Abstract
Background
The embryonic temporal regulator FUSCA3 (FUS3) plays major roles in the establishment of embryonic leaf identity and the regulation of developmental timing. Loss-of-function mutations of this B3 domain transcription factor result in replacement of cotyledons with leaves and precocious germination, whereas constitutive misexpression causes the conversion of leaves into cotyledon-like organs and delays vegetative and reproductive phase transitions.
Results
Herein we show that activation of FUS3 after germination dampens the expression of genes involved in the biosynthesis and response to the plant hormone ethylene, whereas a loss-of-function fus3 mutant shows many phenotypes consistent with increased ethylene signaling. This FUS3-dependent regulation of ethylene signaling also impinges on timing functions outside embryogenesis. Loss of FUS3 function results in accelerated vegetative phase change, and this is again partially dependent on functional ethylene signaling. This alteration in vegetative phase transition is dependent on both embryonic and vegetative FUS3 function, suggesting that this important transcriptional regulator controls both embryonic and vegetative developmental timing.
Conclusion
The results of this study indicate that the embryonic regulator FUS3 not only controls the embryonic-to-vegetative phase transition through hormonal (ABA/GA) regulation but also functions postembryonically to delay vegetative phase transitions by negatively modulating ethylene-regulated gene expression.
Keywords: Arabidopsis, embryonic development, phase transition, FUSCA3, hormones, ethylene
Background
Spatial patterning in most multicellular organisms requires genes to both establish regions of cell differentiation and specify cellular fate. In the early Drosophila embryo, for example, cells are organized into boundaries by the pair rule and segment polarity genes, then they acquire distinct fates through homeotic gene expression [1]. Homeotic genes are also required to establish boundaries during temporal patterning, whereas heterochronic genes define the timing of the cell fate decisions within those boundaries [2]. One challenge in developmental biology is to identify and understand the overall developmental role of genes involved in the temporal patterning of genetic programs.
Higher plants are well-suited for identifying genes involved in developmental timing because they continually produce easily distinguishable organs throughout the life cycle, whose fates are dependent on the time of emergence [3]. The types of leaves that emerge over time often show distinctive developmental changes that allow them to be classified into juvenile and adult leaves. Later, when a plant enters reproductive development, the vegetative meristematic region switches to an inflorescence meristem that produces flower bracts with floral meristems in their axils [4]. Genetic analysis in Arabidopsis thaliana has identified a myriad of genes that converge to control the juvenile to adult leaf transitions and the switch of the vegetative meristem to reproductive development [5].
Unlike flowers and leaves, which form from a shoot apical meristem, the developmental relationship between embryonic leaves (or cotyledons) and adult foliar organs is complicated by cotyledon formation during embryonic patterning. Furthermore, in many plants such as Arabidopsis, cotyledons switch from a storage organ to a more leaflike photosynthetic organ soon after germination. Despite these complexities, single loss-of-function mutations in Arabidopsis have been identified in three genes, LEAFY COTYLEDON1 (LEC1), LEAFY COTYLEDON2 (LEC2) and FUSCA3 (FUS3), whose mutations result in the replacement of cotyledons with organs more similar to vegetative leaves [6-9]. In lec1 and fus3 mutants, genes that encode markers of late embryogenesis are reduced or missing [9,10]. By contrast, germination markers that normally proceed late embryogenesis are precociously activated. These expression patterns suggest that LEC1 and FUS3 may establish temporal boundaries. Although little is known about how these genes contribute to temporal patterns, it is known that FUS3 regulates and is regulated itself by the synthesis of two terpenoid hormones, abscisic acid (ABA) and gibberellins (GA) [10-12]. The ratio of these two hormones contributes to proper cotyledon patterning by regulating the rates of cell cycling [11].
Although extensive analyses of LEC1, LEC2 and FUS3 gene action have been carried out with respect to embryogenesis, the effects of these mutations on vegetative leaf development have not been studied extensively [11,13,14]. It has been shown that after germination, the first juvenile leaves of lec1 seedlings are shifted toward later leaf identities; however, this shift is not maintained, and successive leaves and flowering time were corrected back to a wild-type pattern [15]. This suggests that embryonic leaf development can have a restricted impact on future vegetative leaf identities. What remains unclear, however, is how cotyledon development impinges on later vegetative development, which is temporally and spatially distinct.
To address such questions, we decided to use a combination of controlled FUS3 activation during vegetative development with whole-genome transcript profiling. Using this approach, we discovered that FUS3 downregulates a collection of genes involved in ethylene biosynthesis and signaling. Consistent with this finding, loss-of-function fus3 mutants show ectopic ethylene responses at both the developmental and molecular levels. The fus3 plants also show precocious vegetative phase change; however, unlike the lec1 mutants, this change is not corrected at later adult stages. More importantly, the accelerated vegetative phase transition can be suppressed by inhibiting ethylene action either genetically or pharmacologically. Thus it appears that this previously defined embryonic regulator also has roles in vegetative development. One role of FUS3 during early seedling growth is to dampen ethylene action, which in turn contributes to a slowing of subsequent vegetative phase transitions. These results add ethylene to the list of hormones that contribute to temporal patterning in Arabidopsis.
Results
Downstream effectors of FUS3
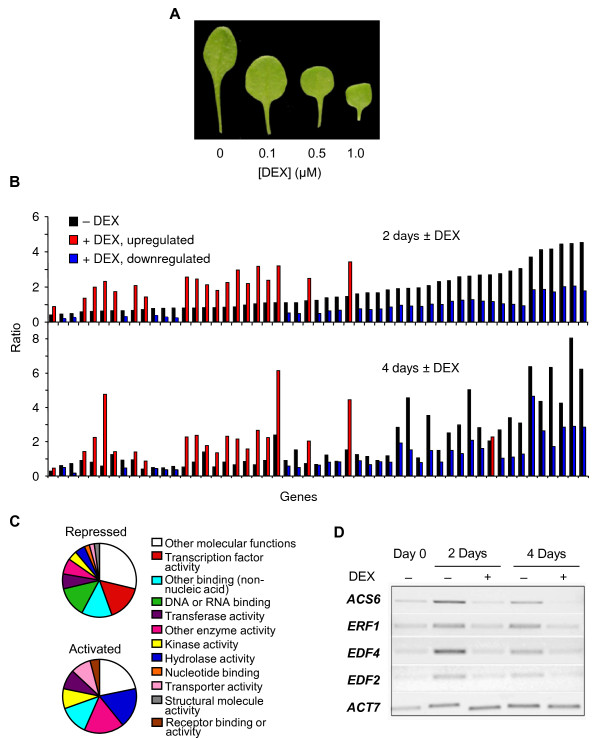
The discovery that FUS3 misexpression outside embryogenesis can influence vegetative leaf identity suggests that potential FUS3-dependent downstream effectors can be identified through whole-genome microarray analysis [11]. We constructed an inducible misexpression system by transforming the fus3-3 (fus3) mutant with a FUS3-glucocorticoid receptor (GR) translational fusion under the control of the epidermal specific AtML1 promoter (fus3 AtML1:FUS3-GR) [11]. Transgenic seeds were germinated and grown in minimal medium (Murashige and Skoog, MS) for 5 days, then plantlets were transferred to various concentrations of dexamethasone (DEX) to determine the minimal amount of activation needed to influence leaf identity. All concentrations of DEX tested had an effect on vegetative leaf shape; increasing concentrations of DEX resulted in a leaf with an increased paddle-like shape and a progressively shorter petiole (Figure 1A). At higher than 0.5 μM DEX concentrations, the appearance of trichomes on the upper or adaxial surface of the leaves was completely inhibited (data not shown). On the basis of these conditions, 1.0 μM DEX was chosen as the FUS3-activating condition.
Figure 1.
