linkage groups within one homoeologous group. This is the case with the QTL for FD/FL detected two consecutive years, 1 and 2 on LGIIIa and LGIIId (FD/FL_IIIa_m123/f23 and FD/FL_IIId_m12), suggesting that two alleles of the same gene on different linkage groups (homoeoalleles) could be involved in the control of strawberry fruit shape at that locus; (3) the control of one trait by homoeo-QTL could be year-dependent as suggested by the FD QTL detected during two consecutive years on LGIIIa (FD_IIIa_m12) and the third year on LGIIIc (FD_IIIc_m3), where it co-localised with a fruit weight QTL. That situation is particularly interesting since it suggests that the expression/function of the homoeo-alleles controlling fruit growth at this particular locus are under environmental control.
Discussion
The major aim of that study was first to give insights into the genetic and physiological control of major fruit quality traits in a polyploid fleshy fruit and, eventually, to provide tools for breeding strawberry varieties with improved fruit quality, for example elite varieties showing high sugar and acid contents associated with a specific fruit size and shape. Therefore, parents of the cultivated strawberry population studied were specifically chosen for their contrasting characteristics for several major fruit quality traits (Moing et al. 2001): the cv. ‘Capitola’ produces large fruits with low sugar and high acidity while the breeding line ‘CF1116’ produces small fruits with high sugar and lower acidity.
Comparison of the trait correlation values (Fig. 2, Online Resource 1) obtained for the population studied with those previously published in strawberry indicated that, in general, correlations between fruit quality traits documented in this study are in agreement with those previously reported in strawberry (see e.g. Shaw 1988, for between and within sugar- and acid-related traits, firmness and colour parameters, as well as Galletta and Maas 1990, for fruit development traits and Zorrilla-Fontanesi et al. 2011 for some fruit quality traits). For instance, pH is positively correlated with soluble-solids and sugar concentration [SSC, FRU, GLU and SUCR, P < 0.0001 or P <0.05 depending on the trait (Online Resource 1)], as in Shaw (1988) and Zorrilla-Fontanesi et al. (2011) for the correlation between pH and SSC. In contrast, large discrepancies with published values were observed for the heritability of several fruit quality traits. For example, while the heritability value obtained for titratable acidity (0.65) is comparable to the broad sense heritability of 0.78 previously reported by Shaw et al. (1987), the heritability value of 0.55 obtained for sucrose concentration is much higher than the value of 0.0004 reported by Ohtsuka et al. (2004). We also found a high heritability value of 0.53 for organic acids but, in that case, no published data are available for comparison in the octoploid strawberry. Conversely, a moderate heritability (0.35) was determined by Shaw et al. (1987) for soluble-solid content while we detected no significant variation for this trait in our progeny, though the parents displayed significant differences (Table 1). The study of several populations by Shaw et al. (1987) instead of one population in our case, or the environmental effects may explain this discrepancy.
For all the traits studied, the segregating population displayed a large range of values consistent with the expectations (Fig. 1). In addition, extreme characteristics were clearly observed in this study and significant differences between segregants and parents were detected for most of the traits. In total, more than 73% of the traits presented transgressive segregation (Table 1). Combinations of alleles from both parents that have effects in the same direction (complementary gene action) are probably responsible for the large number of transgressive segregations observed in our study and for the presence of extreme genotypes. Transgressive segregation is commonly detected in QTL studies (reviewed in Rieseberg et al. 2003) and has been documented at both interspecific [e.g. in Solanum (de Vicente and Tanksley 1993), Prunus (Quilot et al. 2004) and Oryza (Gutierrez et al. 2010)] and intraspecific levels [e.g. in Prunus cerasus (Wang et al. 2000), Cucumis melo (Monforte et al. 2004), Oryza sativa (Mao et al. 2011), Pisum sativum (Ubayasena et al. 2011), and Lolium perenne (Pfender et al. 2011)], as well as for polyploid species [e.g. in Saccharum (Ming et al. 2001) and Triticum durum (Maccaferri et al. 2011)]. In strawberry, this phenomenon might be even stronger due to the high ploidy level of the species. In these species, dominance/over-dominance effects, which can be a form of genomic expression dominance (Flagel and Wendel 2009), may have contributed to the presence of transgressive segregants for fruit quality. In addition, the presence of a large number of alleles, which can interact epistatically, may also provide an explanation for the formation of these transgressive segregants (Coelho et al. 2007).
Each of the detected QTL explains a low percentage of the phenotypic variation of the trait
Though QTL were detected for all the traits, the percentages of phenotypic variation explained by each QTL for a given trait were low to moderate (4.8–17.3%). These low or moderate values could be linked to the relative large size of population compare to other studies on the cultivated strawberry (Weebadde et al. 2008; Zorrilla-Fontanesi et al. 2011). Similar low values have already been reported in another complex polyploid, the sugar cane, in which the percentages of phenotypic variation explained by each QTL for sugar content varied from 3 to 9% (Hoarau et al. 2002; Aitken et al. 2006). In the case of polyploidy species, this can be explained by a multiple loci control of these genetic traits. As has been previously documented (Doerge and Craig 2000; Wang et al. 2000; Ming et al. 2001; Hoarau et al. 2002), the efficiency of QTL mapping using single-dose marker strategy in polyploids is limited, regardless of the chromosomal pairing mechanisms, due to the large number of possible alleles at each locus. This large number of alleles increases the possible number of allele combinations and ultimately restricts the analysis to the detection of only the most significant alleles contributing to the traits. The situation in strawberry might be better since the genetic base of the cultivated strawberry is reduced (Sjulin and Dale 1987; Hancock and Luby 1995), except if favourable alleles have been concentrated through breeding in the modern varieties.
One-fourth of all fruit quality QTL are located on two linkage groups
An important finding was also that, in the population analysed, the fruit quality QTL were non-randomly distributed across the chromosomes as 28% of the QTL were located on two LGs (LGIII-a and LGVI-a) (Table 3). QTL co-location is one result of this preferential distribution on two linkage groups. Cluster of QTL is often reported in the literature (e.g. in cotton for lint fibre development (Rong et al. 2007), in tomato for fruit development and organoleptic quality (Fulton et al. 1997; Saliba-Colombani et al. 2001), and in pepper for fruit traits (Ben Chaim et al. 2001)). In strawberry similar clusters have been reported both for agronomical traits such as yield and fruit number as well as for fruit quality traits (Zorrilla-Fontanesi et al. 2011). It may reflect either pleiotropic effects or the presence of tightly co-ordinately regulated genes. In the present study, QTL clusters often mimicked the level of correlation observed between the traits. Another hypothesis of the concentration of QTL on some linkage groups could be genomic expression dominance, as observed in the allotetraploid cotton where the expression of the D genome parent occurred more often than it did for the A genome parent (Rapp et al. 2009). Clusters of QTL may also result from domestication of the cultivated strawberry, which is a recent species from about 250 years (Davis et al. 2007). When favourable alleles are grouped in cluster or when genomic region has positive pleiotropic effect on traits, breeding for these traits using molecular markers will be facilitated. Interestingly, in the subdioecious Fragaria virginiana, one of the parental species of the cultivated strawberry, Spigler et al. (2011) reported a QTL cluster located on LGVI which is orthologous to one of the LGs belonging to HGVI in the present study, based on several common SSR markers. The characterised LG encompasses regions controlling male and female fertility as well as QTL for sexually dimorphic traits such as fruit and seed set or anther number for examples. The authors suggest that linked and/or pleiotropic reproductive genes have been located on the chromosome 6 before polyploidization and the acquisition of male and female sterility mutations in this species.
