Circadian rhythm is regulated through multiple transcriptional/translational feedback loops. In this study, Kim et al. identify a novel role for the O-GlcNAcylation post-translation modification in controlling clock timing. Drosophila PERIOD (dPER) protein is found to be O-GlcNAcylated in a temporally regulated manner, and this modification determines clock speed. O-GlcNAcylation controls the timing of dPER translocation into the nucleus to carry out its transcription-repressive activity, thus acting as an interval timer in regulating periodic length and preventing premature dPER entry into the nucleus.
Keywords: O-GlcNAcylation, PERIOD protein, O-GlcNAc transferase (OGT), circadian rhythms, Drosophila, nuclear entry
Abstract
Post-translational modifications of one or more central “clock” proteins, most notably time-of-day-dependent changes in phosphorylation, are critical for setting the pace of circadian (≅24 h) clocks. In animals, PERIOD (PER) proteins are the key state variable regulating circadian clock speed and undergo daily changes in abundance and cytoplasmic–nuclear distribution that are partly driven by a complex phosphorylation program. Here, we identify O-GlcNAcylation (O-GlcNAc) as a critical post-translational modification in circadian regulation that also contributes to setting clock speed. Knockdown or overexpression of Drosophila O-GlcNAc transferase (ogt) in clock cells either shortens or lengthens circadian behavioral rhythms, respectively. The Drosophila PERIOD protein (dPER) is a direct target of OGT and undergoes daily changes in O-GlcNAcylation, a modification that is mainly observed during the first half of the night, when dPER is predominantly located in the cytoplasm. Intriguingly, the timing of when dPER translocates from the cytoplasm to the nucleus is advanced or delayed in flies, wherein ogt expression is reduced or increased, respectively. Our results suggest that O-GlcNAcylation of dPER contributes to setting the correct pace of the clock by delaying the timing of dPER nuclear entry. In addition, OGT stabilizes dPER, suggesting that O-GlcNAcylation has multiple roles in circadian timing systems.
Circadian rhythms are daily oscillations in behavioral, physiological, and biochemical processes that persist with ~24-h periods even in the absence of environmental time cues, such as the daily changes in light and temperature (Dunlap 1999). These rhythms are driven by cellular pacemakers based on a small set of core clock proteins that are conserved within a particular kingdom. The overall molecular underpinnings governing circadian rhythms show remarkable similarities from microorganisms to humans (Young and Kay 2001). A shared feature of circadian clocks is that they are based on transcriptional/translational feedback loops (TTFLs) that have both negative and positive elements. Drosophila has served as a pivotal model system in deciphering the cellular and molecular bases underlying circadian rhythms.
In Drosophila, the core transcriptional feedback loop involves the central clock proteins dPERIOD (dPER), TIMELESS (TIM), dCLOCK (dCLK), and CYCLE (CYC) (Hardin 2011). dCLK and CYC are transcription factors that form a heterodimer that binds E-box DNA elements to activate expression of dper and tim, in addition to other clock and nonclock gene targets. After a time delay, dPER and TIM interact and transit from the cytoplasm to the nucleus, where dPER participates with other factors in repressing dCLK–CYC-mediated transcription. Eventually, the levels of dPER and TIM decline in the nucleus, enabling another round of dCLK–CYC-mediated transcription. The phase-specific inhibition of dCLK–CYC activity by dPER is the basis for generating daily rhythms in gene expression. A very similar core transcriptional feedback loop involving PER proteins (mPER1–3) and a homologous pair of transcription factors (CLOCK–BMAL1) operates in the mammalian clockworks (Zhang and Kay 2010).
Although TTFLs constitute the overall architecture of circadian timing mechanisms, post-translational modifications of clock proteins are essential to generate 24-h rhythms (Bae and Edery 2006; Merrow et al. 2006; Gallego and Virshup 2007). Most notably, time-of-day-dependent changes in the phosphorylated state of one or more key clock proteins, which involves the regulated balance between kinases and protein phosphatases, have been shown to be critical for setting the pace of circadian rhythms in a wide variety of organisms. In animal clocks, PER is the main component setting the pace of the clock, and temporal phosphorylation plays a vital role in this function by regulating its daily stability, timing of nuclear entry, duration in the nucleus, and possibly potency as a repressor (Bae and Edery 2006).
Besides phosphorylation, the hydroxyl groups of Ser/Thr residues on proteins can also be modified with O-GlcNAcylation (O-GlcNAc) (Torres and Hart 1984; Holt and Hart 1986). O-GlcNAcylation is a highly conserved post-translational modification that has been shown to modulate diverse protein functions, including protein–protein interactions, protein turnover, subcellular localization, and changes in activity (Wells et al. 2001; Hanover et al. 2010). Two enzymes mediate reversible addition of the β-N-acetylglucosamine moiety to the hydroxyl side chains of Ser/Thr residues of protein substrates; namely, β-N-acetylglucosaminyltransferase (O-GlcNAc transferase; OGT) and β-N-acetylglucosaminidase (O-GlcNAcase; OGA) (Dong and Hart 1994; Kreppel et al. 1997). O-GlcNAcylation is highly dynamic, and numerous findings reveal a complex interplay between phosphorylation and O-GlcNAcylation (Butkinaree et al. 2010; Zeidan and Hart 2010). The ogt gene in Drosophila has recently been reported to be the Polycomb group (PcG) gene super sex combs (sxc) (Gambetta et al. 2009; Sinclair et al. 2009). This finding also suggests that O-GlcNAc modifications play key roles in PcG-mediated epigenetic silencing.
Here, we demonstrate that dPER is O-GlcNAcylated in flies and that this modification is temporally regulated. Remarkably, genetic manipulation of O-GlcNAc levels by either down-regulating or up-regulating OGT in clock cells speeds up or slows down the pace of circadian behavioral rhythms, respectively. The timing of dPER nuclear translocation in key brain pacemaker neurons is advanced in ogt knockdown flies and delayed in ogt-overexpressing flies, providing an attractive mechanism for the behavioral observations. Because O-GlcNAcylation of dPER mainly occurs during the first half of the night, when it is retained in the cytoplasm, our results suggest that O-GlcNAcylation gates the timing of when dPER translocates from the cytoplasm to the nucleus. Moreover, O-GlcNAcylation also stabilizes dPER, revealing multiple functions for this post-translational modification in the clockworks. Together, our findings demonstrate a prominent role for O-GlcNAcylation in circadian timing mechanisms and indicate that setting clock speed involves a complex network of diverse post-translational modifications.
Results
dPER is O-GlcNAcylated in Drosophila cultured cells
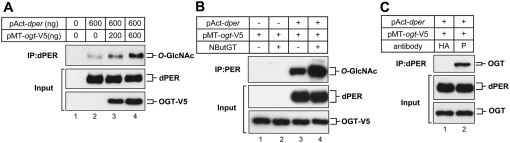
To examine whether circadian clock proteins can be modified by O-GlcNAcylation, we first used a simplified Drosophila tissue culture system. Schneider 2 (S2) cells were cotransfected with pAct-per, pAct-tim, pAct-HA-Clk, or pAct-cyc-3F, along with increasing amounts of pMT-ogt-V5. Cu2+ was added to the medium to induce ectopic expression of OGT via the inducible pMT promoter. O-GlcNAcylation of proteins was measured using an O-GlcNAc-specific antibody (HGAC-85) to probe immunoprecipitated clock proteins (Turner et al. 1990; Gambetta et al. 2009; Sinclair et al. 2009). Among the clock proteins tested, only dPER was strongly modified with O-GlcNAc, with signal intensity increasing in a dose-dependent manner (Fig. 1A). Other clock proteins, such as TIM, CYC, and dCLK, did not manifest significant levels of O-GlcNAc modification under the same conditions (Supplemental Fig. S1). Another O-GlcNAc-specific antibody, RL2 (Snow et al. 1987), also detected dPER (Fig. 1B, lanes 3,4). Moreover, the intensity of O-GlcNAc staining is significantly increased by treating cells with 100 μM NButGT (1,2-dideoxy-2′-propyl-α-D-glucopyranoso-[2,1-D]-Δ2′-thiazoline) (Macauley et al. 2005; Park et al. 2011), which inhibits OGA activity (Fig. 1B, cf. lanes 3 and 4). Furthermore, coexpression of both ogt and dper in S2 cells revealed that dPER can stably interact with OGT (Fig. 1C).
