Abstract
Every year more than 13 million deaths worldwide are due to environmental pollutants, and approximately 24% of diseases are caused by environmental exposures that might be averted through preventive measures. Rapidly growing evidence has linked environmental pollutants with epigenetic variations, including changes in DNA methylation, histone modifications and microRNAs.
Environ mental chemicals and epigenetic changes All of these mechanisms are likely to play important roles in disease aetiology, and their modifications due to environmental pollutants might provide further understanding of disease aetiology, as well as biomarkers reflecting exposures to environmental pollutants and/or predicting the risk of future disease. We summarize the findings on epigenetic alterations related to environmental chemical exposures, and propose mechanisms of action by means of which the exposures may cause such epigenetic changes. We discuss opportunities, challenges and future directions for future epidemiology research in environmental epigenomics. Future investigations are needed to solve methodological and practical challenges, including uncertainties about stability over time of epigenomic changes induced by the environment, tissue specificity of epigenetic alterations, validation of laboratory methods, and adaptation of bioinformatic and biostatistical methods to high-throughput epigenomics. In addition, there are numerous reports of epigenetic modifications arising following exposure to environmental toxicants, but most have not been directly linked to disease endpoints. To complete our discussion, we also briefly summarize the diseases that have been linked to environmental chemicals-related epigenetic changes.
Keywords: Environmental chemicals, epigenetics, disease susceptibility
Background
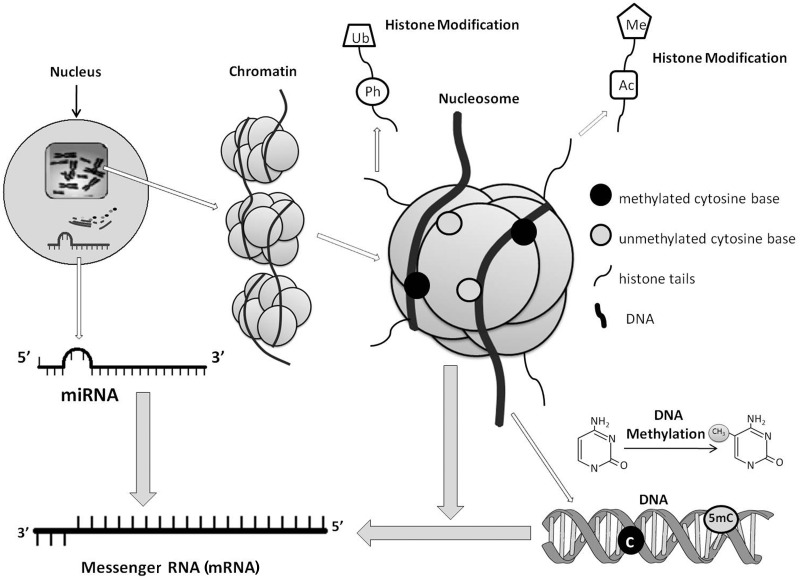
More than 13 million deaths every year are due to environmental pollutants, and as much as 24% of diseases are estimated to be caused by environmental exposures that can be averted.1 In a screening promoted by the United States Center for Disease Control and Prevention, 148 different environmental chemicals were found in the blood and urine from the US population, indicating the extent of our exposure to environmental chemicals.2 Growing evidence suggests that environmental pollutants may cause diseases via epigenetic mechanism-regulated gene expression changes.3,4 Dynamic chromatin remodelling is required for the initial steps in gene transcription, which can be achieved by altering the accessibility of gene promoters and regulatory regions.5 Epigenetic factors, including DNA methylation, histone modifications and microRNAs (miRNAs) (Figure 1), participate in these regulatory processes, thus controlling gene expressions.6,7 Changes in these epigenetic factors have been shown to be induced by exposure to various environmental pollutants, and some of them were linked with different diseases.8–10 In this review, we summarize the findings linking environmental chemical exposures with epigenetic alterations, provide some evidence linking such epigenetic changes with diseases (Table 1), and discuss the challenges and opportunities of environmental epigenomics in epidemiologic studies.
Figure 1.
Transcriptional regulation at the epigenetic level. Epigenetic mechanisms, including DNA methylation, histone modifications and miRNAs, regulate chromatin compaction and gene expression. DNA methylation at CpG sites usually suppresses gene expression. Histones are globular proteins that undergo posttranslational modifications, such as Ac, methylation and phosphorylation, thus influencing chromatin structure and gene expression. Active genes are usually characterized by low DNA methylation and highly acetylated chromatin configuration that allow access to transcription factors. miRNAs are a set of small, non-protein-coding RNAs that negatively regulate expression of target genes at the posttranscriptional level by binding to 3′-untranslated regions of target mRNAs
Table 1.
Effects of environmental chemicals on epigenetic changes
| Environmental chemicals | Epigenetic changes | In vitro/in vivo | Tissue/species | Example of diseases potentially associated with the observed changes in epigenetic changes |
|---|---|---|---|---|
| Arsenic | DNA methylation | |||
| Global hypomethylation | In vitro | Human HaCaT keratinocytes,80 human prostate epithelial cell line RWPE-1,81,82 TRL 1215 rat liver epithelial cell line,83 V79-Cl3 Chinese hamster cells226 | Various cancers227–230 and schizophrenia231 | |
| Global hypomethylation | In vivo | 129/SvJ mice,84 fisher 344 Rat,86 homozygous Tg.AC mice,87 goldfish,232 human PBL233 | Various cancers227–230 and schizophrenia231 | |
| Global hypomethylation and c-Ha-ras hypomethylation | In vivo | C57BL/6J mice85 | Various cancers227–230 and schizophrenia231 | |
| Global hypermethylation | In vivo | Human PBL88,89 | Colorectal cancer,234–236 renal cell carcinoma,237 acute lymphoblastic leukaemia238 and bladder urothelial cell carcinoma239 | |
| DAPK hypermethylation | In vitro | Human uroepithelial SV-HUC-1 cells90 | Various cancers240–251 | |
| P16 hypermethylation | In vitro | Human myeloma cell line U26691 | Various cancers241,248,250,252–257 | |
| DBC1, FAM83A, ZSCAN12 and C1QTNF6 hypermethylation | In vitro | Human UROtsa cells92 | Bladder cancer,258 breast cancer259 and malignant lymphoproliferative neoplasms260 | |
| P53 hypermethylation | In vitro | Human lung adenocarcinoma A549 cells93 | Breast cancer261 and hepatoblastoma262 | |
| C-myc hypomethylation | In vitro | TRL 1215 rat liver epithelial cells94 | Gastric cancer,263,264 colon cancer,263 liver cancer,207,265,266 kidney cancer207 and bladder cancer267 | |
| C-myc and c-Ha-ras hypomethylation | In vitro | Syrian hamster embryo cells95 | Gastric cancer,263,264 colon cancer,263 liver cancer,207,265,266 kidney cancer207 and bladder cancer267 | |
| P16 and RASSF1 hypermethylation | In vivo | A/J mice96 | Various cancers241,248,250,252–257,268,269 | |
| Global hypomethylation and ER-alpha hypomethylation | In vivo | C3H mice97 | Various cancers97,227–230 and schizophrenia231 | |
| P53 and P16 hypermethylation | In vivo | Human PBL98 | Various cancers241,248,250,252–257,261,262 | |
| DAPK hypermethylation | In vivo | Human bladder, kidney and ureter99 | Various cancers240–251 | |
| RASSF1A and PRSS3 hypermethylation | In vivo | Human bladder100 | Lung cancer and prostate cancer268,269 | |
| P16 hypermethylation | In vivo | Human PBL270 | Various cancers241,248,250,252–257 | |
| P53 hypermethylation | In vivo | Human basal cell carcinoma102 | Breast cancer261 and hepatoblastoma262 | |
| Both hypomethylation and hypermethylation of VHL | In vitro | Human kidney cells271 | Renal cell carcinoma271 | |
| Histone modification | ||||
| ↓H3 acetylation | In vitro | UROtsa and URO-ASSC cells92 | Renal cell carcinomas272 | |
| ↓H4K16 acetylation | In vitro | UROtsa cells104 | Bladder cancer273 | |
| ↑H3K14 acetylation | In vitro | NB4 cells105 | Diabetic nephropathy274 | |
| ↑H3S10 phosphorylation | ||||
| ↑H3 phosphorylation | In vitro | WI-38 human diploid fibroblast cells106 | Diabetic nephropathy274 | |
| ↑H3K9 acetylation | In vitro | HepG2 hepatocarcinoma cells107 | Diabetic nephropathy274 | |
| ↓H3, H4, H2a, H2b acetylation ↓H3 and H4 methylation | In vitro | Drosophila melanogaster tissue culture cell line KC161103 | Heart disease275 and traumatic brain injury276 | |
| ↑H2b methylation | ||||
| ↑H3K36 trimethylation | In vitro | Human lung carcinoma A549 cells110 | Diabetic nephropathy,274 multiple myeloma277 and prostate cancer278 | |
| ↓H3K36 dimethylation | ||||
| ↑H3K4 dimethylation | ||||
| ↑H3K9 dimethylation | In vitro | Human lung carcinoma A549 cells110,279 | Prostate cancer,278 kidney cancer,278 lung cancer,280 HCC281 and AML282 | |
| ↓H3K27 trimethylation | ||||
| ↑H3K4 trimethylation | ||||
| ↑H2AX phosphorylation | In vitro | RPMI7951 melanoma cells112 | Ataxia telangiectasia283 | |
| ↓H3K18 acetylation | In vitro | 1470.2 cell line derived from the mouse adenocarcinoma parent line284 | Prostate cancer278 and colon cancer285 | |
| ↓H3R17 methylation | ||||
| miRNAs | ||||
| ↑miR-222, ↓miR-210 | In vitro | TK6 cell line100 | Various cancers286–290 and AD291 | |
| ↓miR-19a | In vitro | T24 cell line115 | Various cancers292–300 | |
| Nickel | DNA methylation | |||
| ATF-1, HIF-1, gpt and Rb hypermethylation | In vitro | G12 cell line116,117 | Various cancers301–306 | |
| P16 hypermethylation | In vivo | Mouse histiocytomas119 | Various cancers241,248,250,252–257 | |
| Histone modification | ||||
| ↑H3K9 methylation | In vitro | Human lung carcinoma A549 cells123,307 | Heart disease275 and traumatic brain injury276 | |
| ↓Ac at all four core histones | ||||
| ↑H3K9 dimethylation | In vitro | Human lung carcinoma A549 cells,122,124 G12 cells,116,123,126,128,279 1HAEo- cell line,120,121 human (HAE) and rat (NRK) cells,125 Chinese hamster cell line127 | Lung cancer,308 heart disease,275 chronic glomerular disease309 and traumatic brain injury276 | |
| ↑H2a, H2b ubiquitylation | ||||
| ↓H3K4 methylation | ||||
| ↓H3K4 acetylation | ||||
| ↓H2a, H2b, H3, H4 acetylation | ||||
| ↓H4K5, H4K8, H4K12, H4K16 acetylation | In vivo | Human lung carcinoma A549 cells130 | Ataxia telangiectasia310 | |
| ↓H2A, H2B, H3, H4 acetylation (especially in H2BK12 and H2BK20) | In vitro | Human airway epithelial 1HAEo- (HAE) cell line131 | Heart disease275 and traumatic brain injury276 | |
| ↑H3 phosphorylation | In vitro | Human lung carcinoma A549 cells132 | Diabetic nephropathy274 | |
| Cadmium | DNA methylation | |||
| Global DNA hypomethylation | In vitro | K562 cell133 | Colorectal cancer,234–236 renal cell carcinoma,237 acute lymphoblastic leukaemia,238 bladder urothelial cell carcinoma239 | |
| Initially induces DNA hypomethylation, prolonged exposure results in DNA hypermethylation | In vitro | TRL1215 rat liver cells134 | Not applicable | |
| miRNAs | ||||
| ↓miR-146a | In vivo | Human PBL137 | Various cancers311–313 | |
| Chromium | DNA methylation | |||
| P16 and hMLH1 hypermethylation | In vivo | Human lung143,144 | Various cancers241,248,250,252–257,314–316 | |
| Gpt hypermethylation | In vitro | G12 cell line317 | Not applicable | |
| Histone modification | ||||
| ↓H3S-10 phosphorylation | In vitro | Human lung carcinoma A549 cells279 | Type 2 diabetes,274 heart disease275 and traumatic brain injury276 | |
| ↓H3K4 trimethylation | ||||
| ↓H3 and H4 acetylation ↑Dimethylation and trimethylation of H3K9 and H3K4 | ||||
| ↓H3K27trimethylation and H3R2 dimethylation | ||||
| Aluminum | miRNAs | |||
| ↑miR-146a | In vitro | HN cells149 | AD,318,319 cardiac hypertrophy320 and various cancers321–328 | |
| ↑miR-9, -128, -125b | In vitro | HN cells329 | AD,330 neurodegeneration331 and various cancers332–335 | |
| Mercury | DNA methylation | |||
| Global hypomethylation | In vivo | Brain tissues in polar bear139 | Neurological disorders336,337 and various cancer338 | |
| Rnd2 hypermethylation | In vitro | Mouse embryonic stem cells140 | neuronal migration defect339 | |
| Lead | DNA methylation | |||
| Global hypomethylation | In vivo | Human PBL,141 newborn umbilical cord blood samples142 | Various cancers227–230 and schizophrenia231 | |
| Pesticides | DNA methylation | |||
| P53 hypermethylation | In vitro | Human lung adenocarcinoma A549 cells93 | Breast cancer261 and hepatoblastoma262 | |
| Alter DNA methylation in the germ line | In vivo | Rat testis154–156 | Potential effects in the offspring | |
| Hypomethylation of c-jun and c-myc | In vivo | Mouse liver158,159 | Gastric cancer,263,264 colon cancer,263 liver cancer,207,265,266 kidney cancer207 and bladder cancer267 | |
| Global hypomethylation (Alu) | In vivo | Human PBL161,162 | Various cancers227–230 and schizophrenia231 | |
| Both hypomethylation and hypermethylation of VHL | In vitro | Human kidney cells271 | Renal cell carcinoma271 | |
| Histone modification | ||||
| ↑Ac of H3 and H4 | In vitro and in vivo | Immortalized rat mesencephalic/dopaminergic cells (N27 cells)169 | Parkinson’s disease169 | |
| Air pollution | DNA methylation | |||
| Global hypomethylation | In vivo | Human PBL8 | Various cancers227–230 and schizophrenia231 | |
| iNOS hypomethylation | In vivo | Human PBL173 | Lung cancer340 | |
| Global hypermethylation | In vivo | C57BL/CBA mice sperm174 | Colorectal cancer,234–236 renal cell carcinoma237, acute lymphoblastic leukaemia238 and bladder urothelial cell carcinoma239 | |
| Hypermethylation of IFNg and hypomethylation of IL4 | In vivo | CD4+ T lymphocytes175 | Asthma175 | |
| Histone modification | ||||
| ↑H3K4 dimethylation and H3K9 acetylation | In vivo | Human PBL177 | Diabetic nephropathy274 | |
| Global hypomethylation (Alu, LINE-1) | In vivo | Human buffy coat317 | Various cancers227–230 and schizophrenia231 | |
| miRNAs | ||||
| ↑miR-222 | In vivo | Human PBL137 | Various cancers286–288 | |
| ↑miR-21 | In vivo | Human PBL137 | Various cancers299,341–347 | |
| Benzene | DNA methylation | |||
| Global hypomethylation (Alu, LINE-1) | In vivo | Human PBL8 | Various cancers227–230 and schizophrenia231 | |
| P15 hypermethylation and melanoma antigen-1 (MAGE-1) hypomethylation | In vivo | Human PBL165–168,186 | Psoriasis348 and various cancers349–360 | |
| Global DNA hypomethylation | In vitro | Human lymphoblastoid cell line TK6187 | Various cancers227–230 and schizophrenia231 | |
| Hypermethylation of poly (ADP-ribose) polymerases-1 (PARP-1) | In vitro | Lymphoblastoid cell line F32188 | Various cancers188 | |
| Bisphenol A | DNA methylation | |||
| Hypomethylation of the Agouti gene and CabpIAP | In vivo | Mouse embryo192 | Mice with hypomethylation of the Agouti gene are obese, diabetic and exhibit increased cancer rates361,362 | |
| Hypomethylation of the homeobox gene Hoxa10 | In vivo | CD-1 mice194 | Not applicable | |
| Hypermethylation of LAMP3. | In vitro | Breast epithelial cells195 | Breast cancer195 | |
| miRNAs | ||||
| ↑miR-146a | In vitro | 3A placental cells196 | Cardiac hypertrophy,320 AD318,319 and various cancers321–328 | |
| Dioxin | DNA methylation | |||
| Igf2 hypomethylation | In vivo | Rat liver198 | Russell–Silver syndrome363–365 and various cancers366–370 | |
| Alterations in DNA methylation at multiple genomic regions | In vivo | Splenocyte of mice199 | Not applicable | |
| miRNAs | ||||
| ↑miR-191 | In vivo | Rat liver200 | Breast cancer,342 colorectal cancer321,371 and gastric cancer372 | |
| RDX | miRNAs | |||
| ↑let-7, miR-15, -16, -26, -181 ↓miR-10b | In vivo | Mouse brain and liver202 | Various cancers325,373–380 | |
| ↑miR-206, -30, -195 | In vivo | Mouse brain and liver202 | Various cancers342,381–385 | |
| DES | miRNAs | |||
| ↓miR-9-3 | In vitro | Breast epithelial cells205 | Breast cancer205 | |
| Drinking water | DNA methylation | |||
| Global hypomethylation c-myc hypomethylation | In vivo | Mice liver207,208 | Gastric cancer,263,264 colon cancer,263 liver cancer,207,265,266 kidney cancer207 and bladder cancer267 |
PBL, peripheral blood leucocytes; HCC, hepatocellular carcinoma; AML, acute myeloid leukaemia; AD, Alzheimer’s disease; HN cells, human neural cells; RDX, hexahydro-1,3,5-trinitro-1,3,5-triazine; DES, diethylstilbestrol.
