Abstract
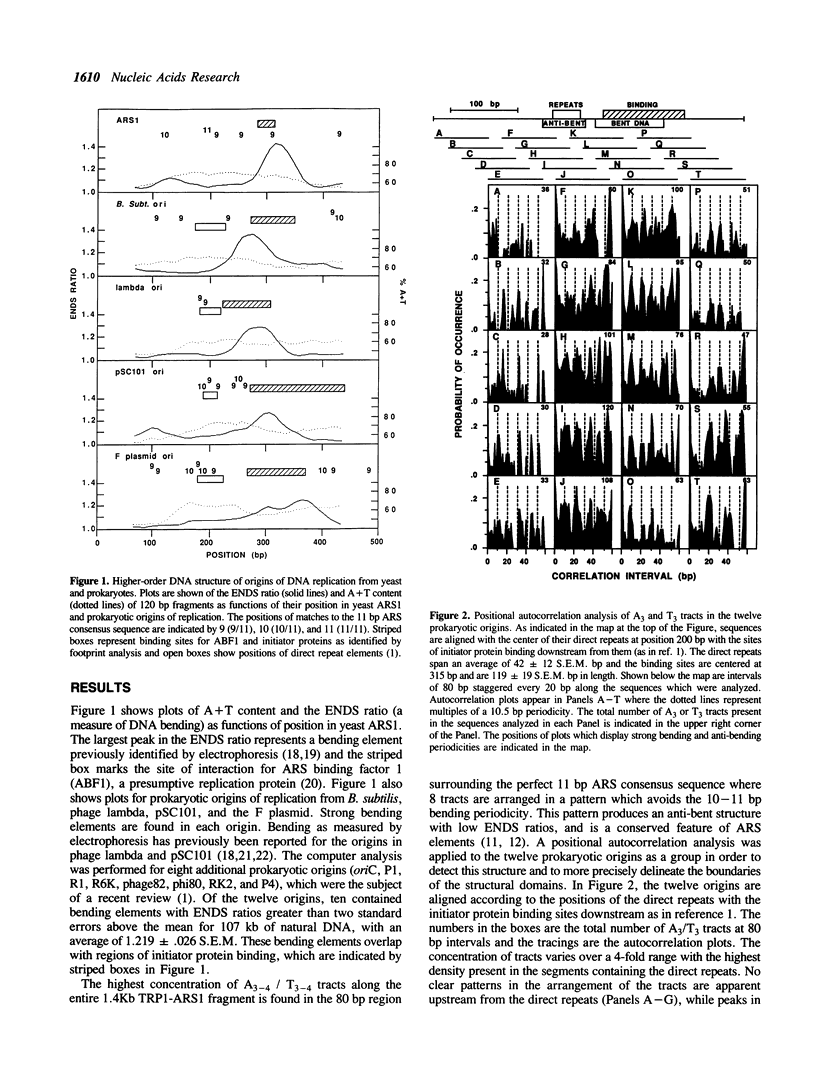
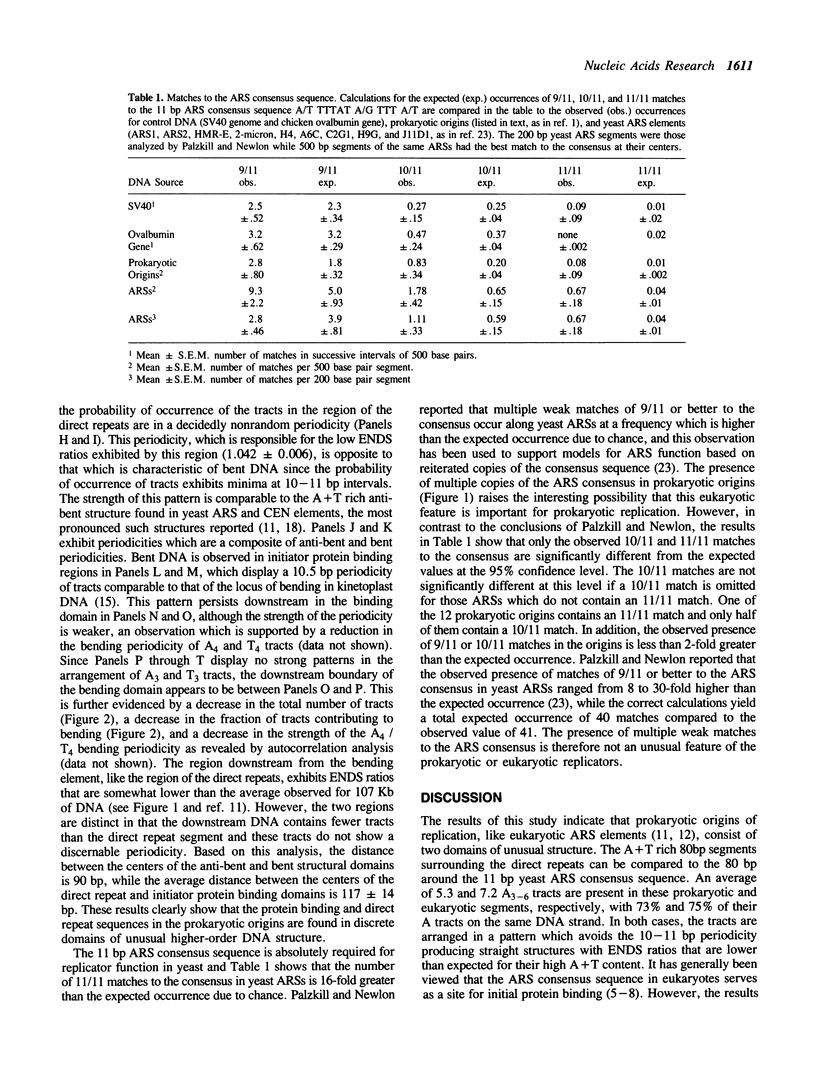
According to the model of Bramhill and Kornberg, initiation of DNA replication in prokaryotes involves binding of an initiator protein to origin DNA and subsequent duplex opening of adjacent direct repeat sequences. In this report, we have used computer analysis to examine the higher-order DNA structure of a variety of origins of replication from plasmids, phages, and bacteria in order to determine whether these sequences are localized in domains of altered structure. The results demonstrate that the primary sites of initiator protein binding lie in discrete domains of DNA bending, while the direct repeats lie within well-defined boundaries of an unusual anti-bent domain. The anti-bent structures arise from a periodicity of A3 and T3 tracts which avoids the 10-11 bp bending periodicity. Since DNA fragments which serve as replicators in yeast also contain these two conserved structural elements, the results provide new insight into the universal role of conserved DNA structures in DNA replication.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson J. N. Detection, sequence patterns and function of unusual DNA structures. Nucleic Acids Res. 1986 Nov 11;14(21):8513–8533. doi: 10.1093/nar/14.21.8513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aymami J., Coll M., Frederick C. A., Wang A. H., Rich A. The propeller DNA conformation of poly(dA).poly(dT). Nucleic Acids Res. 1989 Apr 25;17(8):3229–3245. doi: 10.1093/nar/17.8.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. A model for initiation at origins of DNA replication. Cell. 1988 Sep 23;54(7):915–918. doi: 10.1016/0092-8674(88)90102-x. [DOI] [PubMed] [Google Scholar]