Microarray analysis of seedlings ectopically expressing FUS3. (A) Images showing the development of leaf 4 in 5-day-old fus3 ML1:FUS3-GR seedlings transferred to different concentrations of dexamethasone (DEX). (B) Genes that change at least twofold in expression in fus3 ML1:FUS3-GR seedlings treated with 1 μM DEX. The upper graph (2 days ± DEX) shows the ratios of fold changes in gene expression of 5-day-old seedlings grown for 2 days in the absence of DEX (black bars) or in the presence of DEX (red bars represent upregulated genes, and blue bars represent downregulated genes). The same order of genes is represented in the lower graph (4 days ± DEX), where 5-day-old seedlings were grown for 4 days in the absence of DEX (black bars) or in the presence of DEX (red and blue bars). (C) The proportions of genes associated with various molecular functions (gene ontology) are represented in the pie chart. (D) RT-PCR verification of ethylene-related genes identified by microarray analysis downregulated by FUS3 activation. Expression of four ethylene-related genes and an ACTIN7 (ACT7) control in the absence (-DEX) or presence (+DEX) of FUS3 activation.
Putative FUS3 targets involved in vegetative phase transitions were identified by examining the transcriptome of seedlings that transiently activate FUS3 using the AtML1:FUS3-GR DEX-inducible system [11]. To do this, transgenic seeds from AtML1:FUS3-GR were germinated and grown for 5 days in MS. After this time, half of the seedlings were transferred to MS media supplemented with 1.0 μM DEX (+DEX), and the other half were transferred to MS media supplemented with dimethyl sulfoxide (DMSO) (-DEX or control) for 2 or 4 days (Figure 1B). Putative FUS3 targets were chosen on the basis of the following criteria. Genes that increased (red bars) or decreased (blue bars) at least twofold in expression after two days of FUS3 activation (Figure 1B; +DEX) compared to the control (Figure 1B; -DEX) were considered to represent candidate FUS3 targets involved in phase transition. Utilizing this regime and the average of two replicate experiments, 19 genes increased (Additional file 1, Table S1) and 34 genes decreased (Additional file 2, Table S2) in expression (at least twofold) as the direct result of 2-day FUS3 activation (2 days +DEX) compared to the control (2 days -DEX). To independently verify the expression changes of the 2-day DEX induction experiment, a temporal replicate experiment was performed in which FUS3 activation was extended by transferring 5-day-old seedlings onto DEX for 4 days (4 days +DEX). All but 1 of the 53 genes selected by the 2-day experiment exhibited similar expression ratios in the 4-day experiment, demonstrating that activation of FUS3 results in changes in expression levels of a relatively small but reproducible gene set.
Gene ontology (GO) was utilized to categorize the activated and repressed genes by function (Figure 1C). Thirty-eight percent of genes upregulated by FUS3 activation appear to be involved in enzymatic activity, whereas a number of repressed genes are annotated as having roles in hormone synthesis or action. For example, the CYP707A3 gene that encodes the cytochrome p450 monooxygenase that catabolizes ABA is repressed more than twofold by FUS3 activation (Additional file 2, Table S2) [16]. This observation, in conjunction with the lack of FUS3-dependent induction of known ABA biosynthetic genes, suggests the increased ABA levels observed previously in FUS3 misexpression lines are due to decreased ABA catabolism [11]. A second class of hormone-related genes that are repressed by FUS3 activation at all time points and in both replicates are either involved in ethylene biosynthesis (ACS6, ACC synthase 6) or ethylene response (ERF1, ETHYLENE RESPONSE FACTOR1; ERF104, ETHYLENE RESPONSE FACTOR104; ESE3/EREBP, ETHYLENE AND SALT INDUCIBLE3; EDF4, ETHYLENE RESPONSE DNA BINDING FACTOR4). ERFs and EREBPs are transcription factors which contain an AP2 DNA binding domain, whereas EDF (or RAV) transcription factors contain both an AP2 and a B3 DNA-binding domain. Indeed, transcription factors constitute a dominant category (17%) of FUS3-repressed genes (Additional file 2, Table S2).
Potential downstream targets of FUS3 would be expected to contain the RY promoter element motif CATGCA, to which the B3 domain of FUS3 binds [17,18]. In fact, the RY sequence was found to be statistically enriched in both the up- and downregulated gene set compared to a randomized sample (p-value of 1.77 × 10-4 and 6.06 × 10-3 for 1, 000 bp upregulated and downregulated genes, respectively; Additional files 1 and 2, Tables S1 and S2). Together, these data strongly suggest that our experimental conditions identified a small gene set that is responsive to FUS3. Interestingly, this set includes only a few genes that typically mark seed maturation and late embryogenesis, such as seed storage proteins and late embryogenesis abundant proteins (LEA).
FUS3 negatively regulates a subset of genes that are responsive to ethylene
A closer inspection of our microarray data yielded additional ethylene-induced transcription factors (ERF2, EDF2 and EDF1) whose expression levels were repressed by FUS3 (Additional file 2, Table S2). These genes did not quite meet the stringent criteria of exhibiting a twofold decrease following both 2- and 4-day periods of FUS3 activation. Of the 14 AP2/EREBP/ERF and RAV/EDF genes that are known to be induced by ethylene exposure, six genes (ERF1, ERF2, ERF104, EDF2, EDF4 and EDF1) were dampened by FUS3 activation, suggesting that FUS3 plays a role in reducing the expression of genes involved in ethylene action [19]. This was verified by RT-PCR performed on ACS6, ERF1, EDF4 and EDF2 at 2 and 4 days of DEX induction (Figure 1D).
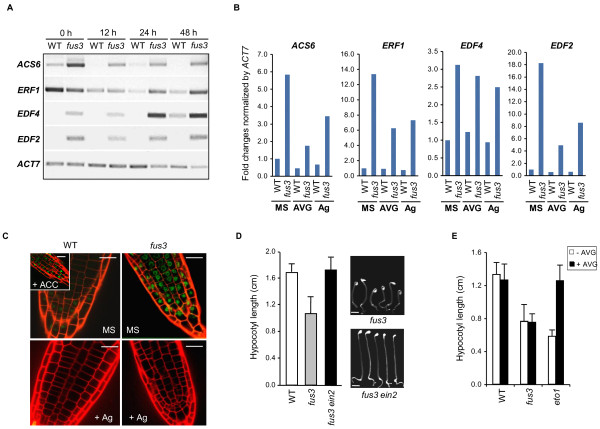
FUS3-mediated repression of genes involved in ethylene synthesis and action predict that fus3 loss-of-function mutants would exhibit an inverse expression pattern of these genes. Indeed, expression of ACS6, EDF2 and EDF4 genes were consistently higher in germinating fus3 versus wild type at all time points surveyed after imbibition, whereas ERF1 expression increased after 24 hours (Figure 2A). These results again support a negative role for FUS3 in regulating the expression of these ethylene responsive genes during germination. The presence of the RY element in the promoters of this gene subset may mean FUS3 directly downregulates their expression. Alternatively, FUS3 may act through ethylene synthesis or signaling to modulate their expression. To test these possibilities, we repeated the RT-PCR analysis of ACS6, ERF1, EDF4 and EDF2 on 2-day old wild-type and fus3 seedlings germinated in the presence of either an ethylene biosynthesis inhibitor, aminoethoxyvinylglycine (AVG), or an ethylene signaling inhibitor, silver ions (AgNO3). In both cases, the inhibitors partially suppressed the increased levels of the gene transcripts in fus3 seedlings compared to untreated controls (Figure 2B). Furthermore, a key transcriptional regulator of ethylene signaling, EIN3 [20,21], showed increased protein stability in the emerging root cells of fus3 compared to wild type, and this stability was eliminated by the addition of silver ions (Figure 2C). Although these results do not exclude a direct role for FUS3 on ethylene-responsive gene transcription, it does suggest that at least part of the aberrant gene expression observed in fus3 mutants does require functional ethylene synthesis or signaling.
Figure 2.
