Abstract
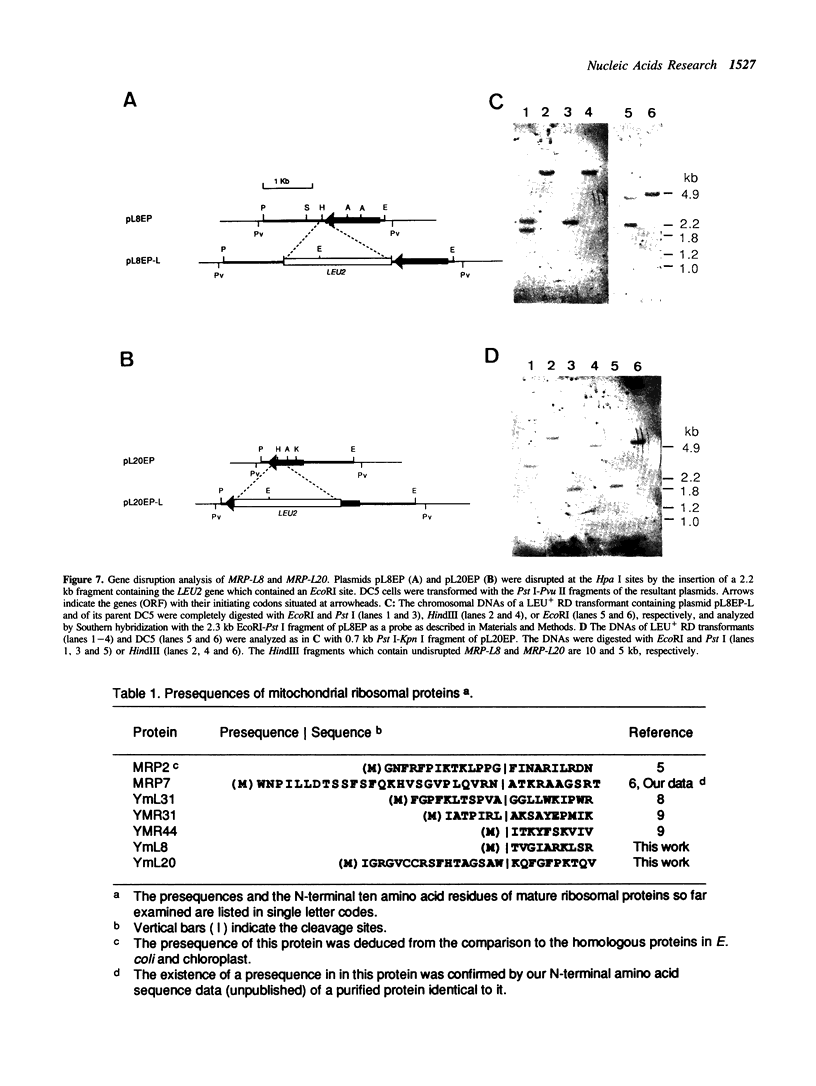
The genes for two large subunit proteins, YmL8 and YmL20, of the mitochondrial ribosome of Saccharomyces cerevisiae were cloned by hybridization with synthetic oligonucleotide mixtures corresponding to their N-terminal amino acid sequences. They were termed MRP-L8 and MRP-L20, respectively, and their nucleotide sequences were determined using a DNA sequencer. The MRP-L8 gene was found to encode a 26.8-kDa protein whose deduced amino acid sequence has a high degree of similarity to ribosomal protein L17 of Escherichia coli. The gene MRP-L20 was found to encode a 22.3-kDa protein with a presequence consisting of 18 amino acid residues. By Southern blot hybridization to the yeast chromosomes separated by field-inversion gel electrophoresis, the MRP-L8 and MRP-L20 genes were located on chromosomes X and XI, respectively. Gene disruption experiments indicate that their products, YmL8 and YmL20 proteins, are essential for the mitochondrial function and the absence of these proteins causes instability of the mitochondrial DNA.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. Codon selection in yeast. J Biol Chem. 1982 Mar 25;257(6):3026–3031. [PubMed] [Google Scholar]
- Bland M. M., Levings C. S., 3rd, Matzinger D. F. The tobacco mitochondrial ATPase subunit 9 gene is closely linked to an open reading frame for a ribosomal protein. Mol Gen Genet. 1986 Jul;204(1):8–16. doi: 10.1007/BF00330180. [DOI] [PubMed] [Google Scholar]
- Bonen L. The mitochondrial S13 ribosomal protein gene is silent in wheat embryos and seedlings. Nucleic Acids Res. 1987 Dec 23;15(24):10393–10404. doi: 10.1093/nar/15.24.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Grivell L. A. Mitochondrial ribosomes. FEBS Lett. 1971 Feb 19;13(2):73–88. doi: 10.1016/0014-5793(71)80204-1. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Strathern J. N., Hicks J. B. Transformation in yeast: development of a hybrid cloning vector and isolation of the CAN1 gene. Gene. 1979 Dec;8(1):121–133. doi: 10.1016/0378-1119(79)90012-x. [DOI] [PubMed] [Google Scholar]
- Burton F. H., Loeb D. D., McGraw R. A., Edgell M. H., Hutchison C. A., 3rd A directed nucleotide-sequencing approach for single-stranded vectors based on recloning intermediates of a progressive DNA synthesis reaction. Gene. 1988 Jul 30;67(2):159–168. doi: 10.1016/0378-1119(88)90393-9. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Frank M., Olson M. V. Electrophoretic separations of large DNA molecules by periodic inversion of the electric field. Science. 1986 Apr 4;232(4746):65–68. doi: 10.1126/science.3952500. [DOI] [PubMed] [Google Scholar]