Figure 1.
dPER is O-GlcNAcylated in S2 cells. (A) S2 cells were transiently transfected with 600 ng of pAct-dper either singly or in combination with increasing amounts of pMT-ogt-V5 (200 and 600 ng). (B,C) S2 cells were transiently cotransfected with pAct-dper and 600 ng of pMT-ogt-V5, as indicated above panels. (B) To inhibit OGA activity, 100 μM NButGT was added as indicated (+). Expression of OGT was induced 24 h after transfection by adding 500 μM (final) CuSO4 to the medium. Cells were harvested 24 h after induction, and protein extracts were directly analyzed by immunoblotting (Input) or subjected to immunoprecipitation with anti-dPER antibody (gp339) or anti-HA antibody (12CA5) as a negative control. Immune complexes were analyzed for O-GlcNAc modification using the HGAC-85 antibody (A) or RL2 antibody (B).
Changing the expression levels of ogt in clock cells alters the pace of circadian rhythms in Drosophila
To investigate the physiological significance of O-GlcNAc modification in circadian timing systems, we examined circadian behavioral rhythms of flies wherein expression of ogt (CG10392) or oga (CG5871) is altered in clock cells. We exploited the binary GAL4/UAS system (Brand and Perrimon 1993) to specifically knock down ogt or oga in clock cells (Dietzl et al. 2007). This was accomplished by crossing transgenic UAS-RNAi lines from the Vienna-based Drosophila stock center (Vienna Drosophila RNAi Center [VDRC]) with flies carrying a tim(UAS)-gal4 driver line (Blau and Young 1999). To enhance RNAi, we coexpressed dicer2 with the UAS-RNAi lines (Dietzl et al. 2007). Flies were synchronized under standard conditions of 12-h light followed by 12-h dark cycles (12:12LD; whereby zeitgeber time 0 [ZT0] is lights-on and ZT12 is lights-off) at 25°C, followed by several days in complete darkness (DD), and the clock-controlled locomotor activity rhythm was analyzed.
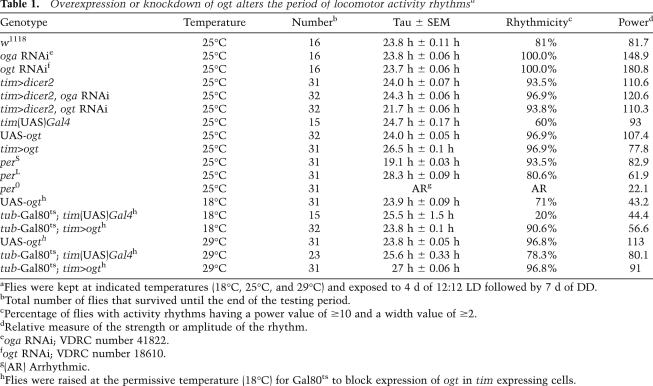
Control flies expressing dicer2 in tim-expressing cells manifested strong rhythms with the normal ~24-h period (Table 1). In sharp contrast, flies expressing ogtRNAi in tim-expressing cells (tim>dicer2/ogt RNAi, referred to here as ogt knockdown flies) manifested robust rhythms but with significantly shortened periods (~21.7 h) (Table 1). Under standard synchronization conditions of 12:12LD at 25°C, flies manifest a bimodal activity pattern, with “morning” and “evening” peaks centered around ZT0 and ZT12, respectively. The evening bout of activity begins earlier in the day in ogt knockdown flies (Fig. 2; Supplemental Fig. S3), consistent with the shorter free-running period (Table 1). Down-regulation of oga expression in the same genetic context as ogt (tim>dicer2/oga RNAi, referred to here as oga knockdown flies) did not alter locomotor activity periods (Table 1). Reduced levels of OGT and OGA were observed for both ogt and oga knockdown flies (Fig. 4A; Supplemental Fig. S2). It is possible that the level of oga reduction is not sufficient to render it rate-limiting with regard to circadian periodicity.
Table 1.
Overexpression or knockdown of ogt alters the period of locomotor activity rhythmsa
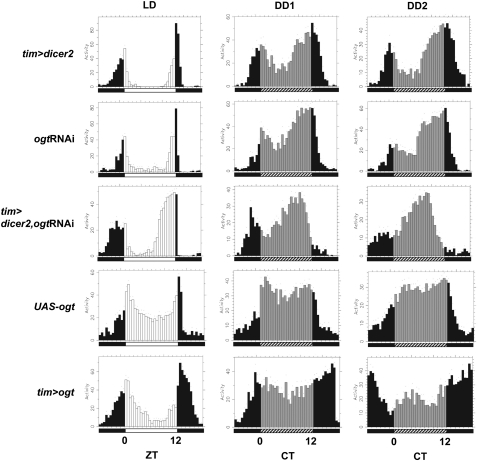
Figure 2.
The daily distribution of activity is altered in ogt knockdown and ogt-overexpressing flies. Each panel represents the average activity (in arbitrary units) of male flies for a given genotype for a consecutive 24-h period during the last day of 1212LD entrainment (LD) followed by the first (DD1) or second (DD2) day of DD. Genotypes are indicated at the left side of each panel. During LD, lights-on is indicated by white vertical bars, and lights-off is indicated by black vertical bars. During DD, subjective day is indicated by gray vertical bars, and subjective dark phase is indicated by black vertical bars. Each vertical bar represents relative activity levels during a 30-min bin. Standard error of the mean is indicated as dots above each bar. (CT) Circadian time. Note that in LD, flies exhibit a bimodal distribution of activity, with a “morning” peak centered around ZT0 and an “evening” peak centered around ZT12. Relative to control flies, the timing of the evening activity component is advanced in ogt knockdown flies (tim>dicer2, ogtRNAi), whereas it is delayed in ogt overexpression flies (tim>ogt).
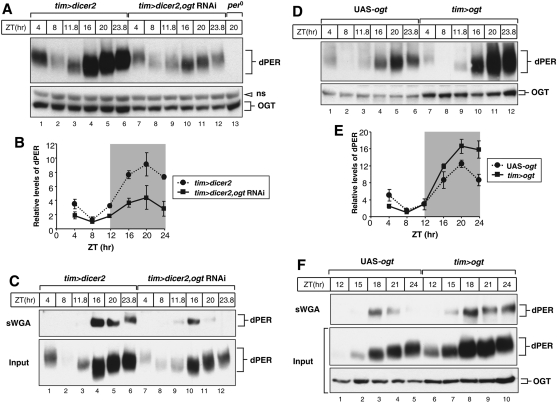
Figure 4.