Genetic control of fruit quality traits
The complexity of the biological processes or metabolic pathways underlying the fruit quality traits studied may provide a likely explanation for the variation in the number of loci contributing to a given trait. Indeed, more QTL were detected for traits such as fruit length (7), firmness (7) anthocyanin content (5) or citrate (5) than for other traits like fructose or sucrose contents (2) (Table 2). Fruit size and shape, for which a large number of QTL have been detected in the strawberry progeny studied (Table 2, Online Resource 2) is under complex genetic and physiological control. In the well-studied fleshy fruit model tomato, up to 30 QTL for fruit size and shape have been detected, among which several loci (<10) controlling fruit patterning and fruit growth are associated with domestication (Tanksley 2004; Paran and van der Knaap 2007). Likewise, fruit firmness at harvest is a very complex trait, depending not only on cell wall degradation during ripening as shown in strawberry (Santiago-Doménech et al. 2008) but also, as reported on tomato, on cell wall biosynthesis at early stages of fruit development (Gilbert et al. 2009), on tissue characteristics including cell number and size (Chaïb et al. 2007) and on water status of the fruit (Saladié et al. 2007). Fruit colour in strawberry is a compound trait in which anthocyanin content, the major contributing factor, is controlled at multiple levels involving transcription factors and structural genes (Carbone et al. 2009). Fruit acidity generally depends on a large number of factors involving several metabolic pathways for the synthesis, transport, storage and degradation of the organic acids and for the transport of the potassium counter-ion (Etienne et al. 2002a; Lobit et al. 2006; Sweetman et al. 2009). Similar findings could be expected for sugar QTL but, actually, fewer numbers of QTL were detected. The large differences in total sugar content observed between the parental lines and within the progeny (Table 1) may reflect an increased assimilate supply from plant photosynthesis or an increased fruit sink strength due to enhanced sugar transport, as previously shown in tomato (Fridman et al. 2000; Schauer et al. 2006). More unexpected is the contrast between the number of QTL found for sucrose (2), glucose (5) and fructose (2). All these sugars may enter complex biochemical pathways, besides being stored in the fruit cell vacuoles. Possible explanation for these findings is that multiple QTL with low effects contributing to the overall phenotypic variation of the trait remained undetected (QTL value < threshold level) for these compounds. A QTL for fructose which was detected at a LOD of 2.68 (i.e. under the threshold level of 2.86) in the year 2 on the LGIIIa of the female map is a good example. It might as well explain that despite the high correlation between fructose and glucose no QTL colocation was observed for these two traits.
Homoeo-QTL may contribute to the genetic control of fruit quality in the octoploid cultivated strawberry
An additional layer of complexity, not present in diploid fleshy fruit species such as tomato, is provided by the contribution of putative homoeo-QTL to the control of quantitative fruit quality traits in the cultivated strawberry. In the present study, about 23% of the QTL were detected at likely homoeologous locations. These QTL were named homoeo-QTL since their orthologous positions on homoeologous linkage groups belonging to the same homoeologous group suggests the presence of allelic forms of the same gene responsible for the variation of the quantitative traits. In the strawberry progeny studied, major fruit quality traits including fruit shape, firmness, glucose and malate contents and pH were controlled by homoeo-QTL (Table 4). To date, few homoeo-QTL have been characterised in other plant species [but see in bread wheat (Kumar et al. 2007, 2009)], though the polyploid species constitute the majority of edible crop plants and have, therefore, been largely studied. Homoeo-QTL have been recently reported in the cultivated strawberry for yield and plant width (Zorrilla-Fontanesi et al. 2011). In strawberry, the presence of homoeoalleles for genes located in orthologous positions on the four homoeologous chromosomes could be due to the structural conservation of the genomes along the polyploidization process. This conservation is supported by results from comparative mapping between diploid and octoploid linkage maps (Rousseau-Gueutin et al. 2008), which revealed high levels of conserved macrosynteny and colinearity within each of the seven homo(eo)logous linkage groups and between the octoploid homoeologous groups and their corresponding diploid linkage groups. Therefore, no major chromosomal rearrangements in genomes involved in Fragaria × ananassa has been triggered along the polyploidization in the Fragaria genus (Rousseau-Gueutin et al. 2008). However, some chromosomal rearrangements have been observed on the homoeologous group VI on other Fragaria species (Goldberg et al. 2010; Spigler et al. 2010). Such structural rearrangements could help restoring the fecundity of the species as hypothesised by Soltis and Soltis (1999) and Spigler et al. (2011).
For various traits (FD, FIRM, GLU and pH), preferential location of homoeo-QTL was observed on the two linkage groups LGIIIa and LGIIIc belonging to the homoeology group HGIII (Table 4). Although the allocation of the linkage groups to the two ancestral genomes of Fragaria × ananassa (Bringhurst 1990; Rousseau-Gueutin et al. 2009) remains unknown, we can address the question if the preferential homoeo-QTL locations on one of the two ancestral genomes observed in this study could reveal a preferential expression of one of these two ancestral genomes. Though more data are needed to study the impact of each ancestral genome of Fragaria × ananassa on fruit quality trait, similar hypothesis has been proved in the polyploidy cotton for fibre quality whereby compiling QTL mapping results from many published studies, Rong et al. (2007) showed that both tetraploid A- (from an ancestor that does produce spinnable fibre) and D-subgenomes (from an ancestor that does not produce spinnable fibre) contribute to QTL for fibre development at largely non-homoeologous locations (see also Paterson et al. 2003; Jiang et al. 1998; Saranga et al. 2001; Chee et al. 2005) suggesting divergent selection acting on many corresponding genes before and/or after polyploidy event. A-subgenome contained favourable alleles prior to polyploid formation whereas D-subgenome would have come under selection after polyploid formation and harbours greater allelic diversity among tetraploid forms (Paterson et al. 2003). On the contrary, in sugarcane, Aitken et al. (2006) identified QTL for sugar-related traits on linkage groups belonging to the same homoeology group. In our study, the preferential location of homoeo-QTL on the homoeologous groups III and VI rather than on the other homoeologous groups suggests a different evolution of the seven homoeologous groups. Such a differential expression was largely shown for the sex chromosomes, which evolve rapidly in plant genomes (Liu et al. 2004), and LGVI being characterised as proto-sex chromosome in Fragaria virginiana (Spigler et al. 2011) might follow such pattern.
