Abstract
Purpose
To improve the significance of insulin-like growth factor binding protein 5 (IGFBP-5) as a prognostic and potentially predictive marker in breast cancer patients.
Experimental Design
Increased IGFBP-5 expression was identified in MCF-7 cells resistant (MCF-7R4) to the IGF-1R/InsR inhibitor, BMS-536924 and its role examined by targeted knockdown and overexpression in multiple experimental models. Protein expression of IGFBP-5 was measured by immunohistochemistry in a cohort of 76 breast cancer patients to examine correlative associations with invasive tumor fraction and outcome. The utility of a combined IGFBP-5/IGFBP-4 (BPR) expression ratio was applied to predict anti-IGF-1R/InsR response in a panel of breast cancer lines and outcome in multiple breast tumor cohorts.
Results
IGFBP-5 knockdown decreased BMS-536924 resistance in MCF-7R4 cells, while IGFBP-5 overexpression in MCF-7 cells conferred resistance. When compared to pathologically normal reduction mammoplasty tissue, IGFBP-5 expression levels were upregulated in both invasive and histologically normal adjacent breast cancer tissue. In both univariate and multivariate modeling, metastasis-free survival (MFS), recurrence free survival (RFS), and overall survival (OS) were significantly associated with high IGFBP-5 expression. Prognostic power of IGFBP-5 was further increased with the addition of IGFBP-4 where tumors were ranked based upon IGFBP-5/IGFBP-4 expression ratio (BPR). Multiple breast cancer cohorts confirm that BPR (high vs. low) was a strong predictor of RFS and OS.
Conclusion
IGFBP-5 expression is a marker of poor outcome in breast cancer patients. An IGFBP-5/IGFBP-4 expression ratio may serve as a surrogate biomarker of IGF pathway activation and predict sensitivity to anti-IGF-1R-targeting.
Keywords: IGFBP, breast cancer, poor outcome
INTRODUCTION
The IGF system is critical to the malignant progression of breast cancer and aberrant pathway activation has been linked to tumors that have acquired resistance to standard therapeutic intervention (1). As targeted therapies directed against IGF-1 receptor (IGF-1R) progress through clinical trials, the success of these agents is largely dependent upon defining IGF-sensitive patient subgroups. While a number of IGF biomarkers have been identified in breast cancer, the clinical utility of gene expression signatures is often limited by the lack of feasibility to qualitatively and quantitatively assess expression patterns in patient tumors (2). As a result, robust markers of IGF pathway activation are needed.
In normal human physiology, IGF signaling is tightly regulated by multiple factors, which are typically altered in cancer (3). For example, increased IGF ligand expression (IGF-I, IGF-II) has been linked to malignant transformation and resistance when overtly present in the breast epithelial and stromal tissue compartments (4, 5). Ligand-mediated IGF-1R activation then results in the recruitment of adaptor molecules (e.g. Grb2, p85, Shc, IRS-1, IRS-2) to the membrane-bound receptor and subsequent downstream signaling initiation (e.g. MAPK, PI3K). However, the IGF binding proteins (IGFBPs), of which six have been identified and found to be expressed in human tissues, play a predominant role in dictating IGF-1R activation through IGF ligand sequestration, localization, and half-life extension (6). In breast cancer, IGFBPs have been shown to be upregulated and play a critical role in tumor cell growth and motility (3, 7). Although IGFBPs share similar function in regulating IGF bioactivity, in the context of cancer, distinct differences are evident. For example, elevated serum levels of IGFBP-2 have been reported in patients presenting with prostate and brain tumors (8). Conversely, IGFBP-3 overexpression correlates to a decreased risk of colorectal cancer development (9). The opposing function may be endocrine-related as IGFBP-3 sequesters ligand away from the tumor via the circulation while IGFBP-2 binds IGFs for local delivery at the level of the tumor microenvironment (10). Despite correlative differences in IGFBP levels and the risk of neoplastic development, a clear link between IGFBP expression and breast cancer prognosis remains undefined.
We previously employed comparative microarray analysis to identify surrogate markers of resistance resulting from prolonged IGF-1R/InsR inhibitor (BMS-536924) exposure in MCF-7 breast cancer cells (11). Data presented here reveal that IGFBP-5 was significantly overexpressed and highly localized to the cellular membrane in MCF-7R4 cells. Transient overexpression of IGFBP-5 sufficiently attenuated drug-induced growth inhibition in parental MCF-7 cells, while targeted knockdown restored BMS-536924 sensitivity in MCF-7R4 cells. Assessment of IGFBP levels in normal breast tissue and invasive breast carcinoma revealed that IGFBP-5 expression correlated directly with disease progression in a manner independent of the other IGFBPs. We then analyzed the association of IGFBP-5 expression at the transcript and protein level with patient metastasis-free survival (MFS), recurrence-free survival (RFS), and overall survival (OS) in both private and publicly available breast tumor cohorts. The further addition of IGFBP-4 to these analyses drastically improved both prognostic and predictive power, where breast cancer cells and patient tumors with an increased ratio of IGFBP-5 to IGFBP-4 expression (BPR High) exhibited decreased sensitivity to anti-IGF-1R/InsR agents and decreased RFS and OS across multiple patient cohorts. These data implicate the IGFBPs as both diagnostic and prognostic indices in breast cancer patients.
