Abstract
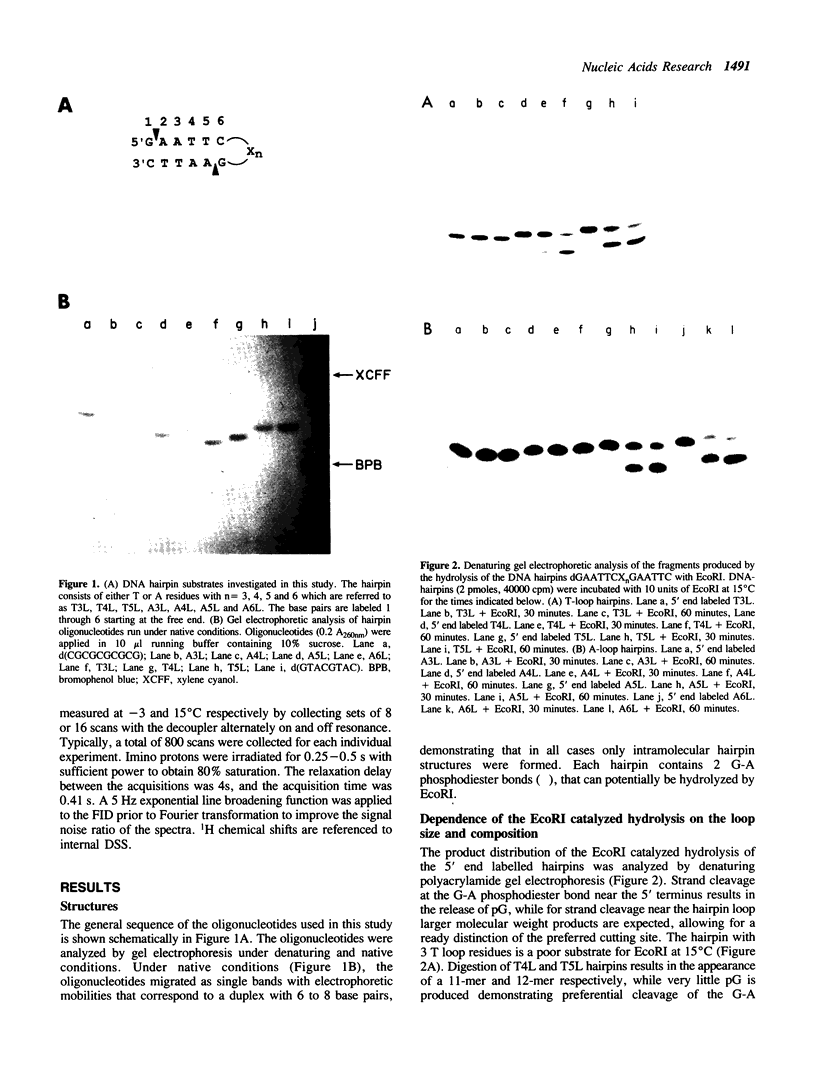
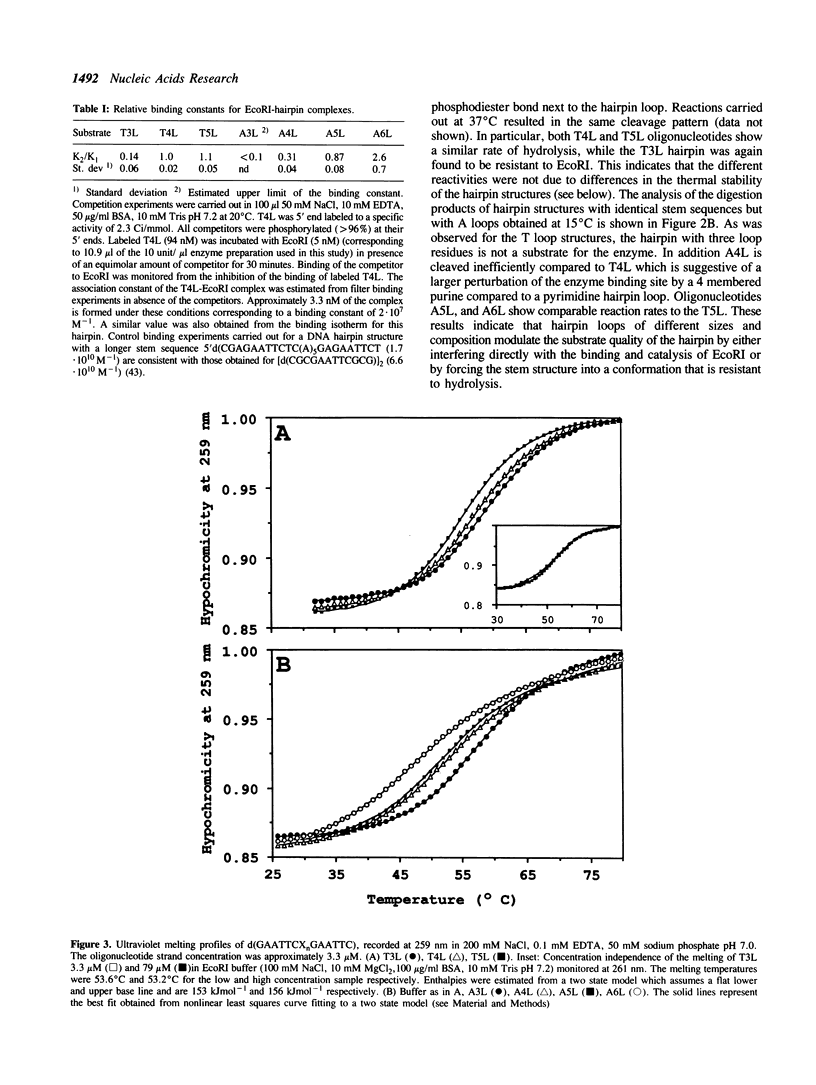
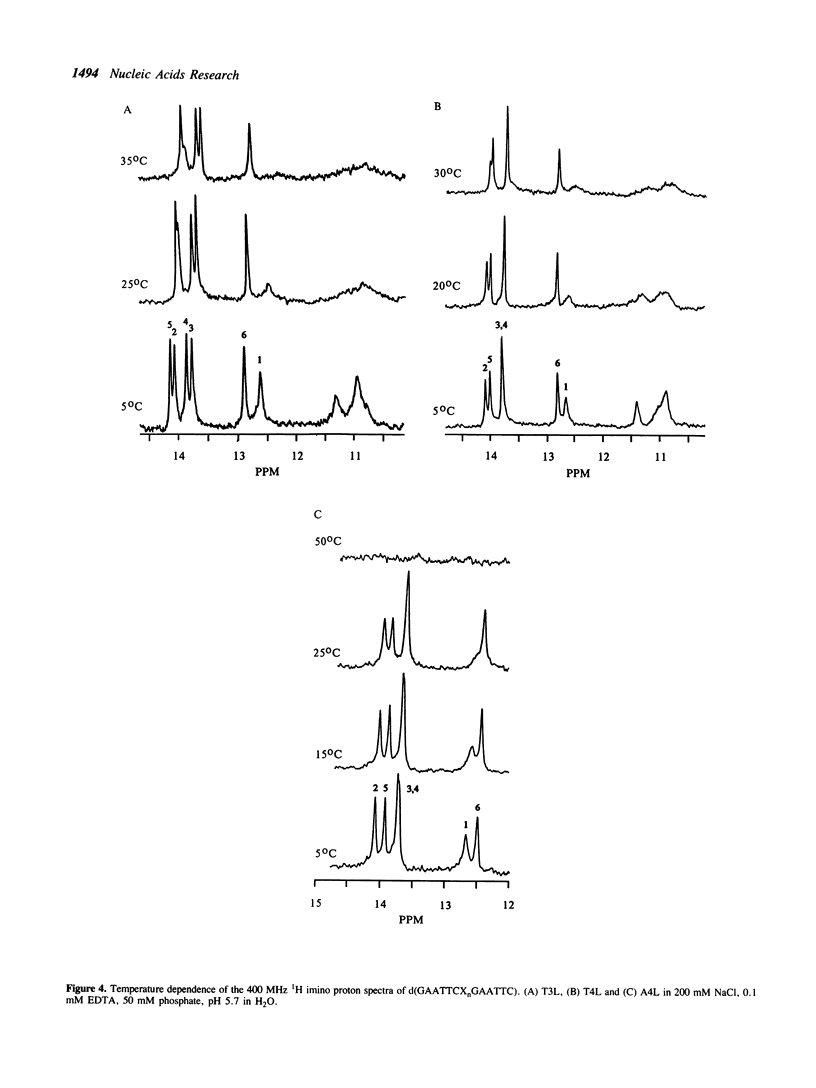
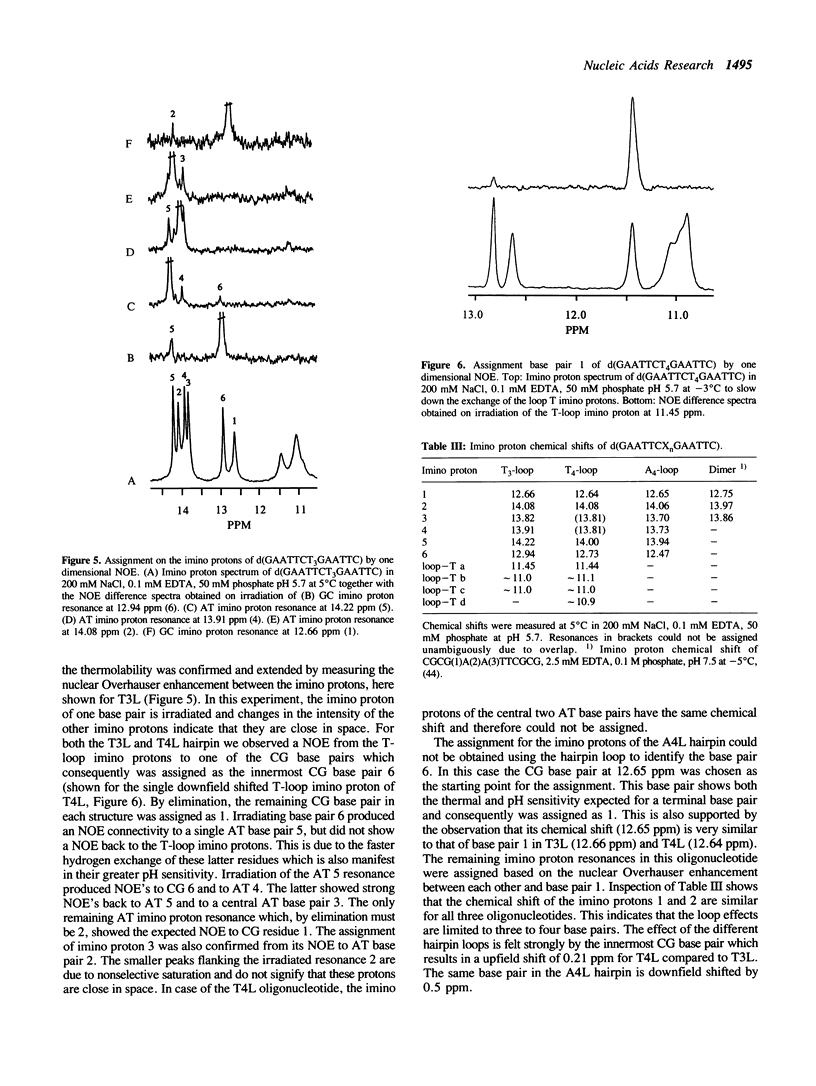
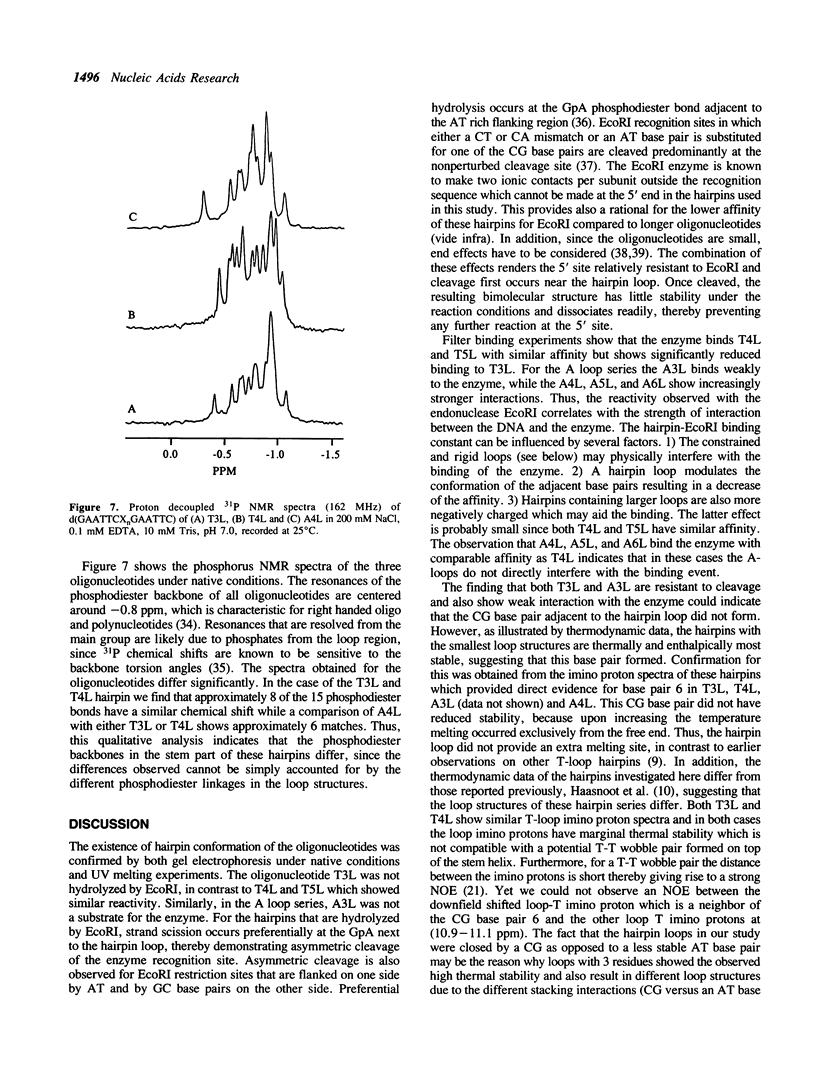
We have investigated loop-induced structural perturbation of the stem structure in hairpins d(GAATTCXnGAATTC) (X = A, T and n = 3, 4, 5 and 6) that contain an EcoRI restriction site in close proximity to the hairpin loop. Oligonucleotides containing either a T3 or a A3 loop were not hydrolyzed by the restriction enzyme and also showed only weak binding to EcoRI in the absence of the cofactor Mg2+. In contrast, hairpins with larger loops are hydrolyzed by the enzyme at the scission site next to the loop although the substrate with a A4 loop is significantly more resistant than the oligonucleotide containing a T4 loop. The hairpin structures with 3 loop residues were found to be thermally most stable while larger hairpin loops resulted in structures with lower melting temperatures. The T-loop hairpins are thermally more stable than the hairpins containing the same number of A residues in the loop. As judged from proton NMR spectroscopy and the thermodynamic data, the base pair closest to the hairpin loop did form in all cases studied. The hairpin loops did, however, affect the conformation of the stem structure of the hairpins. From 31P and 1H NMR spectroscopy we conclude that the perturbation of the stem structure is stronger for smaller hairpin loops and that the extent of the perturbation is limited to 2-3 base pairs for hairpins with T3 or A4 loops. Our results demonstrate that hairpin loops modulate the conformation of the stem residues close to the loop and that this in turn reduces the substrate activity for DNA sequence specific proteins.