However, most of the detected QTL were likely not homoeo-QTL since they were located on only one of the four homoeologous linkage groups in Fragaria × ananassa. The most striking example is that of fruit colour, for which QTL for all the colour-related traits (L, a, b and ANTH) were almost exclusively located on a single linkage group (Fig. 3b and Online Resource 3). There are several, not mutually exclusive, possible explanations for these findings. As gene duplications in polyploid species result in functional redundancy, differential selective pressure may act on the duplicated copies as a process of diploidization whereby gene redundancy is reduced via gene silencing, sequence elimination and rearrangement, demethylation of retroelements and relaxation of imprinting (see reviews in Osborn et al. 2003 as well as in Chen 2007). In a phylogenetical study conducted in three allo-octopolyploid Fragaria species with two nuclear genes, GBSSI-2 and DHAR, we recently found that homoeologous sequences within each octoploid species seemed to evolve faster than orthologous sequences between Fragaria species (Rousseau-Gueutin et al. 2009). In addition, we observed deletion events in alleles of DHAR gene which led to the formation of a pseudogene and of a truncated gene for F. virginiana and F. chiloensis, respectively. Such reduction of genomic redundancy may have contributed to the diploidization process during the evolution of the polyploids (Paterson 2005; Comai 2005), including Fragaria × ananassa, which mainly displays a meiotic disomic behaviour (Rousseau-Gueutin et al. 2008). In addition, we may hypothesise that the alleles conferring to strawberry fruit a valuable sensorial quality such as improved fruit colour may have been in the past under strong selection pressure, first along the domestication process of F. chiloensis and possibly F. virginiana, the parents of Fragaria × ananassa (Darrow 1966) and, more recently, during the breeding process of cultivated strawberry. As a result, the QTL detected on a single linkage group in one homoeology group may reflect the expression of the stronger allele controlling that trait within this homoeology group, which was selected along the domestication/breeding process.
Homoeo-QTL may contribute to the control of fruit quality traits in varying environmental conditions
The differential expression of the loci across years observed in our study is in accordance with previous agronomical studies showing the impact of the environment on strawberry culture (Shaw 1988). Similar results were also reported in a recent study characterising QTL associated with fruit quality traits where about 36% of the QTL were stable over the years (Zorrilla-Fontanesi et al. 2011). Annual variations in temperature and hygrometry have a considerable effect on fruit development and ripening. For example, relatively warm temperatures and high humidity result in softer fruits than cool temperatures and low humidity (Scott and Lawrence 1975). Moreover, elevated temperatures have a negative effect on fruit size and quality (Hancock 1999). Previous work has documented annual differences in the relative expression of sugars and acids in strawberry (Morrow et al. 1958; Sweeney et al. 1970). Variability across years in QTL expression is a rather common phenomenon (see for examples Etienne et al. 2002b and Quilot et al. 2004, as well as Hoarau et al. 2002 and Shi et al. 2009 for polyploid species), but Paterson et al. (2003) observed that even if the genetic control of cotton fibre quality is affected by growing seasons, a basal set of QTL that are relatively unaffected by environmental parameters exists. In our study, developmental and colour traits seemed less affected by environmental factors since 32.1 and 36.8% of the QTL for these traits were detected more than once compared with 16.7, 8.3 and 4.8% of the QTL for texture, sugar-related traits and acid-related traits, respectively.
There are clear indications that homoeo-QTL may be involved in the environmental control of several fruit quality traits, e.g. fruit weight (Fig. 3a) or fruit glucose content (Fig. 3d). In that respect, the fruit shape and weight QTL located on linkage group LGIII provide an interesting example. In years 1 and 2, most fruit development traits including fruit diameter (FD) and length (FL) and FD/FL ratio were located on LGIIIa and LGIIId while new QTL for the highly correlated FD and FW (Online Resource 2) were detected on LGIIIc only on year 3. A clear year effect was indeed detected for FW, which was significantly lower in year 3 (mean FW ~14 g) than in years 1 and 2 (~18 and 22 g, respectively) (data not shown). Fruit size and weight are known to be profoundly affected by environmental cues which regulate the processes of cell division and expansion in the fruit through modifications of photoassimilate supply and hormonal signalling (Baldet et al. 2006). Accordingly, our results suggest the presence on LGIIIa and LGIIIc of homoeoalleles controlling fruit diameter and, consequently, fruit weight, which are sensitive to annual climate variations and/or cultural conditions. It is important to note that climate records in the strawberry production area indicated a cold and rainy weather in spring of year 3, i.e. during the period of strawberry early fruit growth by cell division/cell expansion. As indicated below, such results may have major consequences on the breeding strategy chosen. They also point out the importance of taking into account in physiological studies of strawberry fruit the homoeoallele variability of candidate genes performing the same function.
Strawberry breeding
One goal of strawberry breeders is to combine large size with high fruit quality. Many characteristics of the parents in the present study, including sweetness, aromatic properties, good acid/sugar balance, early maturity and firmness in ‘CF1116’, and large fruit size in ‘Capitola’, fulfil breeders’ requirements (present study and Moing et al. 2001).
The present paper provides new information on the genetic architecture of fruit quality traits relevant for strawberry breeding, while most genetic studies of fruit quality have been carried out so far on the diploid species, F. vesca, examining traits such as fruit colour (Deng and Davis 2001) and seasonal flowering (Cekic et al. 2001; Albani et al. 2004). Only recently, Weebadde et al. (2008) showed that day-neutrality in strawberry is under polygenic control and characterised eight QTL among which one was shared across several locations. In addition, QTL for fruit quality traits (Zorrilla-Fontanesi et al. 2011) and for sexual dimorphism (Spigler et al. 2011) have been also reported. Our study revealed a large number of QTL controlling sweetness and acidity concentrations in the octoploid Fragaria × ananassa, revealing the potential to improve the traits independently. However, since large numbers of QTL co-locations were observed, caution should be applied when trying to modify the alleles present at a locus and the direction of the allele effect should be considered. The QTL cluster on LGVIa-m of ‘CF1116’ is particularly interesting in terms of strawberry breeding. ‘CF1116’ provides favourable alleles for soluble-solids and sucrose, and negative alleles for malate. Therefore, this chromosomal region may have the potential to improve sweetness and acidity. In addition, transgressive individuals resulting from complementary gene action can be regarded as an interesting source of breeding material. The QTL for fruit development and colour present in this chromosomal region have to be considered to have a complete view of the fruit quality at harvest. The differential expression of the genes linked to the QTL across years will also have to be taken into account in attempts to exploit these loci in breeding programs. As example, for breeding genotypes that produce large fruits in unfavourable conditions, it may prove interesting to select the homoeoallele controlling the homoeo-QTL linked to fruit weight located on LGIIIc. A positive effect on fruit weight of this homoeoallele was detected on year 3, a year during which the mean fruit weight and sugar content were low due to bad weather with low temperatures and high precipitations in March and April when the fruit is in expansion. Additional years of evaluation are required to validate these results. Such experiments are planned for fruit size and fruit weight. Finally increasing the number of repetitions per additional year of evaluation would also improve the power of detection.
