Abstract
Protocadherins are transmembrane proteins exhibiting homophilic adhesive activities through their extracellular domain. Pcdh12 is expressed in angiogenic endothelial cells, mesangial cells of kidney glomeruli and glycogen cells of the mouse placenta. To get insights into the role of this protein in vivo, we analyzed PCDH12-deficient mice and investigated their placental phenotype. The mice were alive and fertile, however placental and embryonic sizes were reduced compared to wild types. We observed defects in placental layer segregation and a decreased vascularization of the labyrinth associated with a reduction in cell density in this layer. To understand the molecular events responsible for the phenotypic alterations observed in Pcdh12−/− placentas, we analyzed the expression profile of E12.5 mutant placentas in comparison with wild types, using pangenomic chips. 2,289 genes exhibited statistically significant changes in expressed levels due to loss of PCDH12. Functional grouping of modified genes was obtained by GoMiner software. Gene clusters that contained most of the differentially expressed genes were those involved in tissue morphogenesis and development, angiogenesis, cell-matrix adhesion and migration, immune response and chromatin remodeling. Our data show that loss of PCDH12 leads to morphological alterations of the placenta and to notable changes in its gene expression profile. Specific genes emerging from the microarray screen support the biological modifications observed in PCDH12-deficient placentas.
Keywords: Animals; Animals, Newborn; Cadherins; deficiency; metabolism; Cell Adhesion; Cell Movement; Decidua; cytology; metabolism; Female; Gene Expression Profiling; Glycogen; metabolism; Mice; Morphogenesis; Organ Size; Placenta; cytology; embryology; metabolism; Pregnancy; Reverse Transcriptase Polymerase Chain Reaction
Keywords: knockout mice, gene profiling, trophoblasts, angiogenesis
INTRODUCTION
The placenta constitutes a physical and functional connection between the mother and the developing embryo. It establishes an exchange system for numerous soluble compounds between maternal and fetal bloods. Additionally, it produces hormones that promote the maternal response to pregnancy and seems to play important roles in triggering delivery. The placenta is itself a developing organ with successive morphological and functional modifications adapted to the different gestation phases.
In the mouse, the placenta originates from the ectoplacental cone and the extraembryonic ectoderm. The endothelial cells derive from the allantois (8, 16, 37). From embryonic day (E) 10, the placenta is divided in three layers associated with maternal decidual cells. The labyrinth, located on the fetal side, is composed of an intricate array of fetal and maternal vessels that constitute a selective barrier between the two circulation systems (1, 9). The giant cells are located next to the uterine cells and, until E12, constitute the outermost fetal cell layer. Between the labyrinth and the giant cells, there is a third layer, the junctional zone, also called the “spongy layer” because of its numerous cavities. The junctional zone has been shown to produce several hormones but its general function remains elusive. Nevertheless, there is an absolute requirement of this layer as mutations inducing its disruption are not compatible with embryonic survival (17, 41). The junctional zone is composed of two types of trophoblast: the spongiotrophoblasts and the glycogen cells, recognizable by their high glycogen contents. The glycogen cells form islets within the junctional zone that migrate from E12.5 into the maternal decidua, beyond the giant cell line (4, 16).
Protocadherins constitute a large family of transmembrane proteins exhibiting calcium-dependent homophilic adhesive properties (15, 22). As opposed to the classical cadherins, protocadherins do not or only weakly associate with the actin cytoskeleton. Although some properties of individual protocadherins have been reported, a comprehensive view of their functions is missing.
Protocadherin 12 (PCDH12), previously called VE-cadherin 2, was initially identified in mouse endothelial cells (43). We recently showed that its endothelial expression was more specifically detected in angiogenic endothelium (35). In addition, PCDH12 is abundantly expressed in placental glycogen cells and mesangial cells of renal glomeruli (35).
To gain insight into PCDH12 biological activity in vivo, we produced PCDH12-deficient mice. The mice were alive and fertile. Furthermore, Pcdh12+/− intercrosses produced a normal Mendelian distribution at birth (35). In this paper, we show that PCDH12-deficient placentas and embryos are smaller than their wild type counterparts and we observed two major morphological modifications in the mutant placentas, namely decreased vascular and cell densities in the labyrinth and a missegregation of the labyrinthine and the junctional zone. These general alterations of placental development prompted us to examine whether the placental transcriptional profile was modified in absence of PCDH12. Strikingly, the expression of 2,289 genes was significantly altered in the mutant compared to wild type placentas. Our data show that transcriptional changes occurred in functional groups of genes involved in tissue morphogenesis, angiogenesis, cell migration, immune response and chromatin remodeling. Expression changes of specific genes are discussed in relation with the biological alterations observed in PCDH12-deficient placentas.
MATERIALS AND METHODS
Animals and placenta preparation
All protocols in this study were conducted in strict accordance with the French guidelines for the care and use of laboratory animals. Agreement 38-13 was granted by the veterinary services of the French government to P. H. to perform the in vivo experiments described herein. PCDH12-deficient mice were established on CD1 genetic background. For wild type and Pcdh12−/− (35) placenta production, mice were mated and the day on which a vaginal plug was found was designated 0.5. Pregnant females were killed by cervical dislocation and conceptuses were dissected in phosphate buffer saline (PBS). Placentas and embryos were weighed after rapid liquid draining on paper towels. Placenta volume was calculated from measurements of diameter (D) and thickness (T) with a caliper, using the formula: π/6 × D2 × T. Genotypes were performed on embryonic DNA as previously described (35).
Histology
For immunolocalization, tissues were snap-frozen in OCT compound and sectioned at 10-μm with a cryomicrotome (Leica Microsystems, Wetzlar, Germany). Sections were permeabilized in paraformaldehyde 4%, Triton 0.5%-PBS for 3 min, fixed in 4% paraformaldehyde-PBS for 20 min, saturated with 2% BSA/PBS and incubated with anti-CD31 (45) and either peroxydase-conjugated anti-rat immunoglobulin (Biorad, Marnes-la-Coquette, France) or alexa 488-conjugated anti-rat immunoglobulin (Invitrogen, Cergy-Pontoise, France) antibodies, all at room temperature, using standard procedures. Peroxidase was revealed with diaminobenzidine (DakoCytomation, Trappes, France), followed by nuclear staining with Harris hematoxylin (Sigma-Aldrich, St-Quentin-Fallavier, France). Nuclei in fluorescent images were stained with Hoechst 33258 (Sigma-Aldrich).
For periodic acid-Schiff (PAS)/hematoxylin (both from Sigma-Aldrich) staining, paraffin sections were prepared using standard procedures, from paraformaldehyde prefixed tissues. Slides were mounted in Entellan or Aquatex (VWR International, Fontenay-sous-Bois, France) and observed with a Zeiss Axioplan microscope. Pictures were made with a digital camera (Spot-RT, Diagnostic Instruments, Sterling Heights, Michigan) and were used to calculate the layer surface and cell density using ImageJ software (NIH, Bethesda, http://rsb.info.nih.gov/ij/) and the number of the spongiotrophoblast layer islets in the labyrinth.
To quantify capillary length, images were processed for morphometric analysis using Image J software. A macrocommand was edited to give the total vessel length after binarization, skeletonization and pixel count of the CD31 staining fluorescent image.
Extraction of glycogen and measurement of glycogen content
Glycogen content of placentas was measured by the method of Lo et al. (28). Each placenta was incubated for 30 min at 100°C with 0.5 ml of 30% KOH saturated with Na2SO4. The solution was cooled and the glycogen was precipitated with 0.55 mL of 100% ethanol at 0°C for 30 min. Glycogen was pelleted by centrifugation at 1,000 g for 30 min at 4°C. The pellet was dissolved in 3 mL H2O. Aliquots were incubated for 20 min at 30°C with 0.7% phenol and 70% H2SO4 and absorbance was measured at 490 nm. Mussel glycogen (Roche Biochemicals, Meylan, France) was used as standard. Data were expressed as mg of glycogen per g of wet tissue or per total placenta.
Microarray processing and analysis
Total RNAs were extracted from E12.5 placentas (5 placentas of each genotype, from 3 female and 2 male embryos, were derived from 3 litters) using Tri Reagent (Euromedex, Souffelweyersheim, France), treated with DNase I and purified using NucleoSpin RNA Clean up (Macherey-Nagel, Hoerdt, France). RNA concentration and integrity were tested with an Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Massy, France).
Affymetrix chip hybridization was performed by the Institute of Genetics and Molecular and Cellular Biology (Strasbourg, France). The amplified RNAs were labeled and hybridized to Affymetrix mouse 430 2.0 gene chips containing over 45,000 probe sets and representing more than 34,000 characterized transcripts. The arrays were scanned with a confocal laser GeneChip scanner 3000 7G System. The resulting images were captured using GeneChip Operating Software (all on Affymetrix instruments). The background adjustment and the data normalization have been processed with MAS 5.0 software (Affymetrix). The scaling factors for all arrays formed a homogeneous interval concentrated around 2.33. The exploratory data analysis provided by the Affy package of Bioconductor (www.bioconductor.org) showed no experimental artifacts. Boxplots, densities of log intensities, RNA digestion plots and RNA degradation parameters are provided as supplementary data.