Epigenetic factors
DNA methylation
DNA methylation, a naturally occurring modification that involves the addition of a methyl group to the 5′ position of the cytosine ring, is the most commonly studied and best understood epigenetic mechanism.11 In the human genome, it predominantly occurs at cytosine–guanine dinucleotide (CpG) sites, and serves to regulate gene expression and maintain genome stability.12
Environmental studies have shown distinct DNA methylation abnormalities. One commonly reported alteration is an overall genome-wide reduction in DNA methylation content (global hypomethylation) that may lead to reactivation of transposable elements and alter the transcription of otherwise silenced adjacent genes.13,14 Global hypomethylation is associated with genomic instability and an increased number of mutational events.15–18 There are approximately 1.4 million Alu repetitive elements (sequences containing a recognition site for the restriction enzyme AluI)19 and a half a million long interspersed nucleotide (LINE-1) elements in the human genome that are normally heavily methylated.20 More than one-third of DNA methylation occurs in repetitive elements.20 Because of their high representation throughout the genome, LINE-1 and Alu have been used as global surrogate markers for estimating the genomic DNA methylation level in cancer tissues,20–22 although recent data show lack of correlation with global methylation in normal tissues, such as peripheral blood.23 Other types of abnormalities that can be induced by environmental pollutants are hyper- or hypo-methylation of specific genes or regions, potentially associated with aberrant gene transcription.24–27 DNA methylation alterations that directly affect gene expression often occur in the CpG sites located in the promoter regions of the genes. Recent evidence has shown that differentially methylated sites in various cancer tissues are enriched in sequences, termed ‘CpG island shores’, up to 2 kb distant from the transcription start site.28 However, to date, gene-specific DNA methylation alterations induced by environmental exposures have been mostly investigated in gene promoter regions. CpG island shores are clearly worthy of further investigation in relation to environmental exposures, but whether they hold such importance in a non-cancer setting remains to be determined.
Histone modifications
In humans, protection and packaging of the genetic material are largely performed by histone proteins, which also offer a mechanism for regulating DNA transcription, replication and repair.29 Histones are nuclear globular proteins that can be covalently modified by acetylation (Ac), methylation, phosphorylation, glycosylation, sumoylation, ubiquitination and adenosine diphosphate (ADP) ribosylation,30,31 thus influencing chromatin structure and gene expression.32,33 The most common histone modifications that have been shown to be modified by environmental chemicals are Ac and methylation of lysine residues in the amino terminal of histone 3 (H3) and H4. Histone Ac, with only a single acetyl group added to each amino acid residue usually, increases gene transcriptional activity;34–37 whereas histone methylation (Me), found as mono (Me), di-methyl (Me2), and tri-methyl (Me3) group states38 can inhibit or increase gene expression depending on the amino acid position that is modified.39–41
miRNAs
miRNAs are short single-stranded RNAs of approximately 20–24 nucleotides in length that are transcribed from DNA but not translated into proteins. miRNAs negatively regulate expression of target genes at the post-transcriptional level by binding to 3′-untranslated regions of target mRNAs.42 Each mature miRNA is partially complementary to multiple target mRNAs and directs the RNA-induced silencing complex (RISC) to identify the target mRNAs for inactivation.43 miRNAs are initially transcribed as longer primary transcripts (pri-miRNAs) and processed first by the RNase enzyme complex, and then by Dicer, leading to incorporation of a single strand into the RISC. miRNAs guide RISC to interact with mRNAs and determine post-transcriptional repression. miRNAs are involved in the regulation of gene expression through the targeting of mRNAs during cell proliferation, apoptosis, control of stem cell self renewal, differentiation, metabolism, development and tumour metastasis.44,45 Compared with other mechanisms involved in gene expression, miRNAs act directly before protein synthesis and may be more directly involved in fine-tuning of gene expression or quantitative regulation.46,47 Moreover, miRNAs also play key roles in modifying chromatin structure and participating in the maintenance of genome stability.48 miRNAs can regulate various physiological and pathological processes, such as cell growth, differentiation, proliferation, apoptosis and metabolism.42,49 More than 10 000 miRNAs have been reported in animals, plants and viruses by using computational and experimental methods in miRNA-related public databases. The aberrant expression of miRNAs has been linked to various human diseases, including Alzheimer’s disease, cardiac hypertrophy, altered heart repolarization, lymphomas, leukaemias, and cancer at several sites.50–66
Environmental pollutants and epigenetic alterations
Metals
Heavy metals are widespread environmental contaminants and have been associated with a number of diseases, such as cancer, cardiovascular diseases, neurological disorders and autoimmune diseases.67,68 In recent years, there has been an increasing appreciation of the roles of molecular factors in the aetiology of heavy metal-associated diseases.69–71 Several studies showed that metals act as catalysts in the oxidative deterioration of biological macromolecules.72 Metal ions induce reactive oxygen species (ROS), and thus lead to the generation of free radicals.72,73 ROS accumulation can affect epigenetic factors.74–79 Growing data have linked epigenetic alterations with heavy metal exposure.
Arsenic
Evidence has been rapidly increasing that exposure to arsenic (As) alters DNA methylation both globally and in the promoter regions of certain genes. Upon entering the human body, inorganic As is methylated for detoxification. This detoxification process uses S-adenosyl methionine (SAM), which is a universal methyl donor for methyltransferases including DNA methyltransferases (DNMTs) that determine DNA methylation. Thus, it has been shown that As exposure leads to SAM insufficiency and decreases the activity of DNMTs due to the reduction of their substrate. In addition, As has also been shown to decrease DNMT gene expression.80 These As-induced processes may all contribute to global DNA hypomethylation. Arsenic exposure was shown to induce global hypomethylation in a dose-dependent manner in several in vitro studies.80–83 Further, rats and mice exposed to As for several weeks exhibited global hypomethylation in hepatic DNA.84–87 Nonetheless, evidence in humans is still limited and not completely consistent. In a cross-sectional study of 64 subjects, As level in contaminated water was associated with global DNA hypermethylation in blood mononuclear cells.88 A global dose-dependent hypermethylation of blood DNA was observed in Bangladeshi adults with chronic As exposure.89
Arsenic exposure has also been associated with gene-specific hyper- or hypo-methylation in both experimental settings and human studies.85,90–101 As exposure has been shown to induce dose-dependent promoter hypermethylation of several tumour suppressor genes, such as p15, p16, p53 and DAPK, in vitro and in vivo.91,93,98,101,102 Furthermore, As exposure-related up-regulation of ER-alpha, c-myc and Ha-ras1 gene expression was linked to their promoter hypomethylation in cell lines94,95 and animal studies.84,85,97 Evidence in humans is rapidly growing. Toenail As concentration was positively associated with RASSF1A and PRSS3 promoter methylation levels in bladder tumours.100 Promoter hypermethylation in these two genes was associated with As-induced invasive lung tumours compared with non-invasive tumours.100 Promoter hypermethylation of DAPK was observed in human uroepithelial cells exposed to As,90 as well as in tumours from 13 of 17 patients living in As-contaminated areas relative to 8 of 21 patients living in As non-contaminated areas.99 Increased DNA methylation of the p16 promoter was observed in arseniasis patients when compared with people with no history of As exposure.101
Arsenic exposure has also been shown to cause alterations in histone modifications. The earliest evidence on As-induced histone acetylation reductions was in Drosophila.103 Trivalent As has recently been linked to reduced H3 and H4 lysine 16 (H4K16) acetylation in human bladder epithelial cells.104 On the other hand, trivalent As exposure has also been shown to increase histone acetylation, which was shown to up-regulate genes related to apoptosis or cell stress response.105,106 Ramirez et al. have reported that As could cause global histone acetylation by inhibiting the activity of histone deacetylases (HDACs).107 Together, these studies provide evidence that histone acetylation can be dysregulated by As exposure. Early in 1983, As was also shown to induce methylation changes in H3 and H4 in Drosophila.103 Similar results on H3 were seen in Drosophila Kc 111 cell several years later.108,109 In recent years, in mammalian cells, arsenite (AsIII) exposure has been associated with increased H3 lysine 9 dimethylation (H3K9me2) and H3 lysine 4 trimethylation (H3K4me3), and decreased H3 lysine 27 trimethylation (H3K27me3).110,111 As was shown to induce apoptosis by up-regulation of phosphorylated H2AX112 and cause H3 phosphorylation, which may play important roles in the up-regulation of the oncogenes.106
Exposure of human lymphoblast cell line TK-6 to arsenite exhibited global increases in miRNA expression.113 Arsenic trioxide (As2O3) has been used as a pharmacological treatment in acute promyelocytic leukaemia.114 Cao et al.115 demonstrated that numerous miRNAs were up-regulated or down-regulated in T24 human bladder carcinoma cells exposed to As2O3. In particular, miRNA-19a was substantially decreased, resulting in cell growth arrest and apoptosis. The As-related changes in miRNA expression were shown to be reversible when the exposure was removed.115
Nickel
Nickel has been proposed to increase chromatin condensation and trigger de novo DNA methylation of critical tumour suppressor or senescence genes.116 In Chinese hamster G12 cells transfected with the Escherichia coli guanine phosphoribosyl transferase (gpt) gene, nickel was shown to induce hypermethylation and inhibit the expression of the transfected gpt gene.117 An animal study has further shown that nickel induced DNA hypermethylation, altered heterochromatin states and caused gene inactivation, eventually leading to malignant transformation.118 Govindarajan et al.119 have observed DNA hypermethylation of p16 in nickel-induced tumours of wild-type C57BL/6 mice, as well as in mice heterozygous for the tumour suppressor p53 gene injected with nickel compound.
Nickel may cause diseases also via affecting histone modifications. Evidence on nickel-induced histone modifications includes increases of H3K9 dimethylation, loss of histone acetylation in H2A, H2B, H3 and H4, and increases of the ubiquitination in H2A and H2B.116,120–127 An increase in H3K9 dimethylation and a decrease in H3K4 methylation and histone acetylation was found in the promoter of the gpt transgene in G12 cells exposed to nickel.116,123,128 In mouse PW cells and human cells treated with the HDAC inhibitor trichostatin A, nickel showed a lower capacity to induce malignant transformation.129 This finding suggested that gene silencing mediated by histone deacetylation may play a critical role in nickel-induced cell transformation.129 In addition, nickel has also been shown to induce a loss of histone methylation in vivo and decreased activity of histone H3K9 demethylase in vitro.123 Nickel also suppresses histone H4 acetylation in vitro in both yeast and mammalian cells.130,131 Nickel can induce H3 phosphorylation, specifically in serine 10 (H3S10) via activation of the c-jun N-terminal kinase/stress-activated protein kinase pathway.132
Cadmium
Cadmium (Cd) has been shown to alter global DNA methylation.133 Takiguchi et al.134 demonstrated that Cd inhibits DNMTs and initially induces global DNA hypomethylation in vitro (TRL1215 rat liver cells). However, prolonged exposure was shown to lead to DNA hypermethylation and enhanced DNMTs activity in the same experiment.134 Cd can also decrease DNA methylation in proto-oncogenes and promote oncogenes expression that can result in cell proliferation.133,134
Transcriptional and post-transcriptional gene regulation is critical in responses to Cd exposure, in which miRNAs may play an important role.135,136 Bollati et al.137 have recently demonstrated that increased expression of miR-146a in peripheral blood leucocytes from steel workers was related to inhalation of Cd-rich air particles. miRNA-146a expression is regulated by the transcription factor nuclear factor-kappa B, which represents an important causal link between inflammation and carcinogenesis.138
Other metals
Mercury (Hg) is widely present in various environmental media and foods at levels that can adversely affect humans and animals. Exposure to Hg has been associated with brain tissue DNA hypomethylation in the polar bear.139 Arai et al.140 have studied the effects of Hg on DNA methylation status in mouse embryonic stem cells. After 48 or 96 h of exposure to the chemical, they observed hypermethylation of Rnd2 gene in Hg-treated mouse embryonic stem cells.
Lead is among the most prevalent toxic environmental metals, and has substantial oxidative properties. Long-term exposure to lead was shown to alter epigenetic marks. In the Normative Aging Study, LINE-1 methylation levels were examined in association with patella and tibia lead levels, measured by K-X-Ray fluorescence. Patella lead levels were associated with reduced LINE-1 DNA methylation. The association between lead exposure and LINE-1 DNA methylation may have implications for the mechanisms of action of lead on health outcomes, and also suggests that changes in DNA methylation may represent a biomarker of past lead exposure.141 In addition, Pilsner et al.142 characterized genomic DNA methylation in the lower brain stem region from 47 polar bears hunted in central East Greenland between 1999 and 2001. They have reported an inverse association between cumulative lead measures and genomic DNA methylation level.
Hexavalent chromium [Cr(VI)] is a mutagen and carcinogen that has been linked to lung cancer and other adverse health effects in occupational studies. Kondo et al.143 found p16 and hMLH1 hypermethylation in lung cancer patients with past chromate exposure.144 In vitro experiments on cells exposed to binary mixtures of benzo[a]pyrene (B[a]P) and chromium have shown that B[a]P activates Cyp1A1 transcriptional responses mediated by the aryl hydrocarbon receptor (AhR), whereas chromium represses B[a]P-inducible AhR-mediated gene expression145,146 by inducing cross-links of histone deacetylase 1–DNA methyltransferase 1 (HDAC1–DNMT1) complexes to the Cyp1A1 promoter chromatin and inhibit histone marks, including phosphorylation of histone H3 Ser-10, trimethylation of H3 Lys-4 and various acetylation marks in histones H3 and H4. HDAC1 and DNMT1 inhibitors or depletion of HDAC1 or DNMT1 with siRNAs blocked the chromium-induced transcriptional repression by decreasing the interaction of these proteins with the Cyp1A1 promoter and allowing histone acetylation to proceed. By inhibiting Cyp1A1 expression, chromium stimulate the formation of B[a]P DNA adducts. These findings may link histone modifications to chromium-associated developmental and carcinogenic outcomes.147 Chromate exposure of human lung A549 cells has been shown to increase the global levels of di- and tri-methylated histone H3 lysine 9 (H3K9) and lysine 4 (H3K4), but decrease tri-methylated histone H3 lysine 27 (H3K27) and di-methylated histone H3 arginine 2 (H3R2). Most interestingly, H3K9 dimethylation was enriched in the human MLH1 gene promoter following chromate exposure, and this was correlated with decreased MLH1 mRNA expression. Chromate exposure increased the protein as well as mRNA levels of G9a, a histone methyltransferase that specifically methylates H3K9. This Cr(VI)-induced increase in G9a may account for the global elevation of H3K9 dimethylation. Furthermore, supplementation with ascorbate, the primary reductant of Cr(VI) and also an essential cofactor for the histone demethylase activity, partially reversed the H3K9 dimethylation induced by chromate. These results suggest that Cr(VI) may target histone methyltransferases and demethylases, which in turn affect both global and gene promoter-specific histone methylation, leading to the silencing of specific tumour suppressor genes.148
Recent investigations have demonstrated that aluminum exposure can alter the expression of a number of miRNAs. miR-146a in human neural cells was up-regulated after treatment with aluminium sulphate. Up-regulation of miR-146a corresponded to the decreased expression of complement factor H, a repressor of inflammation.149 In addition, a study on aluminium-sulphate-treated human neural cells in primary culture has shown increased expression of a set of miRNAs, including miR-9, miR-125b and miR-128.150 The same miRNAs were also found to be up-regulated in brain cells of Alzheimer patients, suggesting that aluminum exposure may induce genotoxicity via miRNA-related regulatory elements.150
Pesticides
Growing evidence suggests that epigenetic events can be induced by pesticide exposures.28,151–153 Animal models have shown that exposure to some pesticides, such as vinclozolin and methoxyclor, induces heritable alterations of DNA methylation in male germline associated with testis dysfunction,154–156 or affects ovarian function via altered methylation patterns.157 Decreased methylation in the promoter regions of c-jun and c-myc and increased levels of their mRNAs and proteins were found in livers of mice exposed to dichloro- and trichloro-acetic acid.158,159 Dichlorvos has been demonstrated to induce DNA methylation in multiple tissues in an animal toxicity study.160 DNA methylation in repetitive elements in blood DNA was inversely associated with increased levels of plasma pesticide residues and other persistent organic pollutants in an Arctic population,161 a finding later confirmed in a similar study in a Korean population.162 Whether aberrant DNA methylation represents the link between pesticides and risks of pesticide-related disease, including the excess of cancer risk observed in some epidemiology studies,163–168 remains to be determined.