Mapping of the codominant SSR markers and candidate genes using the Fragaria genome sequencing (Shulaev et al. 2011) helped to partly elucidate the complexity of the genome by identifying homoeologous chromosomes and thereby homoeo-QTL. A further step would be to identify to which donor species each linkage group belongs across homoeology groups. This is crucial to increase our comprehension of the evolution of both genomes in cultivated strawberry and the impact of domestication and breeding on quantitative traits in this species.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors gratefully acknowledge funding from Région Aquitaine, the ANR (No ANR-08-KBBE-007-01) and the European Community (FEDER funds) and S. Courlit, A. Darnaud, D. Camy, J. Bariac, E. Martin for their participation in fruit phenotyping. The authors thank Dr. Brigitte Mangin for discussion on the statistics.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aharoni A, Giri AP, Verstappen FWA, Bertea CM, Sevenier R, Sun ZK, Jongsma MA, Schwab W, Bouwmeester HJ. Gain and loss of fruit flavor compounds produced by wild and cultivated strawberry species. Plant Cell. 2004;16:3110–3131. doi: 10.1105/tpc.104.023895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aitken KS, Jackson PA, McIntyre CL. Quantitative trait loci identified for sugar related traits in a sugarcane (Saccharum spp.) cultivar × Saccharum officinarum population. Theor Appl Genet. 2006;112:1306–1317. doi: 10.1007/s00122-006-0233-2. [DOI] [PubMed] [Google Scholar]
- Aitken KS, Hermann S, Karno K, Bonnett GD, McIntyre LC, Jackson PA. Genetic control of yield related stalk traits in sugarcane. Theor Appl Genet. 2008;117:1191–1203. doi: 10.1007/s00122-008-0856-6. [DOI] [PubMed] [Google Scholar]
- Albani MC, Battey NH, Wilkinson MJ. The development of ISSR-derived SCAR markers around the seasonal flowering locus (SFL) in Fragaria vesca. Theor Appl Genet. 2004;109:571–579. doi: 10.1007/s00122-004-1654-4. [DOI] [PubMed] [Google Scholar]
- Aljanabi SM, Parmessur Y, Kross H, Dhayan S, Saumtally S, Ramdoyal K, Autrey LJC, Dookun-Saumtally A. Identification of a major quantitative trait locus (QTL) for yellow spot (Mycovellosiella koepkei) disease resistance in sugarcane. Mol Breed. 2007;19:1–14. [Google Scholar]
- Atkinson CJ, Dodds PAA, Ford YY, Le Miere J, Taylor JM, Blake PS, Paul N. Effects of cultivar, fruit number and reflected photosynthetically active radiation on Fragaria × ananassa productivity and fruit ellagic acid and ascorbic acid concentrations. Ann Bot. 2006;97:429–441. doi: 10.1093/aob/mcj046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldet P, Hernould M, Laporte F, Mounet F, Just D, Mouras A, Chevalier C, Rothan C. Expression of cell proliferation-related genes in early developing flower is affected by fruit load reduction in tomato plants. J Exp Bot. 2006;57:961–970. doi: 10.1093/jxb/erj082. [DOI] [PubMed] [Google Scholar]
- Basten CJ, Weir BS, Zeng Z-B. QTL Cartographer: a reference manual and tutorial for QTL mapping. Raleigh: Department of Statistics, North Carolina State University; 1997. [Google Scholar]
- Battino M, Beekwilder J, Denoyes-Rothan B, Laimer M, McDougall GJ, Mezzetti B. Bioactive compounds in berries relevant to human health. Nutr Rev. 2009;67(S1):145–150. doi: 10.1111/j.1753-4887.2009.00178.x. [DOI] [PubMed] [Google Scholar]
- Ben Chaim A, Paran I, Grube RC, Jahn M, van Wijk R, Peleman J. QTL mapping of fruit-related traits in pepper (Capsicum annuum) Theor Appl Genet. 2001;102:1016–1028. [Google Scholar]
- Bowers JE, Chapman BA, Rong JK, Paterson AH. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Bringhurst RS. Cytogenetics and evolution in American Fragaria. HortScience. 1990;25:879–881. [Google Scholar]
- Carbone F, Preuss A, De Vos RC, D’Amico E, Perrotta G, Bovy AG, Martens S, Rosati C. Developmental, genetic and environmental factors affect the expression of flavonoid genes, enzymes and metabolites in strawberry fruits. Plant Cell Environ. 2009;32:1117–1131. doi: 10.1111/j.1365-3040.2009.01994.x. [DOI] [PubMed] [Google Scholar]
- Cekic C, Battey NH, Wilkinson MJ. The potential of ISSR-PCR primer-pair combinations for genetic linkage analysis using the seasonal flowering locus in Fragaria as a model. Theor Appl Genet. 2001;103:540–546. [Google Scholar]
- Chaïb J, Devaux MF, Grotte MG, Robini K, Causse M, Lahaye M, Marty I. Physiological relationships among physical, sensory, and morphological attributes of texture in tomato fruits. J Exp Bot. 2007;58:1915–1925. doi: 10.1093/jxb/erm046. [DOI] [PubMed] [Google Scholar]
- Chee P, Draye X, Jiang C-X, Decanini L, Delmonte TA, Bredhauer R, Smith CW, Paterson AH. Molecular dissection of interspecific variation between Gossypium hirsutum and Gossypium barbadense (cotton) by a backcross-self approach: I. Fiber elongation. Theor Appl Genet. 2005;111:757–763. doi: 10.1007/s00122-005-2063-z. [DOI] [PubMed] [Google Scholar]
- Chen ZJ. Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann Rev Plant Biol. 2007;58:377–406. doi: 10.1146/annurev.arplant.58.032806.103835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Qian N, Guo WZ, Song QP, Li BC, Deng FJ, Dong CG, Zhang TZ. Using three overlapped RILs to dissect genetically clustered QTL for fiber strength on Chro.D8 in Upland cotton. Theor Appl Genet. 2009;119:605–612. doi: 10.1007/s00122-009-1070-x. [DOI] [PubMed] [Google Scholar]
- Coelho CM, Wu S, Li YC, Hunter B, Dante RA, Cui Y, Wu R, Larkins BA. Identification of quantitative trait loci that affect endoreduplication in maize endosperm. Theor Appl Genet. 2007;115:1147–1162. doi: 10.1007/s00122-007-0640-z. [DOI] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005;6:836–846. doi: 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Coman MS, Popescu AN (1997) Inheritance of some strawberry quantitative traits. In: van der Sheer HAT, Lieten F, Dijkstra J (eds) Proceedings of Third International Strawberry Symposium, pp 81–88