MATERIALS AND METHODS
Cell culture reagents and cell line construction
Reagents were obtained from the following suppliers: Fetal bovine serum, PBS and trypsin-EDTA from Gibco/Invitrogen (Grand Island, NY); IGF-I, LR3-IGF-I, and IGFBP-5 polyclonal antibody from Gro Pep (Thebarton, SA, Australia); DMEM medium, sodium pyruvate, penicillin/streptomycin from Cellgro/MediaTech (Lawrence, KS); CellTiter 96 Non-Radioactive Cell Proliferation Assay Kit from Promega (Madison, WI). Enhanced chemiluminescence (ECL) kits were purchased from Amersham Biosciences/GE Healthcare (Piscataway, NJ). Antibodies against phosphorylated and total IGF1Rβ, InsRβ, IRS-1, Akt, and MAPK were purchased from Cell Signaling Technology (Danvers, MA). MCF-7 cells were obtained and grown as previously described (12) and subsequent to these works, tested and authenticated as MCF-7 cells (11). The following cell lines were purchased within the last 6 months from American Type Culture Condition (ATCC): BT-474, HCC1937, HCC1954, HS578T, MDA-MB-231, SK-BR-3, T47D, ZR-75-1, and ZR-75-30. All lines were maintained as according to ATCC guidelines and reagents. MCF-7R4 cells were generated by step-wise growth in increasing concentrations of BMS-554417. At a concentration of 10 μM, ~40-fold greater than the determined IC50 of the parental cells, individual clones were selected. MCF-7R4 represented a clone isolated that proliferated under continued selection with BMS-554417, a first-generation IGF-1R/Insulin Receptor (InsR) inhibitor at 10 μM and is cross-resistant to the IG-1R/InsR inhibitor BMS-536924, a structurally related analog of BMS-554417. MCF-7/IGFBP-5 stable transfectants were generated by electroporating parental MCF-7 cells with pcDNA3.0 encoding wild-type IGFBP-5 at 240 V for 10 ms using a BTX820 square wave electroporator, selecting for stable transfectants in 800 μg/mL of geneticin, isolating individual clones using cloning rings, and screening for expression by immunoblotting with anti-IGFBP-5 antibody. Empty vector controls were generated similarly using the pcDNA 3.0 vector.
siRNA
Stealth Select RNAi siRNA targeting IGFBP5 was reverse transfected into MCF-7R4 cells according to manufacturer guidelines (Invitrogen). BLOCK-iT Alexa Fluor Red Fluorescent Control and Stealth RNAi™ siRNA Negative Control were included as controls. Cells were exposed to control or siRNA for 48 hours, assessed for both transfection, knockdown efficiency, and growth response to BMS-536924 (10uM) was assessed by MTS assay at Day 5.
Immunoblot
Upon reaching 70% confluency, cells were placed on ice, washed twice with ice-cold PBS, and lysed with lysis buffer of 50 mm Tris-Cl (pH 7.4), 1% Nonidet P-40, 2 mm EDTA (pH 8.0), 100 mm NaCl, 10 mm sodium orthovanadate, 1 mm phenylmethylsulfonyl fluoride, 20 μg/ml leupeptin, and 20 μg/ml aprotinin). Lysates were clarified by centrifugation at 12,000 × g for 15 min at 4 C. Protein concentrations were determined using the bicinchoninic acid protein assay reagent kit (Pierce). Cellular protein (40 μg) was separated by SDS-PAGE, transferred onto Immuno-Blot polyvinylidene difluoride (PVDF), and immunoblotted according to manufacturer guidelines (BioRad).
Immunofluorescence
MCF-7 and MCF-7R4 cells were grown on glass cover slips to 50% confluence, washed with BRB80 buffer (80 mM PIPES, pH6.9; 1 mM MgCl2; 1 mM EGTA) and fixed with BRB80 containing 0.3% glutaraldehyde. Fixed cells were permeablized with 1% Triton X-100 in PBS pH8.0 and treated with 1 mg/mL NaBH4 in PBS to remove endogenous background fluorescence. Cells were blocked with 5% goat serum and stained with 125pg/mL of rabbit anti-IGFBP5 antibody (GroPep Bioreagents Pty Ltd) or normal rabbit control serum (Jackson ImmunoResearch) followed by goat anti rabbit Cy5 (Jackson ImmunoResearch). Coverslips were mounted with prolong gold with DAPI (Invitrogen) and images captured with an LSM 510 Confocal Laser Scanning Microscope (Carl Zeiss MicroImaging, Inc).
qPCR
Cells were plated at a density of 1 × 106 in 100-mm-diameter dishes, allowed to equilibrate overnight and cellular RNA isolated using the RNeasy Plus Mini Kit according to the manufacturer (Qiagen). Forward and reverse primers were designed to target the following transcripts: IGFBP4, IGFBP5, IGFBP6, ACTB, GAPDH, GUSB, RPLP0, GAPDH. RNA (2ug) was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit and quantitative PCR was performed using the SYBR Green PCR Mastermix on an ABI Prism 7900HT machine according to the manufacturer’s recommended protocol (Applied Biosystems). Relative mRNA concentration was calculated using cycle threshold values that were derived from a standard curve and normalized to ribosomal protein, large, PO as an internal control.
Tumor samples & tissue microarray
Primary breast cancer tumor samples were obtained from 76 patients who underwent surgery at the Mayo Clinic between 1997 and 2006. For inclusion in this study, all of the tumors were invasive ductal breast carcinoma. The mean age at diagnosis was 54 years (range, 30-86 years). The following histopathologic variables were determined: T stage, estrogen receptor (ER), and progesterone receptor (PR). Patients that received adjuvant hormonal therapy, chemotherapy and/or radiation therapy were also included. Immediately following surgical resection, tissues were formalin-fixed and paraffin-embedded. FFPE tissue blocks were used to construct tissue microarray (TMA) specimens for IGFBP-5 assessment as previously described (13). Normal human kidney was included as a positive control. Each tumor was sampled in triplicate from representative areas of tumor blocks with 0.6 mm diameter punch cores.
Immunohistochemistry
Immunohistochemical expression analysis was performed as previously described (13). Briefly, a standard indirect immunoperoxidase procedure (ABC-Elite; Vector Laboratories, Burlingame, CA) was employed and TMA slides were stained with an isoform-specific IGFBP-5 antibody (Santa Cruz Biotechnology, CA). A minimum of three sections from each tumor sample were scored and averaged to derive a highly reproducible and corresponding immunoscore. IGFBP-5 staining assessment was blinded to all clinical parameters and the study endpoint.