Dieldrin, a widely used organochlorine pesticide, has been shown to increase acetylation of core histones H3 and H4 in a time-dependent manner. Histone acetylation was induced within 10 min of dieldrin exposure, suggesting that histone hyperacetylation is an early event in dieldrin-induced diseases. Treatment with anacardic acid, a histone acetyltransferase inhibitor, decreased dieldrin-induced histone acetylation.169 Dieldrin was further shown to induce histone hyperacetylation in the striatum and substantia nigra in mouse models, suggesting the roles for histone hyperacetylation in dieldrin-induced dopaminergic neuronal degeneration.170
Air pollution
Exposure to particulate matter (PM) of ambient air pollution has been associated with increased morbidity and mortality related to cardiovascular and respiratory diseases.171,172 Black carbon, a component of PM derived from vehicular traffic, has been linked to decreased DNA methylation in LINE-1 repetitive elements in 1097 blood DNA samples of elderly men in the Boston area. Additional evidence for PM effects on DNA methylation stemmed from an investigation of workers in a steel plant with well-characterized exposure to PM with diameters of <10 µm (PM10). Methylation of inducible nitric oxide synthase gene promoter region was decreased in blood samples of individuals exposed to PM10 after 3 days of work in the foundry when compared with baseline.173 In the same study, methylation of Alu and LINE-1 was negatively related to long-term exposure to PM10.173 In contrast, an animal experiment on mice exposed to air particles collected from a steel plant showed global DNA hypermethylation in sperm genomic DNA, a change that persisted after removal of environmental exposure.174 Inhaled diesel exhaust particles’ exposure and intranasal Aspergillus fumigatus induced hypermethylation of several sites of the interferon gamma (IFNγ) promoter and hypomethylation at a CpG site of the IL-4 promoter in mice. Altered methylation of promoters of both genes was correlated with changes in IgE levels.175,176
We recently also associated PM exposure with histone modifications in the above-mentioned steel workers with high exposure level to PM.177 In this study, exposure duration (years of work in the foundry) was associated with increased H3K4me2 and H3K4ac in blood leucocytes.177 In the same study, we showed that exposure to metal-rich PM induced rapid changes in the expression of two inflammation-related miRNAs, i.e. miR-21 and miR-222, measured in peripheral blood leucocytes.178 Using microarray profiling, Jardim et al.172 have shown extensive alterations of miRNA expression profiles in human bronchial epithelial cells treated with diesel exhaust particles. Out of 313 detected miRNAs, 197 were either up- or down-regulated by at least 1.5-fold.172
Benzene
Benzene is an environmental chemical that has been associated with increased risk of haematological malignancies, particularly with acute myeloid leukaemia and acute nonlymphocytic leukaemia.179–184 Benzene ranks among the top 20 chemicals for production volume in USA.185 Our results from a study of police officers and gas-station attendants have shown that low-dose exposure to airborne benzene is associated with alterations in DNA methylation in blood DNA of healthy subjects that resemble those found in haematological malignancies,165–168,186 including hypomethylation of LINE-1 and Alu repetitive elements, hypermethylation of p15 tumour suppressor gene and hypomethylation of MAGEA1 (melanoma-associated antigen 1 gene). Consistently, reductions of global DNA methylation has been recently shown in human lymphoblastoid cells treated with benzene metabolites.187 In vitro experiments have also shown that benzene exposure induces hypermethylation of poly (ADP-ribose) polymerases-1 (PARP-1), a gene involved in DNA repair.188
Bisphenol A
Bisphenol A (BPA) is an endocrine disruptor with potential reproductive effects, as well as a weak carcinogen associated with increased cancer risk in adult life through fetal exposures.189,190 BPA is widely used as an industrial plasticizer in epoxy resins for food and beverage containers, baby bottles and dental composites.191 Dolinoy et al.192 reported that periconceptional exposure to BPA shifted the coat colour distribution of the viable yellow agouti (Avy) mouse offspring toward yellow by decreasing CpG methylation in an intracisternal A particle (IAP) retrotransposon upstream of the Agouti gene.193 In this animal model, the yellow-coat phenotype is associated with increased cancer rates, as well as with obesity and insulin resistance. In the same set of experiments, maternal dietary supplementation, with either methyl donors like folic acid or the phytoestrogen genistein, blunted the effect of BPA on IAP methylation and prevented the coat colour change caused by BPA exposure.192 In pregnant CD-1 mice treated with BPA, Bromer et al.194 found decreased methylation and increased expression of the homeobox gene Hoxa10, which controls uterine organogenesis. In breast epithelial cells treated with low-dose BPA, gene expression profiling identified 170 genes with expression changes in response to BPA, of which expression of lysosomal-associated membrane protein 3 (LAMP3) was shown to be silenced due to DNA hypermethylation in its promoter.195
In a recent study by Avissar-Whiting et al.,196 an elevated expression of miR-146a was observed in BPA-treated placental cell lines and miR-146a expression was associated with slower cell proliferation and higher sensitivity to the bleomycin-induced DNA damage.
Dioxin
Dioxin is a compound that has been classified as a human carcinogen by the International Agency for Research on Cancer. As dioxin is only a weak mutagen, extensive research has been conducted to identify potential mechanisms contributing to carcinogenesis. One proposed pathway to carcinogenesis is related to the powerful dioxin-induced activation of microsomal enzymes, such as CYP1B1, that might activate other procarcinogen compounds to active carcinogen. The capability of dioxin to induce CYP1B1 has been recently shown in vitro to depend on the methylation state of the CYP1B1 promoter.197 Also, dioxin was shown to reduce the DNA methylation level of Igf2 in rat liver.198 Recently, alterations in DNA methylation at multiple genomic regions were identified in splenocytes of mice treated with dioxin, a finding potentially related to dioxin immunotoxicity.199 In a xenograft mouse model of hepatocellular carcinoma, Elyakim et al.200 have also found that dioxin up-regulated miR-191. In the same study, inhibition of miR-191 inhibited apoptosis and decreased cell proliferation, suggesting that increased miR-191 expression may contribute to determine dioxin-induced carcinogenicity.
Hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX, also known as hexogen or cyclonite)
Hexahydro-1,3,5-trinitro-1,3,5-triazine (commonly known as RDX, the British code name for Royal Demolition Explosive) is an explosive polynitramine and common ammunition constituent used in military and civil activities. Although most of this environmental pollutant is found in soils, RDX and its metabolites are also found in water sources.201 Exposure to RDX and its metabolites could cause neurotoxicity, immunotoxicity and cancers.202 Zhang et al.202 have recently evaluated the effects of RDX on miRNA expression in mouse brain and liver. In this study, out of 113 miRNAs, 10 were up-regulated and 3 were down-regulated. Most of the miRNAs that showed altered expression, including let-7, miR-17-92, miR-10b, miR-15, miR-16, miR-26 and miR-181, were found to regulate toxicant-metabolizing enzymes, as well as genes related to carcinogenesis and neurotoxicity.202
Diethylstilbestrol
Diethylstilbestrol (DES) is a synthetic oestrogen that was used to prevent miscarriages in pregnant women between the 1940s and the 1960s.203 A moderate increase in breast cancer risk has been shown both in daughters of women who were treated with DES during pregnancy, as well as in their daughters.204 Hsu et al.205 have demonstrated that the expression of 82 miRNAs (9.1% of the 898 miRNAs evaluated) were altered in breast epithelial cells when exposed to DES. In particular, the suppression of miR-9-3 expression was accompanied by promoter hypermethylation of the miR-9-3 coding gene in DES-treated epithelial cells.205
Chemicals in drinking water
Chlorination by-products are formed as a result of the water chlorination for anti-fouling purposes. Various chlorination by-products in drinking water, such as triethyltin,206 chloroform207 and trihalomethanes,208 have been questioned for potential adverse health effects.209 These chemicals have been shown to induce certain epigenetic changes. Rats that were chronically intoxicated with triethyltin in drinking water showed development of cerebral oedema as well as an increase of phosphatidylethanolamine-N-methyltransferase activities. This increased methylation might be a compensatory mechanism for counteracting the membrane damages induced by triethyltin.206 Chloroform, dichloroacetic acid (DCA) and trichloroacetic acid (TCA), three liver and kidney carcinogens, are by-products of chlorine disinfection found in drinking water.210,211 Mice treated with DCA, TCA and chloroform show global hypomethylation and increased expression of c-myc, a proto-oncogene involved in liver and kidney tumours.207 Trihalomethanes (chloroform, bromodichloromethane, chlorodibromomethane and bromoform) are regulated organic contaminants in chlorinated drinking water. In female B6C3F1 mouse liver, trihalomethanes demonstrated carcinogenic activity. Chloroform and bromodichloromethane decreased the level of 5-methylcytosine in hepatic DNA. Methylation in the promoter region of the c-myc gene was reduced by the trihalomethanes, consistent with their carcinogenic activity.208
Environmental epigenomics: challenges and opportunities for epidemiologic studies
The studies reviewed in this article have demonstrated the potential effects of environmental pollutants on the epigenome. Several of the epigenomic changes observed in response to environmental exposures might be mechanistically associated with susceptibility to diseases (Table 1). Further studies of epigenetic mechanisms in disease pathogenesis, including the role of epigenetics in the developmental origins of health and disease, their relationships with environmental exposures and the pathways associated with the disease phenotype may help develop preventive and therapeutic strategies.
Epigenetics and developmental origins of health and disease
During embryogenesis, epigenetic patterns change dynamically to adapt embryos to be fit for further differentiation.7 Two waves of epigenetic reprogramming, which take place at the zygote stage and during primordial germ cells formation, accompany mammalian development.212
Experiments on mice carrying the Avy have demonstrated that embryo life is a window of exquisite sensitivity to the environment. In viable yellow (Avy/a) mice, transcription originating in a IAP retrotransposon inserted upstream of the agouti gene (A) causes ectopic expression of agouti protein, resulting in yellow fur, obesity, diabetes and increased susceptibility to tumours.213 BPA is a high-production-volume chemical used in the manufacture of polycarbonate plastic. In utero or neonatal exposure to BPA is associated with higher body weight, increased breast and prostate cancer and altered reproductive function.
Additional experimental studies have suggested epigenetic mechanisms as potential intermediates for the effects of prenatal exposures to pesticides such as vinclozolin and methoxyclor,154 as well as of other conditions such as nutritional supplies of methyl donors.192 Evidence has also been accumulating in humans. Investigations of candidate loci among individuals prenatally exposed to poor nutrition during the Dutch famine in 1944–45 indicate that epigenetic changes induced by prenatal exposures may be common in humans, although they appear to be relatively small and greatly dependent on the timing of the exposure during gestation.214,215 Based on findings of changes in DNA methylation in subjects exposed to the Dutch famine, Heijmans et al.216 have suggested that the epigenome may represent a molecular archive of the prenatal environment, via which the in-utero environment may produce serious ramifications on health and disease later in life. Terry et al.217 found that prenatal exposure to cigarette smoke was associated with increased overall blood DNA methylation level in adulthood. Other examples include decreased LINE-1 and Sat 2 methylation level in adults and children prenatally exposed to smoking,218 and global DNA hypomethylation in newborns with utero exposures of maternal smoking.219 In addition to these DNA methylation changes, Maccani et al.220 have recently observed that miR-16, miR-21 and miR-146a were down-regulated in cigarette smoke-exposed placentas compared to controls.
Additional well-conducted epigenetic studies are now warranted to generate a catalogue of regions that are sensitive to the prenatal environment and may reflect developmental influences on human disease.
Can we develop epigenomic biosensors of past exposures?
An important property of epigenomic signatures is that, because they can be propagated through cell division even in cells with high turnover, they can persist even after the exposure is removed. In addition, as discussed above, an individual’s epigenome may also reflect his/her prenatal environmental exposure experience. Thus, epigenomic profiling of individuals exposed to environmental pollutants might provide biosensors or molecular archives of one’s past or even prenatal environmental exposures. Using epigenomics, exposure assessment might be brought to research investigations and preventive settings where repeated collections of exposure data might be unfeasible or exceedingly expensive. Further research is needed to establish how rapid are the changes induced by environmental pollutants, as well as whether they accumulate in response to repeated or continuous exposure and how long they persist after the exposure is removed.
What are suitable study designs and approaches for environmental epigenomics?
The field of environmental epigenetics has evolved rapidly in the past several years. As research applications grow, investigators will be facing several difficulties and challenges. Some studies have produced inconsistent results on same pollutants. Several factors may contribute to the inconsistencies. Epigenetic alterations are tissue specific.221 It is conceivable that the same environmental pollutant may produce different epigenetic changes in different tissues, and even within the same tissue on different cell types. Larger studies with well-defined exposure information that allows examining epigenetic changes across different tissues are needed. Different study design, small sample size and different laboratory methods may also be major causes for the inconsistency. Replicating results and identifying the sources of variability across studies is a major challenge for epigenetic investigations. Because epigenetic markers change over time, disease outcomes are prone to reverse causation, i.e. an association between a disease and an epigenetic marker may be determined by an influence of the disease on the epigenetic patterns, rather than vice versa.222 Although epigenetic alterations that were found to be induced by or associated with environmental pollutants were also found in various diseases, almost no study has examined the sequence of exposures, epigenetic alterations and diseases.
Longitudinal studies with prospective collection of objective measures of exposure, biospecimens for epigenetic analyses and preclinical and clinical disease outcomes are needed to appropriately establish causality. Existing prospective epidemiology investigations might provide resources for mapping epigenomic changes in response to specific chemicals. However, cohort studies in which biospecimens have been previously collected for genetic or biochemical studies might pose several challenges. Most studies have collected biospecimens, such as blood, urine or buccal cells, which might not necessarily participate in the aetiology of the disease of interest. Methods of collection and processing (e.g. whole blood vs buffy coat) might modify the cell types stored, thus potentially impacting on epigenetic marks. In addition, high-coverage methods providing high-dimensional data on DNA methylation, histone modifications and miRNA expression are increasingly used in human investigations.
Albeit epigenetic mechanisms have properties that make them ideal molecular intermediates of environmental effects, the proportion of the effects of any individual environmental exposure that might be mediated through epigenetic mechanisms is still undetermined. Epidemiology and statistical approaches, including well-designed prospective studies and advanced statistical methods for causal inference are urgently needed. Similarly to genomic studies,223 epidemiological causal reasoning in epigenomics should include careful consideration of knowledge, data, methods and techniques from multiple disciplines.
The potential interactions between different forms of epigenetic modification
Most studies in environmental epigenetics have separately evaluated only one of the types of the epigenetic marks, i.e. DNA methylation, histone modifications or miRNA expression. However, epigenetic marks are related by an intricate series of interactions that may generate a self-reinforcing cycle of epigenetic events directed to control gene expression.224 For instance, histone deacetylation and methylation at specific amino acid residues contribute to the establishment of DNA methylation patterns. miRNA expression is controlled by DNA methylation in miRNA encoding genes, and, in turn, miRNAs have been shown to modify DNA methylation.225 Future studies that include comprehensive investigations of multiple epigenetic mechanisms might help elucidate the timing and participation of DNA methylation, histone modifications and miRNAs to determine environmental effects on disease development.
Can epigenomics be used for prevention?
One major objective of epidemiology investigations is to provide the groundwork for future preventive interventions. Numerous clinical and preclinical studies showed that most of the epigenetic changes are reversible, which offers novel insights to develop new preventive and therapeutic strategies that might take advantage of molecules that modify the activities of epigenetic enzymes, such as DNMTs and HDACs, as well as of the growing field of RNAi therapeutics. Drugs have been designed and developed that produce functional effects, such as histone acetylation and DNA hypomethylation that might be used to restore the normal transcription level of genes. Future epidemiology studies have a unique opportunity to evaluate whether the effects of environmental exposures on the epigenome are mitigated by positive changes in lifestyles, or worsened by the interaction with other risk factors. Future epigenomic research may provide information for developing preventive strategies, including exposure reduction, as well as pharmacological, dietary or lifestyle interventions.
Funding
Our work is partially supported by grants from the HSPH-NIEHS Center for Environmental Health New Investigator Fund (P30ES000002) and NIH award 1RC1ES018461-01.
Conflict of interest: None declared.