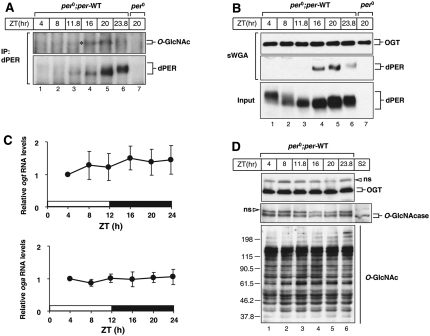
Knockdown and overexpression of OGT in clock cells have opposite effects on the overall daily abundance of dPER. Adult flies of the indicated genotypes (above the panels) were collected at the indicated ZT. Head extracts were prepared and analyzed either directly by immunoblotting (A,D; and C,F, Input) or following sWGA pull-down (C,F, top panels). sWGA complexes were probed by immunoblotting in the presence of anti-dPER antibodies (dPER-Rb1). Anti-OGT (H300) antibody was used to visualize OGT. (ns) Nonspecific band. (B,E) Quantification of dPER levels was performed with NIH ImageJ software. dPER levels at ZT8 in ogt knockdown flies were set to 1, and all other values were normalized. Shown are the average values from two independent experiments. Error bars denote SEM.
Next, we sought to increase the extent of O-GlcNAcylation in clock cells by overexpressing Drosophila ogt (tim>ogt) (Sinclair et al. 2009). Intriguingly, while daily locomotor activity rhythms remain robust in tim>ogt flies, there is a significant increase in the behavioral period (26.5 h) (Table 1). As expected with longer free-running periods, the timing of the evening activity component is delayed in ogt-overexpressing flies (Fig. 2; Supplemental Fig. S3). These results clearly indicate that the expression levels of ogt in clock cells and, presumably, the extent of O-GlcNAc modification play an important role in setting the pace of the circadian timing system in Drosophila.
Given that O-GlcNAc modification of proteins might modulate many cellular events throughout the life cycle of the fly, we wondered whether the behavioral phenotype manifested by changing the expression levels of ogt in clock cells might be due to developmental effects. To test this possibility, we used the temperature-sensitive Gal80ts TARGET system (McGuire et al. 2003; Smith et al. 2008) to restrict ectopic expression of ogt during adulthood. Flies of the appropriate genotype were kept at the permissive temperature (18°C) until they were adults, and one group was evaluated at this temperature, whereas another group was placed at the restrictive temperature (29°C). At the permissive temperature, wherein Gal80ts inhibits GAL4-driven ogt expression in tim-expressing cells, the activity period was ~24 h (Table 1). On the other hand, at the restrictive temperature, flies manifested significantly longer behavioral rhythms of ~27 h (Table 1). These results strongly suggest that the period-altering effects on circadian rhythms by manipulation of OGT levels are due to an active requirement in clock function and not to secondary effects on development.
O-GlyNAcylation of dPER is temporally regulated in flies
We next sought to examine whether O-GlcNAc modification of dPER occurs in flies. Control transgenic flies expressing the wild-type dper transgene in a wper0 genetic background—i.e., the only functional copy of dper is transgene-derived (Ko et al. 2007, 2010; Sun et al. 2010)—were entrained to 12:12LD and collected at different times during a daily cycle, and head extracts were processed for immunoprecipitation with anti-dPER antibodies. Prior work showed that dPER undergoes daily changes in abundance, whereby de novo synthesized dPER is first observed during the early night (ZT12–ZT16), peaks at approximately ZT20, and begins to rapidly decline in abundance beginning at approximately ZT0 (Edery et al. 1994). In addition, dPER undergoes daily changes in electrophoretic mobility that are largely due to progressive increases in phosphorylation, beginning as newly synthesized hypophosphorylated isoforms and slowly transitioning to hyperphosphorylated variants by the late night/early morning, which acts as a trigger for enhanced degradation (Edery et al. 1994). O-GlcNAcylation of dPER was first detected at ZT16, which increased in signal intensity at ZT20 and significantly decreased beginning at approximately ZT24, where the signal remained low or absent for the remainder of dPER's daily life cycle (Fig. 3A). Because the relative change in the intensity of the dPER O-GlcNAcylation signal does not follow the same temporal profile as that of its abundance (e.g., although dPER levels are relatively higher at ZT23.8 compared with ZT16, there is more O-GlcNAcylation signal at ZT16) (Fig. 3A, cf. lanes 4 and 6), the results suggest that the extent of O-GlcNAcylation of dPER is temporally regulated.
Figure 3.
dPER O-GlcNAcylation is temporally regulated in flies. (A) Adult flies of the indicated genotype (above the panels) were collected at the indicated ZT, and head extracts were prepared and processed for immunoprecipitation with anti-dPER antibodies. Immune complexes were analyzed for either O-GlcNAc modification with HGAC-85 antibody or total dPER with antip-dPER antibody. (*) O-GlcNAcylated dPER isoforms. (B) Head extracts were prepared and analyzed either directly by immunoblotting (Input) or following pull-down with sWGA-agarose (sWGA). (C) Adult per0;per-WT transgenic flies were collected at the indicated ZT. RNA was extracted from fly heads, and quantitative real-time RT–PCR was used to measure the relative levels of total ogt or oga mRNAs. Shown are the average values from three independent experiments. (D) Adult per0;per-WT transgenic flies were collected at the indicated ZT, and head extracts were prepared for immunoblotting. In the case of S2 cells (lane denoted S2), they were transiently transfected with pMT-oga, and expression of oga was induced by the addition of 500 μM (final) CuSO4. OGT was visualized with H300 antibody (top panel), OGA was visualized with anti-OGA antibody, and O-GlcNAc modification of total proteins was monitored with RL2 antibody (bottom panel). (ns) Nonspecific bands.
That O-GlcNAcylation of dPER is regulated in a time-of-day-specific manner is further supported by results obtained using succinylated wheat germ agglutinin (sWGA) pull-down assays (Fig. 3B). sWGA is a modified lectin that specifically binds O-GlcNAc on proteins (Love and Hanover 2005; Park et al. 2010). We used sWGA-agarose beads to purify O-GlcNAc-modified proteins from head extracts prepared from flies collected throughout a daily cycle. As previously shown, OGT is auto-GlcNAcylated (Fig. 3B; Tai et al. 2004). The extent of auto-GlcNAcylation of OGT is constant throughout a day (Fig. 3B, top panel). Similar with results obtained using an antibody to O-GlcNAcylation (Fig. 3A), sWGA pull-down also revealed that O-GlcNAcylation of dPER is first detected around ZT16, peaks at ZT20, and is greatly reduced thereafter (Fig. 3B, middle panel). Thus, the time window for maximal O-GlcNAc modification of dPER (i.e., ZT16–ZT20) occurs some 4–8 h prior to maximal phosphorylation (i.e., ZT0–ZT4).
The O-GlcNAcylation status of a protein is likely a balance between the opposing activities of OGT and OGA. In this regard, we tested the possibility that the expression of either ogt or oga is oscillating during a daily cycle, which might contribute to rhythmic O-GlcNAcylation of dPER. However, the mRNA levels of both ogt and oga in total head extracts are constant during an LD cycle (Fig. 3C). Consistent with constitutive expression, OGT and OGA protein levels do not manifest any changes throughout a daily cycle (Fig. 3D, top and middle panels). We also tested the possibility that the activity of OGT and/or OGA undergoes daily changes by monitoring the global pattern of O-GlcNAcylation modification of proteins in fly heads (Fig. 3D, bottom panel). We did not detect any significant changes in the intensities of different bands, suggesting that temporal changes in the extent of dPER O-GlcNAcylation is not an attribute generally shared by other proteins modified by O-GlcNAcylation. However, at this stage, we cannot rule out possible daily fluctuations in the levels/activity of OGT and/or OGA in specific subsets of cells in the adult fly head, such as clock cells.