Clonogenic assays & monolayer proliferation
Clonogenic assays were performed as previously described (14). For monolayer proliferation, cells were plated in 24-well plates at a density of 5,000 cells/well, allowed to equilibrate overnight, and were either starved in SFM for 24 h prior to treatment or remained in full medium for the duration of the assay. Growth was assessed using the MTS proliferation assay according to the manufacturers instructions (CellTiter 96 Aqueous, Promega, Madison, WI). Experiments were performed in triplicate and on three separate occasions.
ELISA
Culture medium and whole cell lysates collected from MCF-7 and MCF-7R4 cells were used to quantify IGFBP-5 expression by ELISA assay (Cat. No. DY875, R&D Systems). Results were similar by triplicate replication (both biological and technical).
Microarray analysis
Comparative microarray studies of MCF-7 and MCF-7R4 cells were performed as previously described (11). IGFBP-5 was identified as one of the most different regulated genes based on fold-induction (≥ 4-fold) and p-value (≤ 0.05) cutoffs. For analysis of public microarray datasets, normalized gene expression data was obtained from the Gene Expression Omnibus (GEO) database for the following independent studies: GSE7390, GSE6523, GSE1456, GSE18229, GSE17705, GSE2034, GSE4823, GSE9014. Data for the NKI-295 was retrieved from the following public repository: http://microarray-pubs.stanford.edu/wound_NKI/explore.html. Patient tumors within each cohort were independently ranked according to both IGFBP-5 and IGFBP-4 expression. The ratio of IGFBP-5/IGFBP-4 (BPR) was determined for each tumor and the following cutoffs implemented for stratification (BPR High ≥ 2.0, BPR Low ≤ 0.5).
Statistical analysis
Statistical significance between two groups was tested using Student’s t test or Mann-Whitney test as indicated and analysis of variance (ANOVA) with Bonferroni’s post hoc test was used for multiple-comparison analysis using Prism 5.0 (GraphPad Software, La Jolla, CA). Univariate survival analyses were performed using the Kaplan-Meier method and corresponding log-rank test for intergroup differences. Multivariate survival analyses were performed with the Cox proportional hazards model. All analyses were conducted using JMP 9.0 software. Error bars represent standard deviation and results are representative of at least three independent experiments.
RESULTS
IGFBP-5 confers resistance to IGF-1R/InsR inhibition
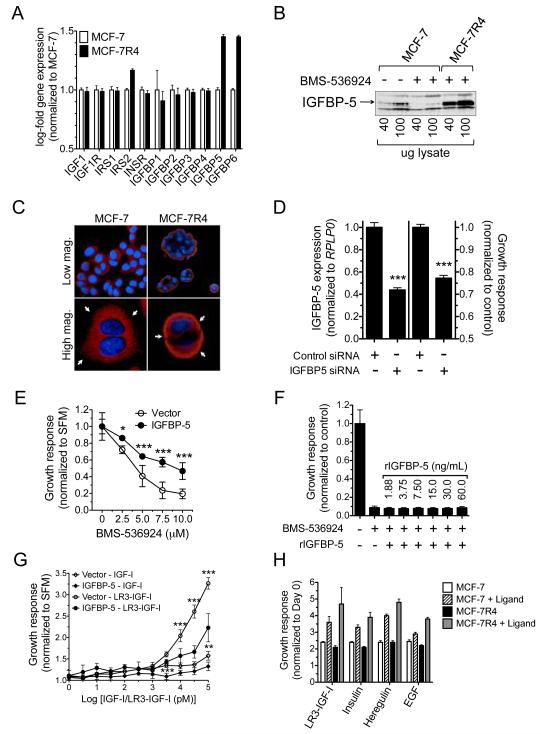
To highlight differences in MCF-7R4 vs. parental MCF-7 cells and illustrate the significance of IGFBP-5 upregulation, comparative gene expression analysis was employed (S1A.). Applying stringent criteria to these analyses (p=<0.001 and fold-regulation >5.0) distinguished IGFBP-5 as the most significantly regulated target gene. Analysis of IGF pathway genes revealed that IGFBP-5 and IGFBP-6 were the only significantly upregulated IGFBP family members (Fig. 1A.). However, validation by qPCR revealed that IGFBP-5 expression was significantly higher (>3-fold) than IGFBP-6 (S1B.). Further characterization of downstream IGF signaling components (IGF-1Rβ/InsRβ, Akt, MAPK) suggested no major differences were present (S1C.).
Figure 1.
IGFBP-5 confers resistance to IGF-1R/InsR inhibition. (A) Comparison of IGF pathway genes in MCF-7 (white bars) vs MCF-7R4 (black bars) cells (B) Western Blot analysis of IGFBP-5 expression in MCF-7 vs. MCF-7R4 cells (C) Immunohistochemistry staining for the nucleus (blue) and IGFBP-5 (red) in MCF-7 (left column) and MCF-7R4 (right column) cells. Colonies (top row) and single cells (bottom row) are shown at different magnifications. (D) Depiction of IGFBP-5 knockdown as measured by qPCR (left) and resultant growth (right) in MCF-7R4 cells. (E) MCF-7 cells overexpressing empty vector (open circles) or IGFBP-5 (closed circles) were exposed to increasing concentrations of BMS-536924 and assessed for colony outgrowth. (F) Growth assessment of MCF-7 cells in response to BMS-536924 (10uM) treatment and addition of recombinant IGFBP-5. (G) Dose response to IGF-1 and LR3-IGF-1 and resulting growth response of MCF-7/Vector (open) vs. MCF-7/IGFBP-5 (closed). (H) Growth response of MCF-7 vs. MCF-7R4 cells to LR3-IGF-1 (10nM), insulin (10nM), Heregulin (10nM), and EGF (10nM). Error bars represent standard deviation, asterisks denote statistical significance; *, P < 0.05; **, P < 0.01; ***, P < 0.001, and results are representative of at least three independent replicates.