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alves J., Pingoud A., Haupt W., Langowski J., Peters F., Maass G., Wolff C. The influence of sequences adjacent to the recognition site on the cleavage of oligodeoxynucleotides by the EcoRI endonuclease. Eur J Biochem. 1984 Apr 2;140(1):83–92. doi: 10.1111/j.1432-1033.1984.tb08069.x. [DOI] [PubMed] [Google Scholar]
- Baumann U., Frank R., Blöcker H. Conformational analysis of hairpin oligodeoxyribonucleotides by a single-strand-specific nuclease. Eur J Biochem. 1986 Dec 1;161(2):409–413. doi: 10.1111/j.1432-1033.1986.tb10460.x. [DOI] [PubMed] [Google Scholar]
- Blommers M. J., Walters J. A., Haasnoot C. A., Aelen J. M., van der Marel G. A., van Boom J. H., Hilbers C. W. Effects of base sequence on the loop folding in DNA hairpins. Biochemistry. 1989 Sep 5;28(18):7491–7498. doi: 10.1021/bi00444a049. [DOI] [PubMed] [Google Scholar]
- Chaconas G., van de Sande J. H. 5'-32P labeling of RNA and DNA restriction fragments. Methods Enzymol. 1980;65(1):75–85. doi: 10.1016/s0076-6879(80)65012-5. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R., Ikuta S., Grzeskowiak K., Dickerson R. E. X-ray structure of a DNA hairpin molecule. Nature. 1988 Jul 14;334(6178):175–179. doi: 10.1038/334175a0. [DOI] [PubMed] [Google Scholar]