Full data sets have been deposited in Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO series accession number GSE7676.
Bioinformatics analysis
Results were annotated using information provided by Affymetrix. The full data set was reduced to 18,542 genes by discarding genes with “EST” and “unknown” annotation labels. To generate the list of genes classified by gene ontology (GO), we used the High-Throughput version of GoMiner (http://discover.nci.nih.gov/gominer/, (49, 50)). We then performed a Gene Set Enrichment Analysis (GSEA) (40) for selected gene sets according to GO biological process categories. The GSEA procedure allowed an interpretation of the gene expression profiles, by using predefined gene sets and ranks of genes to identify significant biological changes in microarray data sets. The family wise error rate (FWER) was used for multiple testing corrections. It provides the probability to get a non zero false discovery rate. The heat maps obtained from this analysis are presented as supplementary material.
Statistical analysis
For Fig. 1–5, the statistical significance was analyzed using Student’s t test. Sample number is indicated in figure legends. We used the “significance analysis of the microarray” (SAM) score (44) to investigate the differentially expressed genes. The estimation of the false discovery rate has been obtained using the q-value package available under the R language (www.r-project.org). The computation of the q-values has been done with the default tuning parameters as input with the exception of the pi0.method entry for which we choose the “smoother “ method. The output value of the pi0 - the estimate of the proportion of null hypothesis - was 0.65351.
Figure 1. Placental, embryonic and postnatal weights of PCDH12-deficient mice.
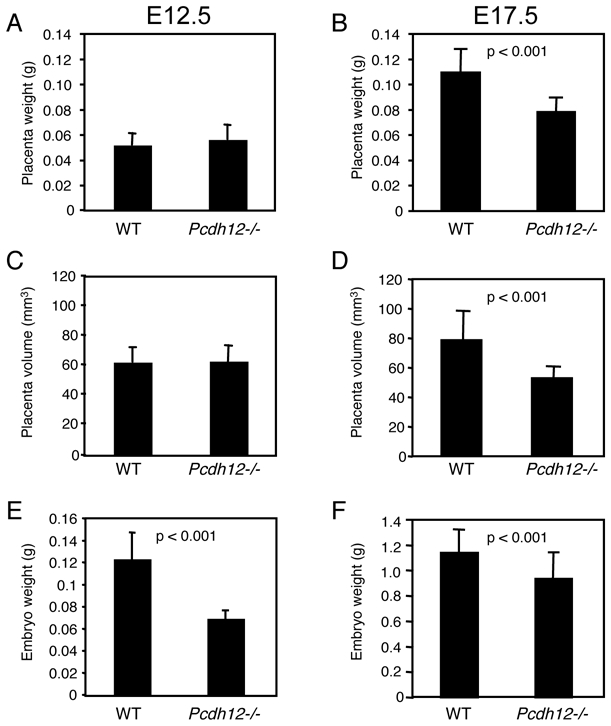
Wild type (WT) and Pcdh12−/− placentas and embryos were dissected at E12.5 (A, C, E) (n = 33 and 42, for WT and Pcdh12−/− individuals, respectively) and at E17.5 (B, D, F) (n = 46 and 33 for WT and Pcdh12−/− individuals, respectively). Placentas were weighed and their volume were calculated after measurement with a caliper, as described in Materials and Methods. Embryos were weighed after elimination of placenta and yolk sac. Histograms show (A, B) placenta weights, (C, D) placenta volumes, and (E, F) embryonic weights. Litter-mates from heterozygous X heterozygous matings were weighed from 2 to 8 weeks of postnatal age. Histograms in (G, H) represent postnatal weights of males (G) (n = 5 and 7, for WT and PCDH12-deficient mice, respectively) and females (H) (n = 7 and 6, for WT and PCDH12-deficient mice, respectively). Data represent the mean with SD. p-values are indicated when statistically significant differences were present.
Figure 5. Glycogen content of Pcdh12−/− placentas.
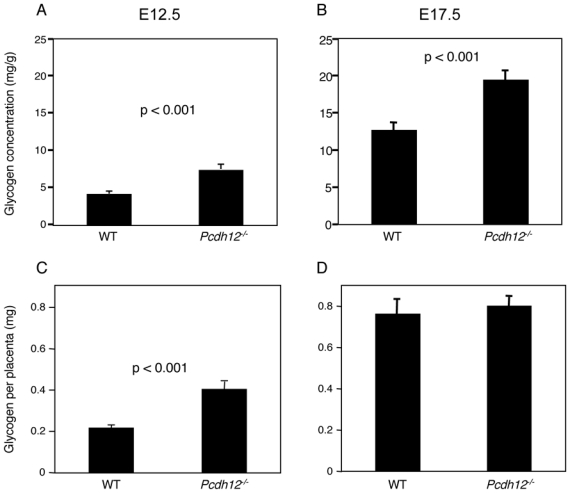
E12.5 (A, C) and E17.5 (B, D) placentas were weighed and glycogen was extracted as described in Materials and Methods. Glycogen content was measured by a colorimetric assay using mussel glycogen as standard. Glycogen concentration (A, B) and glycogen amount (C, D) at E12.5 (n = 41 and 32 for wild type and Pcdh12−/− placentas, respectively) and at E17.5 (n = 31 and 33 for wild type and Pcdh12−/− placentas, respectively) are shown. Data represent the mean with SE.
Real-time RT-PCR
First-strand cDNAs were generated by reverse transcription from 0.5 μg of total RNA in a total volume of 20μl with the SuperScript First strand Synthesis System (Invitrogen) using random primers and SuperScript II Reverse transcriptase.
Real-time PCR was performed using LightCycler apparatus (Roche Diagnostics, Meylan, France) and FastStart DNA master SYBR Green I ready to use PCR mix according to the manufacturer’s protocol (Roche). CDNAs (25 ng) were amplified in 10 mL total volume PCR reaction with specific primers. Gene expression was normalized using values obtained for the housekeeping gene Gapdh as the reference gene.
Primers used for PCR amplification: Phdal2, 5′-gctgtttttccactccatcc and 5′-gtttcacggacccagagc; Gapdh, 5′-tgtgatgggtgtgaaccacgagaa and 5′-aagcatccaaccacatcaca ; Angiopoietin 2, 5′-tacacactgacctcttccccaac and 5′-agtccacaccgccatcttctc; Tek, 5′-gaacaccgaggctatttgtac and 5′-agtgtggaagctgtagtgttgg; Tie1, 5′-ggcagcttccagagtatggt and 5′-tggccagcaatgttaagtca; Gja1, 5′-gagcccgaactctccttttc and 5′-ccatgtctgggcacctct ; Angiogenin2, 5′-tcagcactatgatgccaagc and 5′-tcctttgtgtgtgcaagtgg ; Decysin, 5′-aagcatccaaccacatcaca and 5′-tgtataggtgcacaggataggc. Primers were designed by using Integrated DNA Technologies software (available on line at http://www.idtdna.com/Scitools/Applications/PrimerQuest) or Primer 3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and using sequence data from the GenBank database.
RESULTS
PCDH12 deficiency alters placenta and embryo growths
Absence of PCDH12 does not alter embryo viability, as 25.1% of homozygotes are present at birth upon mating of heterozygous mice (35). Nonetheless, we noticed that Pcdh12−/− placentas and embryos were smaller than their wild-type counterparts. This feature prompted us to investigate several parameters of placental growth and organization. In this study, placentas were examined at two ages: E12.5, when placenta is in a very active phase of organogenesis and E17.5, when placenta is mature. In all following experiments, we compared the Pcdh12−/− mice to wild types. However, it is noteworthy that heretozygotes behaved like the wild types (not shown).
We first measured placenta weight and volume at both embryonic ages. At E17.5, but not at E12.5, Pcdh12−/− placenta weights were indeed significantly (p < 0.001) lower than those of wild types (Fig. 1A, B). Similarly, the volumes of E17.5 PCDH12-deficient placentas were significantly (p < 0.001) reduced, whereas they were similar at E12.5 (Fig. 1C, D). The volume reduction at E17.5 concerns both the diameter and the width of placentas (not shown). The data show that Pcdh12−/− placentas did not grow significantly after E12.5, while the wild types were still in an active phase of expansion. Furthermore, Pcdh12−/− embryo weights were significantly lower at both ages (Fig. 1E, F). Therefore, we conclude that PCDH12 deficiency alters both placenta and embryo growths. However, the average weight of mice of both genotypes aged from 2 weeks to 2 months was identical (Fig. 1G, H), thereby indicating that Pcdh12-deficient mice recovered after birth.