IGFBP-5 expression was then measured in MCF-7R4 cells by Western blot analysis (Fig. 1B.) and ELISA (S1D.) to ensure transcript and protein levels were directly correlative. IGFBP-5 overexpression was not selectively compartmentalized as marked increases were detected both intracellularly via whole cell lysates and extracellularly in the surrounding medium. Furthermore, acute (48 hours) exposure to BMS-536924 (10uM) suppressed IGFBP-5 expression in MCF-7 parental cells. Immunofluoresence analysis of both single cells and colony outgrowths revealed that IGFBP-5, while diffusely expressed in parental cells, was highly localized to the cellular membrane in MCF-7R4 cells in a punctated fashion (Fig. 1C.).
Multiple strategies were then employed to determine if IGFBP-5 is causally or secondarily related to anti-IGF-1R/InsR resistance. First, MCF-7R4 cells were transiently transfected with IGFBP-5-directed siRNA and knockdown confirmed by qPCR (Fig. 1D.) Second, MCF-7 cells were stably transfected with a vector containing full-length IGFBP-5 and overexpression confirmed by ELISA (Fig. 1E.). As an aside, MCF-7/IGFBP-5 and vector-transfected cells exhibited similar patterns of basal growth (S1E.) and signaling (S1F.) with the exception of increased IGF-1Rβ levels in IGFBP-5-overexpressing cells. In both scenarios, modulated IGFBP-5 expression significantly altered growth in response to BMS-536924, where knockdown resensitized (modest as BCRP is also know to play a role in resistance) previously resistant cells and overexpression conferred resistance in sensitive cells. However, exogenous supplementation of IGFBP-5 to the surrounding medium of MCF-7 cells did not overcome BMS-536924 sensitivity, suggesting that IGFBP-5 may act in an autocrine fashion to facilitate resistance (Fig. 1F.). It should be noted that the IGFBP-5 dosing scheme, while supraphysiologic, was designed to overlap with and exceed endogenous MCF-7R4 IGFBP-5 levels (~30ng/mL). Interestingly, MCF-7/IGFBP-5 cell growth in response to IGF ligand stimulation (Fig. 1G.) was opposite to ligand (IGF, Insulin, Heregulin, and EGF) response in MCF-7R4 cells (Fig. 1H).
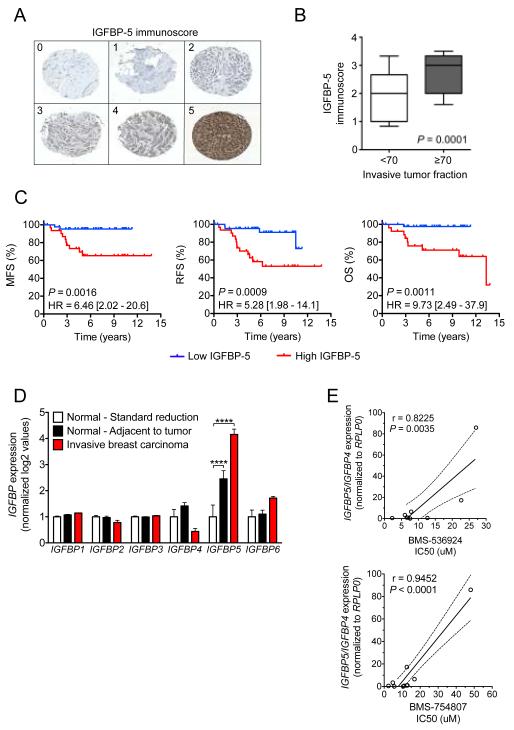
IGFBP-5 expression correlates with increased invasion and poor overall survival
As a preliminary method to explore the range of IGFBP5 expression in breast cancer tumors, a cohort of patients diagnosed with invasive ductal carcinoma that underwent subsequent surgical removal at the Mayo Clinic was assembled (n = 76). Immunohistochemistry (IHC) analysis was employed in tissue microarray (TMA) sections of patient tumors. Stained slides were then assigned an appropriate IGFBP-5 immunoscore (Fig. 2A.). Initially, tumors with highly invasive disease (≥ 70% of the tumor was pathologically infiltrating) were compared to those with less invasive disease (< 70% invasive tumor fraction) and IGFBP-5 was found to be significantly higher (P = 0.0001) in the more invasive disease fractions (Fig. 2B.). To examine IGFBP-5 as a prognostic biomarker, patients were stratified by high (≥ 3) vs. low (<3) IGFBP-5 immunoscore and univariate Kaplan-Meier analysis was performed (Fig. 2C.). There was a significant difference in MFS (HR = 6.46, 95% CI = 2.02 - 20.6, P = 0.0016), RFS (HR = 5.28, 95% CI = 1.98 - 14.1, P = 0.0009), and OS (HR = 9.73, 95% CI = 2.49 - 37.9, P = 0.0011) between patients with high versus low IGFBP-5 expression. To assess the contribution of IGFBP-5 as a prognostic indicator while accounting for additional survival factors (age, estrogen receptor expression (ER), progesterone receptor expression (PR), chemotherapy, radiation therapy, and hormone therapy) multivariate Cox-regression analysis was performed (Table 1). When treated as a continuous variable, IGFBP-5 outperformed all clinical variables in predicting MFS (HR = 3.17, 95% CI = 1.37 - 10.0, P = 0.0050), RFS (HR = 2.67, 95% CI = 1.36 - 6.19, P = 0.0035), and OS (HR = 3.62, 95% CI = 1.37 - 13.7, P = 0.0069). In addition, univariate and multivariate regression analysis was performed in the NKI-295 and confirmed that patient tumors with high levels of IGFBP-5 expression exhibited significantly reduced MFS, RFS and OS (Supplementary Tables S1 & S2).
Figure 2.