Increased expression of ethylene signaling and biosynthetic genes in fus3 mutant, and ethylene-related fus3 phenotypes. (A) RT-PCR of ethylene-related genes in wild-type (WT) and fus3 seeds germinated for 12, 24 and 48 hours in minimal medium (MS). ACTIN7 (ACT7) served as a control. (B) RT-PCR of ethylene-related genes in wild-type and fus3 seeds germinated for 48 hours in MS media or MS media containing 10 μM aminoethoxyvinylglycine (AVG) or 100 μM AgNO3 (Ag). Fold change in gene expression was normalized to ACTIN7, which served as a control. Similar trends were seen in two independent experiments. (C) GFP-EIN3 fluorescence in wild-type and fus3 roots incubated for 48 hours in minimal medium with (+) or without (-) 100 μM AgN03 (Ag). The inset shows the GFP-EIN3 fluorescence in wild-type roots exposed to the ethylene precursor 1-aminocyclopropane-1-carboxylic acid (ACC). Scale bar = 15.2 μm. (D) Images (right panels) and quantification (left panel) of hypocotyls length of 5-day-old fus3 and fus3 ein2-1 seedlings grown in the dark in the air. Scale bar = 0.2 cm. Averages from triplicate experiments ± SD are shown. fus3, n = 24; fus3 ein2-1, n = 21. (E) Quantification of hypocotyls length of 5-day-old wild-type, fus3 and eto1-1 seedlings grown in the dark in MS in the absence (-) or in the presence (+) of the ethylene synthesis inhibitor AVG. The eto1-1 seed, which overaccumulates ethylene gas, was used as a positive control to demonstrate rescue by AVG addition. Averages from triplicate experiments ± SD are shown. Wild type, n = 25; fus3, n = 25; eto1-1, n = 24.
To further probe the connection of FUS3 with ethylene action during germination, we studied the growth of dark-grown fus3 loss-of-function seedlings. When wild-type seedlings are germinated in the dark in the presence of ethylene, they show an exaggerated apical hook, a shortening of the hypocotyl and reduced root growth [22]. Termed the "triple response, " this developmental assay has been extremely useful in genetically dissecting the role of various genes involved in ethylene synthesis or signaling in Arabidopsis [23]. Dark-germinated fus3 plantlets grown in the absence of ethylene exhibited shorter hypocotyl growth and hooked cotyledon development (Figure 2D), which is consistent with their ectopic ethylene gene expression. Moreover, these phenotypes were alleviated by the introduction of a mutation (ein2-1) that reduces ethylene signaling into the fus3 genetic background (Figure 2D). To further differentiate ethylene synthesis and response, we repeated the experiment in the presence of the ethylene biosynthesis inhibitor AVG. In contrast to the ein2 mutation, AVG did not restore the hypocotyl length of fus3, which suggests that the ectopic ethylene responses observed in dark-grown fus3 seedlings were not due to increased ethylene synthesis (Figure 2E).
FUS3 functions to repress ethylene action during germination
The heightened ethylene responses observed in fus3 after germination suggest that loss of this gene function might have other, uncharacterized effects on vegetative development. Because FUS3 is a regulator of embryonic leaf identity, we decided to examine the vegetative leaf identity and phase transitions of the fus3 loss-of-function mutant more closely. From the emergence of the first leaf after germination through to flowering, each wild-type leaf adopts a unique identity based on size and shape, and this graded growth variation is often referred to as the "leaf heteroblastic series" [23]. Although rosette leaves always produce trichomes on the adaxial side of the wild-type leaf, trichomes on the abaxial side begin to appear on leaves only at later nodes (leaf 5 in wild type). Leaves that produce only adaxial trichomes are considered juvenile, whereas later leaves that show trichomes on both sides are regarded as adult [24]. Also, the ratio of leaf blade-to-petiole lengths is generally lower in early juvenile leaves and becomes higher as the plant transitions to adult leaves and flowering [25].
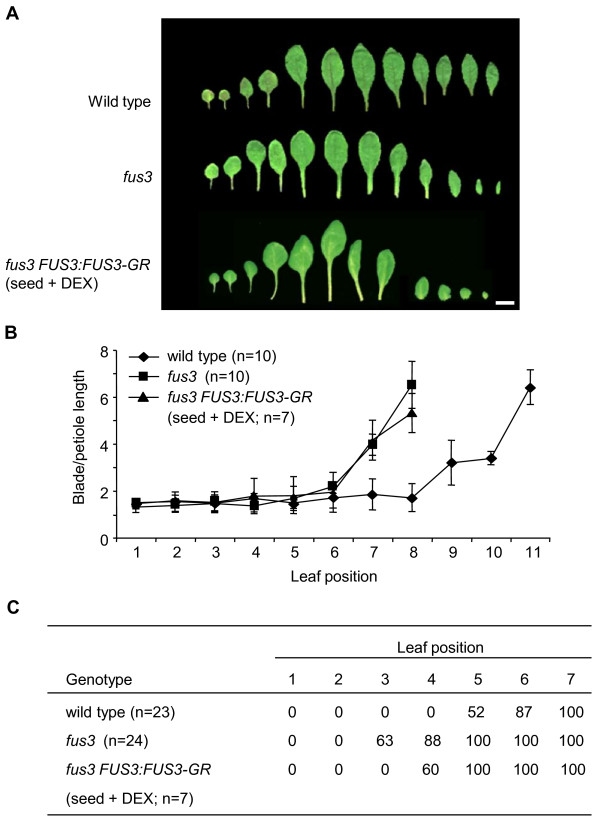
Comparisons of leaf profiles between the fus3 mutant and wild-type plants showed that the first two leaves from the mutant plant were more similar in shape and size to the third and fourth leaves of the wild type, and that this shift continued throughout the heteroblastic series (Figure 3A). This precocious shift was reflected in the blade-to-petiole length ratio of each individual leaf (Figure 3B) and in the leaf trichome distribution (Figure 3C). Abaxial trichomes frequently (63%) appeared on leaf 3 in fus3 mutant plants, approximately two leaves earlier than the wild type.
Figure 3.
Contributions of embryonic FUS3 to leaf identity and phase transitions. (A) Rosette leaf morphology of wild-type (top row), fus3 (middle row) and fus3 plants transformed with the FUS3:FUS3-GR construct (bottom row). fus3 FUS3:FUS3-GR parent plants were sprayed with 30 μM dexamethasone (DEX) during seed production. Leaves from ten plants were dissected, and representative profiles are shown (scale bar = 1 cm). (B) Ratios of blade-to-petiole lengths of individual rosette leaves in wild-type and fus3 and embryonically rescued fus3 FUS3:FUS3-GR plants. (C) Percentage of wild-type, fus3 and embryonically rescued fus3 FUS3:FUS3-GR (seed + DEX) rosettes that developed an abaxial trichome at each leaf position.
FUS3 has functions outside embryogenesis
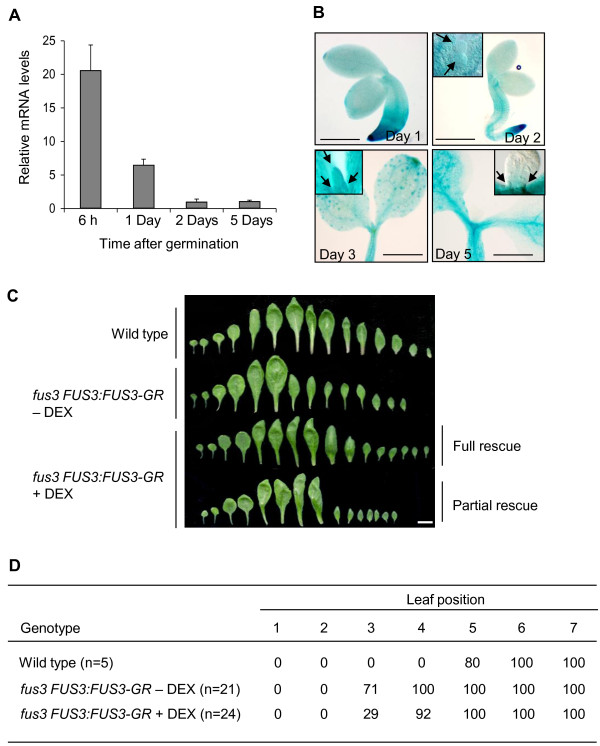
There are two explanations for the vegetative phenotypes of the fus3 mutant. Possibly the altered cotyledon development during embryogenesis in the mutant advances the progression of vegetative leaf identities after germination. Alternatively, FUS3 may function in the developing vegetative meristem. We tested these possibilities directly by measuring FUS3 transcript levels using quantitative RT-PCR (qRT-PCR). In wild-type seeds, FUS3 transcript is detected 6 hours postimbibition and declines over time, as previously shown [26]; it is still detectable 5 days after germination (Figure 4A). To further clarify the source tissue of postembryonic FUS3 expression, a transgenic line containing a sensitive FUS3:GUS transcriptional fusion was germinated and sampled over time for β-glucuronidase (GUS)-dependent blue histochemical staining [27]. The FUS3:GUS line used in these experiments was previously confirmed to reliably report embryonic FUS3 expression patterns based on FUS3 in situ hybridization [27]. Whole-mount preparations of seedlings showed blue staining in emerging leaf primordia from 2- to 5-day-old seedlings (Figure 4B). A similar expression pattern was found using a FUS3:GFP reporter previously described [26] (data not shown).