We thus suspected that feto-maternal interactions were altered in absence of PCDH12. Because embryo growth is dependent upon optimal placental activity, we focused our studies on the placenta.
Histological analysis of Pcdh12−/− placentas
We first examined the histological organization of the different placental layers. E12.5 and E17.5 placenta sections were prepared and labeled with the endothelial-specific anti-CD31 antibody. In the mouse placenta, the labyrinth is highly vascularized, the junctional zone is avascular and the decidua contains large vessels; the limits of the different placental layers are therefore clearly visualized by this technique, as illustrated in Fig. 2A. The layer surface areas, as well as total placenta area were measured on parasagittal sections; the average proportions are shown in Fig. 2B, C. Our results indicate that the proportions were similar between Pcdh12−/− and wild types at both ages, thereby suggesting that growth of all three layers was reduced in the E17.5 Pcdh12−/− placenta. The giant cell line was unmodified (not shown).
Figure 2. Histomorphological analysis of Pcdh12−/− placentas and deciduas.

(A) Parasagittal sections of placentas (shown here at E17.5) were stained with anti-CD31 antibody (brown) and hematoxylin (blue) to visualize the anatomical layers: the labyrinth (L), the junctional zone (J) and the decidua (D). Lines indicate layer separations. At this magnification, individual vessels are not visible, but vascular zones appear brownish. (B, C) The surfaces of the 3 layers, as well as total surfaces, were measured on 3 parasagittal sections (separated by approximately 50 μm) for each placenta at E12.5 (B) and E17.5 (C). The mean values were used to calculate the mean surface proportion (SD) of layers from wild-type (n = 8 and 12, for E12.5 and E17.5 placentas, respectively) and Pcdh12−/− (n = 8 and 10 for E12.5 and E17.5 placentas, respectively) placentas. (D, E) For each placenta at E12.5 (D) and E17.5 (E), nuclei were counted on three different areas at high magnification to calculate the cell density. Data represent the mean with SD. A significant difference was observed for labyrinthine cell density in both E12.5 and E17.5 placentas.
In each layer, the cell density was evaluated after nuclear labeling and counting (Fig. 2D, E). Interestingly, the PCDH12-deficient labyrinths contained significantly less cells per section area than the wild types at both E12.5 and E17.5.
The labyrinth contains a very dense capillary network. As the cell density was lower in the labyrinth, we wondered whether this network was altered in the mutant. As illustrated in Fig. 3A, B, we indeed noticed a decrease in vascular density in the mutant labyrinthine layer. We thus measured the total capillary length in CD31 immunofluorescent images. As shown in Fig. 3C–D, the skeletonized image obtained after processing by ImageJ software superimposed with the CD31-labelled vessels. Our quantitative data show that capillaries were much less developed in the Pcdh12−/− labyrinth at both E12.5 and E17.5 (Fig. 3F, G). This result might be in direct correlation with the lower cell density of the Pcdh12−/− labyrinth and may also have a functional impact on embryonic development, as optimal materno-fetal exchanges are required throughout gestation. In contrast, the vascular pattern of maternal deciduas was similar in wild type and Pcdh12−/− placentas.
Figure 3. Measurement of total capillary length in placenta histological sections.

Histological sections of wild type and Pcdh12−/− E12.5 (n = 5 for each) and E17.5 (n = 6 for each) placentas were labeled with anti-CD31 antibody and visualized by immunofluorescence microscopy at high magnification. Labyrinthine vascular density was decreased in Pcdh12−/− placentas compared to wild types, as shown for E17.5 placentas (A, B). For quantifications, three different images of labyrinth were acquired for each placenta. Images were binarized and skeletonized and total capillary length was measured as described in Material and Methods. (C) Initial CD 31 fluorescent image. (D) Processed image. (E) Merge. (F, G) The data show the average length (SD) of the capillary network per labyrinth area for each genotype at E112.5 (F) and E17.5 (G). CD31-labeled deciduas for which mother and embryo were of same genotype, i.e., wild type or Pcdh12−/−. Vascular patterns of wild type (H) and Pcdh12−/− (I) E12.5 deciduas were similar.
PCDH12 is an homophilic adhesive protein with high expression level in glycogen cells. We thus wondered whether loss of PCDH12 would alter cell interactions during development. E12.5 placentas were chosen to observe glycogen cells before their massive dissemination in the decidua. As Pcdh12-labeling is not useable to identify the Pcdh12−/− glycogen cells, placenta sections were labeled with PAS, which reacts with glycogen and thus highly stains glycogen cells. The junctional zones of wild type and Pcdh12−/− mice contained a similar pattern of glycogen cells assembled into islets with no sign of cell dissociation in the mutant (Fig. 4A,B). Thus, PCDH12 activity is not required for glycogen cell assembly and tissue integrity.
Figure 4. Glycogen cell islets in the junctional zone and the labyrinth of Pcdh12−/− placentas.

PAS staining and hematoxylin counterstaining of wild type (A,C) and Pcdh12−/− (B,D,E) placenta sections. Glycogen cells are stained in deep purple by PAS because of their high glycogen contents. (A,B) E12.5 placenta section images are focused on the junctional zone. Glycogen cell islets are circled and indicated with (*). Note that glycogen cells appear partially empty because of glycogen solubilization occuring during section preparation. (C,D) E17.5 total placenta sections are shown. At this age, the labyrinth constitutes the major part of the placenta; the junctional zone and the decidua form a cap at the placenta margin. (E) is an enlargement of (D) showing that junctional zone islets (circled) are composed of both cell types. Sp, spongiotrophoblast; GC, glycogen cells. (F) The number of islets (pegs) was averaged in sections from wild type (n = 9) and Pcdh12−/− (n = 12) placentas. The error bars represent the SE.
Projections or islets (also called “pegs”) of the junctional zone in the labyrinth may be observed in normal placentas, even in late gestation phase (E17.5). However, we noticed that this feature was enhanced in PCDH12-deficient placentas. Figures 4C and D show two representative placenta sections of WT and Pcdh12−/− genotypes, respectively. The mutant placenta exhibits numerous projections or islets within the labyrinthine zone. Enlargement of one of this peg (Fig. 4E) shows that these structures are composed of both glycogen cells (in dark purple) and spongiotrophoblasts (in light purple). The number of independent islets was counted in E17.5 placentas of each genotype and the results showed a significant difference (Fig. 4F, p < 0.001). A similar alteration was observed at E12.5 (not shown). We conclude that segregation of the junctional zone and the labyrinthine did not occur properly in absence of PCDH12.
Glycogen metabolism in Pcdh12-deficient placentas
One of the major roles of the placenta is to behave as an energy supplier for the embryo. Glycogen cells have the capacity to store huge amounts of glycogen, which can be broken down into glucose after glucagon stimulation (6). Glycogen was extracted from whole placentas and its concentration was measured. Glycogen concentration was significantly higher in Pcdh12−/− placentas at E12.5 and E17.5 (Fig. 5A, B). Correspondingly, the total amount of glycogen per placenta was increased in E12.5 Pcdh12−/− placenta (Fig. 5C); however, glycogen amounts reached a similar level in placentas of both genotypes at E17.5 (Fig. 5D). This feature can be explained by the weight difference between wild type and mutant placentas at this age. Thus, although the glycogen stores reached a similar level in late gestation, glycogen production was accelerated or its accumulation was more efficient in glycogen cells of Pcdh12−/− placentas.
Altogether, these data show that PCDH12 deficiency leads to a complex phenotype affecting several aspects of placental development.
Gene expression profiling of PCDH12-deficient placentas
To understand the molecular events responsible for the phenotypic alterations observed in Pcdh12−/− placentas, we analyzed the genome-wide expression profile of E12.5 mutant placentas in comparison with wild types. RNAs from 5 placentas of each genotype were analyzed with Affymetrix chips. The placentas derived from 3 litters. RNA quality was examined prior to hybridization and expression data were analyzed using several experimental and statistical criteria and gave satisfactory results (see supplemental data).
Remarkably, from the total number of 45,101 probes, 2,289 were assigned as differentially expressed since they present a SAM score superior to 0.75. The SAM threshold of 0.75 used herein corresponds to the 0.95 quantile for both upregulated and downregulated genes. A higher proportion of genes were upregulated (1,620 vs 669). Genes with unknown function were excluded from the initial set of 45,101 probes and the bioinformatic analysis was performed on a set of 18,542 probes.
Expression profiles obtained with DNA chips were examined by qRT-PCR for some specific genes showing different variation levels and different signal intensities (Table 1). Fold variations may be slightly different between the two technical approaches when the signals were at background levels (e.g., decysin in the wild types); however, the qRT-PCR results broadly confirm the DNA chip data.
Table 1.