IGFBP-5 expression correlates with increased invasion and poor outcome. (A) Breast cancer tumor sections were stained for IGFBP-5 in a cohort of patients from the Mayo Clinic (n=76), and an immunoscore assigned (0 = no expression to 5 = highest expression). (B) Box and whisker plot comparing tumors with high (≥70%) vs. low (<70%) invasive fractions. Statistical analysis was performed by Mann-Whitney test and p-value included. (C) Univariate Kaplan-Meier analysis of metastasis-free survival (MFS), recurrence-free survival (RFS), and overall survival (OS) in the Mayo cohort stratified by high (≥3, red line) vs. low (≤3, blue line) IGFBP-5 immunoscore. (D) IGFBP transcript expression in breast tissues obtained from standard reduction surgery (white bars), invasive breast carcinoma (red bars), and histologically normal adjacent breast tissue (black bars). (E) IGFBP-5/IGFBP-4 expression directly correlates to anti-IGF1R/InsR activity in a panel of breast cancer cell lines. IC50 is plotted alongside the IGFBP-5/IGFBP-4 expression value for each cell line. Pearson correlation shows there is a significant correlation for both BMS-536924 (r = 0.8225, P = 0.0035) and BMS-754807 (r = 0.9452, P < 0.0001) with a higher expression ratio being associated with a decreased response to both agents. Error bars represent standard deviation, asterisks denote statistical significance; ***, P < 0.01; ****, P < 0.0001.
Table 1.
Multivariate Cox regression analysis of MFS, RFS, and OS in the Mayo cohort.
| MFS | RFS | OS | ||||
|---|---|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | HR (95% CI) | P | |
| Age | 1.13(0.56-2.32) | 0.7218 | 0.96(0.49-1.80) | 0.9011 | 1.21 (0.59-2.50) | 0.5822 |
| ER | 1.63(0.25-12.0) | 0.6209 | 1.93(0.42-10.4) | 0.4088 | 0.91 (0.14-6.97) | 0.9232 |
| PR | 0.70(0.12-4.84) | 0.7061 | 0.71 (0.17-3.38) | 0.6623 | 1.03(0.12-10.3) | 0.9772 |
| Chemotherapy | 2.76 (0.22-39.4) | 0.4225 | 0.49(0.05-4.17) | 0.5193 | 6.22(0.38-192) | 0.2029 |
| Radiation | 3.21 (0.40-74.6) | 0.3008 | 16.5(2.12-402) | 0.0044 | 2.27 (0.30-54.2) | 0.4706 |
| Hormone therapy | 0.43(0.09-2.17) | 0.2915 | 0.36(0.09-1.52) | 0.1590 | 0.54 (0.09-3.54) | 0.5065 |
| IGFBP-5 | 3.17(1.37-10.0) | 0.0050 | 2.67(1.36-6.19) | 0.0035 | 3.62(1.37-13.7) | 0.0069 |
To further understand how IGFBP-5 regulation is important in tumorigenesis, IGFBP expression was assessed in patients undergoing surgery for breast reduction mammoplasty or breast cancer (15, 16). In comparison to the other IGFBPs, IGFBP-5 expression was markedly upregulated in both the invasive and adjacent histologically normal breast compartments versus normal reduction mammoplasty tissue (Fig. 2D). In contrast, IGFBP-4 expression decreased (albeit not significantly under these statistical parameters) in normal versus invasive disease. While beyond the scope of these works, current efforts are underway to further characterize the role of IGFBP-5 and IGFBP-4 during breast tumorigenesis and disease progression. However, as a preliminary means of exploring the prognostic and potentially predictive value of IGFBP-5 and IGFBP-4 as breast tumor biomarkers, the following breast cancer cell lines were selected: BT-474, HCC1937, HCC1954, HS578T, MCF-7, MDA-MB-231, SK-BR-3, T47D, ZR-75-1, and ZR-75-30. Under basal growth conditions, IGFBP-5 and IGFBP-4 expression values were determined by qPCR and BMS-536924 IC50 values calculated experimentally. While IGFBP-5 and IGFBP-4 alone did not significantly correlate to anti-IGF-1R/InsR sensitivity, the ratio of IGFBP-5/IGFBP-4 was significantly correlated to both BMS-536924 and BMS-754807 resistance (Fig. 2E.). As a confirmatory observation, the ratio of IGFBP-5/IGFBP-4 expression was found to be increased nearly 10-fold in MCF-7R4 vs. MCF-7 cells (data not shown).
IGFBP-5/IGFBP-4 ratio (BPR) correlates with poor outcome across multiple cohorts
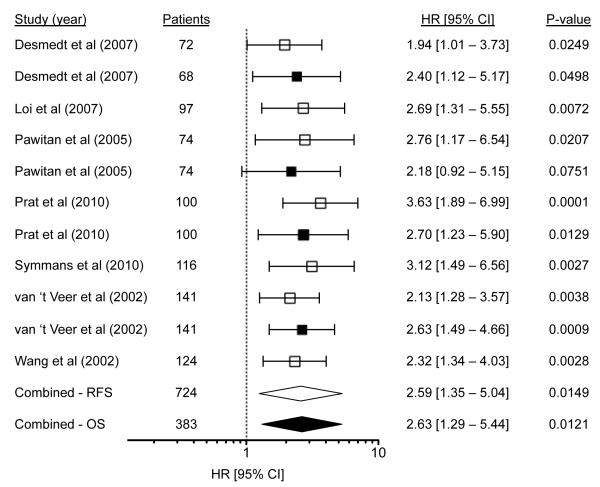
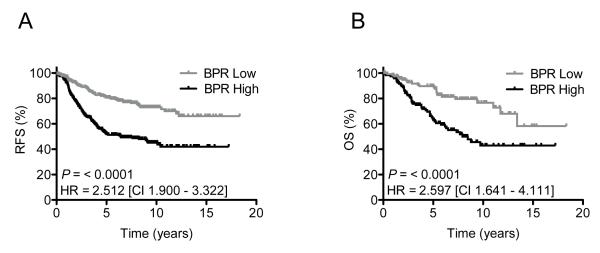
IGFBP-4 and IGFBP-5 expression in breast tumors have both independently been linked to outcome in prior studies (17-19). However, the prognostic power of either IGFBP-4 alone or IGFBP-5 alone is limited to specific cohorts of patients and does not reproduce in a robust manner across multiple datasets. In an effort to increase the prognostic value of IGFBP-5 and based upon the observation that invasive tumors displayed marked reductions in IGFBP-4 expression when compared to normal breast tissue (Fig. 2D.), IGFBP-5 and IGFBP-4 expression was combined to form a ratio. Specifically, patient tumors were independently ranked according to IGFBP-5 and IGFBP-4 expression and an IGFBP-5/IGFBP-4 expression ratio (BPR) was assigned to each sample. Cutoffs were set to distinguish tumors that were BPR High (IGFBP-5/IGFBP-4 expression ratio ≥ 2.0) versus BPR Low (IGFBP-5/IGFBP-4 expression ratio ≤ 0.5). BPR was calculated in multiple breast tumor cohorts and univariate HRs with corresponding 95% CIs for RFS and/or OS are depicted in a forest plot (Fig. 3). These data demonstrate that BPR High patients have poor outcomes compared to BPR Low patients in terms of RFS (HR = 2.59, 95% CI = 1.35 - 5.04, P = 0.0149) and OS (HR = 2.63, 95% CI = 1.29 - 5.44, P = 0.0121). Univariate analysis of the combined groups (all tumors meeting the aforementioned cutoffs) yielded highly significant differences in RFS (HR = 2.52, 95% CI = 1.98 - 3.21, P = <0.0001) and OS (HR = 2.48, 95% CI = 1.73 - 3.54, P = <0.0001) (Fig. 4). In addition, 5- and 10-year estimates of recurrence and survival are presented (Tables 2 & 3). At 5 years, patients with BPR High tumors had shorter recurrence (71%) and survival (73%) rates than patients with BPR Low tumors (29% and 27%) and similar results are reported at 10 years.