- Darrow GM. The strawberry: history, breeding and physiology. New York: Holt, Rinehart, and Winston; 1966. [Google Scholar]
- Davis T, Denoyes-Rothan B, Lerceteau-Köhler E. Strawberry. In: Kole C, editor. Genome mapping and molecular breeding in plants: fruits and nuts. Berlin: Springer; 2007. pp. 189–206. [Google Scholar]
- de Boishebert V, Urruty L, Giraudel JL, Montury M. Assessment of strawberry aroma through solid-phase microextraction-gas chromatography and artificial neuron network methods. Variety classification versus growing years. J Agric Food Chem. 2004;52:2472–2478. doi: 10.1021/jf035376l. [DOI] [PubMed] [Google Scholar]
- de Vicente MC, Tanksley SD. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics. 1993;134:585–596. doi: 10.1093/genetics/134.2.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demmig-Adams B, Adams WW. Antioxidants in photosynthesis and human nutrition. Science. 2002;298:2149–2153. doi: 10.1126/science.1078002. [DOI] [PubMed] [Google Scholar]
- Deng C, Davis TM. Molecular identification of the yellow fruit color (c) locus in diploid strawberry: a candidate gene approach. Theor Appl Genet. 2001;103:316–322. [Google Scholar]
- Doerge RW, Craig BA. Model selection for quantitative trait mocus analysis in polyploids. Proc Natl Acad Sci USA. 2000;97:7951–7956. doi: 10.1073/pnas.97.14.7951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Ann Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Etienne C, Moing A, Dirlewanger E, Raymond P, Monet R, Rothan C. Isolation and characterization of six peach cDNAs encoding key proteins in organic acid metabolism and solute accumulation: involvement in regulating peach fruit acidity. Physiol Plant. 2002;114:259–270. doi: 10.1034/j.1399-3054.2002.1140212.x. [DOI] [PubMed] [Google Scholar]
- Etienne C, Rothan C, Moing A, Plomion C, Bodénès C, Svanella-Dumas L, Cosson P, Pronier V, Monet R, Dirlewanger E. Candidate genes and QTL for sugar and organic acid content in peach [Prunus persica (L.) Batsch] Theor Appl Genet. 2002;105:145–159. doi: 10.1007/s00122-001-0841-9. [DOI] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Fridman E, Pleban T, Zamir D. A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc Natl Acad Sci USA. 2000;97:4718–4723. doi: 10.1073/pnas.97.9.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton TM, Beck-Bunn T, Emmatty D, Eshed Y, Lopez J, Pétiard V, Uhlig J, Zamir D, Tanksley SD. QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTL found in the other wild species. Theor Appl Genet. 1997;95:881–894. [Google Scholar]
- Galletta GJ, Maas JL. Strawberry genetics. HortScience. 1990;25:871–879. [Google Scholar]
- Gilbert L, Alhagdow M, Nunes-Nesi A, Quemener B, Guillon F, Bouchet B, Faurobert M, Gouble B, Page D, Garcia V, Petit J, Stevens R, Causse M, Fernie AR, Lahaye M, Rothan C, Baldet P. GDP-d-mannose 3, 5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009;60:499–508. doi: 10.1111/j.1365-313X.2009.03972.x. [DOI] [PubMed] [Google Scholar]
- Goldberg MT, Spigler RB, Ashman TL. Comparative genetic mapping points to different sex chromosomes in sibling species of wild strawberry (Fragaria) Genetics. 2010;186:1425–1433. doi: 10.1534/genetics.110.122911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green A. Soft fruits. In: Hulme AC, editor. The biochemistry of fruits and their products. London: Academic Press; 1971. pp. 375–409. [Google Scholar]
- Gutierrez AG, Carabali SJ, Giraldo OX, Martinez CP, Correa F, Prado G, Tohme J, Lorieux M. Identification of a rice stripe necrosis virus resistance locus and yield component QTLs using Oryza sativa × O. glaberrima introgression lines. BMC Plant Biol. 2010;10:6. doi: 10.1186/1471-2229-10-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JE. Fruiting and postharvest physiology. Strawberries. Wallingford: CABI Publishing; 1999. [Google Scholar]
- Hancock J, Luby J. Adaptative zones and ancestry of the most important North American strawberry cultivars. Fruit Varieties J. 1995;49:85–90. [Google Scholar]
- Hoarau J-Y, Grivet L, Offmann B, Raboin L-M, Diorflar J-P, Payet J, Hellmann M, D’Hont A, Glaszmann J-C. Genetic dissection of a modern sugarcane cultivar (Saccharum spp.). II. Detection of QTL for yield components. Theor Appl Genet. 2002;105:1027–1037. doi: 10.1007/s00122-002-1047-5. [DOI] [PubMed] [Google Scholar]
- Howard T, Rejab NA, Griffiths S, Leigh F, Leverington-Waite M, Simmonds J, Uauy C, Trafford K. Identification of a major QTL controlling the content of B-type starch granules in Aegilops. J Exp Bot. 2011;62:2217–2228. doi: 10.1093/jxb/erq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummer KE, Nathewet P, Yanagi T. Decaploidy in Fragaria iturupensis (Rosaceae) Am J Bot. 2009;96:713–716. doi: 10.3732/ajb.0800285. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, Vezzi A, Legeai F, Hugueney P, Dasilva C, Horner D, Mica E, Jublot D, Poulain J, Bruyère C, Billault A, Segurens B, Gouyvenoux M, Ugarte E, Cattonaro F, Anthouard V, Vico V, Del Fabbro C, Alaux M, Di Gaspero G, Dumas V, Felice N, Paillard S, Juman I, Moroldo M, Scalabrin S, Canaguier A, Le Clainche I, Malacrida G, Durand E, Pesole G, Laucou V, Chatelet P, Merdinoglu D, Delledonne M, Pezzotti M, Lecharny A, Scarpelli C, Artiguenave F, Pè ME, Valle G, Morgante M, Caboche M, Adam-Blondon AF, Weissenbach J, Quétier F, Wincker P. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang C-X, Wright RJ, El-Zik KM, Paterson AH. Polyploid formation created unique avenues for response to selection in Gossypium (cotton) Proc Natl Acad Sci USA. 1998;95:4419–4424. doi: 10.1073/pnas.95.8.4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader AA. Quality and its maintenance in relation to the post-harvest physiology of strawberry. In: Luby JJ, Dale A, editors. Quality and its maintenance in relation to the post-harvest physiology of strawberry. Portland: Timber press; 1991. pp. 145–152. [Google Scholar]