Figure 4.
FUS3 is expressed and functions postembryonically. (A) Relative expression of the FUS3 gene at various time points after germination. Quantitative RT-PCR was performed on germinating wild-type seeds imbibed for 6 hours, 1 day, 2 days and 5 days in minimal medium (MS). Transcript levels were normalized using ACTIN7 as an internal control. Results from triplicate samples are shown with error bars (SD). Experiments were repeated twice with similar results. Data from one of these replicates are shown. (B) Histochemical staining of FUS3:GUS seedlings at 1, 2, 3 and 5 days after germination in MS. Arrows indicate emerging leaf primordia showing β-glucuronidase (GUS) staining. Scale bars = 0.2 mm (day 1) and 0.4 mm (days 2, 3 and 5). (C) Wild-type and fus3 FUS3:FUS3-GR seeds germinated in MS without dexamethasone (DEX) (-DEX) or on 10 μM DEX (+DEX) for 2 days, then transferred to soil until bolting. Of the plants analyzed, 8.3% showed a full rescue of the rosette leaf morphology, and an example is shown (full rescue). A partial rescue was obtained for 62.5% of the plants, and an example is shown (partial rescue). The morphologies of the first six to eight fus3 FUS3:FUS3-GR leaves were rescued, but those of subsequent leaves were variable. Twenty-one to twenty-four plants were analyzed, constituting a representative profile. Scale bar = 1.0 cm. (D) Percentage of abaxial trichomes in wild-type rosettes and in fus3 FUS3:FUS3-GR rosettes of plants grown with (+) or without (-) DEX.
To functionally determine the effect of FUS3 from embryonic and vegetative tissues on the precocious vegetative phase transition, we constructed transgenic plants in which FUS3 could be activated either in the embryo or in vegetative tissue. fus3 was transformed with a FUS3-GR translational fusion construct that was under the control of the native FUS3 promoter (fus3 FUS3:FUS3-GR). Seeds produced from these lines in the absence of DEX application were phenotypically indistinguishable from fus3 loss-of-function mutants, with approximately 95% of the seed being desiccation-intolerant after 6 weeks of storage (data not shown). Of the small number of seeds that did survive desiccation, all produced ectopic trichomes on their cotyledons, as expected. Thus, in the absence of DEX application, the FUS3-GR fusion protein cannot rescue any of the known embryonic fus3 phenotypes. By contrast, in two independent experiments, approximately 50% of the seed produced by transgenic plants sprayed with 30 μM DEX during flowering showed desiccation tolerance after 6 weeks of storage. Of these, approximately 85% lacked ectopic trichomes on the cotyledons. The large reduction of fus3 seed phenotypes indicates that DEX application during flowering is sufficient to rescue mutant embryos.
From the transgenic seeds that were rescued, seven independent plants were randomly selected and grown to maturity in the absence of DEX, and leaf profiles were performed. Leaf profiles of all these lines still displayed accelerated heteroblastic development similar to that observed in fus3 leaf profiles (Figure 3A). Furthermore, the lines showed the same advanced vegetative phase transition as fus3 with respect to both individual leaf blade-to-petiole ratios (Figure 3B). The abaxial trichomes, however, did show a partial reversion to a wild-type profile (Figure 3C). Together, these results suggest that specifically removing embryonic FUS3 has a limited influence on the development of the first two juvenile leaves of Arabidopsis, but not on later leaves.
The inability of embryonic FUS3 to fully rescue the precocious vegetative phase transitions of fus3 suggests that this gene may function outside seed development. We tested this possibility by using the fus3 FUS3:FUS3-GR transgenic lines. Unlike in the previous experiment, however, we allowed transgenic lines to produce seed in the absence of DEX application and then germinated the seed in the presence of 10 μM DEX for 2 days. This short DEX application did not influence the heteroblastic phase change in wild-type plants as measured by either the blade-to-petiole transition or the appearance of abaxial trichomes in comparison to untreated plants (Additional file 3, Figure S1). By contrast, exposure of germinating fus3 FUS3:FUS3-GR transgenic seeds to DEX for 2 days resulted in a partial (62.5%) or full (8.3%) shift of leaf heteroblasty back toward a wild-type profile (Figure 4C). This rescue was partially reflected in trichome appearance (Figure 4D) and in full-leaf measurements; in wild type, the largest leaf was leaf 7, reaching a size of 3.9 ± 0.2 cm (n = 24). Similarly, the largest leaf in fus3 FUS3:FUS3-GR plants treated with DEX was leaf 7, which reached a size of 3.6 ± 0.3 cm (n = 23). In contrast, leaf 6 was the largest leaf in fus3 FUS3:FUS3-GR untreated plants (3.0 ± 0.3 cm; n = 23). In conclusion, a short pulse of FUS3 during the first 2 days of postembryonic growth can influence leaf identity in juvenile leaves.
FUS3-dependent vegetative phase variation requires functional ethylene signaling
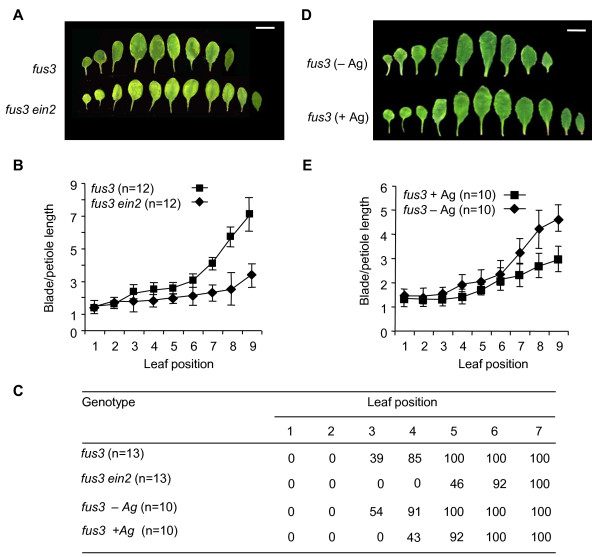
Loss-of-function fus3 mutants show advanced vegetative phase transition, increased expression of ethylene-regulated genes and phenotypes that are characteristic of increased ethylene signaling. To test if these phenotypes are related, a mutation conferring ethylene insensitivity was introduced into the fus3 background. The introduction of the ein2 mutation did suppress defects in vegetative phase change compared to the fus3 single mutant as measured by leaf profiles, blade-to-petiole ratios and abaxial trichome appearance (Figures 5A to 5C). These results suggest that increased ethylene signaling does contribute to the advanced vegetative phase transition phenotype observed in fus3.
Figure 5.
Influence of ethylene signaling on fus3 vegetative phase transition. (A) Mature rosette leaf morphologies of fus3 and fus3 ein2-1 plants. Leaves from 12 plants were dissected, and a representative profile is shown. Scale bar = 1.0 cm. (B) Ratios of blade-to-petiole lengths of mature individual rosette leaves from fus3 and fus3 ein2-1 plants. (C) Percentage of rosettes displaying trichomes on the abaxial surface at each leaf position. (D) Rosette leaf morphology of fus3 germinated for 2 days in minimal medium (MS) in the presence (+) or absence (-) of 100 μM AgNO3 (Ag) and then transferred to soil. Leaves from ten plants were dissected, and a representative profile is shown. Scale bar = 1.0 cm. (E) Ratios of blade-to-petiole lengths of individual fus3 rosette leaves of plants germinated in MS with (+) or without (-) 100 μM AgNO3 (Ag).
To further refine the ethylene contribution, we germinated fus3 seeds on 100 μM AgNO3 for 2 days and transferred the resulting seedlings to soil. Again, the fus3 phase transition phenotypes in the presence of AgNO3 were similar to those seen in the fus3 ein2 double-mutant (Figures 5C to 5E). Because in this experiment fus3 embryonic development occurred in the absence of the ethylene signaling inhibitor AgNO3, any alterations in fus3 vegetative phase transition were due to the transient inhibition of ethylene signaling during germination. In conclusion, the inhibition of ethylene signaling during early germination is able to partially suppress the premature vegetative phase transitions observed in the fus3 mutant.