Figure 3.
Forest plot of RFS (open) and OS (filled) by BPR expression in multiple breast tumor cohorts, showing HRs (BPR Low/High) and 95% CIs where a HR >1 implies a higher risk of recurrence and mortality in the BPR High group.
Figure 4.
Kaplan Meier analysis of the combined cohorts depicting RFS (A) and OS (B) in the BPR Low (gray line) vs. BPR High (black line) groups.
Table 2.
Odds ratios of the combined cohorts depicting RFS at 5 and 10 years in the BPR Low vs. BPR High groups.
| RFS | No. of Patients |
BPR High |
BPR Low |
Odds Ratio (95% CI) |
P |
|---|---|---|---|---|---|
| ≤5yrs | 221 | 71% | 29% | 2.91 | <0.0001 |
| >5yrs | 94 | 46% | 54% | (1.77-4.79) | |
| ≤10yrs | 261 | 67% | 33% | 3.23 | <0.0001 |
| > 10yrs | 352 | 39% | 61% | (2.31 - 4.52) |
Table 3.
Odds ratios of the combined cohorts depicting OS at 5 and 10 years in the BPR Low vs. BPR High groups
| OS | No. of Patients |
BPR High |
BPR Low |
Odds Ratio (95% CI) |
P |
|---|---|---|---|---|---|
| ≤5yrs | 79 | 73% | 27% | 2.57 | 0.0159 |
| >5yrs | 54 | 52% | 48% | (1.24–5.33) | |
| ≤10yrs | 116 | 69% | 31% | 3.11 | <0.0001 |
| > 10yrs | 199 | 42% | 58% | (1.91 - 5.04) |
DISCUSSION
The function of the IGF pathway in breast cancer cells is complex and incompletely understood. IGF-1R activation leads to increased tumor cell proliferation, motility and survival in both in vitro and in vivo model systems. As shown here, the IGF pathway is also important in response to targeted therapeutics, where prolonged exposure to the IGF-1R/InsR inhibitor BMS-536924 resulted in resistant outgrowth. From a mechanistic standpoint, a number of signaling molecules are likely involved. However, striking alterations in IGFBP-5 expression prompted further investigation and revealed highly localized levels of IGFBP-5 in the extracellular and membrane compartment of MCF-7R4 cells. Transient overexpression of IGFBP-5 in parental MCF-7 cells and IGFBP-5 knockdown in MCF-7R4 cells effectively recapitulated resistance and parental sensitivity, further indicating that IGFBP-5 plays a central role as a both means of escape and marker of resistance. This is important, as a number of IGFBPs have been shown to function in the tumorigenesis of primary and secondary breast malignancies.
These findings suggest that the role of IGFBP-5 in resistance appears to be contextual. For example, in MCF-7 cells the presence of functional IGF-1R/InsR coupled with chronic IGFBP-5 overexpression conferred resistance to anti-IGF-1R/InsR targeting and decreased sensitivity to IGF-induced growth. These results correlate with previous reports of IGFBP-5 overexpression effectively inhibiting cell growth (20). Alternatively, chronic IGF-1R/InsR inhibition by BMS-536924 induced IGFBP-5 overexpression and membrane localization in MCF-7R4 cells, likely as a means of preserving IGF pathway function. As a result, MCF-7R4 cells were highly sensitive to IGF-dependent and independent ligand-induced growth. While increased IGFBP-5 represents the common denominator in both aforementioned scenarios, the discordant growth factor response profile reveals contextual differences in biological function. Acute exposure to exogenous rIGFBP-5 did not affect sensitivity to BMS-536924, further supporting the differing roles of IGFBP-5 depending on context.