- Kalt W, Prange RK, Lidster PD. Postharvest color development of strawberries: influence of maturity, temperature and light. Can J Plant Sci. 1993;73:541–548. [Google Scholar]
- Kumar N, Kulwal PL, Balyan HS, Gupta PK. QTL mapping for yield and yield contributing traits in two mapping populations of bread wheat. Mol breed. 2007;19:163–177. [Google Scholar]
- Kumar A, Kumar J, Singh R, Garg T, Chhuneja P, Balyan HS, Gupta PK. QTL analysis for grain colour and pre-harvest sprouting in bread wheat. Plant Sci. 2009;177:114–122. [Google Scholar]
- Lerceteau-Köhler E, Guérin G, Laigret F, Denoyes-Rothan B. Characterization of mixed disomic and polysomic inheritance in the octoploid strawberry (Fragaria × ananassa) using AFLP mapping. Theor Appl Genet. 2003;107:619–628. doi: 10.1007/s00122-003-1300-6. [DOI] [PubMed] [Google Scholar]
- Liu Z, Moore PH, Ma H, Ackerman CM, Ragiba M, Yu Q, Pearl HM, Kim MS, Charlton JW, Stiles JI, Zee FT, Paterson AH, Ming R. A primitive Y chromosome in papaya marks incipient sex chromosome evolution. Nature. 2004;427:348–352. doi: 10.1038/nature02228. [DOI] [PubMed] [Google Scholar]
- Lobit P, Genard M, Soing P, Habib R. Modelling malic acid accumulation in fruits: relationships with organic acids, potassium, and temperature. J Exp Bot. 2006;57:1471–1483. doi: 10.1093/jxb/erj128. [DOI] [PubMed] [Google Scholar]
- Lundergan CA, Moore JN. Inheritance of ascorbic acid content and color intensity in fruits of strawberry (Fragaria × ananassa Duch) J Am Soc Hortic Sci. 1975;100:633–635. [Google Scholar]
- Ma J, Li HB, Zhang CY, Yang XM, Liu YX, Yan GJ, Liu CJ. Identification and validation of a major QTL conferring crown rot resistance in hexaploid wheat. Theor Appl Genet. 2010;120:1119–1128. doi: 10.1007/s00122-009-1239-3. [DOI] [PubMed] [Google Scholar]
- Maccaferri M, Ratti C, Rubies-Autonell C, Vallega V, Demontis A, Stefanelli S, Tuberosa R, Sanguineti MC. Resistance to Soil-borne cereal mosaic virus in durum wheat is controlled by a major QTL on chromosome arm 2BS and minor loci. Theor Appl Genet. 2011;123:527–544. doi: 10.1007/s00122-011-1605-9. [DOI] [PubMed] [Google Scholar]
- Mao DH, Liu TM, Xu CG, Li XH, Xing YZ. Epistasis and complementary gene action adequately account for the genetic bases of transgressive segregation of kilo-grain weight in rice. Euphytica. 2011;180:261–271. [Google Scholar]
- Masterson J. Stomatal size in fossil plants—evidence for polyploidy in majority of angiosperms. Science. 1994;264:421–424. doi: 10.1126/science.264.5157.421. [DOI] [PubMed] [Google Scholar]
- McCord PH, Sosinski BR, Haynes KG, Clough ME, Yencho GC. QTL mapping of internal heat necrosis in tetraploid potato. Theor Appl Genet. 2011;122:129–142. doi: 10.1007/s00122-010-1429-z. [DOI] [PubMed] [Google Scholar]
- Ming R, Wang Y-W. Molecular dissection of complex traits in autopolyploids: mapping QTL affecting sugar yield and related traits in sugarcane. Theor Appl Genet. 2002;105:332–345. doi: 10.1007/s00122-001-0861-5. [DOI] [PubMed] [Google Scholar]
- Ming R, Liu SC, Moore PH, Irvine JE, Paterson AH. QTL analysis in a complex autopolyploid: genetic control of sugar content in sugarcane. Genome Res. 2001;11:2075–2084. doi: 10.1101/gr.198801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moing A, Renaud C, Gaudillere M, Raymond P, Roudeillac P, Denoyes-Rothan B. Biochemical changes during fruit development of four strawberry cultivars. J Am Soc Hortic Sci. 2001;126:394–403. [Google Scholar]
- Monforte AJ, Oliver M, Gonzalo MJ, Alvarez JM, Dolcet-Sanjuan R, Arús P. Identification of quantitative trait loci involved in fruit quality traits in melon (Cucumis melo L.) Theor Appl Genet. 2004;108:750–758. doi: 10.1007/s00122-003-1483-x. [DOI] [PubMed] [Google Scholar]
- Morrow EB, Comstock RE, Kelleher T. Genetic variances in strawberry varieties. Proc Natl Soc Hort Sci. 1958;72:170–185. [Google Scholar]
- Ohno S (1970) Evolution by gene duplication, Springer, New York
- Ohtsuka Y, Kibe H, Hakoda N, Shimura I, Ogiwara I. Heritability of sugar contents in strawberry fruit in the F-1 populations using a common pollen parent. J Jpn Soc Hort Sci. 2004;73:31–35. [Google Scholar]
- Osborn TC, Pires JC, Birchler JA, Auger BDL, Chen ZJ, Lee H, Comai L, Madlung A, Doerge RW, Colot V, Martienssen RA. Understanding mechanisms of novel gene expression in polyploids. Trends Genet. 2003;19:141–147. doi: 10.1016/s0168-9525(03)00015-5. [DOI] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. J Exp Bot. 2007;58:3841–3852. doi: 10.1093/jxb/erm257. [DOI] [PubMed] [Google Scholar]
- Paterson AH. Polyploidy, evolutionary opportunity, and crop adaptation. Genetica. 2005;123:191–196. doi: 10.1007/s10709-003-2742-0. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Saranga Y, Menz M, Jiang C-X, Wright RJ. QTL analysis of genotype × environment interaction affecting fiber quality. Theor Appl Genet. 2003;106:384–396. doi: 10.1007/s00122-002-1025-y. [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA. Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA. 2004;101:9903–9908. doi: 10.1073/pnas.0307901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z, Fahima T, Krugman T, Abbo S, Yakir D, Korol AB, Saranga Y. Genomic dissection of drought resistance in durum wheat x wild emmer wheat recombinant inbreed line population. Plant Cell Environ. 2009;32:758–779. doi: 10.1111/j.1365-3040.2009.01956.x. [DOI] [PubMed] [Google Scholar]
- Pfender WF, Saha MC, Johnson EA, Slabaugh MB. Mapping with RAD (restriction-site associated DNA) markers to rapidly identify QTL for stem rust resistance in Lolium perenne. Theor Appl Genet. 2011;122:1467–1480. doi: 10.1007/s00122-011-1546-3. [DOI] [PubMed] [Google Scholar]