- Connolly B. A., Eckstein F. Assignment of resonances in the 31P NMR spectrum of d(GGAATTCC) by regiospecific labeling with oxygen-17. Biochemistry. 1984 Nov 6;23(23):5523–5527. doi: 10.1021/bi00318a022. [DOI] [PubMed] [Google Scholar]
- Cornelis A. G., Haasnoot J. H., den Hartog J. F., de Rooij M., van Boom J. H., Cornelis A. Local destabilisation of a DNA double helix by a T--T wobble pair. Nature. 1979 Sep 20;281(5728):235–236. doi: 10.1038/281235a0. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Schoenwaelder K. H., van de Sande J. H. Right- and left-handed (Z) helical conformations of the hairpin d(C-G)5T4(C-G)5 monomer and dimer. Biochemistry. 1985 Oct 8;24(21):5698–5702. doi: 10.1021/bi00342a002. [DOI] [PubMed] [Google Scholar]
- Germann M. W., Vogel H. J., Pon R. T., van de Sande J. H. Characterization of a parallel-stranded DNA hairpin. Biochemistry. 1989 Jul 25;28(15):6220–6228. doi: 10.1021/bi00441a013. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B., Borer P. N., Kan L. S., Ts'o P. O. Ring-current effects in the Nmr of nucleic acids: a graphical approach. Biopolymers. 1976 Nov;15(11):2277–2286. doi: 10.1002/bip.1976.360151114. [DOI] [PubMed] [Google Scholar]
- Goppelt M., Pingoud A., Maass G., Mayer H., Köster H., Frank R. The interaction of the EcoRI restriction endonuclease with its substrate. A physico-chemical study employing natural and synthetic oligonucleotides and polynucleotides. Eur J Biochem. 1980 Feb;104(1):101–107. doi: 10.1111/j.1432-1033.1980.tb04405.x. [DOI] [PubMed] [Google Scholar]