Altering the levels of OGT leads to changes in the daily abundance of dPER
As an initial means to examine the effect of O-GlcNAcylation on the central clock mechanism, head extracts were prepared from ogt knockdown and ogt-overexpressing flies, and the daily biochemical cycles in dPER abundance and overall phosphorylation were determined. Decreased expression levels of OGT protein in the ogt knockdown flies were confirmed by immunoblotting endogenous OGT (Fig. 4A, bottom panel). It should be noted that the actual reduction of OGT in clock cells using RNAi is almost certainly more severe compared with the results shown here for total head extracts, as tim expression is spatially limited in the head. Peak levels of dPER were attained at approximately ZT20 for ogt knockdown flies, similar to the control situation (Fig. 4A,B). Although we did not observe a major change in the timing of relative changes in the levels of dPER in ogt knockdown flies during an LD cycle, it is possible that the 4-h time-point resolution is not sufficient. Nonetheless, the overall daily levels of dPER in ogt knockdown flies were significantly lower compared with those observed in control flies (Fig. 4A,B). In sharp contrast, the abundance of the clock protein CYC was not altered in ogt knockdown flies (Supplemental Fig. S4B), indicating that the decrease in dPER levels observed in ogt knockdown flies was not due to nonspecific effects such as cellular toxicity or global inhibition of protein synthesis.
As expected based on down-regulating ogt and the accompanying reductions in the overall daily levels of dPER, the dPER O-GlcNAcylation signal in ogt knockdown flies was significantly decreased (Fig. 4C). This was also the case when we detected immunoprecipitated dPER with an O-GlcNAc-specific antibody (Supplemental Fig. S4A). Nonetheless, whereas O-GlcNAcylated dPER was first observed at ZT16 in control flies, it was detected at earlier times (e.g., at ZT11.8) in the ogt knockdown flies (Fig. 4C, top panel, cf. lanes 3 and 9). This observation is consistent with the shorter behavioral periods manifested by ogt knockdown flies (Table 1) and further suggests that the timing of dPER O-GlcNAcylation is tightly linked to the endogenous pace of the clock (Fig. 4C).
Next, we examined daily dPER protein levels in ogt-overexpressing flies (Fig. 4D). Increased expression of OGT protein was confirmed by immunoblotting for total OGT in ogt-overexpressing flies (Fig. 4D, bottom panel). Although the phase in the daily abundance cycles of dPER proteins is similar between control and ogt-overexpressing flies, as in the case for ogt knockdown flies, the overall daily levels of dPER are higher in ogt-overexpressing flies (Fig. 4D,E). The increased abundance of OGT is accompanied by increases in the dPER O-GlcNAcylation signal, which was prolonged later into the night (e.g., Fig. 4F, top panel, cf. lanes 5 and 10). Thus, whereas decreasing ogt expression in clock cells reduces the overall daily abundance of dPER, increasing ogt expression leads to higher dPER levels. Taken together, our results demonstrate a critical link between O-GlcNAcylation and the levels of dPER protein.
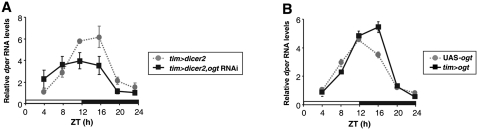
OGT modulates the daily cycle in dper mRNA levels
We examined whether the changes in dPER protein levels in flies wherein ogt expression is altered might derive from a transcriptional contribution by measuring dper mRNA levels throughout a daily cycle. There are several reproducible differences between the daily cycle in dper mRNA levels between control flies and ogt knockdown flies. Most notably, peak levels are attained ~4 h earlier and are ~35% lower in ogt knockdown flies (Fig. 5A). Conversely, when ogt is overexpressed, peak levels in dper mRNA are reached later and are slightly higher compared with the control situation (Fig. 5B). The changes in the timing of when peak levels of dper mRNA are attained in flies wherein ogt expression is genetically manipulated are consistent with the period changes in locomotor activity rhythms (Fig. 2; Table 1). In our experience, measuring dper mRNA cycles offers a more reliable index for assaying changes in the timing of circadian molecular oscillations compared with the dPER protein abundance rhythm (data not shown). Although changes in the levels of dper mRNA might contribute to the observed changes in the overall daily levels of dPER protein (Fig. 4), the more subtle effects on transcript levels suggest that OGT also directly regulates dPER stability. Indeed, the results discussed below provide evidence that OGT has a primary effect on dPER stability (Fig. 7).
Figure 5.
Knockdown and overexpression of OGT in clock cells alter the daily cycles in dper RNA levels. Adult flies of the indicated genotypes were collected at the indicated ZT. RNA was extracted from fly heads, and quantitative real-time RT–PCR was used to measure the relative levels of total dper mRNA. RNA levels from control flies (tim>dicer2 [A] and UAS-ogt [B]) at ZT4 were set to 1, and all other values were normalized. Shown are the average values from three independent experiments. Error bars denote SEM.
Figure 7.
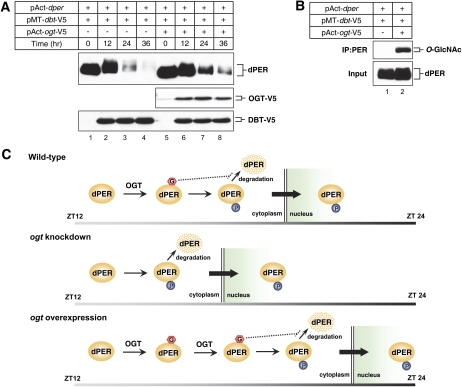
Overexpression of OGT delays DBT-dependent degradation of dPER in S2 cells. (A,B) S2 cells were transiently cotransfected with 600 ng of pAct-dper and 200 ng of pMT-dbt-V5, or in combination with 600 ng of pAct-ogt-V5. To inhibit OGA activity, 200 μM NButGT was added. Expression of DBT was induced 24 h after transfection by adding 500 μM (final) CuSO4 to the medium. Cells were harvested at the indicated times (A) or 12 h (B) after induction, and protein extracts were directly analyzed by immunoblotting (A, Input) or subjected to immunoprecipitation with anti-dPER antibodies. (B) Immune complexes were analyzed for O-GlcNAc modification with RL2 antibody. Each experiment was done at least three times, and representative examples are shown. (C) Model for how O-GlcNAcylation might regulate the timing of dPER nuclear entry. Interplay between O-GlcNAcylation and phosphorylation of dPER might set the proper timing of dPER nuclear entry (wild type). When ogt expression is down-regulated, less O-GlcNAcylation of dPER might increase and/or advance phosphorylation at sites that enhance its translocation to the nucleus, provoking earlier nuclear entry of dPER (ogt knockdown). On the other hand, when ogt is overexpressed, prolonged O-GlcNAcylation of PER might hamper nuclear entry-specific dPER phosphorylation, leading to delayed nuclear entry of dPER (ogt overexpression). The dashed line indicates the stabilizing effect of O-GlcNAcylation on dPER protein abundance. The gray-to-black gradient bar on the bottom of each panel represents the dark period of the day from ZT12 to ZT24. (G) GlcNAc; (P) phosphate.