KEY MESSAGES.
Rapidly growing evidence has linked environmental pollutants with epigenetic variations, including changes in DNA methylation, histone modifications and microRNAs.
Some of such epigenetic changes have been associated with various diseases.
Further studies of epigenetic mechanisms in disease pathogenesis, their relationships with environmental exposures and related pathways are needed for the development of preventive and therapeutic strategies.
Future epidemiology studies on environmental pollutants and epigenome face several challenges.
References
- 1.Prüss-Üstün Annette CC. Organization (WHO); 2006. Preventing disease through healthy environments. Towards an estimate of the environmental burden of disease World Health. [Google Scholar]
- 2.Bollati V, Baccarelli A. Environmental epigenetics. Heredity. 2010;105:105–12. doi: 10.1038/hdy.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezek S, Ujhazy E, Mach M, Navarova J, Dubovicky M. Developmental origin of chronic diseases: toxicological implication. Interdiscip Toxicol. 2008;1:29–31. doi: 10.2478/v10102-010-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang WY, Ho SM. Epigenetic reprogramming and imprinting in origins of disease. Rev Endocr Metab Disord. 2007;8:173–82. doi: 10.1007/s11154-007-9042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 6.Grewal SI, Moazed D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- 7.Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–93. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- 8.Baccarelli A, Bollati V. Epigenetics and environmental chemicals. Curr Opin Pediatr. 2009;21:243–51. doi: 10.1097/mop.0b013e32832925cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heightman TD. Therapeutic prospects for epigenetic modulation. Expert Opin Ther Targets. 2011;15:729–40. doi: 10.1517/14728222.2011.561786. [DOI] [PubMed] [Google Scholar]
- 10.Wright RJ. Epidemiology of stress and asthma: from constricting communities and fragile families to epigenetics. Immunol Allergy Clin North Am. 2011;31:19–39. doi: 10.1016/j.iac.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller OJ, Schnedl W, Allen J, Erlanger BF. 5-Methylcytosine localised in mammalian constitutive heterochromatin. Nature. 1974;251:636–37. doi: 10.1038/251636a0. [DOI] [PubMed] [Google Scholar]
- 12.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–40. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 13.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 14.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 15.Chen RZ, Pettersson U, Beard C, Jackson-Grusby L, Jaenisch R. DNA hypomethylation leads to elevated mutation rates. Nature. 1998;395:89–93. doi: 10.1038/25779. [DOI] [PubMed] [Google Scholar]
- 16.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 17.Gaudet F, Hodgson JG, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–92. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 18.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;(Spec No 1):R65–76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 19.Houck CM, Rinehart FP, Schmid CW. A ubiquitous family of repeated DNA sequences in the human genome. J Mol Biol. 1979;132:289–306. doi: 10.1016/0022-2836(79)90261-4. [DOI] [PubMed] [Google Scholar]
- 20.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weisenberger DJ, Campan M, Long TI, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–36. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int J Epidemiol. 2012;41:126–39. doi: 10.1093/ije/dyq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi JY, James SR, Link PA, et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis. 2009;30:1889–97. doi: 10.1093/carcin/bgp143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gronniger E, Weber B, Heil O, et al. Aging and chronic sun exposure cause distinct epigenetic changes in human skin. PLoS Genet. 2010;6:e1000971. doi: 10.1371/journal.pgen.1000971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deshmukh RS, Ostrup O, Ostrup E, et al. DNA methylation in porcine preimplantation embryos developed in vivo and produced by in vitro fertilization, parthenogenetic activation and somatic cell nuclear transfer. Epigenetics. 2011;6:177–87. doi: 10.4161/epi.6.2.13519. [DOI] [PubMed] [Google Scholar]
- 26.Anier K, Malinovskaja K, Aonurm-Helm A, Zharkovsky A, Kalda A. DNA methylation regulates cocaine-induced behavioral sensitization in mice. Neuropsychopharmacology. 2010;35:2450–61. doi: 10.1038/npp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orta ML, Dominguez I, Pastor N, Cortes F, Mateos S. The role of the DNA hypermethylating agent Budesonide in the decatenating activity of DNA topoisomerase II. Mutat Res. 2010;694:45–52. doi: 10.1016/j.mrfmmm.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 28.Irizarry RA, Ladd-Acosta C, Wen B, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- 30.Suganuma T, Workman JL. Crosstalk among Histone Modifications. Cell. 2008;135:604–07. doi: 10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 31.Zheng YG, Wu J, Chen Z, Goodman M. Chemical regulation of epigenetic modifications: opportunities for new cancer therapy. Med Res Rev. 2008;28:645–87. doi: 10.1002/med.20120. [DOI] [PubMed] [Google Scholar]
- 32.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 33.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 34.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–32. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 35.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–59. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 37.Wang Z, Zang C, Rosenfeld JA, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–18. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- 39.Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005;6:838–49. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 40.Wysocka J, Allis CD, Coonrod S. Histone arginine methylation and its dynamic regulation. Front Biosci. 2006;11:344–55. doi: 10.2741/1802. [DOI] [PubMed] [Google Scholar]
- 41.Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–70. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh SK, Pal Bhadra M, Girschick HJ, Bhadra U. MicroRNAs–micro in size but macro in function. FEBS J. 2008;275:4929–44. doi: 10.1111/j.1742-4658.2008.06624.x. [DOI] [PubMed] [Google Scholar]
- 43.Matkovich SJ, Van Booven DJ, Eschenbacher WH, Dorn GW., 2nd RISC RNA sequencing for context-specific identification of in vivo microrna targets. Circ Res. 2010;108:18–26. doi: 10.1161/CIRCRESAHA.110.233528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams AE. Functional aspects of animal microRNAs. Cell Mol Life Sci. 2008;65:545–62. doi: 10.1007/s00018-007-7355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM, Zhang GZ. Biological functions of microRNAs: a review. J Physiol Biochem. 2011;67:129–39. doi: 10.1007/s13105-010-0050-6. [DOI] [PubMed] [Google Scholar]
- 46.Ying SY, Chang DC, Lin SL. The microRNA (miRNA): overview of the RNA genes that modulate gene function. Mol Biotechnol. 2008;38:257–68. doi: 10.1007/s12033-007-9013-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 48.Guil S, Esteller M. DNA methylomes, histone codes and miRNAs: tying it all together. Int J Biochem Cell Biol. 2009;41:87–95. doi: 10.1016/j.biocel.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 49.Backes C, Meese E, Lenhof HP, Keller A. A dictionary on microRNAs and their putative target pathways. Nucleic Acids Res. 2010;38:4476–86. doi: 10.1093/nar/gkq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho L, Fivecoat H, Wang J, Pasinetti GM. Alzheimer’s disease biomarker discovery in symptomatic and asymptomatic patients: experimental approaches and future clinical applications. Exp Gerontol. 2010;45:15–22. doi: 10.1016/j.exger.2009.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Provost P. Interpretation and applicability of microRNA data to the context of Alzheimer’s and age-related diseases. Aging. 2010;2:166–69. doi: 10.18632/aging.100131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Provost P. MicroRNAs as a molecular basis for mental retardation, Alzheimer’s and prion diseases. Brain Res. 2010;1338:58–66. doi: 10.1016/j.brainres.2010.03.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cheng Y, Zhang C. MicroRNA-21 in cardiovascular disease. J Cardiovasc Transl Res. 2010;3:251–55. doi: 10.1007/s12265-010-9169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Montgomery RL, van Rooij E. MicroRNA regulation as a therapeutic strategy for cardiovascular disease. Curr Drug Targets. 2010;11:936–42. doi: 10.2174/138945010791591368. [DOI] [PubMed] [Google Scholar]
- 55.Shen E, Diao X, Wei C, Wu Z, Zhang L, Hu B. MicroRNAs target gene and signaling pathway by bioinformatics analysis in the cardiac hypertrophy. Biochem Biophys Res Commun. 2010;397:380–85. doi: 10.1016/j.bbrc.2010.05.116. [DOI] [PubMed] [Google Scholar]
- 56.Swynghedauw B, Delcayre C, Samuel JL, Mebazaa A, Cohen-Solal A. Molecular mechanisms in evolutionary cardiology failure. Ann N Y Acad Sci. 2010;1188:58–67. doi: 10.1111/j.1749-6632.2009.05084.x. [DOI] [PubMed] [Google Scholar]
- 57.Fabbri M, Croce CM, Calin GA. MicroRNAs in the ontogeny of leukemias and lymphomas. Leuk Lymphoma. 2009;50:160–70. doi: 10.1080/10428190802535114. [DOI] [PubMed] [Google Scholar]
- 58.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–58. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 59.Olive V, Jiang I, He L. mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int J Biochem Cell Biol. 2010;42:1348–54. doi: 10.1016/j.biocel.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marcucci G, Radmacher MD, Mrozek K, Bloomfield CD. MicroRNA expression in acute myeloid leukemia. Curr Hematol Malig Rep. 2009;4:83–88. doi: 10.1007/s11899-009-0012-7. [DOI] [PubMed] [Google Scholar]
- 61.Motyckova G, Stone RM. The role of molecular tests in acute myelogenous leukemia treatment decisions. Curr Hematol Malig Rep. 2010;5:109–17. doi: 10.1007/s11899-010-0049-7. [DOI] [PubMed] [Google Scholar]
- 62.Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: tiny molecule, great function. Crit Rev Oncol Hematol. 2010;74:149–55. doi: 10.1016/j.critrevonc.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 63.Chen J, Xu X. Diet, epigenetic, and cancer prevention. Adv Genet. 2010;71:237–55. doi: 10.1016/B978-0-12-380864-6.00008-0. [DOI] [PubMed] [Google Scholar]
- 64.Garzon R, Marcucci G, Croce CM. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–89. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lin PY, Yu SL, Yang PC. MicroRNA in lung cancer. Br J Cancer. 2010;103:1144–48. doi: 10.1038/sj.bjc.6605901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mathers JC, Strathdee G, Relton CL. Induction of epigenetic alterations by dietary and other environmental factors. Adv Genet. 2010;71:3–39. doi: 10.1016/B978-0-12-380864-6.00001-8. [DOI] [PubMed] [Google Scholar]
- 67.Howard Hu. Human health and heavy metals exposure. In: McCally M, editor. Life Support: The Environment and Human Health. Boston: Massachusetts Institute of Technology; 2002. [Google Scholar]
- 68.Hemdan NY, Emmrich F, Faber S, Lehmann J, Sack U. Alterations of TH1/TH2 reactivity by heavy metals: possible consequences include induction of autoimmune diseases. Ann NY Acad Sci. 2007;1109:129–37. doi: 10.1196/annals.1398.015. [DOI] [PubMed] [Google Scholar]
- 69.Waalkes MP. Cadmium carcinogenesis. Mutat Res. 2003;533:107–20. doi: 10.1016/j.mrfmmm.2003.07.011. [DOI] [PubMed] [Google Scholar]
- 70.Salnikow K, Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. 2008;21:28–44. doi: 10.1021/tx700198a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang C, Ke Q, Costa M, Shi X. Molecular mechanisms of arsenic carcinogenesis. Mol Cell Biochem. 2004;255:57–66. doi: 10.1023/b:mcbi.0000007261.04684.78. [DOI] [PubMed] [Google Scholar]
- 72.Galaris D, Evangelou A. The role of oxidative stress in mechanisms of metal-induced carcinogenesis. Crit Rev Oncol Hematol. 2002;42:93–103. doi: 10.1016/s1040-8428(01)00212-8. [DOI] [PubMed] [Google Scholar]
- 73.Leonard SS, Bower JJ, Shi X. Metal-induced toxicity, carcinogenesis, mechanisms and cellular responses. Mol Cell Biochem. 2004;255:3–10. doi: 10.1023/b:mcbi.0000007255.72746.a6. [DOI] [PubMed] [Google Scholar]
- 74.Monks TJ, Xie R, Tikoo K, Lau SS. Ros-induced histone modifications and their role in cell survival and cell death. Drug Metab Rev. 2006;38:755–67. doi: 10.1080/03602530600959649. [DOI] [PubMed] [Google Scholar]
- 75.Donaldson K, Stone V, Borm PJ, et al. Oxidative stress and calcium signaling in the adverse effects of environmental particles (PM10) Free Radic Biol Med. 2003;34:1369–82. doi: 10.1016/s0891-5849(03)00150-3. [DOI] [PubMed] [Google Scholar]
- 76.Gilmour PS, Rahman I, Donaldson K, MacNee W. Histone acetylation regulates epithelial IL-8 release mediated by oxidative stress from environmental particles. Am J Physiol Lung Cell Mol Physiol. 2003;284:L533–40. doi: 10.1152/ajplung.00277.2002. [DOI] [PubMed] [Google Scholar]
- 77.Babar IA, Slack FJ, Weidhaas JB. miRNA modulation of the cellular stress response. Future Oncol. 2008;4:289–98. doi: 10.2217/14796694.4.2.289. [DOI] [PubMed] [Google Scholar]
- 78.Sakano K, Inagaki Y, Oikawa S, Hiraku Y, Kawanishi S. Copper-mediated oxidative DNA damage induced by eugenol: possible involvement of O-demethylation. Mutat Res. 2004;565:35–44. doi: 10.1016/j.mrgentox.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Galaris D, Skiada V, Barbouti A. Redox signaling and cancer: the role of “labile” iron. Cancer Lett. 2008;266:21–29. doi: 10.1016/j.canlet.2008.02.038. [DOI] [PubMed] [Google Scholar]
- 80.Reichard JF, Schnekenburger M, Puga A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem Biophys Res Commun. 2007;352:188–92. doi: 10.1016/j.bbrc.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Benbrahim-Tallaa L, Waterland RA, Styblo M, Achanzar WE, Webber MM, Waalkes MP. Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: aberrant genomic DNA methylation and K-ras oncogene activation. Toxicol Appl Pharmacol. 2005;206:288–98. doi: 10.1016/j.taap.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 82.Coppin JF, Qu W, Waalkes MP. Interplay between cellular methyl metabolism and adaptive efflux during oncogenic transformation from chronic arsenic exposure in human cells. J Biol Chem. 2008;283:19342–50. doi: 10.1074/jbc.M802942200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhao CQ, Young MR, Diwan BA, Coogan TP, Waalkes MP. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc Natl Acad Sci USA. 1997;94:10907–12. doi: 10.1073/pnas.94.20.10907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen H, Li S, Liu J, Diwan BA, Barrett JC, Waalkes MP. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: implications for arsenic hepatocarcinogenesis. Carcinogenesis. 2004;25:1779–86. doi: 10.1093/carcin/bgh161. [DOI] [PubMed] [Google Scholar]
- 85.Okoji RS, Yu RC, Maronpot RR, Froines JR. Sodium arsenite administration via drinking water increases genome-wide and Ha-ras DNA hypomethylation in methyl-deficient C57BL/6J mice. Carcinogenesis. 2002;23:777–85. doi: 10.1093/carcin/23.5.777. [DOI] [PubMed] [Google Scholar]
- 86.Uthus EO, Davis C. Dietary arsenic affects dimethylhydrazine-induced aberrant crypt formation and hepatic global DNA methylation and DNA methyltransferase activity in rats. Biol Trace Elem Res. 2005;103:133–45. doi: 10.1385/BTER:103:2:133. [DOI] [PubMed] [Google Scholar]
- 87.Xie Y, Trouba KJ, Liu J, Waalkes MP, Germolec DR. Biokinetics and subchronic toxic effects of oral arsenite, arsenate, monomethylarsonic acid, and dimethylarsinic acid in v-Ha-ras transgenic (Tg.AC) mice. Environ Health Perspect. 2004;112:1255–63. doi: 10.1289/txg.7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Majumdar S, Chanda S, Ganguli B, Mazumder DN, Lahiri S, Dasgupta UB. Arsenic exposure induces genomic hypermethylation. Environ Toxicol. 2010;25:315–18. doi: 10.1002/tox.20497. [DOI] [PubMed] [Google Scholar]
- 89.Pilsner JR, Liu X, Ahsan H, et al. Genomic methylation of peripheral blood leukocyte DNA: influences of arsenic and folate in Bangladeshi adults. Am J Clin Nutr. 2007;86:1179–86. doi: 10.1093/ajcn/86.4.1179. [DOI] [PubMed] [Google Scholar]