- Gorenstein D. G., Schroeder S. A., Fu J. M., Metz J. T., Roongta V., Jones C. R. Assignments of 31P NMR resonances in oligodeoxyribonucleotides: origin of sequence-specific variations in the deoxyribose phosphate backbone conformation and the 31P chemical shifts of double-helical nucleic acids. Biochemistry. 1988 Sep 20;27(19):7223–7237. doi: 10.1021/bi00419a009. [DOI] [PubMed] [Google Scholar]
- Gupta G., Sarma M. H., Sarma R. H., Bald R., Engelke U., Oei S. L., Gessner R., Erdmann V. A. DNA hairpin structures in solution: 500-MHz two-dimensional 1H NMR studies on d(CGCCGCAGC) and d(CGCCGTAGC). Biochemistry. 1987 Dec 1;26(24):7715–7723. doi: 10.1021/bi00398a027. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., Hilbers C. W., van der Marel G. A., van Boom J. H., Singh U. C., Pattabiraman N., Kollman P. A. On loop folding in nucleic acid hairpin-type structures. J Biomol Struct Dyn. 1986 Apr;3(5):843–857. doi: 10.1080/07391102.1986.10508468. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., de Bruin S. H., Berendsen R. G., Janssen H. G., Binnendijk T. J., Hilbers C. W., van der Marel G. A., van Boom J. H. Structure, kinetics and thermodynamics of DNA hairpin fragments in solution. J Biomol Struct Dyn. 1983 Oct;1(1):115–129. doi: 10.1080/07391102.1983.10507429. [DOI] [PubMed] [Google Scholar]
- Haasnoot C. A., den Hartog J. H., de Rooij J. F., van Boom J. H., Altona C. Loopstructures in synthetic oligodeoxynucleotides. Nucleic Acids Res. 1980 Jan 11;8(1):169–181. doi: 10.1093/nar/8.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare D. R., Reid B. R. Three-dimensional structure of a DNA hairpin in solution: two-dimensional NMR studies and distance geometry calculations on d(CGCGTTTTCGCG). Biochemistry. 1986 Sep 9;25(18):5341–5350. doi: 10.1021/bi00366a053. [DOI] [PubMed] [Google Scholar]