Discussion
The FUS3 hormonal framework
Four lines of evidence suggest that FUS3 negatively regulates ethylene action. First, gain-of-function activation of FUS3 dampens the expression of a collection of ethylene-responsive genes, whereas a loss-of-function fus3 allele shows the opposite effect. Second, the ectopic expression of these ethylene-responsive genes in light-grown seedlings is dampened by the addition of ethylene synthesis or signaling inhibitors. Third, loss-of-function fus3 seedlings show common ethylene-response phenotypes in the dark which are dependent on ethylene signaling and also have increased stability of the key positive regulator of ethylene signaling, EIN3. Finally, the precocious vegetative phase variation observed in fus3 mutants is suppressed if ethylene signaling is inhibited genetically or pharmacologically.
Inhibition of ethylene biosynthesis does not rescue fus3 phenotypes in the dark, suggesting that alteration of ethylene signaling in fus3 is not merely a consequence of increased ethylene levels and requires functional ethylene signaling. However, genetic manipulation of FUS3 did influence the expression of ACS6, encoding for an enzyme involved in ethylene biosynthesis. The altered expression of this gene might reflect a feedback response to changes in ethylene signaling. On this note, only a subset of ethylene signaling genes was regulated by FUS3, which suggests that the relationship between FUS3 and ethylene is complex. Many of the ethylene-responsive genes regulated by FUS3 contain RY elements in their promoter, which suggests that this transcription factor may bind these promoters directly. A direct regulation of gene transcription may explain why the addition of ethylene inhibitors did not fully alleviate the ectopic expression of the ethylene-responsive genes. Nevertheless, the negative effect of FUS3 on the regulation of ethylene signaling adds this hormone to ABA and GA as those that are dependent on FUS3 during germination [11].
Coordination of the synthesis and signaling of various hormones is important in regulating overall plant growth and development, and many examples of ethylene, ABA and GA interactions have been reported [28-33]. In one of the best-studied cases, initial submergence of deepwater rice plants resulted in the accumulation of ethylene, which in turn enhanced GA sensitivity of the internodes to promote rapid growth [29]. The addition of ABA had the opposite effect by decreasing the sensitivity of internode tissues to GA [32]. With respect to leaf identity, interactions between ethylene and ABA have also been implicated in semiaquatic plants that exhibit heterophylly [34]. ABA induces terrestrial leaf development and ethylene stimulates the formation of submerged leaves in part by reducing ABA levels [30]. In Arabidopsis, where genetic analysis can be applied, loss of ethylene response does increase ABA levels in leaves, and, conversely, Arabidopsis mutants deficient in ABA synthesis show increased ethylene production [35-38]. The antagonistic and seemingly common relationships between ABA and GA and between ABA and ethylene imply that FUS3 acts as a control point of these multiple hormone pathways.
Suppressing ethylene signaling in fus3 mutants partially rescued the defects in juvenile leaf identities and the accelerated vegetative phase variation of this mutant, but did not rescue the embryonic leaf identity phenotypes. Perhaps this is not surprising, because there are major cellular differences beyond size and shape between embryonic cotyledons and juvenile and adult leaves. For example, genetic programs involved in storage reserves and desiccation appear to be embryo-specific. Related to this is that controlled FUS3 activation outside seed development was not sufficient to upregulate most of the standard seed-specific marker genes such as storage proteins, suggesting that other regulators are also required. The absence of seed-specific marker induction by the controlled activation of FUS3 in seedlings is similar to results obtained by Kagaya et al. [39], who also utilized a transgene-based activation system. Under their study conditions, vegetatively activated FUS3 could induce ectopic seed storage gene expression only with the addition of ABA. These observations are in contrast to constitutive FUS3 misexpression experiments, which result in storage reserve accumulation in vegetative tissues. Genetically reducing ABA levels in plants that constitutively express FUS3 also attenuates the ability of FUS3 to activate embryonic programs ectopically, consistent with an ABA requirement for full FUS3 function [11]. It appears that there is a limited time during germination during which tissues are more sensitized to FUS3 action. The presence of the transcription factor ABI5 has been posited to define a 60-hour developmental window during germination during which ABA can still arrest the growth of the emerging embryo and induce late-embryogenesis gene expression [40]. Whether this 60-hour checkpoint also defines a window in which FUS3 can reactivate late embryogenesis programming needs to be determined, but our expression studies place postembryonic FUS3 within this developmental window.
FUS3 and vegetative phase change
Transitions from one leaf identity to the next depend on the coordination of at least two independent processes: the timing of leaf initiation and a program that determines the duration of a developmental phase [3]. In the latter case, the genes that define the juvenile boundary may repress the expression of adult leaf-promoting genes above a certain threshold. One prediction of this model is that these juvenile boundary regulators will decrease in activity over time to allow adult phase change to occur [41]. FUS3 transcripts are detected after germination, and although they decrease, they can still be detected 5 days postgermination at a time when at least four to five leaf primordia have formed [42]. Because leaf 5 is considered to be the first to show adult characteristics, the decreased temporal domain of FUS3 expression correlates well with decreasing juvenile leaf identity. A simple model would suggest that the presence of FUS3 in leaf primordia contributes to juvenile leaf identity and also that as FUS3 activity drops, the transition to adult leaf phases can occur.
Although we have shown a vegetative role for FUS3, embryonic expression of FUS3 also affects the leaf identity of the first two juvenile leaves. This is similar to studies involving LEC1, which suggests that the identity of the embryonic foliar organs influences the identity of subsequent vegetative organs despite their different developmental origins [15]. Unlike fus3 mutants, however, aberrant vegetative phase transitions in lec1 plants are corrected in later leaves [15]. The lack of a detectable LEC1 signal after germination, as shown in public microarrays [43], may explain the difference between these mutants.
In contrast to animals, plant hormones can function in many different tissues and at various times during development, resulting in various developmental outcomes. One of the mechanisms by which FUS3 prolongs the juvenile phase appears to involve the downregulation of ethylene action, which suggests that this hormone is important in promoting adult-phase transitions (Figure 6). Morphometric analysis of leaf growth of etr1 mutants have shown that this ethylene-insensitive plant produce shorter, broader leaves, which are more akin to a juvenile leaf identity [37]. Consistent with this, the vegetative-to-floral transition in a collection of ethylene-insensitive mutants is delayed compared to the wild type [44]. In agreement with this, ML1:FUS3-GFP plants that constitutively express FUS3 postembryonically also show delayed flowering [11,26]. Using ethylene to quicken vegetative phase transitions is consistent with the role of this hormone in promoting germination [36,38]. Perhaps the short vegetative pulse of FUS3 serves to dampen the action of this hormone after it has stimulated germination so that the leaf identities do not advance too quickly.
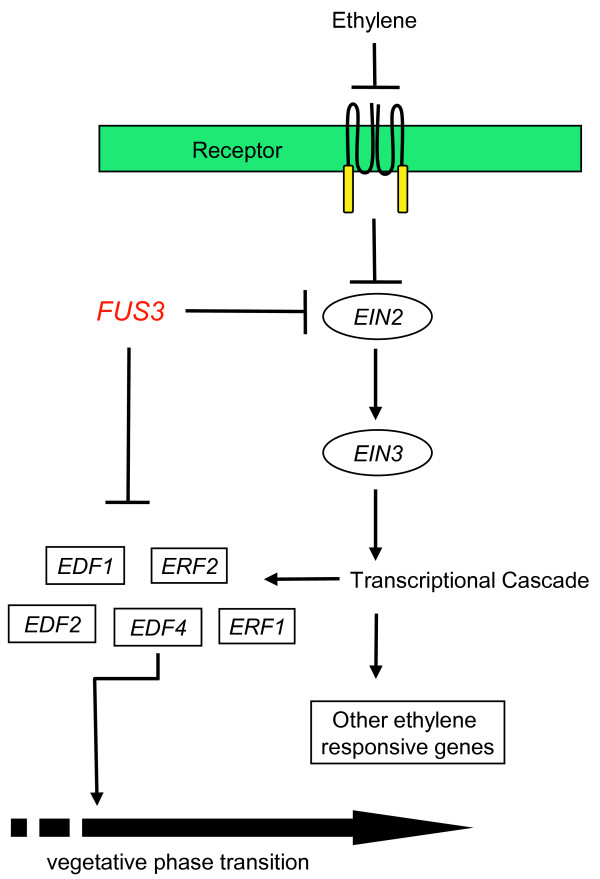
Figure 6.