- Bramhill D., Kornberg A. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell. 1988 Mar 11;52(5):743–755. doi: 10.1016/0092-8674(88)90412-6. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Li Y. Y., Feldman J., Jayaram M., Abraham J., Nasmyth K. A., Hicks J. B. Localization and sequence analysis of yeast origins of DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):1165–1173. doi: 10.1101/sqb.1983.047.01.132. [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Sweder K., Srienc F., Bailey J. E., Campbell J. L. Deletion mutations affecting autonomously replicating sequence ARS1 of Saccharomyces cerevisiae. Mol Cell Biol. 1984 Nov;4(11):2455–2466. doi: 10.1128/mcb.4.11.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckdahl T. T., Anderson J. N. Computer modelling of DNA structures involved in chromosome maintenance. Nucleic Acids Res. 1987 Oct 26;15(20):8531–8545. doi: 10.1093/nar/15.20.8531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goursot R., Goze A., Niaudet B., Ehrlich S. D. Plasmids from Staphylococcus aureus replicate in yeast Saccharomyces cerevisiae. Nature. 1982 Jul 29;298(5873):488–490. doi: 10.1038/298488a0. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao C. L., Carbon J. High-frequency transformation of yeast by plasmids containing the cloned yeast ARG4 gene. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3829–3833. doi: 10.1073/pnas.76.8.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huberman J. A., Spotila L. D., Nawotka K. A., el-Assouli S. M., Davis L. R. The in vivo replication origin of the yeast 2 microns plasmid. Cell. 1987 Nov 6;51(3):473–481. doi: 10.1016/0092-8674(87)90643-x. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Marini J. C., Effron P. N., Goodman T. C., Singleton C. K., Wells R. D., Wartell R. M., Englund P. T. Physical characterization of a kinetoplast DNA fragment with unusual properties. J Biol Chem. 1984 Jul 25;259(14):8974–8979. [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Crothers D. M. Structural basis for DNA bending. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2622–2626. doi: 10.1073/pnas.86.8.2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palzkill T. G., Newlon C. S. A yeast replication origin consists of multiple copies of a small conserved sequence. Cell. 1988 May 6;53(3):441–450. doi: 10.1016/0092-8674(88)90164-x. [DOI] [PubMed] [Google Scholar]