- 90.Chai CY, Huang YC, Hung WC, Kang WY, Chen WT. Arsenic salts induced autophagic cell death and hypermethylation of DAPK promoter in SV-40 immortalized human uroepithelial cells. Toxicol Lett. 2007;173:48–56. doi: 10.1016/j.toxlet.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 91.Fu HY, Shen JZ. Hypermethylation of CpG island of p16 gene and arsenic trioxide induced p16 gene demethylation in multiple myeloma. Zhonghua Nei Ke Za Zhi. 2005;44:411–14. [PubMed] [Google Scholar]
- 92.Jensen TJ, Novak P, Eblin KE, Gandolfi AJ, Futscher BW. Epigenetic remodeling during arsenical-induced malignant transformation. Carcinogenesis. 2008;29:1500–508. doi: 10.1093/carcin/bgn102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Mass MJ, Wang L. Arsenic alters cytosine methylation patterns of the promoter of the tumor suppressor gene p53 in human lung cells: a model for a mechanism of carcinogenesis. Mutat Res. 1997;386:263–77. doi: 10.1016/s1383-5742(97)00008-2. [DOI] [PubMed] [Google Scholar]
- 94.Chen H, Liu J, Zhao CQ, Diwan BA, Merrick BA, Waalkes MP. Association of c-myc overexpression and hyperproliferation with arsenite-induced malignant transformation. Toxicol Appl Pharmacol. 2001;175:260–68. doi: 10.1006/taap.2001.9253. [DOI] [PubMed] [Google Scholar]
- 95.Takahashi M, Barrett JC, Tsutsui T. Transformation by inorganic arsenic compounds of normal Syrian hamster embryo cells into a neoplastic state in which they become anchorage-independent and cause tumors in newborn hamsters. Int J Cancer. 2002;99:629–34. doi: 10.1002/ijc.10407. [DOI] [PubMed] [Google Scholar]
- 96.Cui X, Wakai T, Shirai Y, Hatakeyama K, Hirano S. Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol Sci. 2006;91:372–81. doi: 10.1093/toxsci/kfj159. [DOI] [PubMed] [Google Scholar]
- 97.Waalkes MP, Liu J, Chen H, et al. Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J Natl Cancer Inst. 2004;96:466–74. doi: 10.1093/jnci/djh070. [DOI] [PubMed] [Google Scholar]
- 98.Chanda S, Dasgupta UB, Guhamazumder D, et al. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol Sci. 2006;89:431–37. doi: 10.1093/toxsci/kfj030. [DOI] [PubMed] [Google Scholar]
- 99.Chen WT, Hung WC, Kang WY, Huang YC, Chai CY. Urothelial carcinomas arising in arsenic-contaminated areas are associated with hypermethylation of the gene promoter of the death-associated protein kinase. Histopathology. 2007;51:785–92. doi: 10.1111/j.1365-2559.2007.02871.x. [DOI] [PubMed] [Google Scholar]
- 100.Marsit CJ, Karagas MR, Danaee H, et al. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis. 2006;27:112–16. doi: 10.1093/carcin/bgi172. [DOI] [PubMed] [Google Scholar]
- 101.Zhang AH, Bin HH, Pan XL, Xi XG. Analysis of p16 gene mutation, deletion and methylation in patients with arseniasis produced by indoor unventilated-stove coal usage in Guizhou, China. J Toxicol Environ Health A. 2007;70:970–75. doi: 10.1080/15287390701290808. [DOI] [PubMed] [Google Scholar]
- 102.Boonchai W, Walsh M, Cummings M, Chenevix-Trench G. Expression of p53 in arsenic-related and sporadic basal cell carcinoma. Arch Dermatol. 2000;136:195–98. doi: 10.1001/archderm.136.2.195. [DOI] [PubMed] [Google Scholar]
- 103.Arrigo AP. Acetylation and methylation patterns of core histones are modified after heat or arsenite treatment of Drosophila tissue culture cells. Nucleic Acids Res. 1983;11:1389–404. doi: 10.1093/nar/11.5.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jo WJ, Ren X, Chu F, et al. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol Appl Pharmacol. 2009;241:294–302. doi: 10.1016/j.taap.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li J, Chen P, Sinogeeva N, et al. Arsenic trioxide promotes histone H3 phosphoacetylation at the chromatin of CASPASE-10 in acute promyelocytic leukemia cells. J Biol Chem. 2002;277:49504–10. doi: 10.1074/jbc.M207836200. [DOI] [PubMed] [Google Scholar]
- 106.Li J, Gorospe M, Barnes J, Liu Y. Tumor promoter arsenite stimulates histone H3 phosphoacetylation of proto-oncogenes c-fos and c-jun chromatin in human diploid fibroblasts. J Biol Chem. 2003;278:13183–91. doi: 10.1074/jbc.M300269200. [DOI] [PubMed] [Google Scholar]
- 107.Ramirez T, Brocher J, Stopper H, Hock R. Sodium arsenite modulates histone acetylation, histone deacetylase activity and HMGN protein dynamics in human cells. Chromosoma. 2008;117:147–57. doi: 10.1007/s00412-007-0133-5. [DOI] [PubMed] [Google Scholar]
- 108.Desrosiers R, Tanguay RM. Further characterization of the posttranslational modifications of core histones in response to heat and arsenite stress in Drosophila. Biochem Cell Biol. 1986;64:750–57. doi: 10.1139/o86-102. [DOI] [PubMed] [Google Scholar]
- 109.Desrosiers R, Tanguay RM. Methylation of Drosophila histones at proline, lysine, and arginine residues during heat shock. J Biol Chem. 1988;263:4686–92. [PubMed] [Google Scholar]
- 110.Zhou X, Sun H, Ellen TP, Chen H, Costa M. Arsenite alters global histone H3 methylation. Carcinogenesis. 2008;29:1831–36. doi: 10.1093/carcin/bgn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhou X, Li Q, Arita A, Sun H, Costa M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol Appl Pharmacol. 2009;236:78–84. doi: 10.1016/j.taap.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zykova TA, Zhu F, Lu C, et al. Lymphokine-activated killer T-cell-originated protein kinase phosphorylation of histone H2AX prevents arsenite-induced apoptosis in RPMI7951 melanoma cells. Clin Cancer Res. 2006;12:6884–93. doi: 10.1158/1078-0432.CCR-06-0410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–48. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 114.Binet F, Antoine F, Girard D. Interaction between arsenic trioxide and human primary cells: emphasis on human cells of myeloid origin. Inflamm Allergy Drug Targets. 2009;8:21–27. doi: 10.2174/187152809787582516. [DOI] [PubMed] [Google Scholar]
- 115.Cao Y, Yu SL, Wang Y, Guo GY, Ding Q, An RH. MicroRNA-dependent regulation of PTEN after arsenic trioxide treatment in bladder cancer cell line T24. Tumour Biol. 2011;32:179–88. doi: 10.1007/s13277-010-0111-z. [DOI] [PubMed] [Google Scholar]
- 116.Lee YW, Klein CB, Kargacin B, et al. Carcinogenic nickel silences gene-expression by chromatin condensation and DNA methylation - a new model for epigenetic carcinogens. Mol Cell Biol. 1995;15:2547–57. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lee YW, Klein CB, Kargacin B, et al. Carcinogenic nickel silences gene expression by chromatin condensation and DNA methylation: a new model for epigenetic carcinogens. Mol Cell Biol. 1995;15:2547–57. doi: 10.1128/mcb.15.5.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Klein CB, Conway K, Wang XW, et al. Senescence of nickel-transformed cells by an X chromosome: possible epigenetic control. Science. 1991;251:796–99. doi: 10.1126/science.1990442. [DOI] [PubMed] [Google Scholar]
- 119.Govindarajan B, Klafter R, Miller MS, et al. Reactive oxygen-induced carcinogenesis causes hypermethylation of p16(Ink4a) and activation of MAP kinase. Mol Med. 2002;8:1–8. [PMC free article] [PubMed] [Google Scholar]
- 120.Karaczyn AA, Golebiowski F, Kasprzak KS. Truncation, deamidation, and oxidation of histone H2B in cells cultured with nickel(II) Chem Res Toxicol. 2005;18:1934–42. doi: 10.1021/tx050122a. [DOI] [PubMed] [Google Scholar]
- 121.Karaczyn A, Ivanov S, Reynolds M, Zhitkovich A, Kasprzak KS, Salnikow K. Ascorbate depletion mediates up-regulation of hypoxia-associated proteins by cell density and nickel. J Cell Biochem. 2006;97:1025–35. doi: 10.1002/jcb.20705. [DOI] [PubMed] [Google Scholar]
- 122.Broday L, Peng W, Kuo MH, Salnikov K, Zoroddu M, Costa M. Nickel compounds are novel inhibitors of histone H4 acetylation. Cancer Res. 2000;60:238–41. [PubMed] [Google Scholar]
- 123.Chen H, Ke Q, Kluz T, Yan Y, Costa M. Nickel ions increase histone H3 lysine 9 dimethylation and induce transgene silencing. Mol Cell Biol. 2006;26:3728–37. doi: 10.1128/MCB.26.10.3728-3737.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ke QD, Davidson T, Chen HB, Kluz T, Costa M. Alterations of histone modifications and transgene silencing by nickel chloride. Carcinogenesis. 2006;27:1481–88. doi: 10.1093/carcin/bgl004. [DOI] [PubMed] [Google Scholar]
- 125.Golebiowski F, Kasprzak KS. Inhibition of core histones acetylation by carcinogenic nickel(II) Mol Cell Biochem. 2005;279:133–39. doi: 10.1007/s11010-005-8285-1. [DOI] [PubMed] [Google Scholar]
- 126.Klein CB, Costa M. DNA methylation, heterochromatin and epigenetic carcinogens. Mutat Res-Rev Mutat Res. 1997;386:163–80. doi: 10.1016/s1383-5742(96)00052-x. [DOI] [PubMed] [Google Scholar]
- 127.Klein CB, Conway K, Wang XW, et al. Senescence of nickel-transformed cells by an x-chromosome - possible epigenetic control. Science. 1991;251:796–99. doi: 10.1126/science.1990442. [DOI] [PubMed] [Google Scholar]
- 128.Yan Y, Kluz T, Zhang P, Chen HB, Costa M. Analysis of specific lysine histone H3 and H4 acetylation and methylation status in clones of cells with a gene silenced by nickel exposure. Toxicol Appl Pharmacol. 2003;190:272–77. doi: 10.1016/s0041-008x(03)00169-8. [DOI] [PubMed] [Google Scholar]
- 129.Zhang QW, Salnikow K, Kluz T, Chen LC, Su WC, Costa M. Inhibition and reversal of nickel-induced transformation by the histone deacetylase inhibitor trichostatin A. Toxicol Appl Pharmacol. 2003;192:201–11. doi: 10.1016/s0041-008x(03)00280-1. [DOI] [PubMed] [Google Scholar]
- 130.Broday L, Peng W, Kuo MH, Salnikow K, Zoroddu M, Costa M. Nickel compounds are novel inhibitors of histone H4 acetylation. Cancer Res. 2000;60:238–41. [PubMed] [Google Scholar]
- 131.Golebiowski F, Kasprzak KS. Inhibition of core histones acetylation by carcinogenic nickel(II) Mol Cell Biochem. 2005;279:133–39. doi: 10.1007/s11010-005-8285-1. [DOI] [PubMed] [Google Scholar]
- 132.Ke Q, Li Q, Ellen TP, Sun H, Costa M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis. 2008;29:1276–81. doi: 10.1093/carcin/bgn084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Huang D, Zhang Y, Qi Y, Chen C, Ji W. Global DNA hypomethylation, rather than reactive oxygen species (ROS), a potential facilitator of cadmium-stimulated K562 cell proliferation. Toxicol Lett. 2008;179:43–47. doi: 10.1016/j.toxlet.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 134.Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res. 2003;286:355–65. doi: 10.1016/s0014-4827(03)00062-4. [DOI] [PubMed] [Google Scholar]
- 135.Tellez-Plaza M, Navas-Acien A, Crainiceanu CM, Guallar E. Cadmium exposure and hypertension in the 1999–2004 National Health and Nutrition Examination Survey (NHANES) Environ Health Perspect. 2008;116:51–56. doi: 10.1289/ehp.10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bhatnagar A. Environmental cardiology - Studying mechanistic links between pollution and heart disease. Circulation Res. 2006;99:692–705. doi: 10.1161/01.RES.0000243586.99701.cf. [DOI] [PubMed] [Google Scholar]
- 137.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect. 2010;118:763–68. doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Williams AE, Perry MM, Moschos SA, Larner-Svensson HM, Lindsay MA. Role of miRNA-146a in the regulation of the innate immune response and cancer. Biochem Soc Trans. 2008;36(Pt 6):1211–15. doi: 10.1042/BST0361211. [DOI] [PubMed] [Google Scholar]
- 139.Pilsner JR, Lazarus AL, Nam DH, et al. Mercury-associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay: a sensitive method to study epigenetics in wildlife. Mol Ecol. 2010;19:307–14. doi: 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- 140.Arai Y, Ohgane J, Yagi S, et al. Epigenetic Assessment of environmental chemicals detected in maternal peripheral and cord blood samples. J Reprod Dev. 2011;57:507–17. doi: 10.1262/jrd.11-034a. [DOI] [PubMed] [Google Scholar]
- 141.Wright RO, Schwartz J, Wright RJ, et al. Biomarkers of lead exposure and DNA methylation within retrotransposons. Environ Health Perspect. 2010;118:790–95. doi: 10.1289/ehp.0901429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Pilsner JR, Hu H, Ettinger A, et al. Influence of prenatal lead exposure on genomic methylation of cord blood DNA. Environ Health Perspect. 2009;117:1466–71. doi: 10.1289/ehp.0800497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kondo K, Takahashi Y, Hirose Y, et al. The reduced expression and aberrant methylation of p16(INK4a) in chromate workers with lung cancer. Lung Cancer. 2006;53:295–302. doi: 10.1016/j.lungcan.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 144.Takahashi Y, Kondo K, Hirose T, et al. Microsatellite instability and protein expression of the DNA mismatch repair gene, hMLH1, of lung cancer in chromate-exposed workers. Mol Carcinogenesis. 2005;42:150–58. doi: 10.1002/mc.20073. [DOI] [PubMed] [Google Scholar]
- 145.Frouin H, Fortier M, Fournier M. Toxic effects of various pollutants in 11B7501 lymphoma B cell line from harbour seal (Phoca vitulina) Toxicology. 2010;270:66–76. doi: 10.1016/j.tox.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 146.Apostoli P, Catalani S. Mechanisms of action for metallic elements and their species classified carcinogen R 45 and R 49 by EU. G Ital Med Lav Ergon. 2008;30:382–91. [PubMed] [Google Scholar]
- 147.Schnekenburger M, Talaska G, Puga A. Chromium cross-links histone deacetylase 1-DNA methyltransferase 1 complexes to chromatin, inhibiting histone-remodeling marks critical for transcriptional activation. Mol Cell Biol. 2007;27:7089–101. doi: 10.1128/MCB.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Sun H, Zhou X, Chen H, Li Q, Costa M. Modulation of histone methylation and MLH1 gene silencing by hexavalent chromium. Toxicol Appl Pharmacol. 2009;237:258–66. doi: 10.1016/j.taap.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Pogue AI, Li YY, Cui JG, et al. Characterization of an NF-kappaB-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;103:1591–95. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 150.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–69. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kim MS, Lee J, Sidransky D. DNA methylation markers in colorectal cancer. Cancer Metastasis Rev. 2010;29:181–206. doi: 10.1007/s10555-010-9207-6. [DOI] [PubMed] [Google Scholar]
- 153.Markowitz SD, Bertagnolli MM. Molecular origins of cancer: Molecular basis of colorectal cancer. N Engl J Med. 2009;361:2449–60. doi: 10.1056/NEJMra0804588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–69. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Guerrero-Bosagna C, Settles M, Lucker B, Skinner MK. Epigenetic transgenerational actions of vinclozolin on promoter regions of the sperm epigenome. PLoS One. 2010;5:e13100. doi: 10.1371/journal.pone.0013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Anway MD, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors. Endocrinology. 2006;147(6 Suppl):S43–49. doi: 10.1210/en.2005-1058. [DOI] [PubMed] [Google Scholar]
- 157.Zama AM, Uzumcu M. Fetal and neonatal exposure to the endocrine disruptor methoxychlor causes epigenetic alterations in adult ovarian genes. Endocrinology. 2009;150:4681–91. doi: 10.1210/en.2009-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Hypomethylation and overexpression of c-jun and c-myc protooncogenes and increased DNA methyltransferase activity in dichloroacetic and trichloroacetic acid-promoted mouse liver tumors. Cancer Lett. 2000;158:185–93. doi: 10.1016/s0304-3835(00)00518-8. [DOI] [PubMed] [Google Scholar]
- 159.Tao L, Yang S, Xie M, Kramer PM, Pereira MA. Effect of trichloroethylene and its metabolites, dichloroacetic acid and trichloroacetic acid, on the methylation and expression of c-Jun and c-Myc protooncogenes in mouse liver: prevention by methionine. Toxicol Sci. 2000;54:399–407. doi: 10.1093/toxsci/54.2.399. [DOI] [PubMed] [Google Scholar]
- 160.Hathaway G, Proctor N, Hughes J, Fischman M. Proctor And Hughes’ Chemical Hazards of the Workplace. New York: Van Nostrand Reinhold; 1991. [Google Scholar]
- 161.Rusiecki JA, Baccarelli A, Bollati V, Tarantini L, Moore LE, Bonefeld-Jorgensen EC. Global DNA hypomethylation is associated with high serum-persistent organic pollutants in Greenlandic Inuit. Environ Health Perspect. 2008;116:1547–52. doi: 10.1289/ehp.11338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Kim KY, Kim DS, Lee SK, et al. Association of low-dose exposure to persistent organic pollutants with global DNA hypomethylation in healthy koreans. Environ Health Perspect. 2010;118:370–74. doi: 10.1289/ehp.0901131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Alavanja MC, Bonner MR. Pesticides and human cancers. Cancer Invest. 2005;23:700–11. doi: 10.1080/07357900500360008. [DOI] [PubMed] [Google Scholar]