Working model of phase change regulation by FUS3. During late embryogenesis and early germination, FUS3 negatively influences a number of factors, including ethylene signaling. As a consequence, the EIN3 protein, a key positive regulator of ethylene signaling, is reduced, thereby causing a decrease in the expression of downstream transcription factors such as ERFs and EDFs. A reduction of these ethylene-dependent transcription factors prevents the premature transition from the juvenile to the adult phase of development. In the loss-of-function fus3 mutant, ethylene signaling increases, which in turn accelerates vegetative phase transitions. The lack of full restoration of altered gene expression by ethylene inhibitors and the observation that only a subset of ethylene responsive genes are affected by FUS3 also suggest that FUS3 may have ethylene-independent effects. Consistent with this hypothesis, many ethylene-responsive genes contain FUS3-binding RY sequences in their promoter elements. It is therefore possible that FUS3 also influences these genes and some aspects of phase transitions through ethylene-independent mechanisms.
Conclusions
Herein we provide evidence that the embryonic regulator FUS3 also plays an important role postembryonically by negatively regulating vegetative phase transitions through repression of ethylene action. This study also implicates a role for ethylene in temporal patterning. Together with previous findings showing that FUS3 controls the embryonic-to-vegetative phase transition by modulating the ABA/GA ratio, this highlights a pivotal role of FUS3 in controlling the timing of expression of embryonic and vegetative programs through hormonal regulation. The pivotal role of FUS3 in regulating hormone levels and responses introduces another dimension to understanding the transitions of leaf identity that occur throughout the plant's life cycle. ABA and ethylene levels are also very dependent on both abiotic and biotic external signals, such as water availability, temperature and pathogen attack. Hence, the phase transitions observed will be dependent not only on developmental regulators such as FUS3 but also on environmental changes that influence hormone concentrations. In the future, it will be interesting to see if some of phenotypic plasticity observed in leaf shape can be linked not only to the relative timing of developmental regulators like FUS3 but also to environmental conditions, both of which impinge on ethylene, ABA and GA synthesis and/or signaling.
Methods
Plant material, growth conditions and leaf profiles
fus3-3 (fus3) [7] and all other strains used in this study were derived from a Columbia (Col) genetic background. For all experiments, plants were grown at 20°C under constant light. Seeds from all genotypes were generated from parent plants grown under identical conditions and stored under the same conditions for the same length of time. All transformations were performed as previously described [45]. DEX (Sigma-Aldrich, St Louis, MO, USA) was dissolved in DMSO. We performed leaf profiles according to Telfer et al. [24], for which 10 to 15 seedlings were germinated and grown in individual 4-inch round pots for approximately 3.5 weeks. To determine if DEX had any off-target effects on leaf heteroblasty, we performed blade-to-petiole measurements and trichome appearance assays on 10 wild-type plants and found that the chemical did not influence either of these markers of phase transition (Additional file 3, Figure S1). For the ein2 fus3 double-mutant construction, ein2-1 homozygotes were identified by screening for an altered triple-response phenotype in a segregating F2 population. Positive ein2-1 plants were grown and assayed for fus3 allele using a cleaved amplified polymorphic sequence marker that marks the mutant polymorphism [11]. For the pharmacological studies, 100 μM of the inhibitor AgNO3 or 10 μM of the inhibitor AVG were added to MS plates.
Cloning and generation of transgenic plants
The EIN3 cDNA was amplified and carboxy-terminally fused to GFP under the control of the 35S promoter in the pEGAD vector using the primers EIN3-BamHI forward 5'-ATA GGA TCC ATG ATG TTT AAT GAG ATG GGA AAT G-3'; EIN3-BamHI reverse 5'-ATA GGA TCC GAA CCA TAT GGA TAC ATC TTG C-3'; and the BamHI enzyme [46] to generate 35S:GFP-EIN3. The GFP-EIN3 translational fusion reporter fully rescues ein3 loss-of-function ethylene-dependent phenotypes (data not shown).
RT-PCR analysis
Thirty PCR cycles were performed with 250 ng of total RNA to amplify ethylene-related genes identified through microarrays using the following primers: ERF1 forward 5'-GTA TCC TCA ACG ACG CCT TTC AC-3'; ERF1 reverse 5'-CTT CAC CGT CAA TCC CTT ATC C-3'; EDF4/RAV1 forward 5'-GTA CAG GTT CCA TCT GTG AAA CC-3'; EDF4/RAV1 reverse 5'-CTC GTC TTC GTC CAT CTT CAC GTC-3'; EDF2/RAV2 forward 5'-GCA TAG ACG AGA TAA GTT CCT CCA C-3'; EDF2/RAV2 reverse 5'-GTT CTT GAA GTT GAC GAC GGC GTC G-3'; ACS6 forward 5'-ACA TCC AGA AGC TTC GAT TTG TAC-3'; and ACS6 reverse 5'-CTC GTC TAC AAA CTC TTC ATC GGA-3'. These primers were also used to check for expression of genes involved in ethylene synthesis and signaling in dry wild-type (Col) and fus3 immature seeds. ACTIN7 was amplified using the following primers: ACT7 forward 5'-GGT GAG GAT ATT CAG CCA CTT GTC TG-3' and ACT7 reverse 5'-TGT GAG ATC CCG ACC CGC AAG ATC-3' and was used as the control. Seeds were imbibed as described for 12, 24 and 48 hours.
Quantitative RT-PCR
RNA extraction and qRT-PCR were performed as previously described [26]. Sequences of gene-specific primers were designed to amplify 190-bp products. These included the following: ACT7190 forward 5'-TCA CAG AGG CAC CTC TTA ACC-3'; ACT7190 reverse, 5'-CCC TCG TAG ATT GGC ACA G-3'; FUS3190 forward, 5'-TGT GAA TGC TCA TGG TCT GC-3'; and FUS3190 reverse, 5'-GGA GGA GAA GAT CGT TAA CCA C-3'.
Histochemistry and microscopy
Detection of GUS activity and sectioning was performed as described by Donnelly et al. [47]. Leaf tissues were cleared in 70% ethanol and 8:2:1 chloral hydrate:glycerol:water, then mounted in the same solution on microscope slides.
Microarray and bioinformatic analysis
Wild-type Columbia seeds containing the ML1:FUS3:GR construct [11] were imbibed for 5 days at 4°C on filter paper placed on MS plates. To minimize the effects of biological variation, we followed a pooling strategy according to that described by Zhu et al. [48] in which approximately 100 individual plants per replicate experiment were harvested and pooled before RNA extraction and hybridization. The seedlings were then transferred to 20°C for 5 days. Shoot tissues from seedlings were collected for RNA isolation (day 5). The remaining seedlings on the filter paper were transferred to either MS plates consisting of 1 μM DEX or the same concentration of DMSO. Two and four days after the transfer, shoot tissue was collected from the seedlings. The above protocol was repeated to obtain a second biological replica.
The standard Affymetrix labeling and hybridization protocols were used on an ATH1 Affymetrix GeneChip microarray encompassing 22, 814 probe sets (Affymetrix Inc, Santa Clara, CA, USA). Data were globally normalized using the MAS5.0 global normalization algorithm with a target value of 500. The resulting data were filtered to eliminate MAS5.0 "marginal" and "absent" calls (Present requires both sets of replicates for treatment and control samples). In addition, the r values for the correlation of the expression values between the two replicate set of experiments was greater than 0.95. The complete data set has been submitted to the EMBL-EBI ArrayExpress database under the accession number E-MEXP-3465 in compliance with MIAME standards.
The average expression value for each gene was calculated for two replicate samples at each time point. The Cluster and TreeView software programs were used for cluster analysis [49]. The data set was filtered to identify genes that exhibited more than a twofold increase or decrease in expression level [50]. Cluster analysis was performed using average linkage hierarchical clustering with the centered Pearson correlation coefficient as the similarity metric. RY motif (CATGCA) and p-values were obtained using the "Motif Analysis" tool on TAIR http://www.arabidopsis.org/tools/bulk/motiffinder/index.jsp.