Confirmation by qRT-PCR of representative gene expression in Pcdh12−/− placenta compared to wild type
| Gene Symbol* | qRT-PCR† |
Microarray ratio | ||
|---|---|---|---|---|
| WT | Pcdh12−/− | WT/Pcdh12−/− ratio | ||
| Phdal2 (Ipl) | 2,188 | 1,230 | 0.56 | 0.50 |
| Gapdh | 360,815 | 355,650 | 0.99 | 1.03 |
| Angiopoietin 2 | 1,021 | 1,017 | 1.00 | 1.08 |
| Tek (Tie 2) | 122 | 171 | 1.40 | 1.58 |
| Tie1 | 177 | 364 | 2.06 | 2.55 |
| Gja1 (Cx43) | 22,436 | 79,117 | 3.53 | 4.29 |
| Angiogenin 2 | 103 | 4,808 | 47 | 102 |
| Decysin | 19 | 9,137 | 491 | 180 |
Genes were chosen for their different expression levels and fold variations according to DNA chips data.
Data are expressed in arbitrary units and represent the mean value for 5 placentas in each case, normalized by Gapdh values, except for Gapdh.
First, a functional classification of differentially expressed genes was performed with the use of the visualization tool GoMiner. On the basis of prior knowledge provided by the gene ontology (GO) biological process category, we focused on classes of interest. Hence, selected gene sets were further analyzed with the GSEA method in order to detect top differential gene expression. In a predefined set, this method yields even small but coordinated changes (40). As shown in Table 2, functional groups that contained most of the differentially expressed genes were those involved in tissue morphogenesis and development, angiogenesis, cell-matrix adhesion and migration, immune response, and chromatin remodeling. All clusters except the chromatin-remodeling group contained only up-regulated genes. The heat maps, representing these variations in each cluster for individual samples, are shown in Supplemental Fig. 1–5.
Table 2.
Changes in gene expression categorized by gene ontology
| Gene Cluster* | Total | Changes
|
FWER value† | |
|---|---|---|---|---|
| Under | Over | |||
| Development and tissue morphogenesis | 485 | 0 | 144 | 0.010 |
| Cell adhesion and migration, matrix proteins | 80 | 0 | 62 | 0.010 |
| Immune response | 216 | 0 | 39 | 0.000 |
| Angiogenesis | 47 | 0 | 16 | 0.040 |
| Chromatin | 70 | 9 | 0 | 0.040 |
Gene grouping was obtained through GoMiner software and genes were further gathered in 5 clusters.
The detection of over and under expressed genes for each cluster and the corresponding Family Wise Error Rate was obtained with Gene Set Enrichment Analysis procedure (see Materials and Methods).
Independently, we examined the variations of specific genes that have previously been involved in placenta morphogenesis or secretion, cell-cell junction and angiogenesis (Table 3). As expected, Pcdh12 expression signals reached background levels
Table 3.
Changes in expression of specific placental, junctional or angiogenic genes*
| Gene Symbol | Gene Product† | Affymetrix ID | WT Signal Average‡ | Pcdh12−/− Signal Average‡ | Pcdh12−/− WT Signal Ratio | SAM Score | Q-Value |
|---|---|---|---|---|---|---|---|
| Placental genes | |||||||
| Phlda2 | Pleckstrin homology-like domain, family A, member 2 (IPL, TSSC3) | 1417837_at | 2252.22 | 1132.68 | 0.50 | 2.75 | 0.059 |
| Tpbpa | Trophoblast specific protein alpha | 1415808_at | 17589.14 | 15863.9 | 0.90 | 0.36 | 0.285 |
| Ascl2 | Achaete-scute complex homolog-like 2 (MASH2) | 1422396_s_at | 178.82 | 134.86 | 0.75 | 0.35 | 0.212 |
| Plac1 | Placental specific protein 1 | 1417553_at | 2345.84 | 1780.32 | 0.76 | 0.48 | 0.156 |
| Pem | Placentae and embryos oncofetal gene | 1423429_at | 4379.28 | 3564.46 | 0.81 | 0.61 | 0.105 |
| Prlpb | Prolactin-like protein B | 1420571_at | 4874.54 | 2649.18 | 0.54 | 0.84 | 0.143 |
| Prlpc | Prolactin-like protein C (PLP-H) | 1418418_a_at | 588.28 | 729.88 | 1.24 | 0.39 | 0.317 |
| Prlpcb | Prolactin-like protein C beta | 1448532_at | 13424.94 | 12016.8 | 0.90 | 0.48 | 0.309 |
| Prlpcg | Prolactin-like protein C-gamma (PLP-D) | 1424387_at | 2249.24 | 2006.56 | 0.89 | 0.16 | 0.532 |
| Upar2 | Urokinase-type plasminogen activator receptor,type 2 | 1452521_a_at | 176.28 | 205.1 | 1.16 | 0.33 | 0.173 |
| Epo | Erythropoietin 1 | 1433508_at | 545.18 | 576.22 | 1.06 | 0.16 | 0.524 |
| Igf2 | Insulin-like growth factor 2 | 1415931_at | 71.14 | 112.06 | 1.58 | 0.58 | 0.100 |
| Igf2r | Insulin-like growth factor 2 receptor | 1424112_at | 254.76 | 387.14 | 1.52 | 0.68 | 0.175 |
| Ptgs1 | Prostaglandin-endoperoxide synthase 1 (COX1) | 1423414_at | 21.06 | 64.88 | 3.08 | 0.95 | 0.108 |
| Ptgs2 | Prostaglandin-endoperoxide synthase 2 (COX2) | 1417262_at | 60.24 | 66.24 | 1.10 | 0.16 | 0.506 |
| Slc21a2 | Solute carrier family 21 (prostaglandin transporter) | 1420913_at | 507.48 | 1280.96 | 2.52 | 1.74 | 0.046 |
| Psg16 | Pregnancy specific GP16 | 1449238_at | 378.72 | 153.34 | 0.40 | 0.85 | 0.095 |
| Psg19 | Pregnancy specific glycoprotein 19 | 1421418_a_at | 39.78 | 175.06 | 4.40 | 1.28 | 0.068 |
| Cell junction genes | |||||||
| Pcdh12 | Protocadherin 12 | 1450473_at | 64.96 | 6.92 | 0.11 | 1.32 | 0.089 |
| Cdh1 | E-cadherin | 1448261_at | 744.42 | 680.3 | 0.91 | 0.11 | 0.524 |
| Cdh2 | N-cadherin | 1418815_at | 14.24 | 28.36 | 1.99 | 0.69 | 0.182 |
| Cdh3 | P-cadherin | 1426673_at | 499.22 | 775.92 | 1.55 | 0.80 | 0.158 |
| Cdh5 | VE-cadherin | 1433956_at | 496.2 | 1024.34 | 2.06 | 1.38 | 0.083 |
| Gja1 | Gap junction membrane channel protein alpha 1 (CX43) | 1438650_x_at | 706.08 | 3031.44 | 4.29 | 2.17 | 0.063 |
| Gjb2 | Gap junction membrane channel protein beta 2 (CX26) | 1423271_at | 3194.64 | 3498.7 | 1.10 | 0.17 | 0.511 |
| Gjb3 | Gap junction membrane channel protein beta 3 (CX31) | 1416715_at | 349.7 | 290.74 | 0.83 | 0.57 | 0.149 |
| Cldn1 | Claudin 1 | 1437932_a_at | 112.42 | 296.88 | 2.64 | 0.97 | 0.079 |
| Cldn5 | Claudin 5 | 1417839_at | 170.6 | 235.88 | 1.38 | 1.25 | 0.103 |
| Cldn11 | Claudin 11 | 1416003_at | 39.26 | 107.26 | 2.73 | 0.88 | 0.099 |
| Angiogenic genes | |||||||
| Vegfa | Vascular endothelial growth factor A | 1420909_at | 97.88 | 102.56 | 1.05 | 0.06 | 0.581 |
| Pgf | Placental growth factor | 1418471_at | 163.88 | 202.56 | 1.24 | 0.42 | 0.318 |
| Fgfr2 | Fibroblast growth factor receptor 2 | 1433489_s_at | 332.46 | 244.36 | 0.74 | 0.73 | 0.185 |
| Fgfr1 | Fibroblast growth factor receptor 1 | 1424050_s_at | 327.44 | 637.68 | 1.95 | 1.16 | 0.068 |
| Tie1 | Tyrosine kinase receptor 1 | 1416238_at | 35.26 | 89.76 | 2.55 | 0.90 | 0.143 |
| Tek | Endothelial-specific receptor tyrosine kinase | 1418788_at | 69.82 | 110.1 | 1.58 | 1.03 | 0.087 |
| Eng | Endoglin | 1417271_a_at | 108.98 | 201.18 | 1.85 | 1.69 | 0.046 |
| Angpt2 | Angiopoietin 2 | 1448831_at | 789.16 | 855.16 | 1.08 | 0.08 | 0.558 |
| Alk1 | Activin A receptor, type II-like 1 | 1451604_a_at | 103.24 | 160.12 | 1.55 | 0.82 | 0.143 |
| Tgfb1 | Transforming growth factor, beta 1 | 1420653_at | 86.22 | 191.68 | 2.22 | 0.95 | 0.102 |
| Tgfb2 | Transforming growth factor, beta 2 | 1438303_at | 34.78 | 95.78 | 2.75 | 1.51 | 0.057 |
| Tgfbr1 | Transforming growth factor, beta receptor I | 1420895_at | 255.16 | 275.84 | 1.08 | 0.25 | 0.417 |
| Hif1a | Hypoxia-inducible factor one alpha | 1427418_a_at | 1340.2 | 1431.92 | 1.07 | 0.22 | 0.399 |
| sFlt1 | Soluble FMS-like tyrosine kinase 1 | 1451756_at | 976.78 | 1673.66 | 1.71 | 0.71 | 0.143 |
| Flt1 | FMS-like tyrosine kinase 1 | 1419300_at | 197.68 | 294.3 | 1.49 | 0.51 | 0.143 |
| Kdr | Kinase insert domain protein receptor | 1449379_at | 75.66 | 130.66 | 1.73 | 0.87 | 0.103 |
Genes of these categories were selected as follows: lowest average signal value above 15 (except Pcdh12).