The timing of dPER nuclear entry in key brain pacemaker neurons is regulated by OGT in a manner consistent with clock speed
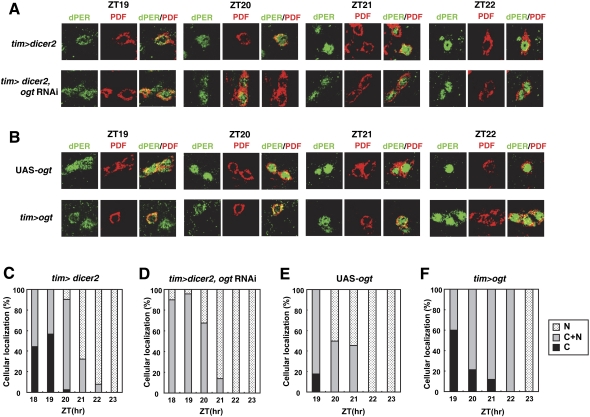
To further understand how OGT might regulate the pace of the clock, we examined the timing of dPER nuclear entry in key pacemaker neurons in the Drosophila brain. Several neuronal clusters in the Drosophila brain drive behavioral rhythms in locomotor activity. Among those, the small ventral lateral neurons (s-LNvs) are central for the maintenance of ~24-h locomotor activity rhythms in constant dark conditions (Nitabach and Taghert 2008). Moreover, prior work showed that the timing of dPER nuclear entry in the s-LNvs is gated, such that it occurs around ZT19–ZT20 (Curtin et al. 1995). Flies were collected at different times in a daily cycle, and whole-brain mounts were probed with anti-dPER antibodies. In addition, antibodies against pigment-dispersing factor (PDF), a circadian relevant neuropeptide, were used to label the cytoplasm of the LNvs (Fig. 6; Renn et al. 1999).
Figure 6.
Manipulating the levels of OGT in clock cells leads to changes in dPER nuclear entry that are consistent with the period of activity rhythms. Adult flies of the indicated genotypes were collected at the indicated times in LD, and isolated brains were processed for whole-mount immunohistochemistry. (A,B) Shown are representative staining patterns obtained for the s-LNvs. (C–F) For each genotype, the subcellular localization of dPER at different times in a daily cycle was quantified for the s-LNvs from at least five flies. dPER was visualized with anti-dPER antibodies (shown in green). PDF was visualized with an anti-PDF antibody (shown in red) and serves as a convenient cytoplasmic marker for the s-LNvs. Note that nuclear entry time of dPER is advanced in ogt knockdown flies, whereas it is delayed in ogt-overexpressing flies. (A,C,D) Moreover, whereas exclusively cytoplasmic dPER was observed in control flies from ZT18 to ZT20, we did not observe any s-LNvs where dPER was restricted to the cytoplasm in ogt knockdown flies. (B,E,F) On the other hand, in ogt-overexpressing flies, dPER mainly localized to the cytoplasm was still visible even later into the night compared with the control situation.
Consistent with prior work, dPER transitions from a mostly cytoplasmic location at ZT18 and ZT19 to a largely mixed cytoplasmic and nuclear location at ZT20, followed by essentially exclusive nuclear localization by ZT22 in control flies (Fig. 6A [top panel], C). In sharp contrast, in ogt knockdown flies, at ZT18, when dPER started to be clearly detected in our immunohistochemical analysis, most of the neurons already manifested a mixed distribution in both compartments (Fig. 6A [bottom panel], D), and by ZT21, almost all of the LNvs manifested exclusive nuclear staining of dPER. We did not observe any s-LNvs stained with only cytoplasmic dPER in ogt knockdown flies (Fig. 6; data not shown). Thus, our results indicate that when ogt expression is down-regulated in clock cells, the timing of dPER nuclear entry in the s-LNv is advanced. We also examined the nuclear entry of dPER in ogt-overexpressing flies (Fig. 6B,E,F). Intriguingly, in ogt-overexpressing flies, the nuclear entry timing of dPER is clearly delayed, and strong nuclear accumulation was only observed beginning at ZT23 (Fig. 6B [bottom panel], F). Moreover, we detected some s-LNvs with clear cytoplasmic dPER even at ZT21 in ogt-overexpressing flies (Fig. 6F). Thus, the relative changes in the timing of dPER nuclear entry/accumulation by genetically manipulating ogt expression in clock cells are consistent with the changes in the periods of behavioral rhythms. For example, the earlier nuclear accumulation of dPER in ogt knockdown flies likely explains the faster pace of the clock in these flies (Fig. 2; Table 1). In summary, our findings indicate that the timing of dPER nuclear entry in key pacemaker neurons is highly sensitive to the levels of OGT and that altering the timing of this event correlates with the modulation in clock speed.
OGT delays DOUBLE TIME (DBT)-mediated degradation of dPER in S2 cells
Based on the results in flies whereby altering OGT levels is associated with changes in the daily abundance of dPER, we sought to examine in a more direct assay whether OGT can lead to increases in the levels of dPER by using a simplified S2 cell culture system. Prior work has shown that DBT (the Drosophila homolog of casein kinase 1ɛ [CK1ɛ]) is the main kinase controlling dPER stability, a mechanism that involves progressive increases in global dPER phosphorylation until highly phosphorylated isoforms are targeted for rapid degradation (Kloss et al. 1998; Price et al. 1998). A similar observation can be recapitulated in S2 cells by expressing dper under a constitutive promoter and inducing dbt (Fig. 7A, top panel, lanes 1–4; Ko et al. 2002). Coexpression of OGT significantly delayed the DBT-dependent decline in dPER levels (Fig. 7A, top panel, cf. lanes 5–8 and 1–4; see Supplemental Fig. S5A for quantification of results). Ectopic expression of ogt clearly leads to a greater extent of dPER O-GlycNAcylation, even when similar amounts of dPER are compared (Fig. 7B). To more directly measure the stability of dPER, we inhibited translation by adding cycloheximide into the medium at 24 h post-DBT induction and measured dPER levels (Supplemental Fig. S5B,C). When OGT was coexpressed, the rate of decrease in dPER protein levels was significantly less (Supplemental Fig. S5C). Thus, in both flies and S2 cells, overexpressing OGT increases the abundance of dPER.
Discussion
Circadian clocks operate through negative feedback loops wherein positive elements activate negative elements, which in turn repress the activity of the positive elements until the levels of the negative elements decline, enabling another round of activation by the positive elements. To construct such a type of oscillating system, a lag is required before the negative elements can act to inhibit the positive elements (Leloup and Goldbeter 1998; Allada et al. 2001; Stanewsky 2003). In Drosophila, the delayed nuclear entry/accumulation of dPER contributes to the time delay in feedback repression. A complex web of kinases and phosphatases regulates when in a daily cycle dPER participates in repressing dCLK–CYC-mediated transcription by regulating its stability, timing of nuclear entry, and duration in the nucleus. While DBT is the major kinase driving daily cycles in dPER levels, other kinases such as CK2 and GSK-3β/SGG appear to have preferential effects on regulating the translocation of dPER from the cytoplasm to the nucleus (Bao et al. 2001; Lin et al. 2002; Akten et al. 2003; Nawathean and Rosbash 2004; Ko et al. 2010). The association of TIM with dPER in the cytoplasm not only protects dPER from DBT-mediated degradation, but also enhances, yet is not obligatory for, dPER nuclear entry (Shafer et al. 2002). Thus, the regulation of when in a daily cycle dPER translocates from the cytoplasm to the nucleus involves numerous factors. Here, we show that the timing of dPER nuclear entry is more complex than previously thought and identify O-GlcNAcylation as a critical post-translational modification in setting clock speed.