IGF ligand expression correlates with poor prognosis and endocrine resistance in breast cancer (21, 22). However, IGFBP expression does not universally correlate to that of ligand as IGF ligand bioavailability can be increased or decreased depending upon which IGFBP isoform is present. For example, IGFBP-4 inhibits IGF-I activation of IGF-1R when the binding protein/ligand complex is targeted for proteolysis (23). Moreover, protease-resistant IGFBP-4 blocks breast tumor IGF activity, growth, and angiogenesis (24). These data support our observation of an inverse correlation between IGFBP-4 expression and invasive breast cancer. While IGFBP-4 is exclusively inhibitory to IGF action, the role of IGFBP-5 is complex as more than 100 proteins potentially interact with IGFBP-5 and influence novel functional properties (25). In addition, hallmark studies examining the relationship between growth factors and fibroblasts demonstrate that IGFBP-5 has a propensity to accumulate in the extracellular matrix (ECM), thereby avoiding degradation and potentiating the effects of IGF-I ligand on IGF-1R activation (26). Where IGFBP-5 readily binds ECM components such as collagen, laminin, and fibronectin, IGFBP-4 exhibits the reverse phenotype and is not present in the ECM. As these and our data suggest, it is therefore plausible that breast tumor cells inversely regulate IGFBP-5 and IGFBP-4 expression, localization, and bioavailability in an effort to maximize IGF signaling. Furthermore, a number of reports support a role for increased IGFBP-5 expression in breast cancer tumors and formation of secondary metastases (27). While the prognostic significance of IGFBP-5 and IGFBP-4 has been shown in patient breast tumors, applicability is limited to select patient subsets. In this study, IGFBP-5 and IGFBP-4 were combined to form a ratio of expression (BPR) and cutoff parameters were enforced to result in widespread utility of BPR High as a marker of poor outcome in a large (RFS = 724, OS = 383) and diverse (7 patient cohorts) set of tumors. This ratio will be key and may explain why decreased and/or loss of IGFBP-5 expression is directly related to tamoxifen sensitivity (28). It is plausible that while both IGFBP-5 and IGFBP-4 levels are decreased in hormone-refractory patient tumors, the degree of IGFBP-4 downregulation may exceed that of IGFBP-5 to result in increased IGFBP-5 vs IGFBP-4. As a result, current efforts are underway to determine the role of BPR in hormone-refractory breast cancer.
Perhaps the greatest potential for targeting the IGF system hinges upon disrupting crosstalk with other signaling pathways as preclinical work has identified significant antitumor activity when IGF-1R is concordantly targeted with mTOR, ERα, EGFR and HER2. As a result, IGF-1R has become one of the most well studied molecular targets in oncology with over 60 ongoing clinical trials currently examining potential benefit in patients with therapeutically refractive, recurrent and/or metastatic disease (29). In a recent report examining a small cohort of patients with advanced sarcomas and other solid tumors, the pharmacokinetic profile and preliminary antitumor effects of combined IGF-1R/mTOR inhibition are reported (30). While significant antitumor activity and maintenance of stable disease was reported in select patients, the authors note that a primary limitation of the study was a lack of biomarkers to assess the status of IGF pathway activation. These findings highlight the salient need for IGF pathway patient stratifiers and, as was the case in the development of trastuzumab, further recapitulate the power of patient selection through appropriate biomarker development. The current study utilized IGF-1R/InsR-resistant breast cancer cells to delineate potential markers of acquired resistance and found that IGFBP-5 was significantly associated with poor outcome in patient tumors. Addition of BPR further improved the prognostic power and is presented here as a potentially predictive clinical application.
Supplementary Material
TRANSLATIONAL RELEVANCE.
Herein, we demonstrate that IGFBP-5 expression is a marker of poor prognosis in breast cancer patients. Increased IGFBP-5 was highly correlative to invasive disease, distant metastasis, recurrence, and overall survival. In comparison to pathologically normal reduction mammoplasty tissue, IGFBP-5 levels were upregulated in both invasive and histologically normal adjacent breast cancer tissue, indicating that IGFBP-5 may serve as a marker of early carcinogenesis and progression. While deregulated IGFBP expression was specific to IGFBP-5, the ratio of IGFBP-5 to IGFBP-4 expression (BPR) was a strong predictor of both anti-IGF-1R/InsR response in a panel of breast tumor lines and outcome across multiple breast tumor cohorts. These data demonstrate that BPR predicts disease recurrence and patient survival and provide compelling evidence that IGFBPs may serve as surrogate markers of IGF pathway activation to improve IGF-targeted therapeutics.
Acknowledgments
Supported in part by the Mayo Clinic Breast SPORE (CA116201-03), NIH K12 (CA090628-05) and the Mayo Clinic Cancer Center (CA15083).
GRANT SUPPORT
Supported in part by the Mayo Clinic Breast SPORE (CA116201-03), NIH K12 (CA090628-05) and the Mayo Clinic Cancer Center (CA15083).