Other designations are indicated in parenthesis.
In arbitrary units.
For placental genes, the most significant variations were for the pleckstrin homology-like domain, family A, member 2 (also called Ipl or Tssc3), the pregnancy-specific GP16 and 19, the prostaglandin-endoperoxide synthase 1 (coding for cyclooxygenase 1), the solute carrier family 21 (a prostaglandin transporter) genes. The products of these genes have pivotal roles in placenta morphogenesis, modulation of the maternal immune system and fetal delivery (5, 7, 14, 39).
The expression of three classes of cell junction genes was altered in the mutant placenta: the cadherin (N, P and VE), the connexin (Gja1 coding for connexin 43) and the claudin (1 and 11) families, whose products are located in adherens, gap and tight junctions, respectively. Genes encoding for angiogenic factors and their receptors, such as the fibroblast growth factor receptor 1, Tie 1, the transforming growth factors beta 1 and 2, and soluble Flt1, were significantly upregulated in the mutant placentas.
Functional grouping of differentially expressed genes revealed that a large number of matrix-related and integrin transcripts were significantly upregulated in the mutant placenta and, for some of them, with a high Pcdh12−/−/wild type signal ratio. Table 4 shows selected genes of this category with lowest signal value above 15 and SAM score above 0.50. Genes coding for several collagen chains, or involved in collagen synthesis, for laminin chains, fibrillin 1 and two fibulins are present. Most selected genes encoding matrix proteins are expressed at high levels in the placenta. Six genes encoding integrins or proteins involved in cell-matrix adhesion and cell migration were significantly upregulated. Cell migration also requires matrix and integrin proteases and indeed a number of metalloproteinase family genes were upregulated. One member, the disintegrin metalloprotease (also called decysin) gene exhibited a 180-fold overexpression in the mutant placenta.
Table 4.
Changes in expression of genes involved in cell-matrix adhesion and migration*
| Gene Symbol | Gene Product | Affymetrix ID | WT Signal Average | Pcdh12−/− Signal Average | Pcdh12−/− WT Signal Ratio | SAM Score | Q-Value |
|---|---|---|---|---|---|---|---|
| Matrix and matrix-related genes | |||||||
| Col1a1 | Procollagen, type I, alpha-1 | 1423669_at | 119.52 | 364.06 | 3.05 | 1.15 | 0.115 |
| Col1a2 | Procollagen, type I, alpha 2 | 1450857_a_at | 359.84 | 996.74 | 2.77 | 1.53 | 0.087 |
| Col4a2 | Procollagen, type IV, alpha 2 | 1424051_at | 904.66 | 2044.74 | 2.26 | 1.16 | 0.096 |
| Col5a1 | Procollagen, type V, alpha 1 | 1416740_at | 90.32 | 306.74 | 3.40 | 1.75 | 0.078 |
| Col5a2 | Procollagen, type V, alpha 2 | 1450625_at | 153.46 | 551.64 | 3.59 | 1.27 | 0.068 |
| Col6a3 | Procollagen type VI, alpha 3 | 1424131_at | 38.94 | 131.54 | 3.38 | 1.27 | 0.095 |
| Col15a1 | Procollagen, type XV, alpha 1 | 1426955_at | 67.24 | 150.2 | 2.23 | 1.35 | 0.058 |
| Plod2 | Procollagen lysine, 2-oxoglutarate 5-dioxygenase 2 | 1416687_at | 316.70 | 990.40 | 3.13 | 1.15 | 0.066 |
| Lox | Lysyl oxidase | 1416121_at | 337.24 | 893.10 | 2.65 | 0.78 | 0.112 |
| Lama1 | Laminin, alpha 1 | 1418153_at | 113.22 | 349.60 | 3.09 | 0.77 | 0.165 |
| Lama5 | Laminin, alpha 5 | 1427009_at | 312.54 | 706.98 | 2.26 | 1.83 | 0.078 |
| Lamb2 | Laminin, beta 2 | 1416513_at | 126.9 | 290.38 | 2.29 | 1.45 | 0.090 |
| Fbn1 | Fibrillin 1 | 1425896_a_at | 66.12 | 182.16 | 2.75 | 0.98 | 0.089 |
| Fbln2 | Fibulin 2 | 1423407_a_at | 131.9 | 324.54 | 2.46 | 1.04 | 0.083 |
| Fbln5 | Fibulin 5 | 1416164_at | 50.04 | 181.10 | 3.62 | 1.26 | 0.061 |
| Integrin genes | |||||||
| Itga3 | Integrin alpha 3 | 1421997_s_at | 49 | 140.4 | 2.87 | 1.45 | 0.082 |
| Itga6 | Integrin alpha 6 | 1422444_at | 205.18 | 292.2 | 1.42 | 1.17 | 0.124 |
| Itgav | Integrin alpha V | 1421198_at | 165.18 | 221.82 | 1.34 | 0.74 | 0.093 |
| Itgb1 | Integrin beta1D | 1452545_a_at | 696.92 | 916.86 | 1.32 | 1.01 | 0.098 |
| Itgb5 | Integrin beta 5 | 1417533_a_at | 755.58 | 1392.96 | 1.84 | 1.17 | 0.075 |
| Itgb7 | Integrin beta 7 | 1418741_at | 22.88 | 46.88 | 2.05 | 0.76 | 0.152 |
| Matrix protease and matrix protease inhibitor genes | |||||||
| Dcsn | Disintegrin metalloprotease (decysin) | 1419476_at | 8.90 | 1598.20 | 179.57 | 1.78 | 0.068 |
| Mmp9 | Gelatinase | 1416298_at | 15.0 | 35.44 | 2.41 | 0.88 | 0.141 |
| Mmp14 | Matrix metalloproteinase 14 | 1448383_at | 189.3 | 396.1 | 2.09 | 0.93 | 0.087 |
| Mmp23 | Matrix metalloproteinase 23 | 1417281_a_at | 14.44 | 43.98 | 3.05 | 0.61 | 0.238 |
| Adam9 | A disintegrin and metalloproteinase domain 9 | 1416094_at | 233.4 | 335.9 | 1.44 | 0.83 | 0.068 |
| Adam12 | A disintegrin and metalloproteinase domain 12 | 1421171_at | 58.06 | 100.96 | 1.74 | 0.50 | 0.189 |
| Adam17 | A disintegrin and metalloproteinase domain 17 | 1421858_at | 208.26 | 262 | 1.26 | 0.92 | 0.098 |
| Adam19 | A disintegrin and metalloprotease domain 19 | 1418403_at | 196.76 | 385.88 | 1.96 | 0.90 | 0.093 |
| Adamts2 | A disintegrin-like and metalloprotease with thrombospondin type 1 motif, 2 | 1457058_at | 248.08 | 562.9 | 2.27 | 0.85 | 0.118 |
| Adamts9 | A disintegrin-like and metalloprotease with thrombospondin type 1 motif, 9 | 1431399_at | 28.76 | 47.94 | 1.67 | 0.77 | 0.143 |
| Timp2 | Tissue inhibitor of metalloproteinase 2 | 1450040_at | 39.44 | 74.6 | 1.89 | 0.81 | 0.103 |
Genes of these categories were selected as follows: lowest signal value above 15, SAM score above 0.50.
In addition, some miscellaneous genes, not classified by GoMiner software, were dramatically upregulated. Table 5 shows genes with Pcdh12−/−/wild type signal ratio above 2 or below 0.5 with highest average value above 100 and a SAM score above 0.8. Strikingly, a gene family for RNases or RNA-binding proteins emerged as highly differentially expressed. For example, genes coding for eosinophil-associated ribonucleases 1 and 2 and angiogenin-related genes were upregulated 238, 96 and 102-fold, respectively. Other unrelated genes including a gene involved in cell proliferation, cyclin E2, exhibited a significant expression variation.