In this study, we show that dPER is a direct target of OGT and is modified by O-GlcNAcylation. This modification occurs in a temporally regulated manner, first detected in the early night, peaking in the middle of the night, and declining/disappearing thereafter for the remainder of the dPER daily life cycle (Fig. 3). At present, it is not clear how O-GlcNAcylation of dPER is temporally regulated. Based on our studies indicating that expression of either ogt or oga is not under circadian regulation (Fig. 3), some other factor(s) might favor O-GlcNAcylation of dPER during the first half of the night. For example, uridine diphosphate-N-acetylglucosamine (UDP-GlcNAc), the donor substrate of OGT, is generated from glucose via the hexosamine biosynthetic pathway, indicating that the extent of protein O-GlcNAcylation can be sensitive to nutrient availability (Walgren et al. 2003; Housley et al. 2008). Thus, it is possible that metabolic cues are affecting the activity of OGT in clock cells, leading to rhythmic activity of OGT.
The physiological significance of O-GlcNAc modification in circadian clock systems was demonstrated by showing that the period of daily activity rhythms is sensitive to the levels of ogt expression in clock-specific cells (Table 1). Remarkably, the periodicity of behavioral rhythms in ogt knockdown flies is shortened, whereas longer periods are observed in flies overexpressing ogt (Table 1). This strongly suggests that O-GlcNAcylation of dPER is a key variable in setting clock speed. We identified the timing in dPER nuclear entry (Fig. 6) as a key event in the clockworks that is altered by changes in ogt expression in a manner consistent with the changes in overt behavioral rhythms. For example, inhibiting endogenous ogt expression in clock cells advanced the time of dPER nuclear entry (Fig. 6), likely underlying the shorter behavioral rhythms.
Alterations in the timing of dPER nuclear entry by genetically manipulating ogt levels might also explain the differences in dper mRNA abundance cycles (Fig. 5). For example, the more rapid nuclear entry of dPER in ogt knockdown flies (Fig. 6A,C,D) could contribute to the earlier decline in dper mRNA levels and its inability to attain maximal peak values (Fig. 5A). Clearly, the earlier nuclear entry of dPER when ogt levels are reduced is not due to an increase in the abundance of dPER (Fig. 4). Rather, it appears that dPER stability is increased by more extensive O-GlyNAcylation. Experiments in S2 cells suggest that OGT has a primary effect on stabilizing dPER against DBT-mediated degradation (Fig. 7; Supplemental Fig. S5). How this occurs is presently not clear. It is also not established whether OGT-mediated changes in the levels of dPER contribute to altering the timing of dPER nuclear entry. However, it should be noted that other factors, such as DBT and TIM, regulate both the stability of dPER and its nuclear entry time (Bae and Edery 2006), so multiple effects of O-GlyNAcylation on both dPER levels and nuclear translocation are not unanticipated. Because O-GlyNAcylation of dPER is mainly observed when it resides in the cytoplasm (Figs. 3, 4), we propose that this modification acts as an interval timer to prevent the premature nuclear entry of dPER (Fig. 7C). While future work will be required to better understand the mechanism for how O-GlcNAcylation regulates dPER nuclear entry, it is possible that O-GlcNAcylation of dPER might attenuate phosphorylation at sites that enhance its translocation to the nucleus (Bao et al. 2001; Akten et al. 2003; Nawathean and Rosbash 2004; Cyran et al. 2005; Muskus et al. 2007; Ko et al. 2010). Indeed, there are numerous reports illustrating complex interplays between phosphorylation and O-GlcNAcylation (Butkinaree et al. 2010; Zeidan and Hart 2010).
It is intriguing that in plants, Spindly (Spy), which has significant similarity to animal OGT, functions in the same pathways with GIGANTE (GI) (Tseng et al. 2004; Olszewski et al. 2010). Like GI, SPY affects circadian rhythms in Arabidopsis. In loss of spy function mutants, the circadian period in cotyledon movement rhythm is lengthened, whereas overexpressing spy shortens the rhythm. Recent work by Durgan et al. (2011) using cardiomyocytes showed diurnal variations in total protein O-GlcNAcylation and identified Bmal1 as an O-GlcNAc-modified protein in mammals. Together with our results, this implies that O-GlcNAc modification has a conserved role in regulating circadian clock pace.
Materials and methods
Fly strains and behavioral assays
UAS-oga RNAi, UAS-ogt RNAi, and UAS-ogt flies have been described (Sinclair et al. 2009; Park et al. 2011), and UAS-ogt flies were kindly provided by Barry M. Honda (Simon Fraser University, Canada). tub-gal80ts flies were kindly provided by Kwang-Min Choe (Yonsei University, Korea).
The locomotor activities of individual flies were measured as previously described using the Drosophila Activity Monitoring system from Trikinetics (Sun et al. 2010). Briefly, young adult flies were used for the analysis and kept in incubators at the indicated temperature (18°C, 25°C, and 29°C), exposed to at least 4 d of 12:12LD (where ZT0 is defined as the time when the light phase begins), and subsequently kept in constant dark (DD) conditions for 7 d. Locomotor activity data for each individual fly were analyzed using the FaasX software (Fly Activity Analysis suite for Mac OS X), which was generously provided by F. Rouyer (CNRS, France). Periods were calculated for each individual fly using χ2 periodogram analysis and pooled to obtain a group average for each independent transgenic line or genotype. Power is a quantification of the relative strength of the rhythm during DD. Individual flies with a power ≥10 and a “width” value of ≥2 (denotes number of peaks in 30-min increments above the periodogram 95% confidence line) were considered rhythmic.
Plasmids and methods for S2 cell-based assays
The pAct-per, pAct-tim, pAct-per-V5, pMT-Clk-V5, and pMT-dbt-V5 plasmids were described previously (Ceriani et al. 1999; Ko et al. 2002; Kim and Edery 2006; Kim et al. 2007). To generate pMT-ogt-V5, coding regions of ogt were amplified by PCR from a cDNA clone and subcloned into pMT/V5-His (Invitrogen) using ApaI and XbaI.
S2 cells were obtained from Invitrogen and transfected using effectene reagent following the manufacturer's protocol (Qiagen). Recombinant proteins under the control of the metallothionein promoter (pMT) were induced by adding CuSO4 to a final concentration of 500 μM in the medium.
Immunoblotting and immunoprecipitation
Protein extracts from S2 cells were prepared as previously described (Kim et al. 2007). Briefly, cells were lysed using modified RIPA buffer (50 mM Tris-HCl at pH 7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) with the addition of protease inhibitor cocktail and PhosSTOP. For protein extracts from flies, flies were collected by freezing at the indicated times in LD, heads were isolated, and total fly head extracts were prepared using modified RIPA buffer as previously described (Kim et al. 2007). Protein extracts from either S2 cells or heads were resolved by SDS-polyacrylamide gel electrophoresis (PAGE), and target proteins were identified by immunoblotting as previously described (Sun et al. 2010). Immunoblots were probed using primary antibodies at the following dilutions: anti-V5 (Invitrogen), 1:10,000; anti-TIM (TR3), 1:3000 (Sidote et al. 1998); and anti-dCLK (GP208), 1:3000 (Kim et al. 2007). We also raised a novel rabbit anti-dPER antiserum termed dPER-Rb1 and dPER-Rb2 (AbFrontier) using the same antigen previously described (Lim et al. 2007), and dPER-Rb1 was used in this study. To detect Drosophila OGT and OGA, anti-OGT Ab (H300, Santa Cruz Biotechnology) and anti-OGA Ab (Kang et al. 2009) were used at the following dilutions: anti-OGT, 1:3000; and anti-OGA, 1:2000. Quantification of immunoblots was performed with NIH ImageJ software.