- Potter D, Eriksson T, Evans RC, Oh S, Smedmark JEE, Morgan DR, Kerr M, Robertson KR, Arsenault M, Dickinson TA, Campbell CS. Phylogeny and classification of Rosaceae. Plant Syst Evol. 2007;266:5–43. [Google Scholar]
- Quilot B, BH WU, Kervella J, Génard M, Foulongne M, Moreau K. QTL analysis of quality traits in an advanced backcross between Prunus persica cultivars and the wild relative species P. davidiana. Theor Appl Genet. 2004;109:884–897. doi: 10.1007/s00122-004-1703-z. [DOI] [PubMed] [Google Scholar]
- Rapp RA, Udall JA, Wendel JF. Genomic expression dominance in allopolyploids. BMC Biol. 2009;7:18. doi: 10.1186/1741-7007-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH, Widmer A, Arntz AM, Burke JM. The genetic architecture necessary for transgressive segregation is common in both natural and domesticated populations. Phil Trans R Soc Lond B. 2003;358:1141–1147. doi: 10.1098/rstb.2003.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong J, Feltus FA, Waghmare VN, Pierce GJ, Chee PW, Draye X, Saranga Y, Wright RJ, Wilkins TA, May OL, Smith CW, Gannaway JR, Wendel JF, Paterson AH. Meta-analysis of polyploid cotton QTL shows unequal contributions of subgenomes to a complex network of genes and gene clusters implicated in lint fiber development. Genetics. 2007;176:2577–2588. doi: 10.1534/genetics.107.074518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Lerceteau-Köhler E, Barrot L, Sargent DJ, Monfort A, Simpson D, Arus P, Guérin G, Denoyes-Rothan B. Comparative genetic mapping between octoploid and diploid Fragaria species reveals a high level of colinearity between their genomes and the essentially disomic behavior of the cultivated octoploid strawberry. Genetics. 2008;179:2045–2060. doi: 10.1534/genetics.107.083840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseau-Gueutin M, Gaston A, Aïnouche A, Aïnouche ML, Olbricht K, Staudt G, Richard L, Denoyes-Rothan B. Tracking the evolutionary history of polyploidy in Fragaria L. (strawberry): New insights from phylogenetic analyses of low-copy nuclear genes. Mol Phylogenet Evol. 2009;51:515–530. doi: 10.1016/j.ympev.2008.12.024. [DOI] [PubMed] [Google Scholar]
- Sagredo B, Balbyshev N, Lafta A, Casper H, Lorenzen J. A QTL that confers resistance to Colorado potato beetle (Leptinotarsa decemlineata [Say]) in tetraploid potato populations segregating for leptine. Theor Appl Genet. 2009;119:1171–1181. doi: 10.1007/s00122-009-1118-y. [DOI] [PubMed] [Google Scholar]
- Saladié M, Matas AJ, Isaacson T, Jenks MA, Goodwin SM, Niklas KJ, Xiaolin R, Labavitch JM, Shackel KA, Fernie AR, Lytovchenko A, O’Neill MA, Watkins CB, Rose JK. A reevaluation of the key factors that influence tomato fruit softening and integrity. Plant Physiol. 2007;144:1012–1028. doi: 10.1104/pp.107.097477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saliba-Colombani V, Causse M, Langlois D, Philouze J, Buret M. Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTL for physical and chemical traits. Theor Appl Genet. 2001;102:259–272. [Google Scholar]
- Santiago-Doménech N, Jiménez-Bemúdez S, Matas AJ, Rose JK, Muñoz-Blanco J, Mercado JA, Quesada MA. Antisense inhibition of a pectate lyase gene supports a role for pectin depolymerization in strawberry fruit softening. J Exp Bot. 2008;59:2769–2779. doi: 10.1093/jxb/ern142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz C, Perez AG, Olias R, Olias JM. Quality of strawberries packed with perforated polypropylene. J Food Sci. 1999;64:748–752. [Google Scholar]
- Saranga Y, Menz M, Jiang CX, Wright RJ, Yakir D, Paterson AH. Genomic dissection of genotype X environment interactions conferring adaptation of cotton to arid conditions. Genome Res. 2001;11:1988–1995. doi: 10.1101/gr.157201. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Fernandéz-Fernandéz F, Ruiz-Roja JJ, Sutherland BG, Passey A, Whitehouse AB, Simpson DW. A genetic linkage map of the cultivated strawberry (Fragaria × ananassa) and its comparison to the diploid Fragaria reference map. Mol Breed. 2009;24:293–303. [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol. 2006;24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- Scott DH, Lawrence FJ. Strawberries. In: Janick J, Moore JN, editors. Advances in fruit breeding. USA: Purdue University Press; 1975. [Google Scholar]
- Shaw DV. Genotypic variation and genotypic correlations for sugars and organic acids of strawberries. J Am Soc Hortic Sci. 1988;113:770–774. [Google Scholar]
- Shaw DV, Bringhurst RS, Voth V. Genetic variation for quality traits in an advanced-cycle breeding population of strawberries. J Am Soc Hortic Sci. 1987;112:699–702. [Google Scholar]
- Shi J, Li R, Qiu D, Jiang C, Long Y, Morgan C, Bancroft I, Zhao J, Meng J. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics. 2009;182:851–861. doi: 10.1534/genetics.109.101642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulaev V, Sargent DJ, Crowhurst RN, Mockler TC, Folkerts O, Delcher AL, Jaiswal P, Mockaitis K, Liston A, Mane SP, Burn P, Davis TM, Slovin JP, Bassil N, Hellens RP, Evans C, Harkins T, Kodira C, Desany B, Crasta OR, Jensen RV, Allan AC, Michael TP, Setubal JC, Celton JM, Rees DJG, Williams KP, Holt SH, Rojas JJR, Chatterjee M, Liu B, Silva H, Meisel L, Adato A, Filichkin S, Troggio R, Viola M, Ashman TL, Wang H, Dharmawardhana P, Elser J, Raja R, Priest HD, Bryant DW, Jr, Fox SE, Givan SA, Wilhelm LJ, Naithan S, Christoffels A, Salama DY, Carter J, Girona EL, Zdepski A, Wang W, Kerstetter RA, Schwab W, Korban SS, Davik J, Monfort A, Denoyes-Rothan B, Arus P, Mittler R, Flinn B, Aharoni A, Bennetzen JL, Salzberg SL, Dickerman AW, Velasco R, Borodovsky M, Veilleux RE, Folta KM. The genome of woodland strawberry (Fragaria vesca) Nat Genet. 2011;43:U109–U151. doi: 10.1038/ng.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sjulin TM, Dale A. Genetic diversity of North American strawberry cultivars. J Am Soc Hortic Sci. 1987;112:375–385. [Google Scholar]