Table 5.
Changes in expression of miscellaneous genes*
| Gene Symbol | Gene Product | Affymetrix ID | WT Signal Average | Pcdh12−/− Signal Average | Pcdh12−/− WT Signal Ratio | SAM Score | Q-Value |
|---|---|---|---|---|---|---|---|
| RNA-related genes | |||||||
| Ear1 | Eosinophil-associated ribonuclease 1 | 1421802_at | 2.90 | 690.48 | 238.10 | 3.78 | 0.046 |
| Ear2 | Eosinophil-associated ribonuclease 2 | 1449846_at | 24.56 | 2367.70 | 96.40 | 2.74 | 0.055 |
| Ear3 | Eosinophil-associated ribonuclease 3 | 1422411_s_at | 161.88 | 1810.56 | 11.18 | 2.03 | 0.065 |
| RNasea4 | RNase A family 4 | 1422603_at | 527.26 | 1475.30 | 2.80 | 0.92 | 0.099 |
| Ang2 | Angiogenin 2, ribonuclease A family | 1422415_at | 2.72 | 278.22 | 102.29 | 1.51 | 0.075 |
| Rbm3 | RNA-binding motif protein 3 | 1422660_at | 3490.62 | 1505.94 | 0.43 | 1.13 | 0.100 |
| Immune response genes | |||||||
| C1i | Complement component 1 inhibitor | 1416625_at | 997.34 | 2616.52 | 2.62 | 0.75 | 0.121 |
| Daf1 | Decay accelerating factor 1 | 1418762_at | 130.74 | 374.96 | 2.87 | 0.87 | 0.068 |
| Ccl6 | Chemokine (C-C motif) ligand 6 | 1417266_at | 47.24 | 107.36 | 2.27 | 1.03 | 0.110 |
| Cxcl1 | Chemokine (C-X-C motif) ligand 1 | 1419209_at | 33.62 | 100.06 | 2.98 | 2.15 | 0.068 |
| H2-Q7 | Histocompatibility 2, Q region locus 7 | 1418536_at | 45.9 | 111.62 | 2.43 | 1.04 | 0.068 |
| Lilrb4 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | 1420394_s_at | 50.46 | 105.52 | 2.09 | 0.84 | 0.153 |
| Lyzs | Lysozyme | 1423547_at | 91.7 | 188.56 | 2.06 | 0.91 | 0.150 |
| Stab1 | Stabilin | 1450199_a_at | 55.82 | 127.52 | 2.28 | 0.92 | 0.106 |
| Unrelated genes | |||||||
| Hmgb2 | High mobility group box 2 | 1437313_x_at | 843.62 | 356.00 | 0.42 | 1.07 | 0.098 |
| Lbp | Lipopolysaccharide binding protein | 1448550_at | 516.62 | 1815.60 | 3.51 | 0.90 | 0.099 |
| Hk2 | Hexokinase 2 | 1422612_at | 260.28 | 721.66 | 2.77 | 1.19 | 0.082 |
| Gsn | Gelsolin | 1456312_x_at | 607.50 | 2428.50 | 4.00 | 1.23 | 0.091 |
| Cyclin E2 | Cyclin E2 | 1422535_at | 415.10 | 170.10 | 0.41 | 0.97 | 0.123 |
DISCUSSION
In this paper, we show that loss of PCDH12 induced several morphological and transcriptional changes in placental development. To our knowledge, this is the first gene profiling study regarding a protocadherin or cadherin gene inactivation.
The embryonic growth defect was attributed to placental anomalies because the mutant mice reached the weights of controls 2 weeks after birth. Thus, this study supports the close link between placental and embryonic developments. As fetal growth was already retarded at E12.5, it is likely that placental functional defects were present much earlier on.
The catch-up of PCDH12-deficient pups after birth is reminiscent of the phenotype of mice deficient in the homeobox gene Esx1, for which placental defects, including missegregation of labyrinthine and spongiotrophoblast layers, were observed (26). Conversely, mice with other genetic ablations leading to placental anomalies and subsequent embryonic growth retardation did not recover after birth the body weight of their wild type littermates (10, 47). This observation may have important consequences in pediatrics for the management of children born small because of abnormal placental function: a close examination of placental phenotype and/or genetic alteration may provide indications on child thriving and may impact medical decisions (21).
The macroscopic morphology of PCDH12-deficient placentas was modified in two aspects: placenta size (in late gestation) and segregation of the labyrinthine and the junctional zone. Furthermore, the labyrinthine vasculature was much less developed; a feature that may be related to the lower cell density in the labyrinth. As the labyrinth is subjected to a major development between E12.5 and E17.5, and because vessel and cell densities were lower in the mutant labyrinth, we first suspected that labyrinth growth was specifically reduced during this time period in the mutant placenta. However, the surface proportions of the three layers were not statistically different between the two genotypes at both ages, indicating that placental growth retardation was general and not specific to the labyrinth. Nevertheless, it is possible that labyrinth development influences or coordinates the growth of the junctional zone and decidua. Alternatively, angiogenesis defects in the labyrinth and alteration of glycogen cell behavior in the junctional zone and decidua may independently decrease the growth rate of these layers.
The histological modifications observed herein are shared with other mouse genetic models. For example, a missegregation of labyrinth and junctional zone was also observed in interspecies hybrids, cloned conceptuses and after Esx1 deletion (26, 42, 48). However, this phenotype may be caused by different molecular mechanisms (38).
Considering Pcdh12−/− histomorphological anomalies, it is striking that gene clustering through GoMiner detected that clusters of genes involved in development, tissue morphogenesis, adhesion and migration, as well as angiogenesis, are significantly modified by loss of PCDH12. The majority of these genes was upregulated. Interestingly, genes of the chromatin-remodeling cluster, mostly silencing genes, were downregulated in the mutant. This may be connected to the general overexpression of regulated genes in PCDH12-deficient placentas. Upregulation of genes of the immune response cluster is more intriguing; it might reflect a modification in the activity of the uterine natural killer cells of the decidua. More generally, it is noteworthy that loss of a protein exhibiting cell-cell adhesive properties has a significant impact on gene transcription.
Most of the modified genes are expressed by PCDH12-positive cells, i.e., glycogen and endothelial cells. For example, Gja1 (CX43), Psg19, Mmp9 or Cox1 are solely or primarily expressed by glycogen cells (5, 6, 23, 36), and TIE1 or KDR are mainly located on endothelial cell surface (11, 33). This suggests intracellular signaling mechanisms between the membrane-linked Pcdh12 and the nuclear transcriptional machinery.
Remarkably, the cytoplasmic domains of γ-protocadherins and protocadherin FAT1 may be cleaved and translocated to the nucleus (18, 29). This opens interesting prospects for protocadherin signaling.
Other modified genes are expressed by cell types that do not express PCDH12. For example, PHLDA2 or the EAR family members are specific of labyrinthine trophoblasts (14) or immune response cells (24), respectively. This feature indicates that PCDH12 activity may influence other cell types of the placenta through intercellular signaling mechanisms that remain to be elucidated.
Angiogenesis
As shown by the histomorphological analysis (Fig. 2), proportions of placenta layers were not altered in Pcdh12-deficient mice. However, labyrinthine vascular density was decreased (Fig. 4), which may be caused by expression variation of several genes: first, increased levels of the VEGF decoy receptor gene Flt1 and moreover of its soluble form sFlt1; both are known for their antiangiogenic properties and are strongly expressed by the junctional zone (11, 13, 20). Similarly, increased expression of Flt1 and sFlt1 was recently shown to be responsible for the vascular defect phenotype observed in Adra2b−/− placentas (30); second, upregulation of Tie1, an orphan receptor regulating endothelial quiescence (34); third, upregulation of Tgfbeta1 and 2, which are known promoters of vessel maturation; fourth, upregulation of numerous matrix protease genes, which may release cryptic matrix or non-matrix derived inhibitors of angiogenesis (see (31), for review); and fifth, upregulation of fibulin 5, which inhibits the ability of endothelial cells to undergo angiogenic sprouting (2). Some of these anti-angiogenic factors may also target the maternal vasculature as well (see below).
Placenta morphogenesis and function
Expression of major placental morphogenetic genes, such as Plac1, Pem or Ascl2 was not significantly modified by loss of PCDH12, with the exception of the imprinted gene Phlda2 (also called Ipl or Tssc3) that was downregulated. Its protein product acts to limit placental growth in mice. The functional consequences of Phlda2 upregulation is at present unclear.