To probe for O-GlcNAc modification on clock proteins, we first performed immunoprecipitation essentially as previously described (Sun et al. 2010). To protein extracts from S2 cells and fly head extracts, we added 3 μL of anti-PER (GP339), anti-CLK(GP208), anti-HA(12CA5), or anti-Flag (M2) antibody, depending on the target protein sought, and incubated with gentle rotation for 3–5 h at 4°C, followed by the addition of 20 μL of Gammabind G-Sepharose (GE Healthcare) with a further incubation of 1–2 h. Beads were collected by light centrifugation, and immune complexes were mixed with 30 μL of 1× SDS-PAGE sample buffer and incubated for 5 min at 95°C; the resulting supernatants were analyzed by immunoblotting. O-GlcNAc-specific antibodies were used at the following dilutions: HGAC-85, 1:1000 (Abcam); and RL-2, 1:2000 (Thermo). sWGA pull-down was performed by adding sWGA-agarose (L-1020S, Vector Laboratories) to the protein extracts, and bound material was analyzed by immunoblotting.
Quantitative real-time RT–PCR
Quantitative real-time RT–PCR was performed as described previously (Sun et al. 2010). Total RNA was isolated from frozen heads using TRI reagent (Molecular Research Center, Inc.). Five-hundred nanograms of total RNA was reverse-transcribed with oligo-dT primer using amfiRivert reverse transcriptase (GenDEPOT), and real-time PCR was performed using the Corbett Rotor Gene 6000 instrument (Corbett Life Science) in the presence of Quantitect SYBR Green PCR Kit (Qiagen). The primer sequences used here for quantitation of dper and tim RNAs were as described in Yoshii et al. (2007) and were as follows: dper forward, 5′-GACCGAATCCCTGCTCAATA-3′; dper reverse, 5′-GTGTCATTGGCGGACTTCTT-3′; tim forward, 5′-CCCTTATACCCGAGGTGGAT-3′; and tim reverse, 5′-TGATCGAGTTGCAGTGCTTC-3′. The primer sequences used here for quantitation of ogt and oga RNAs were as follows: ogt forward, 5′-GCTATACGCCTGGGAACAAA-3′; ogt reverse, 5′-CCTTTGGCATACTGTGAGCA-3′; oga forward, 5′-ACAAGGAGATTTGCCACGAG-3′; and oga reverse, 5′-AGCTGCCCTCTGTGTAGGAA-3′. We also included primers for the noncycling mRNA coding for CBP20 as previously described (Majercak et al. 2004), and sequences were as follows: cbp20 forward, 5′-GTCTGATTCGTGTGGACTGG-3′; and cbp20 reverse, 5′-CAACAGTTTGCCATAACCCC-3′. Results were analyzed with software associated with Rotor Gene 6000, and relative mRNA levels were quantitated using the 2−ΔΔCt method.
Immunohistochemistry
Confocal imaging of adult brains was performed as described (Ko et al. 2007). Briefly, adult fly heads were cut open in ice-cold PBS, and at least five to seven brains were analyzed for each time point in a daily cycle. Heads were fixed in 4% paraformaldehyde and rinsed with PBS containing 1% Triton X-100. Fixed heads were dissected with needles, and isolated brains were incubated for 30 min to several hours in a blocking solution comprised of PBT solution (PBS containing 0.5% Triton X-100) containing 10% horse serum. Primary antibodies were directly added to the blocking solution and incubated overnight at 4°C. The following antibodies and final dilutions were used: (1) anti-dPER antibody, 1:250 (Lim et al. 2007); and (2) anti-PDF antibody (C7), 1:200 (Cyran et al. 2005). Subsequently, brains were washed with PBT, and blocking solution containing secondary antibodies was added and incubated overnight at 4°C. The secondary antibodies used were Alexa488-conjugated anti-rabbit IgG (Invitrogen) or Alexa555-conjugated anti-mouse IgG (Sigma), both at a final dilution of 1:200. After several washes with PBT, brains were incubated in 0.1 M phosphate buffer containing 50% glycerol for 15–30 min, and tissues were transferred onto slides and mounted with Vectashield (Vector Laboratories). Confocal images were obtained with an LSM7 Confocal Microscope (Zeiss) and processed with Zen software (Zeiss).
Acknowledgments
We are very grateful to Barry M. Honda and Kwang-Min Choe for providing flies, Junho Choe for anti-dPER antibody and plasmid for dPER antigen production, Justin Blau for anti-PDF antibody, and Jae Hyup Shin for help in immunoblotting. Work in the laboratory of E.Y.K. was supported by Basic Science Research Program (2009-0067106 and NRF-2008-521-C00220) and Chronic Inflammatory Disease Research Center (R13-2003-019) through the National Research Foundation of Korea (NRF), funded by the Ministry of Education, Science and Technology. Work in the laboratory of J.W.C. was supported by the National Research Foundation (NRF), funded by the Korean Government (2011-0020479 to J.W.C.) and World Class University Program (R31-2008-000-10086-0 to J.W.C.). I.E. is supported by the National Institutes of Health (R01NS34958 and R01NS42088).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.182378.111.
References
- Akten B, Jauch E, Genova GK, Kim EY, Edery I, Raabe T, Jackson FR 2003. A role for CK2 in the Drosophila circadian oscillator. Nat Neurosci 6: 251–257 [DOI] [PubMed] [Google Scholar]
- Allada R, Emery P, Takahashi JS, Rosbash M 2001. Stopping time: The genetics of fly and mouse circadian clocks. Annu Rev Neurosci 24: 1091–1119 [DOI] [PubMed] [Google Scholar]
- Bae K, Edery I 2006. Regulating a circadian clock's period, phase and amplitude by phosphorylation: Insights from Drosophila. J Biochem 140: 609–617 [DOI] [PubMed] [Google Scholar]
- Bao S, Rihel J, Bjes E, Fan JY, Price JL 2001. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J Neurosci 21: 7117–7126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW 1999. Cycling vrille expression is required for a functional Drosophila clock. Cell 99: 661–671 [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N 1993. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118: 401–415 [DOI] [PubMed] [Google Scholar]
- Butkinaree C, Park K, Hart GW 2010. O-linked β-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochim Biophys Acta 1800: 96–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceriani MF, Darlington TK, Staknis D, Mas P, Petti AA, Weitz CJ, Kay SA 1999. Light-dependent sequestration of TIMELESS by CRYPTOCHROME. Science 285: 553–556 [DOI] [PubMed] [Google Scholar]
- Curtin KD, Huang ZJ, Rosbash M 1995. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 365–372 [DOI] [PubMed] [Google Scholar]
- Cyran SA, Yiannoulos G, Buchsbaum AM, Saez L, Young MW, Blau J 2005. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J Neurosci 25: 5430–5437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448: 151–156 [DOI] [PubMed] [Google Scholar]
- Dong DL, Hart GW 1994. Purification and characterization of an O-GlcNAc selective N-acetyl-β-D-glucosaminidase from rat spleen cytosol. J Biol Chem 269: 19321–19330 [PubMed] [Google Scholar]
- Dunlap JC 1999. Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Durgan DJ, Pat BM, Laczy B, Bradley JA, Tsai JY, Grenett MH, Ratcliffe WF, Brewer RA, Nagendran J, Villegas-Montoya C, et al. 2011. O-GlcNAcylation, novel post-translational modification linking myocardial metabolism and cardiomyocyte circadian clock. J Biol Chem 286: 44606–44619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M 1994. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci 91: 2260–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM 2007. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol 8: 139–148 [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325: 93–96 [DOI] [PubMed] [Google Scholar]
- Hanover JA, Krause MW, Love DC 2010. The hexosamine signaling pathway: O-GlcNAc cycling in feast or famine. Biochim Biophys Acta 1800: 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin PE 2011. Molecular genetic analysis of circadian timekeeping in Drosophila. Adv Genet 74: 141–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt GD, Hart GW 1986. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. J Biol Chem 261: 8049–8057 [PubMed] [Google Scholar]
- Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW 2008. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem 283: 16283–16292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Park SY, Ji S, Jang I, Park S, Kim HS, Kim SM, Yook JI, Park YI, Roth J, et al. 2009. O-GlcNAc protein modification in cancer cells increases in response to glucose deprivation through glycogen degradation. J Biol Chem 284: 34777–34784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Edery I 2006. Balance between DBT/CKIɛ kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc Natl Acad Sci 103: 6178–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EY, Ko HW, Yu W, Hardin PE, Edery I 2007. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol Cell Biol 27: 5014–5028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B, Price JL, Saez L, Blau J, Rothenfluh A, Wesley CS, Young MW 1998. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase I&epsiv. Cell 94: 97–107 [DOI] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678 [DOI] [PubMed] [Google Scholar]
- Ko HW, DiMassa S, Kim EY, Bae K, Edery I 2007. Cis-combination of the classic per(S) and per(L) mutations results in arrhythmic Drosophila with ectopic accumulation of hyperphosphorylated PERIOD protein. J Biol Rhythms 22: 488–501 [DOI] [PubMed] [Google Scholar]
- Ko HW, Kim EY, Chiu J, Vanselow JT, Kramer A, Edery I 2010. A hierarchical phosphorylation cascade that regulates the timing of PERIOD nuclear entry reveals novel roles for proline-directed kinases and GSK-3β/SGG in circadian clocks. J Neurosci 30: 12664–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW 1997. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem 272: 9308–9315 [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A 1998. A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J Biol Rhythms 13: 70–87 [DOI] [PubMed] [Google Scholar]
- Lim C, Lee J, Choi C, Kim J, Doh E, Choe J 2007. Functional role of CREB-binding protein in the circadian clock system of Drosophila melanogaster. Mol Cell Biol 27: 4876–4890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Kilman VL, Keegan K, Paddock B, Emery-Le M, Rosbash M, Allada R 2002. A role for casein kinase 2α in the Drosophila circadian clock. Nature 420: 816–820 [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA 2005. The hexosamine signaling pathway: Deciphering the ‘O-GlcNAc code.’ Sci STKE 2005: re13 doi: 10.1126/stke.3122005re13 [DOI] [PubMed] [Google Scholar]
- Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ 2005. O-GlcNAcase uses substrate-assisted catalysis: Kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem 280: 25313–25322 [DOI] [PubMed] [Google Scholar]
- Majercak J, Chen WF, Edery I 2004. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol Cell Biol 24: 3359–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire SE, Le PT, Osborn AJ, Matsumoto K, Davis RL 2003. Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302: 1765–1768 [DOI] [PubMed] [Google Scholar]
- Merrow M, Mazzotta G, Chen Z, Roenneberg T 2006. The right place at the right time: Regulation of daily timing by phosphorylation. Genes Dev 20: 2629–2633 [DOI] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL 2007. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27: 8049–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawathean P, Rosbash M 2004. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol Cell 13: 213–223 [DOI] [PubMed] [Google Scholar]
- Nitabach MN, Taghert PH 2008. Organization of the Drosophila circadian control circuit. Curr Biol 18: R84–R93 doi: 10.1016/j.cub.2007.11.061 [DOI] [PubMed] [Google Scholar]
- Olszewski NE, West CM, Sassi SO, Hartweck LM 2010. O-GlcNAc protein modification in plants: Evolution and function. Biochim Biophys Acta 1800: 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Kim HS, Kim NH, Ji S, Cha SY, Kang JG, Ota I, Shimada K, Konishi N, Nam HW, et al. 2010. Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J 29: 3787–3796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S, Park SH, Baek JY, Jy YJ, Kim KS, Roth J, Cho JW, Choe KM 2011. Protein O-GlcNAcylation regulates Drosophila growth through the insulin signaling pathway. Cell Mol Life Sci 68: 3377–3384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price JL, Blau J, Rothenfluh A, Abodeely M, Kloss B, Young MW 1998. double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 94: 83–95 [DOI] [PubMed] [Google Scholar]
- Renn SC, Park JH, Rosbash M, Hall JC, Taghert PH 1999. A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell 99: 791–802 [DOI] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW 2002. Sequential nuclear accumulation of the clock proteins period and timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci 22: 5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidote D, Majercak J, Parikh V, Edery I 1998. Differential effects of light and heat on the Drosophila circadian clock proteins PER and TIM. Mol Cell Biol 18: 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc Natl Acad Sci 106: 13427–13432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EM, Lin JM, Meissner RA, Allada R 2008. Dominant-negative CK2α induces potent effects on circadian rhythmicity. PLoS Genet 4: e12 doi: 10.1371/journal.pgen.0040012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L 1987. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J Cell Biol 104: 1143–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R 2003. Genetic analysis of the circadian system in Drosophila melanogaster and mammals. J Neurobiol 54: 111–147 [DOI] [PubMed] [Google Scholar]
- Sun WC, Jeong EH, Jeong HJ, Ko HW, Edery I, Kim EY 2010. Two distinct modes of PERIOD recruitment onto dCLOCK reveal a novel role for TIMELESS in circadian transcription. J Neurosci 30: 14458–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai HC, Khidekel N, Ficarro SB, Peters EC, Hsieh-Wilson LC 2004. Parallel identification of O-GlcNAc-modified proteins from cell lysates. J Am Chem Soc 126: 10500–10501 [DOI] [PubMed] [Google Scholar]
- Torres CR, Hart GW 1984. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem 259: 3308–3317 [PubMed] [Google Scholar]
- Tseng TS, Salome PA, McClung CR, Olszewski NE 2004. SPINDLY and GIGANTEA interact and act in Arabidopsis thaliana pathways involved in light responses, flowering, and rhythms in cotyledon movements. Plant Cell 16: 1550–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JR, Tartakoff AM, Greenspan NS 1990. Cytologic assessment of nuclear and cytoplasmic O-linked N-acetylglucosamine distribution by using anti-streptococcal monoclonal antibodies. Proc Natl Acad Sci 87: 5608–5612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walgren JL, Vincent TS, Schey KL, Buse MG 2003. High glucose and insulin promote O-GlcNAc modification of proteins, including α-tubulin. Am J Physiol Endocrinol Metab 284: E424–E434 doi: 10.1152/ajpendo.00382.2002 [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW 2001. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science 291: 2376–2378 [DOI] [PubMed] [Google Scholar]
- Yoshii T, Fujii K, Tomioka K 2007. Induction of Drosophila behavioral and molecular circadian rhythms by temperature steps in constant light. J Biol Rhythms 22: 103–114 [DOI] [PubMed] [Google Scholar]
- Young MW, Kay SA 2001. Time zones: A comparative genetics of circadian clocks. Nat Rev Genet 2: 702–715 [DOI] [PubMed] [Google Scholar]
- Zeidan Q, Hart GW 2010. The intersections between O-GlcNAcylation and phosphorylation: Implications for multiple signaling pathways. J Cell Sci 123: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang EE, Kay SA 2010. Clocks not winding down: Unravelling circadian networks. Nat Rev Mol Cell Biol 11: 764–776 [DOI] [PubMed] [Google Scholar]