- Carle G. F., Olson M. V. An electrophoretic karyotype for yeast. Proc Natl Acad Sci U S A. 1985 Jun;82(11):3756–3760. doi: 10.1073/pnas.82.11.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E. Y., Seeburg P. H. Supercoil sequencing: a fast and simple method for sequencing plasmid DNA. DNA. 1985 Apr;4(2):165–170. doi: 10.1089/dna.1985.4.165. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas M. G., McCammon M. T., Vassarotti A. Targeting proteins into mitochondria. Microbiol Rev. 1986 Jun;50(2):166–178. doi: 10.1128/mr.50.2.166-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faye G., Sor F. Analysis of mitochondrial ribosomal proteins of Saccharomyces cerevisiae by two dimensional polyacrylamide gel electrophoresis. Mol Gen Genet. 1977 Sep 21;155(1):27–34. doi: 10.1007/BF00268557. [DOI] [PubMed] [Google Scholar]
- Fearon K., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP7, a protein of the large subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3636–3646. doi: 10.1128/mcb.8.9.3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graack H. R., Grohmann L., Choli T. Mitochondrial ribosomes of yeast: isolation of individual proteins and N-terminal sequencing. FEBS Lett. 1988 Dec 19;242(1):4–8. doi: 10.1016/0014-5793(88)80975-x. [DOI] [PubMed] [Google Scholar]
- Gray M. W., Doolittle W. F. Has the endosymbiont hypothesis been proven? Microbiol Rev. 1982 Mar;46(1):1–42. doi: 10.1128/mr.46.1.1-42.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grohmann L., Graack H. R., Kitakawa M. Molecular cloning of the nuclear gene for mitochondrial ribosomal protein YmL31 from Saccharomyces cerevisiae. Eur J Biochem. 1989 Jul 15;183(1):155–160. doi: 10.1111/j.1432-1033.1989.tb14907.x. [DOI] [PubMed] [Google Scholar]
- Gualberto J. M., Wintz H., Weil J. H., Grienenberger J. M. The genes coding for subunit 3 of NADH dehydrogenase and for ribosomal protein S12 are present in the wheat and maize mitochondrial genomes and are co-transcribed. Mol Gen Genet. 1988 Dec;215(1):118–127. doi: 10.1007/BF00331312. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hicks J., Fink G. R. Identification of chromosomal location of yeast DNA from hybrid plasmid p Yeleu 10. Nature. 1977 Sep 15;269(5625):265–267. doi: 10.1038/269265a0. [DOI] [PubMed] [Google Scholar]
- Isono K. A computer program package for storing and retrieving DNA/RNA and protein sequence data. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):101–112. doi: 10.1093/nar/12.1part1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M., Chow C. K. The complete amino acid sequences of ribosomal proteins L17, L27, and S9 from Bacillus stearothermophilus. Eur J Biochem. 1984 Mar 1;139(2):225–234. doi: 10.1111/j.1432-1033.1984.tb07998.x. [DOI] [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreader C. A., Langer C. S., Heckman J. E. A mitochondrial protein from Neurospora crassa detected both on ribosomes and in membrane fractions. Analysis of the gene, the message, and the protein. J Biol Chem. 1989 Jan 5;264(1):317–327. [PubMed] [Google Scholar]
- Kuiper M. T., Akins R. A., Holtrop M., de Vries H., Lambowitz A. M. Isolation and analysis of the Neurospora crassa Cyt-21 gene. A nuclear gene encoding a mitochondrial ribosomal protein. J Biol Chem. 1988 Feb 25;263(6):2840–2847. [PubMed] [Google Scholar]