Genes coding for placental hormones (prolactins) or growth factors (IGF2, EPO) did not show major alterations in their expression. However, transcription levels of two genes coding for proteins involved in prostaglandin synthesis and uptake, Ptgs1 (cyclooxygenase 1 or Cox1) and Slc21a2 (coding for a prostaglandin transporter) respectively, were upregulated. Prostaglandins are critical molecules for initiation of parturition (reviewed in (32)) and are currently used to trigger delivery in humans. PCDH12 levels may thus influence the timing of birth through regulation of prostaglandin activity. This feature may be related to the upregulation in the mutant placenta of Mmp9 expression, encoding a matrix protease facilitating tissue remodeling in delivery (25).
We observed altered expression of two members of the pregnancy-specific glycoprotein (Psg) gene family, Psg16 and Psg19. PSGs are the most abundant fetal proteins in the maternal bloodstream in late pregnancy (27). In situ hybridization studies showed that Psg19 was exclusively transcribed by the junctional zone in the mouse (23). The importance of PSGs in the maintenance of pregnancy has been demonstrated by injection of anti-PSG antibodies, which induced abortion in mice (19). Most studies on PSGs are linked to the modulation of the maternal immune system, through cytokine secretion induction, preventing rejection of the fetuses. Consistently, expression of several immune response genes was upregulated in Pcdh12−/− placentas, including interleukin and chemokine ligands and receptors.
Matrix protein and proteases
Various types of collagen, laminin and fibulin were upregulated in absence of PCDH12. Once assembled, these secreted proteins are the major constituents of the extracellular matrix where they have a dual function: they provide tissues with their biomechanical properties and they support cell guidance during migration. In the placenta, the extracellular matrix enables the attachment of fetal and maternal tissues and provides a path for trophoblast invasion.
Pericellular protease activity further facilitates cell migration and invasion. Matrix metalloproteinases (MMPs) and their tissue inhibitors (TIMPs) are involved in morphogenesis of many epitheliomesenchymal organs (review, see (46)). Genes encoding three matrix metalloproteases, MMP9, 14 and 23, and several members of ADAMs (A Disintegrin And Metalloproteinase) or ADAMTS (ADAM with ThromboSpondin repeats) families were upregulated in the PCDH12-deficient placentas. All are involved in cell adhesion, cell migration, membrane protein shedding and proteolysis. The most upregulated gene of this category was decysin. This protein is a new member of the ADAM family. In placenta, decysin is mostly expressed in the junctional zone and around maternal vessels (3), i.e., at sites of PCDH12 expression. However, its substrate specificity is unknown.
Altogether, the transcriptional upregulation of these gene families is consistent with increased cell migration and tissue invasion. Therefore, the fact that PCDH12-deficient placentas showed defects in layer segregation is intriguing. It is possible that the orchestration of the various players of the cell migration process be compromised in the mutant placenta, leading to uncoordinated cell movement.
RNases
Surprisingly, a number of genes coding for secreted RNases, especially those of EAR 1–3 and angiogenin 2, were dramatically upregulated in PCDH12-deficient placentas. Some of these proteins are expressed by neutrophils or macrophages (see (12) and references therein) and possibly uterine natural killer cells. It has been suggested that these enzymes, as most of the RNase family members, may be involved in host defense. As indicated for the other immune response genes whose expression is modified in the mutant placenta, it is possible that PCDH12 modulates the uterine natural killer cell activity by an unknown mechanism.
Conclusions
Our data show that PCDH12 is a participant in placental morphogenesis. In absence of PCDH12, placentas exhibited growth retardation and histomorphological alterations. The gene profiling study comparing wild type and Pcdh12−/− placentas shows expression variations of a surprisingly large number of genes. This study constitutes a basis for the investigation of the multiple signaling pathways in which PCDH12 is involved.
Supplementary Material
Acknowledgments
C. Rampon was supported by the Ligue contre le Cancer and the Fondation pour la Recherche Médicale. This work was supported by recurrent grants from the Institut National de la Santé et de la Recherche Médicale, the Commissariat à l’Energie Atomique and Grenoble University. C. Rampon’s present address : Inserm U770, 94276 Le Kremlin Bicètre, France.
References
- 1.Adamson SL, Lu Y, Whiteley KJ, Holmyard D, Hemberger M, Pfarrer C, Cross JC. Interactions between trophoblast cells and the maternal and fetal circulation in the mouse placenta. Dev Biol. 2002;250:358–373. doi: 10.1016/s0012-1606(02)90773-6. [DOI] [PubMed] [Google Scholar]
- 2.Albig AR, Schiemann WP. Fibulin-5 antagonizes vascular endothelial growth factor (VEGF) signaling and angiogenic sprouting by endothelial cells. DNA and cell biology. 2004;23:367–379. doi: 10.1089/104454904323145254. [DOI] [PubMed] [Google Scholar]
- 3.Baran N, Kelly PA, Binart N. Decysin, a new member of the metalloproteinase family, is regulated by prolactin and steroids during mouse pregnancy. Biol Reprod. 2003;68:1787–1792. doi: 10.1095/biolreprod.102.009761. [DOI] [PubMed] [Google Scholar]
- 4.Bouillot S, Rampon C, Tillet E, Huber P. Tracing the glycogen cells with protocadherin 12 during mouse placenta development. Placenta. 2006;27:882–888. doi: 10.1016/j.placenta.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 5.Burdon C, Mann C, Cindrova-Davies T, Ferguson-Smith AC, Burton GJ. Oxidative Stress and the Induction of Cyclooxygenase Enzymes and Apoptosis in the Murine Placenta. Placenta. 2007 doi: 10.1016/j.placenta.2006.12.001. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coan PM, Conroy N, Burton GJ, Ferguson-Smith AC. Origin and characteristics of glycogen cells in the developing murine placenta. Dev Dyn. 2006;235:3280–3294. doi: 10.1002/dvdy.20981. [DOI] [PubMed] [Google Scholar]
- 7.Cook JL, Shallow MC, Zaragoza DB, Anderson KI, Olson DM. Mouse placental prostaglandins are associated with uterine activation and the timing of birth. Biol Reprod. 2003;68:579–587. doi: 10.1095/biolreprod.102.008789. [DOI] [PubMed] [Google Scholar]
- 8.Cross JC, Baczyk D, Dobric N, Hemberger M, Hughes M, Simmons DG, Yamamoto H, Kingdom JC. Genes, development and evolution of the placenta. Placenta. 2003;24:123–130. doi: 10.1053/plac.2002.0887. [DOI] [PubMed] [Google Scholar]
- 9.Cross JC, Hemberger M, Lu Y, Nozaki T, Whiteley K, Masutani M, Adamson SL. Trophoblast functions, angiogenesis and remodeling of the maternal vasculature in the placenta. Mol Cell Endocrinol. 2002;187:207–212. doi: 10.1016/s0303-7207(01)00703-1. [DOI] [PubMed] [Google Scholar]
- 10.DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345:78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 11.Dumont DJ, Fong GH, Puri MC, Gradwohl G, Alitalo K, Breitman ML. Vascularization of the mouse embryo: a study of flk-1, tek, tie, and vascular endothelial growth factor expression during development. Dev Dyn. 1995;203:80–92. doi: 10.1002/aja.1002030109. [DOI] [PubMed] [Google Scholar]
- 12.Dyer KD, Rosenberg HF. The RNase a superfamily: generation of diversity and innate host defense. Molecular diversity. 2006;10:585–597. doi: 10.1007/s11030-006-9028-2. [DOI] [PubMed] [Google Scholar]
- 13.Ferrara N, Gerber HP, LeCouter J. The biology of VEGF and its receptors. Nat Med. 2003;9:669–676. doi: 10.1038/nm0603-669. [DOI] [PubMed] [Google Scholar]
- 14.Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B. Placental overgrowth in mice lacking the imprinted gene Ipl. Proc Natl Acad Sci U S A. 2002;99:7490–7495. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frank M, Kemler R. Protocadherins. Curr Opin Cell Biol. 2002;14:557–562. doi: 10.1016/s0955-0674(02)00365-4. [DOI] [PubMed] [Google Scholar]
- 16.Georgiades P, Ferguson-Smith AC, Burton GJ. Comparative developmental anatomy of the murine and human definitive placentae. Placenta. 2002;23:3–19. doi: 10.1053/plac.2001.0738. [DOI] [PubMed] [Google Scholar]