- Prunell A. Nucleosome reconstitution on plasmid-inserted poly(dA) . poly(dT). EMBO J. 1982;1(2):173–179. doi: 10.1002/j.1460-2075.1982.tb01143.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnos M., Zahn K., Inman R. B., Blattner F. R. Initiation protein induced helix destabilization at the lambda origin: a prepriming step in DNA replication. Cell. 1988 Feb 12;52(3):385–395. doi: 10.1016/s0092-8674(88)80031-x. [DOI] [PubMed] [Google Scholar]
- Snyder M., Buchman A. R., Davis R. W. Bent DNA at a yeast autonomously replicating sequence. Nature. 1986 Nov 6;324(6092):87–89. doi: 10.1038/324087a0. [DOI] [PubMed] [Google Scholar]
- Stenzel T. T., Patel P., Bastia D. The integration host factor of Escherichia coli binds to bent DNA at the origin of replication of the plasmid pSC101. Cell. 1987 Jun 5;49(5):709–717. doi: 10.1016/0092-8674(87)90547-2. [DOI] [PubMed] [Google Scholar]
- Strich R., Woontner M., Scott J. F. Mutations in ARS1 increase the rate of simple loss of plasmids in Saccharomyces cerevisiae. Yeast. 1986 Sep;2(3):169–178. doi: 10.1002/yea.320020305. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Umek R. M., Kowalski D. Yeast regulatory sequences preferentially adopt a non-B conformation in supercoiled DNA. Nucleic Acids Res. 1987 Jun 11;15(11):4467–4480. doi: 10.1093/nar/15.11.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. S., Eckdahl T. T., Anderson J. N. Bent DNA functions as a replication enhancer in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Jul;8(7):2763–2769. doi: 10.1128/mcb.8.7.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson D. H. The yeast ARS element, six years on: a progress report. Yeast. 1985 Sep;1(1):1–14. doi: 10.1002/yea.320010102. [DOI] [PubMed] [Google Scholar]
- Zahn K., Blattner F. R. Direct evidence for DNA bending at the lambda replication origin. Science. 1987 Apr 24;236(4800):416–422. doi: 10.1126/science.2951850. [DOI] [PubMed] [Google Scholar]