- Lindemann H., Wittmann-Liebold B. Primary structure of protein S13 from the small ribosomal subunit of Escherichia coli. FEBS Lett. 1976 Dec 1;71(2):251–255. doi: 10.1016/0014-5793(76)80944-1. [DOI] [PubMed] [Google Scholar]
- Matsushita Y., Kitakawa M., Isono K. Cloning and analysis of the nuclear genes for two mitochondrial ribosomal proteins in yeast. Mol Gen Genet. 1989 Oct;219(1-2):119–124. doi: 10.1007/BF00261166. [DOI] [PubMed] [Google Scholar]
- Myers A. M., Crivellone M. D., Tzagoloff A. Assembly of the mitochondrial membrane system. MRP1 and MRP2, two yeast nuclear genes coding for mitochondrial ribosomal proteins. J Biol Chem. 1987 Mar 5;262(7):3388–3397. [PubMed] [Google Scholar]
- Myers A. M., Pape L. K., Tzagoloff A. Mitochondrial protein synthesis is required for maintenance of intact mitochondrial genomes in Saccharomyces cerevisiae. EMBO J. 1985 Aug;4(8):2087–2092. doi: 10.1002/j.1460-2075.1985.tb03896.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederacher D., Entian K. D. Isolation and characterization of the regulatory HEX2 gene necessary for glucose repression in yeast. Mol Gen Genet. 1987 Mar;206(3):505–509. doi: 10.1007/BF00428892. [DOI] [PubMed] [Google Scholar]
- Partaledis J. A., Mason T. L. Structure and regulation of a nuclear gene in Saccharomyces cerevisiae that specifies MRP13, a protein of the small subunit of the mitochondrial ribosome. Mol Cell Biol. 1988 Sep;8(9):3647–3660. doi: 10.1128/mcb.8.9.3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratzkin B., Carbon J. Functional expression of cloned yeast DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1977 Feb;74(2):487–491. doi: 10.1073/pnas.74.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roise D., Schatz G. Mitochondrial presequences. J Biol Chem. 1988 Apr 5;263(10):4509–4511. [PubMed] [Google Scholar]
- Röhl R., Nierhaus K. H. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci U S A. 1982 Feb;79(3):729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. M., Dayhoff M. O. Origins of prokaryotes, eukaryotes, mitochondria, and chloroplasts. Science. 1978 Jan 27;199(4327):395–403. doi: 10.1126/science.202030. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Cowe E., Higgins D. G., Shields D. C., Wolfe K. H., Wright F. Codon usage patterns in Escherichia coli, Bacillus subtilis, Saccharomyces cerevisiae, Schizosaccharomyces pombe, Drosophila melanogaster and Homo sapiens; a review of the considerable within-species diversity. Nucleic Acids Res. 1988 Sep 12;16(17):8207–8211. doi: 10.1093/nar/16.17.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Tuohy T. M., Mosurski K. R. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986 Jul 11;14(13):5125–5143. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wahleithner J. A., Wolstenholme D. R. Ribosomal protein S14 genes in broad bean mitochondrial DNA. Nucleic Acids Res. 1988 Jul 25;16(14B):6897–6913. doi: 10.1093/nar/16.14.6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]