- 164.Weichenthal S, Moase C, Chan P. A review of pesticide exposure and cancer incidence in the Agricultural Health Study cohort. Environ Health Perspect. 2010;118:1117–25. doi: 10.1289/ehp.0901731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Alavanja MC, Ward MH, Reynolds P. Carcinogenicity of agricultural pesticides in adults and children. J Agromedicine. 2007;12:39–56. doi: 10.1300/J096v12n01_05. [DOI] [PubMed] [Google Scholar]
- 166.Bassil KL, Vakil C, Sanborn M, Cole DC, Kaur JS, Kerr KJ. Cancer health effects of pesticides: systematic review. Can Fam Physician. 2007;53:1704–11. [PMC free article] [PubMed] [Google Scholar]
- 167.Koutros S, Alavanja MC, Lubin JH, et al. An update of cancer incidence in the Agricultural Health Study. J Occup Environ Med. 2010;52:1098–105. doi: 10.1097/JOM.0b013e3181f72b7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Waggoner JK, Kullman GJ, Henneberger PK, et al. Mortality in the agricultural health study, 1993–2007. Am J Epidemiol. 2011;173:71–83. doi: 10.1093/aje/kwq323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77:621–32. doi: 10.1124/mol.109.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Song C, Kanthasamy A, Anantharam V, Sun F, Kanthasamy AG. Environmental neurotoxic pesticide increases histone acetylation to promote apoptosis in dopaminergic neuronal cells: relevance to epigenetic mechanisms of neurodegeneration. Mol Pharmacol. 2010;77:621–32. doi: 10.1124/mol.109.062174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Baccarelli A, Cassano PA, Litonjua A, et al. Cardiac autonomic dysfunction - effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation. 2008;117:1802–09. doi: 10.1161/CIRCULATIONAHA.107.726067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Jardim MJ, Fry RC, Jaspers I, Dailey L, Diaz-Sanchez D. Disruption of microRNA expression in human airway cells by diesel exhaust particles is linked to tumorigenesis-associated pathways. Environ Health Perspect. 2009;117:1745–51. doi: 10.1289/ehp.0900756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tarantini L, Bonzini M, Apostoli P, et al. Effects of particulate matter on genomic DNA methylation content and iNOS promoter methylation. Environ Health Perspect. 2009;117:217–22. doi: 10.1289/ehp.11898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Yauk C, Polyzos A, Rowan-Carroll A, et al. Germ-line mutations, DNA damage, and global hypermethylation in mice exposed to particulate air pollution in an urban/industrial location. Proc Natl Acad Sci USA. 2008;105:605–10. doi: 10.1073/pnas.0705896105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Liu J, Ballaney M, Al-alem U, et al. Combined inhaled diesel exhaust particles and allergen exposure alter methylation of T helper genes and IgE production in vivo. Toxicol Sci. 2008;102:76–81. doi: 10.1093/toxsci/kfm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Perera F, Tang WY, Herbstman J, et al. Relation of DNA methylation of 5′-CpG island of ACSL3 to transplacental exposure to airborne polycyclic aromatic hydrocarbons and childhood asthma. PLoS One. 2009;4:e4488. doi: 10.1371/journal.pone.0004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cantone L, Nordio F, Hou L, et al. Effects of Inhalable Metal-rich Air Particles on Histone H3K4 Dimethylation and H3K9 Acetylation in a Cross-sectional Study of Steel Workers. Environ Health Perspect. 2011;119:964–9. doi: 10.1289/ehp.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bollati V, Marinelli B, Apostoli P, et al. Exposure to metal-rich particulate matter modifies the expression of candidate micrornas in peripheral blood leukocytes. Environ Health Perspect. 2010 doi: 10.1289/ehp.0901300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Smith MT. Advances in understanding benzene health effects and susceptibility. Annu Rev Public Health. 2010;31:133–48. doi: 10.1146/annurev.publhealth.012809.103646. 2 p following 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Rinsky RA, Hornung RW, Silver SR, Tseng CY. Benzene exposure and hematopoietic mortality: a long-term epidemiologic risk assessment. Am J Ind Med. 2002;42:474–80. doi: 10.1002/ajim.10138. [DOI] [PubMed] [Google Scholar]
- 181.Goldstein BD. Benzene toxicity. Occup Med. 1988;3:541–54. [PubMed] [Google Scholar]
- 182.Whitworth KW, Symanski E, Coker AL. Childhood lymphohematopoietic cancer incidence and hazardous air pollutants in southeast Texas, 1995–2004. Environ Health Perspect. 2008;116:1576–80. doi: 10.1289/ehp.11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Overall evaluations of carcinogenicity: an updating of IARC Monographs volumes 1 to 42. IARC Monogr Eval Carcinog Risks Hum Suppl. 1987;7:1–440. [PubMed] [Google Scholar]
- 184.Richardson DB. Temporal variation in the association between benzene and leukemia mortality. Environ Health Perspect. 2008;116:370–74. doi: 10.1289/ehp.10841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.CDC. Facts About Benzene. 2005 Available from: http://www.bt.cdc.gov/agent/benzene/basics/facts.asp. [Google Scholar]
- 186.Bollati V, Baccarelli A, Hou L, et al. Changes in DNA methylation patterns in subjects exposed to low-dose benzene. Cancer Res. 2007;67:876–80. doi: 10.1158/0008-5472.CAN-06-2995. [DOI] [PubMed] [Google Scholar]
- 187.Ji Z, Zhang L, Peng V, Ren X, McHale CM, Smith MT. A comparison of the cytogenetic alterations and global DNA hypomethylation induced by the benzene metabolite, hydroquinone, with those induced by melphalan and etoposide. Leukemia. 2010;24:986–91. doi: 10.1038/leu.2010.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Gao A, Zuo X, Liu Q, Lu X, Guo W, Tian L. Methylation of PARP-1 promoter involved in the regulation of benzene-induced decrease of PARP-1 mRNA expression. Toxicol Lett. 2010;195:114–18. doi: 10.1016/j.toxlet.2010.03.005. [DOI] [PubMed] [Google Scholar]
- 189.Keri RA, Ho SM, Hunt PA, Knudsen KE, Soto AM, Prins GS. An evaluation of evidence for the carcinogenic activity of bisphenol A. Reprod Toxicol. 2007;24:240–52. doi: 10.1016/j.reprotox.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ho SM, Tang WY, de Frausto JB, Prins GS. Developmental exposure to estradiol and bisphenol A increases susceptibility to prostate carcinogenesis and epigenetically regulates phosphodiesterase type 4 variant 4. Cancer Res. 2006;66:5624–32. doi: 10.1158/0008-5472.CAN-06-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.vom Saal FS, Hughes C. An extensive new literature concerning low-dose effects of bisphenol A shows the need for a new risk assessment. Environ Health Perspect. 2005;113:926–33. doi: 10.1289/ehp.7713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci USA. 2007;104:13056–61. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Waterland RA. Is epigenetics an important link between early life events and adult disease? Horm Res. 2009;71(Suppl 1):13–16. doi: 10.1159/000178030. [DOI] [PubMed] [Google Scholar]
- 194.Bromer JG, Zhou Y, Taylor MB, Doherty L, Taylor HS. Bisphenol-A exposure in utero leads to epigenetic alterations in the developmental programming of uterine estrogen response. FASEB J. 2010;24:2273–80. doi: 10.1096/fj.09-140533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Weng YI, Hsu PY, Liyanarachchi S, et al. Epigenetic influences of low-dose bisphenol A in primary human breast epithelial cells. Toxicol Appl Pharmacol. 2010;248:111–21. doi: 10.1016/j.taap.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Avissar-Whiting M, Veiga KR, Uhl KM, et al. Bisphenol A exposure leads to specific microRNA alterations in placental cells. Reprod Toxicol. 2010;29:401–06. doi: 10.1016/j.reprotox.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Beedanagari SR, Taylor RT, Bui P, Wang F, Nickerson DW, Hankinson O. Role of epigenetic mechanisms in differential regulation of the dioxin-inducible human CYP1A1 and CYP1B1 genes. Mol Pharmacol. 2010;78:608–16. doi: 10.1124/mol.110.064899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wang J, Zhao YY, Liu H, et al. The role of insulin-like growth factor-2 gene differentially methylated regions in TCDD-induced malformation. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2007;24:162–66. [PubMed] [Google Scholar]
- 199.McClure EA, North CM, Kaminski NE, Goodman JI. Changes in DNA methylation and gene expression during 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD)-induced suppression of the lipopolysaccharide (LPS)-stimulated IgM response in splenocytes. Toxicol Sci. 2011;120:339–48. doi: 10.1093/toxsci/kfq396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Elyakim E, Sitbon E, Faerman A, et al. hsa-miR-191 is a candidate oncogene target for hepatocellular carcinoma therapy. Cancer Res. 2010;70:8077–87. doi: 10.1158/0008-5472.CAN-10-1313. [DOI] [PubMed] [Google Scholar]
- 201.Beller HR, Tiemeier K. Use of liquid chromatography/tandem mass spectrometry to detect distinctive indicators of in situ RDX transformation in contaminated groundwater. Environ Sci Techno. 2002;36:2060–66. doi: 10.1021/es0157696. [DOI] [PubMed] [Google Scholar]
- 202.Zhang B, Pan X. RDX induces aberrant expression of microRNAs in mouse brain and liver. Environ Health Perspect. 2009;117:231–40. doi: 10.1289/ehp.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Laitman CJ. DES exposure and the aging woman: mothers and daughters. Curr Womens Health Rep. 2002;2:390–93. [PubMed] [Google Scholar]
- 204.Palmer JR, Hatch EE, Rosenberg CL, et al. Risk of breast cancer in women exposed to diethylstilbestrol in utero: prelimiinary results (United States) Cancer Causes Control. 2002;13:753–58. doi: 10.1023/a:1020254711222. [DOI] [PubMed] [Google Scholar]
- 205.Hsu PY, Deatherage DE, Rodriguez BA, et al. Xenoestrogen-induced epigenetic repression of microRNA-9-3 in breast epithelial cells. Cancer Res. 2009;69:5936–45. doi: 10.1158/0008-5472.CAN-08-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Mages F, Macovschi O, Prigent AF, Fonlupt P. Increased methylation of chloroform extractable products and CTP: cholinephosphate cytidylyltransferase in brain membrane preparations from triethyltin-intoxicated rats. Pharmacol Toxicol. 1989;65:302–05. doi: 10.1111/j.1600-0773.1989.tb01178.x. [DOI] [PubMed] [Google Scholar]
- 207.Pereira MA, Kramer PM, Conran PB, Tao L. Effect of chloroform on dichloroacetic acid and trichloroacetic acid-induced hypomethylation and expression of the c-myc gene and on their promotion of liver and kidney tumors in mice. Carcinogenesis. 2001;22:1511–19. doi: 10.1093/carcin/22.9.1511. [DOI] [PubMed] [Google Scholar]
- 208.Coffin JC, Ge R, Yang S, Kramer PM, Tao L, Pereira MA. Effect of trihalomethanes on cell proliferation and DNA methylation in female B6C3F1 mouse liver. Toxicol Sci. 2000;58:243–52. doi: 10.1093/toxsci/58.2.243. [DOI] [PubMed] [Google Scholar]
- 209.Chlorinated drinking-water. IARC Monogr Eval Carcinog Risks Hum. 1991;52:45–141. [PMC free article] [PubMed] [Google Scholar]
- 210.Uden PC, Miller JW. Chlorinated acids and chloral in drinking water. J Am Water Works Assoc. 1983;75:524–27. [Google Scholar]
- 211.Coleman WE, Munch JW, Kaylor WH, Streicher RP, Ringhand HP, Meier JR. Gas chromatography/mass spectroscopy analysis of mutagenic extracts of aqueous chlorinated humic acid. A comparison of the by-products to drinking water contaminants Environ. Sci. Technol. 1984;18:674–78. [Google Scholar]
- 212.Shi L, Wu J. Epigenetic regulation in mammalian preimplantation embryo development. Reprod Biol Endocrinol. 2009;7:59–69. doi: 10.1186/1477-7827-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Morgan HD, Sutherland HG, Martin DI, Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet. 1999;23:314–18. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 214.Heijmans BT, Tobi EW, Stein AD, et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci USA. 2008;105:17046–49. doi: 10.1073/pnas.0806560105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Tobi EW, Lumey LH, Talens RP, et al. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum Mol Genet. 2009;18:4046–53. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Heijmans BT, Tobi EW, Lumey LH, Slagboom PE. The epigenome: archive of the prenatal environment. Epigenetics. 2009;4:526–31. doi: 10.4161/epi.4.8.10265. [DOI] [PubMed] [Google Scholar]
- 217.Terry MB, Ferris JS, Pilsner R, et al. Genomic DNA methylation among women in a multiethnic New York City birth cohort. Cancer Epidemiol Biomarkers Prev. 2008;17:2306–10. doi: 10.1158/1055-9965.EPI-08-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Flom J, Ferris J, Gonzalez K, Santella R, Terry M. Prenatal tobacco smoke exposure and genomewide methylation in adulthood. Cancer Epidemiol Biomarkers Prev. 2011;20:720–21. doi: 10.1158/1055-9965.EPI-11-0553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Guerrero-Preston R, Goldman LR, Brebi-Mieville P, et al. Global DNA hypomethylation is associated with in utero exposure to cotinine and perfluorinated alkyl compounds. Epigenetics. 2010;5:539–46. doi: 10.4161/epi.5.6.12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Maccani MA, Avissar-Whiting M, Banister CE, McGonnigal B, Padbury JF, Marsit CJ. Maternal cigarette smoking during pregnancy is associated with downregulation of miR-16, miR-21, and miR-146a in the placenta. Epigenetics. 2010;5:583–89. doi: 10.4161/epi.5.7.12762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Minard ME, Jain AK, Barton MC. Analysis of epigenetic alterations to chromatin during development. Genesis. 2009;47:559–72. doi: 10.1002/dvg.20534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Relton CL, Davey Smith G. Epigenetic epidemiology of common complex disease: prospects for prediction, prevention, and treatment. PLoS Med. 2010;7:e1000356. doi: 10.1371/journal.pmed.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Geneletti S, Gallo V, Porta M, Khoury MJ, Vineis P. Assessing causal relationships in genomics: From Bradford-Hill criteria to complex gene-environment interactions and directed acyclic graphs. Emerg Themes Epidemiol. 2011;8:5–22. doi: 10.1186/1742-7622-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–95. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 225.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 226.Sciandrello G, Caradonna F, Mauro M, Barbata G. Arsenic-induced DNA hypomethylation affects chromosomal instability in mammalian cells. Carcinogenesis. 2004;25:413–17. doi: 10.1093/carcin/bgh029. [DOI] [PubMed] [Google Scholar]
- 227.Smith IM, Mydlarz WK, Mithani SK, Califano JA. DNA global hypomethylation in squamous cell head and neck cancer associated with smoking, alcohol consumption and stage. Int J Cancer. 2007;121:1724–28. doi: 10.1002/ijc.22889. [DOI] [PubMed] [Google Scholar]
- 228.Roman-Gomez J, Jimenez-Velasco A, Agirre X, et al. Promoter hypermethylation and global hypomethylation are independent epigenetic events in lymphoid leukemogenesis with opposing effects on clinical outcome. Leukemia. 2006;20:1445–48. doi: 10.1038/sj.leu.2404257. [DOI] [PubMed] [Google Scholar]
- 229.Deng G, Nguyen A, Tanaka H, et al. Regional hypermethylation and global hypomethylation are associated with altered chromatin conformation and histone acetylation in colorectal cancer. Int J Cancer. 2006;118:2999–3005. doi: 10.1002/ijc.21740. [DOI] [PubMed] [Google Scholar]
- 230.Brothman AR, Swanson G, Maxwell TM, et al. Global hypomethylation is common in prostate cancer cells: a quantitative predictor for clinical outcome? Cancer Genet Cytogenet. 2005;156:31–36. doi: 10.1016/j.cancergencyto.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 231.Shimabukuro M, Sasaki T, Imamura A, et al. Global hypomethylation of peripheral leukocyte DNA in male patients with schizophrenia: a potential link between epigenetics and schizophrenia. J Psychiatr Res. 2007;41:1042–46. doi: 10.1016/j.jpsychires.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 232.Bagnyukova TV, Luzhna LI, Pogribny IP, Lushchak VI. Oxidative stress and antioxidant defenses in goldfish liver in response to short-term exposure to arsenite. Environ Mol Mutagen. 2007;48:658–65. doi: 10.1002/em.20328. [DOI] [PubMed] [Google Scholar]