- 17.Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of Mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- 18.Haas IG, Frank M, Veron N, Kemler R. Presenilin-dependent processing and nuclear function of gamma-protocadherins. J Biol Chem. 2005;280:9313–9319. doi: 10.1074/jbc.M412909200. [DOI] [PubMed] [Google Scholar]
- 19.Hau J, Gidley-Baird AA, Westergaard JG, Teisner B. The effect on pregnancy of intrauterine administration of antibodies against two pregnancy-associated murine proteins: murine pregnancy-specific beta 1-glycoprotein and murine pregnancy-associated alpha 2-glycoprotein. Biomedica biochimica acta. 1985;44:1255–1259. [PubMed] [Google Scholar]
- 20.Hirashima M, Lu Y, Byers L, Rossant J. Trophoblast expression of fms-like tyrosine kinase 1 is not required for the establishment of the maternal-fetal interface in the mouse placenta. Proc Natl Acad Sci U S A. 2003;100:15637–15642. doi: 10.1073/pnas.2635424100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jimenez-Chillaron JC, Patti ME. To catch up or not to catch up: is this the question? Lessons from animal models. Current opinion in endocrinology, diabetes, and obesity. 2007;14:23–29. doi: 10.1097/MED.0b013e328013da8e. [DOI] [PubMed] [Google Scholar]
- 22.Junghans D, Haas IG, Kemler R. Mammalian cadherins and protocadherins: about cell death, synapses and processing. Curr Opin Cell Biol. 2005;17:446–452. doi: 10.1016/j.ceb.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Kromer B, Finkenzeller D, Wessels J, Dveksler G, Thompson J, Zimmermann W. Coordinate expression of splice variants of the murine pregnancy-specific glycoprotein (PSG) gene family during placental development. European journal of biochemistry/FEBS. 1996;242:280–287. doi: 10.1111/j.1432-1033.1996.0280r.x. [DOI] [PubMed] [Google Scholar]
- 24.Larson KA, Olson EV, Madden BJ, Gleich GJ, Lee NA, Lee JJ. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc Natl Acad Sci U S A. 1996;93:12370–12375. doi: 10.1073/pnas.93.22.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Unlugedik E, Bocking AD, Challis JR. The role of prostaglandins in the mechanism of lipopolysaccharide-induced proMMP9 secretion from human placenta and fetal membrane cells. Biol Reprod. 2007;76:654–659. doi: 10.1095/biolreprod.106.057034. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Behringer RR. Esx1 is an X-chromosome-imprinted regulator of placental development and fetal growth. Nature genetics. 1998;20:309–311. doi: 10.1038/3129. [DOI] [PubMed] [Google Scholar]
- 27.Lin TM, Halbert SP, Spellacy WN. Measurement of pregnancy-associated plasma proteins during human gestation. J Clin Invest. 1974;54:576–582. doi: 10.1172/JCI107794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo S, Russel J, Taylor A. Determination of glycogen in small tissue samples. J Appl Physiol. 1970;28:234–236. doi: 10.1152/jappl.1970.28.2.234. [DOI] [PubMed] [Google Scholar]
- 29.Magg T, Schreiner D, Solis GP, Bade EG, Hofer HW. Processing of the human protocadherin Fat1 and translocation of its cytoplasmic domain to the nucleus. Exp Cell Res. 2005;307:100–108. doi: 10.1016/j.yexcr.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Muthig V, Gilsbach R, Haubold M, Philipp M, Ivacevic T, Gessler M, Hein L. Upregulation of Soluble Vascular Endothelial Growth Factor Receptor 1 Contributes to Angiogenesis Defects in the Placenta of {alpha}2B-Adrenoceptor Deficient Mice. Circ Res. 2007 doi: 10.1161/CIRCRESAHA.107.151563. [DOI] [PubMed] [Google Scholar]
- 31.Nyberg P, Xie L, Kalluri R. Endogenous inhibitors of angiogenesis. Cancer Res. 2005;65:3967–3979. doi: 10.1158/0008-5472.CAN-04-2427. [DOI] [PubMed] [Google Scholar]
- 32.Olson DM, Zaragoza DB, Shallow MC, Cook JL, Mitchell BF, Grigsby P, Hirst J. Myometrial activation and preterm labour: evidence supporting a role for the prostaglandin F receptor--a review. Placenta. 2003;24 (Suppl A):S47–54. doi: 10.1053/plac.2002.0938. [DOI] [PubMed] [Google Scholar]
- 33.Partanen J, Armstrong E, Makela TP, Korhonen J, Sandberg M, Renkonen R, Knuutila S, Huebner K, Alitalo K. A novel endothelial cell surface receptor tyrosine kinase with extracellular epidermal growth factor homology domains. Mol Cell Biol. 1992;12:1698–1707. doi: 10.1128/mcb.12.4.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Patan S. TIE1 and TIE2 receptor tyrosine kinases inversely regulate embryonic angiogenesis by the mechanism of intussusceptive microvascular growth. Microvascular research. 1998;56:1–21. doi: 10.1006/mvre.1998.2081. [DOI] [PubMed] [Google Scholar]
- 35.Rampon C, Prandini MH, Bouillot S, Pointu H, Tillet E, Frank R, Vernet M, Huber P. Protocadherin 12 (VE-cadherin 2) is expressed in endothelial, trophoblast, and mesangial cells. Exp Cell Res. 2005;302:48–60. doi: 10.1016/j.yexcr.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Reuss B, Hellmann P, Dahl E, Traub O, Butterweck A, Grummer R, Winterhager E. Connexins and E-cadherin are differentially expressed during trophoblast invasion and placenta differentiation in the rat. Dev Dyn. 1996;205:172–182. doi: 10.1002/(SICI)1097-0177(199602)205:2<172::AID-AJA8>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 37.Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol. 2005;284:12–24. doi: 10.1016/j.ydbio.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 38.Singh U, Fohn LE, Wakayama T, Ohgane J, Steinhoff C, Lipkowitz B, Schulz R, Orth A, Ropers HH, Behringer RR, Tanaka S, Shiota K, Yanagimachi R, Nuber UA, Fundele R. Different molecular mechanisms underlie placental overgrowth phenotypes caused by interspecies hybridization, cloning, and Esx1 mutation. Dev Dyn. 2004;230:149–164. doi: 10.1002/dvdy.20024. [DOI] [PubMed] [Google Scholar]
- 39.Snyder SK, Wessner DH, Wessells JL, Waterhouse RM, Wahl LM, Zimmermann W, Dveksler GS. Pregnancy-specific glycoproteins function as immunomodulators by inducing secretion of IL-10, IL-6 and TGF-beta1 by human monocytes. Am J Reprod Immunol. 2001;45:205–216. doi: 10.1111/j.8755-8920.2001.450403.x. [DOI] [PubMed] [Google Scholar]
- 40.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka M, Gertsenstein M, Rossant J, Nagy A. Mash2 acts cell autonomously in mouse spongiotrophoblast development. Dev Biol. 1997;190:55–65. doi: 10.1006/dbio.1997.8685. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka S, Oda M, Toyoshima Y, Wakayama T, Tanaka M, Yoshida N, Hattori N, Ohgane J, Yanagimachi R, Shiota K. Placentomegaly in cloned mouse concepti caused by expansion of the spongiotrophoblast layer. Biol Reprod. 2001;65:1813–1821. doi: 10.1095/biolreprod65.6.1813. [DOI] [PubMed] [Google Scholar]
- 43.Telo P, Breviario F, Huber P, Panzeri C, Dejana E. Identification of a novel cadherin (vascular endothelial cadherin-2) located at intercellular junctions in endothelial cells. J Biol Chem. 1998;273:17565–17572. doi: 10.1074/jbc.273.28.17565. [DOI] [PubMed] [Google Scholar]
- 44.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vecchi A, Garlanda C, Lampugnani MG, Resnati M, Matteucci C, Stoppacciaro A, Schnurch H, Risau W, Ruco L, Mantovani A, Dejana E. Monoclonal antibodies specific for endothelial cells of mouse blood vessels. Their application in the identification of adult and embryonic endothelium. Eur J Cell Biol. 1994;63:247–254. [PubMed] [Google Scholar]
- 46.Visse R, Nagase H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ Res. 2003;92:827–839. doi: 10.1161/01.RES.0000070112.80711.3D. [DOI] [PubMed] [Google Scholar]
- 47.Xiao X, Zuo X, Davis AA, McMillan DR, Curry BB, Richardson JA, Benjamin IJ. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. Embo J. 1999;18:5943–5952. doi: 10.1093/emboj/18.21.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zechner U, Reule M, Orth A, Bonhomme F, Strack B, Guenet, Hameister H, Fundele R. An X-chromosome linked locus contributes to abnormal placental development in mouse interspecific hybrid. Nature genetics. 1996;12:398–403. doi: 10.1038/ng0496-398. [DOI] [PubMed] [Google Scholar]
- 49.Zeeberg BR, Feng W, Wang G, Wang MD, Fojo AT, Sunshine M, Narasimhan S, Kane DW, Reinhold WC, Lababidi S, Bussey KJ, Riss J, Barrett JC, Weinstein JN. GoMiner: a resource for biological interpretation of genomic and proteomic data. Genome biology. 2003;4:R28. doi: 10.1186/gb-2003-4-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zeeberg BR, Qin H, Narasimhan S, Sunshine M, Cao H, Kane DW, Reimers M, Stephens RM, Bryant D, Burt SK, Elnekave E, Hari DM, Wynn TA, Cunningham-Rundles C, Stewart DM, Nelson D, Weinstein JN. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