- Hilbers C. W., Haasnoot C. A., de Bruin S. H., Joordens J. J., van der Marel G. A., van Boom J. H. Hairpin formation in synthetic oligonucleotides. Biochimie. 1985 Jul-Aug;67(7-8):685–695. doi: 10.1016/s0300-9084(85)80156-5. [DOI] [PubMed] [Google Scholar]
- Ikuta S., Chattopadhyaya R., Ito H., Dickerson R. E., Kearns D. R. NMR study of a synthetic DNA hairpin. Biochemistry. 1986 Aug 26;25(17):4840–4849. doi: 10.1021/bi00365a018. [DOI] [PubMed] [Google Scholar]
- Jen-Jacobson L., Kurpiewski M., Lesser D., Grable J., Boyer H. W., Rosenberg J. M., Greene P. J. Coordinate ion pair formation between EcoRI endonuclease and DNA. J Biol Chem. 1983 Dec 10;258(23):14638–14646. [PubMed] [Google Scholar]
- Lilley D. M. Hairpin-loop formation by inverted repeats in supercoiled DNA is a local and transmissible property. Nucleic Acids Res. 1981 Mar 25;9(6):1271–1289. doi: 10.1093/nar/9.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClarin J. A., Frederick C. A., Wang B. C., Greene P., Boyer H. W., Grable J., Rosenberg J. M. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986 Dec 19;234(4783):1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Mizuuchi M., Gellert M. Cruciform structures in palindromic DNA are favored by DNA supercoiling. J Mol Biol. 1982 Apr 5;156(2):229–243. doi: 10.1016/0022-2836(82)90325-4. [DOI] [PubMed] [Google Scholar]