Temporal activation of FUS3
The FUS3:FUS3-GR construct was made by replacing the ML1 promoter in the ML1:FUS3-GR construct with the FUS3 promoter from the FUS3:FUS3-GFP construct previously described [11]. For FUS3:FUS3-GR activation during seed development, DEX was diluted to 30 μM in dH2O containing 0.05% Triton X-100 and sprayed on flowering plants twice weekly until the plants had senesced.
Confocal microscopy
To study EIN3 stability in a fus3 background, we crossed fus3 with an ein3 35S:GFP:EIN3 transgenic line. The stability of the EIN3 protein in the fus3 background was compared to non-fus3 mutant plants from the same F1 plant. At least 20 seedlings were observed for each genotype. A Nikon inverted fluorescence microscope equipped with a Nikon water immersion objective (Nikon Instruments, Inc, Melville, NY, USA) and a Bio-Rad Radiance 2000 confocal head (Bio-Rad Laboratories, Hercules, CA, USA) was used to detect GFP fluorescence. The same confocal settings were used in all experiments.
Abbreviations
bp: base pair; GFP: green fluorescent protein; RT-PCR: reverse transcriptase polymerase chain reaction.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PM and SG designed the experiments and wrote the manuscript. SL and SG performed the biological part of the microarray study and the data analysis of the microarray experiment (Figure 1 and supplementary tables). SL and YT performed RT-PCR. SL performed the experiments shown in Figures 3 and 5. QSL and SG performed the experiments shown in Figure 4. FD and JH constructed and selected 35S:GFP-EIN3 transgenics. YT constructed and selected FUS3-GR constructs. NJP performed cluster analysis. All authors read and approved the final manuscript.
Supplementary Material
Table S1 Genes upregulated by ectopic FUS3 activation. Values for fold changes in expression after 2 and 4 days (d) of FUS3 activation with dexamethasone (+DEX) are the averages of two replicates. The presence of RY promoter motifs (CATGCA) in the 500 bp (0.5 K), 1, 000 bp (1 K) and 3, 000 bp (3 K) upstream regions of each gene is included. p-values are 3.06 × 10-4, 1.77 × 10-4 and 2.57 × 10-2, respectively.
Table S2 Genes downregulated by ectopic FUS3 activation. Values for fold changes in expression after 2 and 4 days (d) of FUS3 activation with dexamethasone (+DEX) are the averages of two replicates. Genes involved in hormone metabolism or response are in bold. Ethylene response genes that were downregulated less than twofold by FUS3 activation at both 2 and 4 days are in italics. The presence of RY promoter motifs (CATGCA) in 500 bp (0.5 K), 1, 000 bp (1 K) and 3, 000 bp (3 K) upstream regions of each gene is included. p-values are 4.27 × 10-2, 6.06 × 10-3 and 3.46 × 10-2, respectively.
Figure S1 Phenotypic analysis of wild-type plants exposed to dexamethasone (DEX). Wild-type seeds were germinated in 10 μM DEX for 2 days and then transferred to soil. The increase in the blade-to-petiole ratio and the appearance of the abaxial trichomes on leaf 5 is comparable to wild-type profiles shown in Figure 3. (A) Ratios of blade-to-petiole lengths of individual wild-type rosette leaves. (B) Percentage of wild-type rosettes that formed abaxial trichomes at each leaf position.
Contributor Information
Shelley Lumba, Email: shelley.lumba@utoronto.ca.
Yuichiro Tsuchiya, Email: yuichiro.tsuchiya@utoronto.ca.
Frederic Delmas, Email: frederic.delmas@bordeaux.inra.fr.
Jodi Hezky, Email: jodi.hezky@gmail.com.
Nicholas J Provart, Email: nicholas.provart@utoronto.ca.
Qing Shi Lu, Email: Mimmie.lu@utoronto.ca.
Peter McCourt, Email: peter.mccourt@utoronto.ca.
Sonia Gazzarrini, Email: gazzarrini@utsc.utoronto.ca.
Acknowledgements
We are grateful to the Gazzarrini and McCourt research groups for discussion and to K Breitkreuz for critical reading of the manuscript. We thank J Kieber (University of North Carolina) for the eto1-1 seed. This work was funded by a Natural Sciences and Engineering Research Council of Canada (NSERC) grant (to SG) and an NSERC Industrial Research Chair grant (to PM).
References
- Akam M. The molecular basis for metameric pattern in the Drosophila embryo. Development. 1987;101:1–22. [PubMed] [Google Scholar]
- Thummel CS. Molecular mechanisms of developmental timing in C. elegans and Drosophila. Dev Cell. 2001;1:453–465. doi: 10.1016/s1534-5807(01)00060-0. [DOI] [PubMed] [Google Scholar]
- Kerstetter RA, Poethig RS. The specification of leaf identity during shoot development. Annu Rev Cell Dev Biol. 1998;14:373–398. doi: 10.1146/annurev.cellbio.14.1.373. [DOI] [PubMed] [Google Scholar]
- Henderson IR, Dean C. Control of Arabidopsis flowering: the chill before the bloom. Development. 2004;131:3829–3838. doi: 10.1242/dev.01294. [DOI] [PubMed] [Google Scholar]
- Poethig RS. Small RNAs and developmental timing in plants. Curr Opin Genet Dev. 2009;19:374–378. doi: 10.1016/j.gde.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäumlein H, Miséra S, Luerßen H, Kölle K, Horstmann C, Wobus U, Müller AJ. The FUS3 gene of Arabidopsis thaliana is a regulator of gene expression during late embryogenesis. Plant J. 1994;6:379–387. [Google Scholar]
- Keith K, Kraml M, Dengler NG, McCourt P. fusca3: a heterochronic mutation affecting late embryo development in Arabidopsis. Plant Cell. 1994;6:589–600. doi: 10.1105/tpc.6.5.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke DW, Franzmann LH, Nickle TC, Yeung EC. Leafy cotyledon mutants of Arabidopsis. Plant Cell. 1994;6:1049–1064. doi: 10.1105/tpc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Matsudaira Yee K, Danao J, Zimmerman JL, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON1 is an essential regulator of late embryogenesis and cotyledon identity in Arabidopsis. Plant Cell. 1994;6:1731–1745. doi: 10.1105/tpc.6.12.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Hayama R, Tsuchiya Y, Nishimura M, Kawaide H, Kamiya Y, Naito S. The role of ABI3 and FUS3 loci in Arabidopsis thaliana on phase transition from late embryo development to germination. Dev Biol. 2000;220:412–423. doi: 10.1006/dbio.2000.9632. [DOI] [PubMed] [Google Scholar]
- Gazzarrini S, Tsuchiya Y, Lumba S, Okamoto M, McCourt P. The transcription factor FUSCA3 controls developmental timing in Arabidopsis through the hormones gibberellin and abscisic acid. Dev Cell. 2004;7:373–385. doi: 10.1016/j.devcel.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Curaba J, Moritz T, Blervaque R, Parcy F, Raz V, Herzog M, Vachon G. AtGA3ox2, a key gene responsible for bioactive gibberellin biosynthesis, is regulated during embryogenesis by LEAFY COTYLEDON2 and FUSCA3 in Arabidopsis. Plant Physiol. 2004;136:3660–3669. doi: 10.1104/pp.104.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotan T, Ohto M, Matsudaira Yee K, West MAL, Lo R, Kwong RW, Yamagishi K, Fischer RL, Goldberg RB, Harada JJ. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Stone SL, Kwong LW, Matsudaira Yee K, Pelletier J, Lepiniec L, Fischer RL, Goldberg RB, Harada JJ. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. Proc Natl Acad Sci USA. 2001;98:11806–11811. doi: 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H, Shoda K, Kim GT, Uchimiya H. Heteroblasty in Arabidopsis thaliana (L.) Heynh. Planta. 2000;210:536–542. doi: 10.1007/s004250050042. [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E. The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO J. 2004;23:1647–1656. doi: 10.1038/sj.emboj.7600121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reidt W, Wohlfarth T, Ellerström M, Czihal A, Tewes A, Ezcurra I, Rask L, Bäumlein H. Gene regulation during late embryogenesis: the RY motif of maturation-specific gene promoters is a direct target of the FUS3 gene product. Plant J. 2000;21:401–408. doi: 10.1046/j.1365-313x.2000.00686.x. [DOI] [PubMed] [Google Scholar]