- Soltis DE, Soltis PS. Polyploidy: recurrent formation and genome evolution. Trends Ecol Evol. 1999;14:348–352. doi: 10.1016/s0169-5347(99)01638-9. [DOI] [PubMed] [Google Scholar]
- Somers DJ, Fedak G, Clarke J, Cao W. Mapping of FHB resistance QTL in tetraploid wheat. Genome. 2006;49:1586–1596. doi: 10.1139/g06-127. [DOI] [PubMed] [Google Scholar]
- Sone K, Mochizuki T, Okimura M, Noguchi Y, Kitadani E. Inheritance of ascorbic acid content in strawberry fruits. J Jpn Soc Hort Sci. 2003;72:141–147. [Google Scholar]
- Spigler RB, Lewers KS, Johnson AL, Ashman T-L (2010) Comparative mapping reveals autosomal origin of sex chromosome in octoploid Fragaria virginiana. J Heredity 101(Suppl 1):S107–117 [DOI] [PubMed]
- Spigler RB, Lewers KS, Ashman T-L. Genetic architecture of sexual dimorphism in a subdioecious plant with a proto-sex chromosome. Evolution. 2011;65:1114–1126. doi: 10.1111/j.1558-5646.2010.01189.x. [DOI] [PubMed] [Google Scholar]
- Staudt G. The species of Fragaria, their taxonomy and geographical distribution. Acta Hortic. 1989;265:23–33. [Google Scholar]
- Staudt G. Strawberry biogeography, genetics and systematics. Acta Hortic. 2009;842:1019. [Google Scholar]
- Sweeney JP, Chapman VJ, Hepner PA. Sugar, acid, and flavor in fresh fruit. J Am Diet Assoc. 1970;57:432–435. [PubMed] [Google Scholar]
- Sweetman C, Deluc LG, Cramer GR, Ford CM, Soole KL. Regulation of malate metabolism in grape berry and other developing fruits. Phytochemistry. 2009;70:1329–1344. doi: 10.1016/j.phytochem.2009.08.006. [DOI] [PubMed] [Google Scholar]
- Tanksley SD. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 2004;16:S181–S189. doi: 10.1105/tpc.018119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Arabidopsis Genome Initiative Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- Tulipani S, Mezzetti B, Capocasa F, Bompadre S, Beekwilder J, de Vos CH, Capanoglu E, Bovy A, Battino M. Antioxidants, phenolic compounds, and nutritional quality of different strawberry genotypes. J Agric Food Chem. 2008;56:696–704. doi: 10.1021/jf0719959. [DOI] [PubMed] [Google Scholar]
- Tulipani S, Mezzetti B, Battino M. Impact of strawberries on human health: insight into marginally discussed bioactive compounds for the Mediterranean diet. Public health nutrition. 2009;12:1656–1662. doi: 10.1017/S1368980009990516. [DOI] [PubMed] [Google Scholar]
- Tuskan GA, DiFazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, Schein J, Sterck L, Aerts A, Bhalerao RR, Bhalerao RP, Blaudez D, Boerjan W, Brun A, Brunner A, Busov V, Campbell M, Carlson J, Chalot M, Chapman J, Chen GL, Cooper D, Coutinho PM, Couturier J, Covert S, Cronk Q, Cunningham R, Davis J, Degroeve S, Dejardin A, Depamphilis C, Detter J, Dirks B, Dubchak I, Duplessis S, Ehlting J, Ellis B, Gendler K, Goodstein D, Gribskov M, Grimwood J, Groover A, Gunter L, Hamberger B, Heinze B, Helariutta Y, Henrissat B, Holligan D, Holt R, Huang W, Islam-Faridi N, Jones S, Jones-Rhoades M, Jorgensen R, Joshi C, Kangasjarvi J, Karlsson J, Kelleher C, Kirkpatrick R, Kirst M, Kohler A, Kalluri U, Larimer F, Leebens-Mack J, Leple JC, Locascio P, Lou Y, Lucas S, Martin F, Montanini B, Napoli C, Nelson DR, Nelson C, Nieminen K, Nilsson O, Pereda V, Peter G, Philippe R, Pilate G, Poliakov A, Razumovskaya J, Richardson P, Rinaldi C, Ritland K, Rouze P, Ryaboy D, Schmutz J, Schrader J, Segerman B, Shin H, Siddiqui A, Sterky F, Terry A, Tsai CJ, Uberbacher E, Unneberg P, Vahala J, Wall K, Wessler S, Yang G, Yin T, Douglas C, Marra M, Sandberg G, Van de Peer Y, Rokhsar D. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray) Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- Ubayasena L, Bett K, Tar’an B, Warkentin T. Genetic control and identification of QTLs associated with visual quality traits of field pea (Pisum sativum L.) Genome. 2011;54:261–272. doi: 10.1139/g10-117. [DOI] [PubMed] [Google Scholar]
- Velterop JS, Vos F. A rapid and inexpensive microplate assay for the enzymatic determination of glucose, fructose, sucrose, l-malate and citrate in tomato (Lycopersicon esculentum) extracts and in orange juice. Phytochem Analysis. 2001;12:299–304. doi: 10.1002/pca.598. [DOI] [PubMed] [Google Scholar]
- Wang D, Karle R, Iezzoni AF. QTL analysis of flower and fruit traits in sour cherry. Theor Appl Genet. 2000;100:535–544. [Google Scholar]
- Weebadde CK, Wang D, Finn CE, Lewers KS, Luby JJ, Bushakra J, Sjulin TM, Hancock JF. Using a linkage mapping approach to identify QTL for day-neutrality in the octoploid strawberry. Plant Breeding. 2008;127:94–101. [Google Scholar]
- Wrolstad RE, Shallenberger RS. Free sugars and sorbitol in fruits—a compilation from the literature. J Assoc Off Anal Chem. 1981;64:91–103. [PubMed] [Google Scholar]
- Wrolstad RE, Putnam TP, Varseveld GW. Color quality of frozen strawberries: effect of anthocyanin, pH, total acidity and ascorbic acid variability. J Food Sci. 1970;35:448–452. [Google Scholar]
- Zeng Z. Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA. 1993;90:10972–10976. doi: 10.1073/pnas.90.23.10972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Z. Precision mapping of quantitative trait loci. Genetics. 1994;136:1457–1468. doi: 10.1093/genetics/136.4.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorrilla-Fontanesi Y, Cabeza A, Domínguez P, Medina-Minguez J, Valpuesta V, Denoyes-Rothan B, Sánchez-Sevilla J, Amaya Saavedra I (2011) Quantitative trait loci and underlying candidate genes controlling agronomical and fruit quality traits in octoploid strawberry (Fragaria × ananassa). Theor Appl Genet. doi10.1007/s00122-011-1624-6 (in press) [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.