- Otting G., Grütter R., Leupin W., Minganti C., Ganesh K. N., Sproat B. S., Gait M. J., Wüthrich K. Sequential NMR assignments of labile protons in DNA using two-dimensional nuclear-Overhauser-enhancement spectroscopy with three jump-and-return pulse sequences. Eur J Biochem. 1987 Jul 1;166(1):215–220. doi: 10.1111/j.1432-1033.1987.tb13504.x. [DOI] [PubMed] [Google Scholar]
- Panayotatos N., Wells R. D. Cruciform structures in supercoiled DNA. Nature. 1981 Feb 5;289(5797):466–470. doi: 10.1038/289466a0. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Broka C., Rice J. A., Itakura K., Breslauer K. J. Premelting and melting transitions in the d(CGCGAATTCGCG) self-complementary duplex in solution. Biochemistry. 1982 Feb 2;21(3):428–436. doi: 10.1021/bi00532a002. [DOI] [PubMed] [Google Scholar]
- Pramanik P., Kanhouwa N., Kan L. S. Hairpin and duplex formation in DNA fragments CCAATTTTGG, CCAATTTTTTGG, and CCATTTTTGG: a proton NMR study. Biochemistry. 1988 Apr 19;27(8):3024–3031. doi: 10.1021/bi00408a054. [DOI] [PubMed] [Google Scholar]
- Pörschke D., Jung M. Stability decrease of RNA double helices by phenylalanine-, tyrosine- and tryptophane-amides. Analysis in terms of site binding and relation to melting proteins. Nucleic Acids Res. 1982 Oct 11;10(19):6163–6176. doi: 10.1093/nar/10.19.6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinkel L. J., van der Marel G. A., van Boom J. H., Altona C. Influence of N6-methylation of residue A(5) on the conformational behaviour of d(C-C-G-A-A-T-T-C-G-G) in solution studied by 1H-NMR spectroscopy. 2. The hairpin form. Eur J Biochem. 1987 Mar 2;163(2):287–296. doi: 10.1111/j.1432-1033.1987.tb10799.x. [DOI] [PubMed] [Google Scholar]