- 233.Pilsner JR, Liu X, Ahsan H, et al. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ Health Perspect. 2009;117:254–60. doi: 10.1289/ehp.11872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Cheetham S, Tang MJ, Mesak F, Kennecke H, Owen D, Tai IT. SPARC promoter hypermethylation in colorectal cancers can be reversed by 5-Aza-2′ deoxycytidine to increase SPARC expression and improve therapy response. British J Cancer. 2008;98:1810–19. doi: 10.1038/sj.bjc.6604377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Alemayehu A, Sebova K, Fridrichova I. Redundant DNA methylation in colorectal cancers of Lynch-syndrome patients. Genes Chrom Cancer. 2008;47:906–14. doi: 10.1002/gcc.20586. [DOI] [PubMed] [Google Scholar]
- 236.Norrie MWA, Hawkins NJ, Todd AV, Meagher AP, O’Connor TW, Ward RL. The role of hMLH1 methylation in the development of synchronous sporadic colorectal carcinomas. Dis Colon Rectum. 2002;45:674–80. doi: 10.1007/s10350-004-6266-1. [DOI] [PubMed] [Google Scholar]
- 237.Minardi D, Lucarini G, Filosa A, et al. Prognostic role of global DNA-methylation and histone acetylation in pT1a clear cell renal carcinoma in partial nephrectomy specimens. J Cell Mol Med. 2009;13:2115–21. doi: 10.1111/j.1582-4934.2008.00482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Schafer E, Irizarry R, Negi S, et al. Promoter hypermethylation in MLL-r infant acute lymphoblastic leukemia: biology and therapeutic targeting. Blood. 2010;115:4798–809. doi: 10.1182/blood-2009-09-243634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Owen HC, Giedl J, Wild PJ, et al. Low frequency of epigenetic events in urothelial tumors in young patients. J Urol. 2010;184:459–63. doi: 10.1016/j.juro.2010.03.131. [DOI] [PubMed] [Google Scholar]
- 240.Qian J, Yao DM, Lin J, et al. Alteration of methylation status of death-associated protein kinase (dapk) gene promoter in patients with acute myeloid leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2010;18:1390–94. [PubMed] [Google Scholar]
- 241.Laytragoon-Lewin N, Chen F, Castro J, et al. DNA content and methylation of p16, DAPK and RASSF1A gene in tumour and distant, normal mucosal tissue of head and neck squamous cell carcinoma patients. Anticancer Res. 2010;30:4643–48. [PubMed] [Google Scholar]
- 242.Paluszczak J, Misiak P, Wierzbicka M, Wozniak A, Baer-Dubowska W. Frequent hypermethylation of DAPK, RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell carcinomas and adjacent normal mucosa. Oral Oncol. 2011;47:104–7. doi: 10.1016/j.oraloncology.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 243.Hafner N, Diebolder H, Jansen L, Hoppe I, Durst M, Runnebaum IB. Hypermethylated DAPK in serum DNA of women with uterine leiomyoma is a biomarker not restricted to cancer. Gynecol Oncol. 2011;121:224–9. doi: 10.1016/j.ygyno.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 244.Li B, Wang B, Niu LJ, Jiang L, Qiu CC. Hypermethylation of multiple tumor-related genes associated with DMNT3b up-regulation served as a biomarker for early diagnosis of esophageal squamous cell carcinoma. Epigenetics. 2011;6:307–16. doi: 10.4161/epi.6.3.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Ben Ayed-Guerfali D, Benhaj K, Khabir A, et al. Hypermethylation of tumor-related genes in tunisian patients with gastric carcinoma: Clinical and biological significance. J Surg Oncol. 2011;103:687–94. doi: 10.1002/jso.21875. [DOI] [PubMed] [Google Scholar]
- 246.Sugita H, Iida S, Inokuchi M, et al. Methylation of BNIP3 and DAPK indicates lower response to chemotherapy and poor prognosis in gastric cancer. Oncol Rep. 2011;25:513–18. doi: 10.3892/or.2010.1085. [DOI] [PubMed] [Google Scholar]
- 247.Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303:21–8. doi: 10.1016/j.canlet.2010.12.011. [DOI] [PubMed] [Google Scholar]
- 248.Hu SL, Kong XY, Cheng ZD, et al. Promoter methylation of p16, Runx3, DAPK and CHFR genes is frequent in gastric carcinoma. Tumori. 2010;96:726–33. doi: 10.1177/030089161009600515. [DOI] [PubMed] [Google Scholar]
- 249.Van der Auwera I, Bovie C, Svensson C, et al. Quantitative methylation profiling in tumor and matched morphologically normal tissues from breast cancer patients. BMC Cancer. 2010;10:97–104. doi: 10.1186/1471-2407-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Zhang C, Jin Y, Xu H, et al. Relationship between promoter methylation of p16, DAPK and RAR beta genes and the clinical data of non-small cell lung cancer. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2011;28:23–28. doi: 10.3760/cma.j.issn.1003-9406.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 251.Peng Z, Shan C, Wang H. Value of promoter methylation of RASSF1A, p16, and DAPK genes in induced sputum in diagnosing lung cancers. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2010;35:247–53. doi: 10.3969/j.issn.1672-7347.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 252.Malhotra P, Kochhar R, Vaiphei K, Wig JD, Mahmood S. Aberrant promoter methylation of p16 in colorectal adenocarcinoma in North Indian patients. World J Gastrointest Oncol. 2010;2:295–303. doi: 10.4251/wjgo.v2.i7.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Poetsch M, Hemmerich M, Kakies C, et al. Alterations in the tumor suppressor gene p16 (INK4A) are associated with aggressive behavior of penile carcinomas. Virchows Arch. 2011;458:221–29. doi: 10.1007/s00428-010-1007-4. [DOI] [PubMed] [Google Scholar]
- 254.Lin HH, Ke HL, Wu WJ, Lee YH, Chang LL. Hypermethylation of E-cadherin, p16, p14, and RASSF1A genes in pathologically normal urothelium predict bladder recurrence of bladder cancer after transurethral resection. Urol Oncol. 2010 doi: 10.1016/j.urolonc.2010.01.002. in press. [DOI] [PubMed] [Google Scholar]
- 255.Wang D, Wang J, Li Y, He Z, Zhang Y. The influence of anthracosis and p16 ink4a gene aberrant methylation on small-sized pulmonary adenocarcinoma. Exp Mol Pathol. 2011;90:131–36. doi: 10.1016/j.yexmp.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 256.Zainuddin N, Kanduri M, Berglund M, et al. Quantitative evaluation of p16(INK4a) promoter methylation using pyrosequencing in de novo diffuse large B-cell lymphoma. Leuk Res. 2011;35:438–43. doi: 10.1016/j.leukres.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 257.Shaw R, Risk JM, Liloglou T. RE: P16 INK41 promoter hypermethylation is associated with invasiveness and prognosis of oral squamous cell carcinoma in an age dependent manner. Su et al. Oral Oncol. 2010;46:734–39. doi: 10.1016/j.oraloncology.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 258.Serizawa RR, Ralfkiaer U, Steven K, et al. Integrated genetic and epigenetic analysis of bladder cancer reveals an additive diagnostic value of FGFR3 mutations and hypermethylation events. Int J Cancer. 2011;129:78–87. doi: 10.1002/ijc.25651. [DOI] [PubMed] [Google Scholar]
- 259.Hill VK, Hesson LB, Dansranjavin T, et al. Identification of 5 novel genes methylated in breast and other epithelial cancers. Mol Cancer. 2010;9:51–63. doi: 10.1186/1476-4598-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Gronbaek K, Ralfkiaer U, Dahl C, et al. Frequent hypermethylation of DBC1 in malignant lymphoproliferative neoplasms. Mod Pathol. 2008;21:632–38. doi: 10.1038/modpathol.2008.27. [DOI] [PubMed] [Google Scholar]
- 261.Radpour R, Barekati Z, Haghighi MM, et al. Correlation of telomere length shortening with promoter methylation profile of p16/Rb and p53/p21 pathways in breast cancer. Mod Pathol. 2010;23:763–72. doi: 10.1038/modpathol.2009.195. [DOI] [PubMed] [Google Scholar]
- 262.Hanafusa T, Shinji T, Shiraha H, et al. Functional promoter upstream p53 regulatory sequence of IGFBP3 that is silenced by tumor specific methylation. BMC Cancer. 2005;5:9–19. doi: 10.1186/1471-2407-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Luo J, Li YN, Wang F, Zhang WM, Geng X. S-adenosylmethionine inhibits the growth of cancer cells by reversing the hypomethylation status of c-myc and H-ras in human gastric cancer and colon cancer. Int J Biol Sci. 2010;6:784–95. doi: 10.7150/ijbs.6.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fang JY, Zhu SS, Xiao SD, et al. Studies on the hypomethylation of c-myc, c-Ha-ras oncogenes and histopathological changes in human gastric carcinoma. J Gastroenterol Hepatol. 1996;11:1079–82. doi: 10.1111/j.1440-1746.1996.tb00040.x. [DOI] [PubMed] [Google Scholar]
- 265.Tsujiuchi T, Tsutsumi M, Sasaki Y, Takahama M, Konishi Y. Hypomethylation of CpG sites and c-myc gene overexpression in hepatocellular carcinomas, but not hyperplastic nodules, induced by a choline-deficient L-amino acid-defined diet in rats. Jpn J Cancer Res. 1999;90:909–13. doi: 10.1111/j.1349-7006.1999.tb00834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Shen L, Qui D, Fang J. Correlation between hypomethylation of c-myc and c-N-ras oncogenes and pathological changes in human hepatocellular carcinoma. Zhonghua Zhong Liu Za Zhi. 1997;19:173–76. [PubMed] [Google Scholar]
- 267.Del Senno L, Maestri I, Piva R, et al. Differential hypomethylation of the c-myc protooncogene in bladder cancers at different stages and grades. J Urol. 1989;142:146–49. doi: 10.1016/s0022-5347(17)38700-1. [DOI] [PubMed] [Google Scholar]
- 268.Rabiau N, Thiam MO, Satih S, et al. Methylation analysis of BRCA1, RASSF1, GSTP1 and EPHB2 promoters in prostate biopsies according to different degrees of malignancy. In Vivo. 2009;23:387–91. [PubMed] [Google Scholar]
- 269.Buckingham L, Penfield Faber L, Kim A, et al. PTEN, RASSF1 and DAPK site-specific hypermethylation and outcome in surgically treated stage I and II nonsmall cell lung cancer patients. Int J Cancer. 2010;126:1630–39. doi: 10.1002/ijc.24896. [DOI] [PubMed] [Google Scholar]
- 270.Zhang A, Feng H, Yang G, et al. Unventilated indoor coal-fired stoves in Guizhou province, China: cellular and genetic damage in villagers exposed to arsenic in food and air. Environ Health Perspect. 2007;115:653–8. doi: 10.1289/ehp.9272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Zhong CX, Mass MJ. Both hypomethylation and hypermethylation of DNA associated with arsenite exposure in cultures of human cells identified by methylation-sensitive arbitrarily-primed PCR. Toxicol Lett. 2001;122:223–34. doi: 10.1016/s0378-4274(01)00365-4. [DOI] [PubMed] [Google Scholar]
- 272.Kanao K, Mikami S, Mizuno R, Shinojima T, Murai M, Oya M. Decreased acetylation of histone H3 in renal cell carcinoma: a potential target of histone deacetylase inhibitors. J Urol. 2008;180:1131–36. doi: 10.1016/j.juro.2008.04.136. [DOI] [PubMed] [Google Scholar]
- 273.Jo WJ, Ren XF, Chu FX, et al. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol Appl Pharmacol. 2009;241:294–302. doi: 10.1016/j.taap.2009.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Sayyed SG, Gaikwad AB, Lichtnekert J, et al. Progressive glomerulosclerosis in type 2 diabetes is associated with renal histone H3K9 and H3K23 acetylation, H3K4 dimethylation and phosphorylation at serine 10. Nephrol Dial Transplant. 2010;25:1811–17. doi: 10.1093/ndt/gfp730. [DOI] [PubMed] [Google Scholar]
- 275.Gaikwad AB, Sayyed SG, Lichtnekert J, Tikoo K, Anders HJ. Renal failure increases cardiac histone h3 acetylation, dimethylation, and phosphorylation and the induction of cardiomyopathy-related genes in type 2 diabetes. Am J Pathol. 2010;176:1079–83. doi: 10.2353/ajpath.2010.090528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Gao WM, Chadha MS, Kline AE, et al. Immunohistochemical analysis of histone H3 acetylation and methylation - evidence for altered epigenetic signaling following traumatic brain injury in immature rats. Brain Res. 2006;1070:31–4. doi: 10.1016/j.brainres.2005.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Zhao F, Chen Y, Zeng L, et al. Role of triptolide in cell proliferation, cell cycle arrest, apoptosis and histone methylation in multiple myeloma U266 cells. Eur J Pharmacol. 2010;646:1–11. doi: 10.1016/j.ejphar.2010.05.034. [DOI] [PubMed] [Google Scholar]
- 278.Seligson DB, Horvath S, McBrian MA, et al. Global Levels of Histone Modifications Predict Prognosis in Different Cancers. Am J Pathol. 2009;174:1619–28. doi: 10.2353/ajpath.2009.080874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Arita A, Costa M. Epigenetics in metal carcinogenesis: Nickel, Arsenic, Chromium and Cadmium. Metallomics. 2009;1:222–28. doi: 10.1039/b903049b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Chen MW, Hua KT, Kao HJ, et al. H3K9 Histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–40. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 281.Yao JY, Zhang L, Zhang X, et al. H3K27 trimethylation is an early epigenetic event of p16(INK4a) silencing for regaining tumorigenesis in fusion reprogrammed hepatoma cells. J Biol Chem. 2010;285:18828–37. doi: 10.1074/jbc.M109.077974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Paul TA, Bies J, Small D, Wolff L. Signatures of polycomb repression and reduced H3K4 trimethylation are associated with p15INK4b DNA methylation in AML. Blood. 2010;115:3098–108. doi: 10.1182/blood-2009-07-233858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Porcedda P, Turinetto V, Brusco A, et al. A rapid flow cytometry test based on histone H2AX phosphorylation for the sensitive and specific diagnosis of ataxia telangiectasia. Cytometry A. 2008;73:508–16. doi: 10.1002/cyto.a.20566. [DOI] [PubMed] [Google Scholar]
- 284.Barr FD, Krohmer LJ, Hamilton JW, Sheldon LA. Disruption of histone modification and CARM1 recruitment by arsenic represses transcription at glucocorticoid receptor-regulated promoters. PLoS One. 2009;4:e6766. doi: 10.1371/journal.pone.0006766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Ashktorab H, Belgrave K, Hosseinkhah F, et al. Global histone H4 acetylation and HDAC2 expression in colon adenoma and carcinoma. Digestive Dis Sci. 2009;54:2109–17. doi: 10.1007/s10620-008-0601-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Felli N, Fontana L, Pelosi E, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–86. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.le Sage C, Nagel R, Egan DA, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Garofalo M, Di Leva G, Romano G, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 289.Mi SL, Lu J, Sun M, et al. MicroRNA expression signatures accurately discriminate acute lymphoblastic leukemia from acute myeloid leukemia. Blood. 2007;110:75a. doi: 10.1073/pnas.0709313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Saumet A, Vetter G, Bouttier M, et al. Transcriptional repression of microRNA genes by PML-RARA increases expression of key cancer proteins in acute promyelocytic leukemia. Blood. 2009;113:412–21. doi: 10.1182/blood-2008-05-158139. [DOI] [PubMed] [Google Scholar]
- 291.Hebert SS, Horre K, Nicolai L, et al. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc Natl Acad Sci USA. 2008;105:6415–20. doi: 10.1073/pnas.0710263105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Takakura S, Mitsutake N, Nakashima M, et al. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008;99:1147–54. doi: 10.1111/j.1349-7006.2008.00800.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Calin GA, Liu CG, Sevignani C, et al. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc Natl Acad Sci USA. 2004;101:11755–60. doi: 10.1073/pnas.0404432101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Arndt GM, Dossey L, Cullen LM, et al. Characterization of global microRNA expression reveals oncogenic potential of miR-145 in metastatic colorectal cancer. BMC Cancer. 2009;9:374–90. doi: 10.1186/1471-2407-9-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Bandres E, Cubedo E, Agirre X, et al. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;5:29–38. doi: 10.1186/1476-4598-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Malzkorn B, Wolter M, Grzendowski M, Stuhler K, Reifenberger G. Identification and functional characterization of microRNAs involved in the malignant progression of astrocytic gliomas. Brain Pathol. 2010;20:539–50. doi: 10.1111/j.1750-3639.2009.00328.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5–15. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Budhu A, Jia HL, Forgues M, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 299.Connolly E, Melegari M, Landgraf P, et al. Elevated expression of the miR-17-92 polycistron and miR-21 in hepadnavirus-associated hepatocellular carcinoma contributes to the malignant phenotype. Am J Pathol. 2008;173:856–64. doi: 10.2353/ajpath.2008.080096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–32. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 301.Chim CS, Fung TK, Liang R. Disruption of INK4/CDK/Rb cell cycle pathway by gene hypermethylation in multiple myeloma and MGUS. Leukemia. 2003;17:2533–35. doi: 10.1038/sj.leu.2403133. [DOI] [PubMed] [Google Scholar]