- Mönke G, Altschmied L, Tewes A, Reidt W, Mock HP, Bäumlein H, Conrad U. Seed-specific transcription factors ABI3 and FUS3: molecular interaction with DNA. Planta. 2004;219:158–166. doi: 10.1007/s00425-004-1206-9. [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, Gadrinab C, Heller C, Jeske A, Koesema E, Meyers CC, Parker H, Prednis L, Ansari Y, Choy N, Deen H, Geralt M, Hazari N, Hom E, Karnes M, Mulholland C, Ndubaku R, Schmidt I, Guzman P, Aguilar-Henonin L, Schmid M, Weigel D, Carter DE, Marchand T, Risseeuw E, Brogden D, Zeko A, Crosby WL, Berry CC, Ecker JR. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- Schaller GE, Kieber JJ. In: The Arabidopsis Book. Somerville C, Meyerowitz E, editor. Vol. 1. Rockville, MD: American Society of Plant Biologists; 2002. Ethylene; p. e0071.http://www.bioone.org/doi/pdf/10.1199/tab.0071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukaya H. In: The Arabidopsis Book. Somerville C, Meyerowitz E, editor. Vol. 1. Rockville, MD: American Society of Plant Biologists; 2002. Leaf development; p. e0072.http://www.bioone.org/doi/pdf/10.1199/tab.0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124:645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- Bollman KM, Aukerman MJ, Park MY, Hunter C, Berardini TZ, Poethig RS. HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development. 2003;130:1493–1504. doi: 10.1242/dev.00362. [DOI] [PubMed] [Google Scholar]
- Lu QS, Dela Paz J, Pathmanathan A, Chiu RS, Tsai AYL, Gazzarrini S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. Plant J. 2010;64:100–113. doi: 10.1111/j.1365-313X.2010.04307.x. [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y, Nambara E, Naito S, McCourt P. The FUS3 transcription factor functions through the epidermal regulator TTG1 during embryogenesis in Arabidopsis. Plant J. 2004;37:73–81. doi: 10.1046/j.1365-313x.2003.01939.x. [DOI] [PubMed] [Google Scholar]
- Cox MCH, Benschop JJ, Vreeburg RAM, Wagemaker CAM, Moritz T, Peeters AJM, Voesenek LACJ. The roles of ethylene, auxin, abscisic acid, and gibberellin in the hyponastic growth of submerged Rumex palustris petioles. Plant Physiol. 2004;136:2948–2960. doi: 10.1104/pp.104.049197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kende H, van der Knaap E, Cho HT. Deepwater rice: a model plant to study stem elongation. Plant Physiol. 1998;118:1105–1110. doi: 10.1104/pp.118.4.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara A, Ikegami K, Koshiba T, Nagata T. Effects of ethylene and abscisic acid upon heterophylly in Ludwigia arcuata (Onagraceae) Planta. 2003;217:880–887. doi: 10.1007/s00425-003-1062-z. [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Jackson MB, Gühl K, Vreeburg RAM, Croker SJ, Peeters AJM, Voesenek LACJ. Contrasting interactions between ethylene and abscisic acid in Rumex species differing in submergence tolerance. Plant J. 2005;44:756–768. doi: 10.1111/j.1365-313X.2005.02563.x. [DOI] [PubMed] [Google Scholar]
- Hoffmann-Benning S, Kende H. On the role of abscisic acid and gibberellin in the regulation of growth in rice. Plant Physiol. 1992;99:1156–1161. doi: 10.1104/pp.99.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazzarrini S, McCourt P. Cross-talk in plant hormone signalling: what Arabidopsis mutants are telling us. Ann Bot. 2003;91:605–612. doi: 10.1093/aob/mcg064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii K, Kuwabara A, Nagata T. Characterization of anisocotylous leaf formation in Streptocarpus wendlandii (Gesneriaceae): significance of plant growth regulators. Ann Bot. 2004;94:457–467. doi: 10.1093/aob/mch160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiwocha SDS, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross ARS, Kermode AR. The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 2005;42:35–48. doi: 10.1111/j.1365-313X.2005.02359.x. [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Nambara E, Cutler S, Kawaide H, Kamiya Y, McCourt P. Regulation of abscisic acid signalling by the ethylene response pathway in Arabidopsis. Plant Cell. 2000;12:1117–1126. doi: 10.1105/tpc.12.7.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeNoble ME, Spollen WG, Sharp RE. Maintenance of shoot growth by endogenous ABA: genetic assessment of the involvement of ethylene suppression. J Exp Bot. 2004;55:237–245. doi: 10.1093/jxb/erh031. [DOI] [PubMed] [Google Scholar]
- Beaudoin N, Serizet C, Gosti F, Giraudat J. Interactions between abscisic acid and ethylene signaling cascades. Plant Cell. 2000;12:1103–1116. doi: 10.1105/tpc.12.7.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagaya Y, Toyoshima R, Okuda R, Usui H, Yamamoto A, Hattori T. LEAFY COTYLEDON1 controls seed storage protein genes through its regulation of FUSCA3 and ABSCISIC ACID INSENSITIVE3. Plant Cell Physiol. 2005;46:399–406. doi: 10.1093/pcp/pci048. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc Natl Acad Sci USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter CA, Aukerman MJ, Sun H, Fokina M, Poethig RS. PAUSED encodes the Arabidopsis exportin-t ortholog. Plant Physiol. 2003;132:2135–2143. doi: 10.1104/pp.103.023309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toufighi K, Brady SM, Austin R, Ly E, Provart NJ. The Botany Array Resource: e-Northerns, Expression Angling, and promoter analyses. Plant J. 2005;43:153–163. doi: 10.1111/j.1365-313X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Ogawara T, Higashi K, Kamada H, Ezura H. Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J Plant Physiol. 2003;160:1335–1340. doi: 10.1078/0176-1617-01129. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation in Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc Natl Acad Sci USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly PM, Bonetta D, Tsukaya H, Dengler RE, Dengler NG. Cell cycling and cell enlargement in developing leaves of Arabidopsis. Dev Biol. 1999;215:407–419. doi: 10.1006/dbio.1999.9443. [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Chen W, Provart N, Chang HS, Guimil S, Su G, Estes B, Zou G, Wang X. Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol J. 2003;1:59–70. doi: 10.1046/j.1467-7652.2003.00006.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 Genes upregulated by ectopic FUS3 activation. Values for fold changes in expression after 2 and 4 days (d) of FUS3 activation with dexamethasone (+DEX) are the averages of two replicates. The presence of RY promoter motifs (CATGCA) in the 500 bp (0.5 K), 1, 000 bp (1 K) and 3, 000 bp (3 K) upstream regions of each gene is included. p-values are 3.06 × 10-4, 1.77 × 10-4 and 2.57 × 10-2, respectively.
Table S2 Genes downregulated by ectopic FUS3 activation. Values for fold changes in expression after 2 and 4 days (d) of FUS3 activation with dexamethasone (+DEX) are the averages of two replicates. Genes involved in hormone metabolism or response are in bold. Ethylene response genes that were downregulated less than twofold by FUS3 activation at both 2 and 4 days are in italics. The presence of RY promoter motifs (CATGCA) in 500 bp (0.5 K), 1, 000 bp (1 K) and 3, 000 bp (3 K) upstream regions of each gene is included. p-values are 4.27 × 10-2, 6.06 × 10-3 and 3.46 × 10-2, respectively.
Figure S1 Phenotypic analysis of wild-type plants exposed to dexamethasone (DEX). Wild-type seeds were germinated in 10 μM DEX for 2 days and then transferred to soil. The increase in the blade-to-petiole ratio and the appearance of the abaxial trichomes on leaf 5 is comparable to wild-type profiles shown in Figure 3. (A) Ratios of blade-to-petiole lengths of individual wild-type rosette leaves. (B) Percentage of wild-type rosettes that formed abaxial trichomes at each leaf position.