- Roy S., Weinstein S., Borah B., Nickol J., Appella E., Sussman J. L., Miller M., Shindo H., Cohen J. S. Mechanism of oligonucleotide loop formation in solution. Biochemistry. 1986 Nov 18;25(23):7417–7423. doi: 10.1021/bi00371a025. [DOI] [PubMed] [Google Scholar]
- Scheffler I. E., Elson E. L., Baldwin R. L. Helix formation by dAT oligomers. I. Hairpin and straight-chain helices. J Mol Biol. 1968 Sep 28;36(3):291–304. doi: 10.1016/0022-2836(68)90156-3. [DOI] [PubMed] [Google Scholar]
- Senior M. M., Jones R. A., Breslauer K. J. Influence of loop residues on the relative stabilities of DNA hairpin structures. Proc Natl Acad Sci U S A. 1988 Sep;85(17):6242–6246. doi: 10.1073/pnas.85.17.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers M. F., Byrd R. A., Gallo K. A., Samson C. J., Zon G., Egan W. Nuclear magnetic resonance and circular dichroism studies of a duplex--single-stranded hairpin loop equilibrium for the oligodeoxyribonucleotide sequence d(CGCGATTCGCG). Nucleic Acids Res. 1985 Sep 11;13(17):6375–6386. doi: 10.1093/nar/13.17.6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Wolk S. K., Hardin C. C., Germann M. W., van de Sande J. H., Tinoco I., Jr Comparison of the B- and Z-form hairpin loop structures formed by d(CG)5T4(CG)5. Biochemistry. 1988 Sep 6;27(18):6960–6967. doi: 10.1021/bi00418a043. [DOI] [PubMed] [Google Scholar]