- 302.Stirzaker C, Millar DS, Paul CL, et al. Extensive DNA methylation spanning the Rb promoter in retinoblastoma tumors. Cancer Res. 1997;57:2229–37. [PubMed] [Google Scholar]
- 303.Chen MN, Mao Q, Liu YH, Mao BY. Methylation and expression analysis of p16(INK4a) and RB genes in meningiomas. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2004;21:277–79. [PubMed] [Google Scholar]
- 304.Zhao YF, Shen SP, Jiang JY, Geng H, Guo JG, Xie LP. Methylation and expression of gene p16INK4a and RB in breast carcinoma. Zhonghua Bing Li Xue Za Zhi. 2010;39:377–81. [PubMed] [Google Scholar]
- 305.Zhao Y, Tian X, Du J, et al. Methylation and expression of gene p16INK4a and RB in gastric carcinomas. Beijing Da Xue Xue Bao. 2003;35:382–85. [PubMed] [Google Scholar]
- 306.Li H, Lu S, Fong L. Study on the status of methylation of Rb gene promoter in human esophageal cancer and effect of NMBzA on Rb gene promoter in monkey esophageal epithelium. Zhonghua Zhong Liu Za Zhi. 1998;20:412–14. [PubMed] [Google Scholar]
- 307.Ke Q, Davidson T, Chen H, Kluz T, Costa M. Alterations of histone modifications and transgene silencing by nickel chloride. Carcinogenesis. 2006;27:1481–88. doi: 10.1093/carcin/bgl004. [DOI] [PubMed] [Google Scholar]
- 308.Chen MW, Hua KT, Kao HJ, et al. H3K9 histone methyltransferase G9a promotes lung cancer invasion and metastasis by silencing the cell adhesion molecule Ep-CAM. Cancer Res. 2010;70:7830–40. doi: 10.1158/0008-5472.CAN-10-0833. [DOI] [PubMed] [Google Scholar]
- 309.Lefevre GM, Patel SR, Kim D, Tessarollo L, Dressler GR. Altering a histone H3K4 methylation pathway in glomerular podocytes promotes a chronic disease phenotype. PLoS Genet. 2010;6:e1001142. doi: 10.1371/journal.pgen.1001142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Kumar R, Hunt CR, Gupta A, et al. Purkinje cell-specific males absent on the first (mMof) gene deletion results in an ataxia-telangiectasia-like neurological phenotype and backward walking in mice. Proc Natl Acad Sci USA. 2011;108:3636–41. doi: 10.1073/pnas.1016524108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Gramantieri L, Ferracin M, Fornari F, et al. Cyclin g1 is a target of miR-122a, a MicroRNA frequently down-regulated in human hepatocellular carcinoma. Cancer Res. 2007;67:6092–99. doi: 10.1158/0008-5472.CAN-06-4607. [DOI] [PubMed] [Google Scholar]
- 312.Jazdzewski K, Murray EL, Franssila K, Jarzab B, Schoenberg DR, de la Chapelle A. Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2008;105:7269–74. doi: 10.1073/pnas.0802682105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Lin SL, Chiang A, Chang D, Ying SY. Loss of mir-146a function in hormone-refractory prostate cancer. Rna-a Publication of the RNA Society. 2008;14:417–24. doi: 10.1261/rna.874808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Gonzalez-Ramirez I, Ramirez-Amador V, Irigoyen-Camacho ME, et al. hMLH1 promoter methylation is an early event in oral cancer. Oral Oncol. 2011;47:22–26. doi: 10.1016/j.oraloncology.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 315.Vasavi M, Kiran V, Ravishankar B, Prabhakar B, Ahuja YR, Hasan Q. Microsatellite instability analysis and its correlation with hMLH1 repair gene hypermethylation status in esophageal pathologies including cancers. Cancer Biomark. 2010;7:1–10. doi: 10.3233/CBM-2010-0135. [DOI] [PubMed] [Google Scholar]
- 316.Ling ZQ, Tanaka A, Li P, et al. Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 2010;297:244–51. doi: 10.1016/j.canlet.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 317.Klein CB, Su L, Bowser D, Leszczynska J. Chromate-induced epimutations in mammalian cells. Environ Health Perspect. 2002;110(Suppl 5):739–43. doi: 10.1289/ehp.02110s5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Lukiw WJ, Zhao YH, Cui JG. An NF-kappa B-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–22. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Pogue AI, Li YY, Cui JG, et al. Characterization of an NF-kappa B-regulated, miRNA-146a-mediated down-regulation of complement factor H (CFH) in metal-sulfate-stressed human brain cells. J Inorg Biochem. 2009;103:1591–95. doi: 10.1016/j.jinorgbio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 320.Cheng YH, Ji RR, Yue JM, et al. MicroRNAs are aberrantly expressed in hypertrophic heart - Do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–40. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappa B-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–86. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Bhaumik D, Scott GK, Schokrpur S, Patil CK, Campisi J, Benz CC. Expression of microRNA-146 suppresses NF-kappa B activity with reduction of metastatic potential in breast cancer cells. Oncogene. 2008;27:5643–5647. doi: 10.1038/onc.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Shen J, Ambrosone CB, DiCioccio RA, Odunsi K, Lele SB, Zhao H. A functional polymorphism in the miR-146a gene and age of familial breast/ovarian cancer diagnosis. Carcinogenesis. 2008;29:1963–66. doi: 10.1093/carcin/bgn172. [DOI] [PubMed] [Google Scholar]
- 325.Calin GA, Ferracin M, Cimmino A, et al. A microRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. New Engl J Med. 2005;353:1793–801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 326.Xu T, Zhu Y, Wei QK, et al. A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis. 2008;29:2126–31. doi: 10.1093/carcin/bgn195. [DOI] [PubMed] [Google Scholar]
- 327.Yanaihara N, Caplen N, Bowman E, et al. Unique MicroRNA molecular profiles in lung cancer diagnosis and prognosis. Toxicol Pathol. 2006;34:1017–18. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 328.Kozaki KI, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- 329.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–69. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lukiw WJ. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport. 2007;18:297–300. doi: 10.1097/WNR.0b013e3280148e8b. [DOI] [PubMed] [Google Scholar]
- 331.Saba R, Goodman CD, Huzarewich RLCH, Robertson C, Booth SA. A miRNA Signature of Prion Induced Neurodegeneration. PLos One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Roehle A, Hoefig KP, Repsilber D, et al. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. British J Haematol. 2008;142:732–44. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 333.Wang Y, Lee ATC, Ma JZI, et al. Profiling MicroRNA expression in hepatocellular carcinoma reveals MicroRNA-224 up-regulation and apoptosis inhibitor-5 as a MicroRNA-224-specific target. J Biol Chem. 2008;283:13205–15. doi: 10.1074/jbc.M707629200. [DOI] [PubMed] [Google Scholar]
- 334.Tan HX, Wang QA, Chen LZ, et al. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol. 2010;27:654–60. doi: 10.1007/s12032-009-9264-2. [DOI] [PubMed] [Google Scholar]
- 335.Veerla S, Lindgren D, Kvist A, et al. MiRNA expression in urothelial carcinomas: Important roles of miR-10a, miR-222, miR-125b, miR-7 and miR-452 for tumor stage and metastasis, and frequent homozygous losses of miR-31. Int J Cancer. 2009;124:2236–42. doi: 10.1002/ijc.24183. [DOI] [PubMed] [Google Scholar]
- 336.Mill J, Tang T, Kaminsky Z, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am J Hum Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 339.Heng JI, Nguyen L, Castro DS, et al. Neurogenin 2 controls cortical neuron migration through regulation of Rnd2. Nature. 2008;455:114–18. doi: 10.1038/nature07198. [DOI] [PubMed] [Google Scholar]
- 340.Pereira MA, Tao L, Liu Y, Li L, Steele VE, Lubet RA. Modulation by budesonide of DNA methylation and mRNA expression in mouse lung tumors. Int J Cancer. 2007;120:1150–53. doi: 10.1002/ijc.22468. [DOI] [PubMed] [Google Scholar]
- 341.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 342.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 343.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–33. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 344.Zhu SM, Si ML, Wu HL, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–36. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 345.Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–36. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–08. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 347.Meng FY, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Zhang K, Zhang R, Li X, Yin G, Niu X. Promoter methylation status of p15 and p21 genes in HPP-CFCs of bone marrow of patients with psoriasis. Eur J Dermatol. 2009;19:141–46. doi: 10.1684/ejd.2008.0618. [DOI] [PubMed] [Google Scholar]
- 349.Furonaka O, Takeshima Y, Awaya H, Ishida H, Kohno N, Inai K. Aberrant methylation of p14(ARF), p15(INK4b) and p16(INK4a) genes and location of the primary site in pulmonary squamous cell carcinoma. Pathol Int. 2004;54:549–55. doi: 10.1111/j.1440-1827.2004.01663.x. [DOI] [PubMed] [Google Scholar]
- 350.Lindberg D, Akerstrom G, Westin G. Evaluation of CDKN2C/p18, CDKN1B/p27 and CDKN2B/p15 mRNA expression, and CpG methylation status in sporadic and MEN1-associated pancreatic endocrine tumours. Clin Endocrinol. 2008;68:271–77. doi: 10.1111/j.1365-2265.2007.03034.x. [DOI] [PubMed] [Google Scholar]
- 351.Kim M, Yim SH, Cho NS, et al. Homozygous deletion of CDKN2A (p16, p14) and CDKN2B (p15) genes is a poor prognostic factor in adult but not in childhood B-lineage acute lymphoblastic leukemia: a comparative deletion and hypermethylation study. Cancer Genet Cytogenet. 2009;195:59–65. doi: 10.1016/j.cancergencyto.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 352.Shimamoto T, Ohyashiki JH, Ohyashiki K. Methylation of p15(INK4b) and E-cadherin genes is independently correlated with poor prognosis in acute myeloid leukemia. Leuk Res. 2005;29:653–59. doi: 10.1016/j.leukres.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 353.Chen W, Wu Y, Zhu J, Liu J, Tan S, Xia C. Methylation of p16 and p15 genes in multiple myeloma. Clin Med Sci J. 2002;17:101–05. [PubMed] [Google Scholar]
- 354.Gallardo F, Esteller M, Pujol RM, Costa C, Estrach T, Servitje O. Methylation status of the p15, p16 and MGMT promoter genes in primary cutaneous T-cell lymphomas. Haematologica. 2004;89:1401–403. [PubMed] [Google Scholar]
- 355.El-Shakankiry NH, Mossallam GI. p15 (INK4B) and E-cadherin CpG island methylation is frequent in Egyptian acute myeloid leukemia. J Egypt Natl Canc Inst. 2006;18:227–32. [PubMed] [Google Scholar]
- 356.Matsuno N, Hoshino K, Nanri T, et al. p15 mRNA expression detected by real-time quantitative reverse transcriptase-polymerase chain reaction correlates with the methylation density of the gene in adult acute leukemia. Leuk Res. 2005;29:557–64. doi: 10.1016/j.leukres.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 357.Wemmert S, Bettscheider M, Alt S, et al. p15 promoter methylation - a novel prognostic marker in glioblastoma patients. Int J Oncol. 2009;34:1743–48. doi: 10.3892/ijo_00000305. [DOI] [PubMed] [Google Scholar]
- 358.Yalcin A, Serin MS, Emekdas G, et al. Promoter methylation of P15(INK4B) gene is possibly associated with parvovirus B19 infection in adult acute leukemias. Int J Lab Hematol. 2009;31:407–19. doi: 10.1111/j.1751-553X.2008.01052.x. [DOI] [PubMed] [Google Scholar]
- 359.Berg T, Guo Y, Abdelkarim M, Fliegauf M, Lubbert M. Reversal of p15/INK4b hypermethylation in AML1/ETO-positive and -negative myeloid leukemia cell lines. Leuk Res. 2007;31:497–506. doi: 10.1016/j.leukres.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 360.Wong TS, Man MW, Lam AK, Wei WI, Kwong YL, Yuen AP. The study of p16 and p15 gene methylation in head and neck squamous cell carcinoma and their quantitative evaluation in plasma by real-time PCR. Eur J Cancer. 2003;39:1881–87. doi: 10.1016/s0959-8049(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 361.Morgan DA, Rahmouni K. Differential effects of insulin on sympathetic nerve activity in agouti obese mice. J Hypertens. 2010;28:1913–19. doi: 10.1097/HJH.0b013e32833c2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Xiang Z, Proneth B, Dirain ML, Litherland SA, Haskell-Luevano C. Pharmacological characterization of 30 human melanocortin-4 receptor polymorphisms with the endogenous proopiomelanocortin-derived agonists, synthetic agonists, and the endogenous agouti-related protein antagonist. Biochemistry. 2010;49:4583–600. doi: 10.1021/bi100068u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 363.Gucev ZS, Tasic V, Jancevska A, Kirovski I. A case of Silver-Russell syndrome (SRS): multiple pituitary hormone deficiency, lack of H19 hypomethylation and favourable growth hormone (GH) treatment response. J Genet. 2009;88:239–43. doi: 10.1007/s12041-009-0033-y. [DOI] [PubMed] [Google Scholar]
- 364.Zeschnigk M, Albrecht B, Buiting K, et al. IGF2/H19 hypomethylation in Silver-Russell syndrome and isolated hemihypoplasia. Eur J Hum Genet. 2008;16:328–34. doi: 10.1038/sj.ejhg.5201974. [DOI] [PubMed] [Google Scholar]
- 365.Chopra M, Amor DJ, Sutton L, Algar E, Mowat D. Russell-Silver syndrome due to paternal H19/IGF2 hypomethylation in a patient conceived using intracytoplasmic sperm injection. Reprod Biomed Online. 2010;20:843–47. doi: 10.1016/j.rbmo.2010.02.025. [DOI] [PubMed] [Google Scholar]
- 366.Dammann RH, Kirsch S, Schagdarsurengin U, et al. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int J Oncol. 2010;36:171–79. [PubMed] [Google Scholar]
- 367.Baba Y, Nosho K, Shima K, et al. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139:1855–64. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Li Y, Meng G, Huang L, Guo QN. Hypomethylation of the P3 promoter is associated with up-regulation of IGF2 expression in human osteosarcoma. Hum Pathol. 2009;40:1441–47. doi: 10.1016/j.humpath.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 369.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–46. [PubMed] [Google Scholar]
- 370.Ito Y, Koessler T, Ibrahim AE, et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–43. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Xi Y, Formentini A, Chien M, et al. Prognostic values of microRNAs in colorectal cancer. Biomark Insights. 2006;2:113–21. [PMC free article] [PubMed] [Google Scholar]
- 372.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–61. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Calin GA, Dumitru CD, Shimizu M, et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–29. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Cimmino A, Calin GA, Fabbri M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–49. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Ambs S, Prueitt RL, Yi M, et al. Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res. 2008;68:6162–70. doi: 10.1158/0008-5472.CAN-08-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Roccaro AM, Sacco A, Thompson B, et al. MicroRNAs 15a and 16 regulate tumor proliferation in multiple myeloma. Blood. 2009;113:6669–80. doi: 10.1182/blood-2009-01-198408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 378.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Yanaihara N, Caplen N, Bowman E, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 380.Sampson VB, Rong NH, Han J, et al. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007;67:9762–70. doi: 10.1158/0008-5472.CAN-07-2462. [DOI] [PubMed] [Google Scholar]
- 381.Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ER alpha) and represses ER alpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21:1132–47. doi: 10.1210/me.2007-0022. [DOI] [PubMed] [Google Scholar]
- 382.Roccaro AM, Jia X, Ngo HT, et al. MicroRNA expression in the biology, prognosis, and therapy of waldenstrom macroglobulinemia. Clin Lymphoma Myeloma. 2009;9:S126. doi: 10.1182/blood-2008-09-178228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Dixon-McIver A, East P, Mein CA, et al. Distinctive patterns of microrna expression associated with karyotype in acute myeloid leukaemia. PLos One. 2008;3:e2141. doi: 10.1371/journal.pone.0002141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA. 2006;103:18255–60. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Sayed D, Hong C, Chen IY, Lypowy J, Abdellatif M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circulation Res. 2007;100:416–24. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]