Footnotes
Authors’ Disclosure of Potential Conflicts of Interests:
Joan M. Carboni and Marco M. Gottardis are employees of Bristol-Myers Squibb
REFERENCES
- 1.Turner BC, Haffty BG, Narayanan L, Yuan J, Havre PA, Gumbs AA, et al. Insulin- like growth factor-I receptor overexpression mediates cellular radioresistance and local breast cancer recurrence after lumpectomy and radiation. Cancer Res. 1997 Aug 1;57(15):3079–83. [PubMed] [Google Scholar]
- 2.Creighton CJ, Casa A, Lazard Z, Huang S, Tsimelzon A, Hilsenbeck SG, et al. Insulin-like growth factor-I activates gene transcription programs strongly associated with poor breast cancer prognosis. J Clin Oncol. 2008 Sep 1;26(25):4078–85. doi: 10.1200/JCO.2007.13.4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuire WL, Jr., Jackson JG, Figueroa JA, Shimasaki S, Powell DR, Yee D. Regulation of insulin-like growth factor-binding protein (IGFBP) expression by breast cancer cells: use of IGFBP-1 as an inhibitor of insulin-like growth factor action. J Natl Cancer Inst. 1992 Sep 2;84(17):1336–41. doi: 10.1093/jnci/84.17.1336. [DOI] [PubMed] [Google Scholar]
- 4.Paik S. Expression of IGF-I and IGF-II mRNA in breast tissue. Breast Cancer Res Treat. 1992;22(1):31–8. doi: 10.1007/BF01833331. [DOI] [PubMed] [Google Scholar]
- 5.Lu Y, Zi X, Zhao Y, Mascarenhas D, Pollak M. Insulin-like growth factor-I receptor signaling and resistance to trastuzumab (Herceptin) J Natl Cancer Inst. 2001 Dec 19;93(24):1852–7. doi: 10.1093/jnci/93.24.1852. [DOI] [PubMed] [Google Scholar]
- 6.Yee D, Rosen N, Favoni RE, Cullen KJ. The insulin-like growth factors, their receptors, and their binding proteins in human breast cancer. Cancer Treat Res. 1991;53:93–106. doi: 10.1007/978-1-4615-3940-7_5. [DOI] [PubMed] [Google Scholar]
- 7.Zhang X, Yee D. Insulin-like growth factor binding protein-1 (IGFBP-1) inhibits breast cancer cell motility. Cancer Res. 2002 Aug 1;62(15):4369–75. [PubMed] [Google Scholar]
- 8.Mehrian-Shai R, Chen CD, Shi T, Horvath S, Nelson SF, Reichardt JK, et al. Insulin growth factor-binding protein 2 is a candidate biomarker for PTEN status and PI3K/Akt pathway activation in glioblastoma and prostate cancer. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5563–8. doi: 10.1073/pnas.0609139104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J, Pollak MN, Giovannucci E, Chan JM, Tao Y, Hennekens CH, et al. Prospective study of colorectal cancer risk in men and plasma levels of insulin-like growth factor (IGF)-I and IGF-binding protein-3. J Natl Cancer Inst. 1999 Apr 7;91(7):620–5. doi: 10.1093/jnci/91.7.620. [DOI] [PubMed] [Google Scholar]
- 10.Pollak M. Insulin and insulin-like growth factor signalling in neoplasia. Nat Rev Cancer. 2008 Dec;8(12):915–28. doi: 10.1038/nrc2536. [DOI] [PubMed] [Google Scholar]
- 11.Hou X, Huang F, Carboni JM, Flatten K, Asmann YW, Ten Eyck C, et al. Drug efflux by breast cancer resistance protein is a mechanism of resistance to the benzimidazole insulin-like growth factor receptor/insulin receptor inhibitor, BMS-536924. Mol Cancer Ther. 2011 Jan;10(1):117–25. doi: 10.1158/1535-7163.MCT-10-0438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haluska P, Carboni JM, Loegering DA, Lee FY, Wittman M, Saulnier MG, et al. In vitro and in vivo antitumor effects of the dual insulin-like growth factor-I/insulin receptor inhibitor, BMS-554417. Cancer Res. 2006 Jan 1;66(1):362–71. doi: 10.1158/0008-5472.CAN-05-1107. [DOI] [PubMed] [Google Scholar]
- 13.Wang H, Zhang W, Fuller GN. Overexpression of IGFBP5, but not IGFBP3, correlates with the histologic grade of human diffuse glioma: a tissue microarray and immunohistochemical study. Technol Cancer Res Treat. 2006 Jun;5(3):195–9. doi: 10.1177/153303460600500303. [DOI] [PubMed] [Google Scholar]
- 14.Haluska P, Carboni JM, TenEyck C, Attar RM, Hou X, Yu C, et al. HER receptor signaling confers resistance to the insulin-like growth factor-I receptor inhibitor, BMS- 536924. Mol Cancer Ther. 2008 Sep;7(9):2589–98. doi: 10.1158/1535-7163.MCT-08-0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finak G, Sadekova S, Pepin F, Hallett M, Meterissian S, Halwani F, et al. Gene expression signatures of morphologically normal breast tissue identify basal-like tumors. Breast Cancer Res. 2006;8(5):R58. doi: 10.1186/bcr1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008 May;14(5):518–27. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 17.Vendrell JA, Robertson KE, Ravel P, Bray SE, Bajard A, Purdie CA, et al. A candidate molecular signature associated with tamoxifen failure in primary breast cancer. Breast Cancer Res. 2008;10(5):R88. doi: 10.1186/bcr2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mita K, Zhang Z, Ando Y, Toyama T, Hamaguchi M, Kobayashi S, et al. Prognostic significance of insulin-like growth factor binding protein (IGFBP)-4 and IGFBP-5 expression in breast cancer. Jpn J Clin Oncol. 2007 Aug;37(8):575–82. doi: 10.1093/jjco/hym066. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Cao X, Zhang W, Feng Y. Expression level of insulin-like growth factor binding protein 5 mRNA is a prognostic factor for breast cancer. Cancer Sci. 2007 Oct;98(10):1592–6. doi: 10.1111/j.1349-7006.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butt AJ, Dickson KA, McDougall F, Baxter RC. Insulin-like growth factor-binding protein-5 inhibits the growth of human breast cancer cells in vitro and in vivo. J Biol Chem. 2003 Aug 8;278(32):29676–85. doi: 10.1074/jbc.M301965200. [DOI] [PubMed] [Google Scholar]
- 21.Huynh HT, Tetenes E, Wallace L, Pollak M. In vivo inhibition of insulin-like growth factor I gene expression by tamoxifen. Cancer Res. 1993 Apr 15;53(8):1727–30. [PubMed] [Google Scholar]
- 22.Peyrat JP, Bonneterre J, Hecquet B, Vennin P, Louchez MM, Fournier C, et al. Plasma insulin-like growth factor-1 (IGF-1) concentrations in human breast cancer. Eur J Cancer. 1993;29A(4):492–7. doi: 10.1016/s0959-8049(05)80137-6. [DOI] [PubMed] [Google Scholar]
- 23.Jones JI, Clemmons DR. Insulin-like growth factors and their binding proteins: biological actions. Endocr Rev. 1995 Feb;16(1):3–34. doi: 10.1210/edrv-16-1-3. [DOI] [PubMed] [Google Scholar]
- 24.Ryan AJ, Napoletano S, Fitzpatrick PA, Currid CA, O’Sullivan NC, Harmey JH. Expression of a protease-resistant insulin-like growth factor-binding protein-4 inhibits tumour growth in a murine model of breast cancer. Br J Cancer. 2009 Jul 21;101(2):278–86. doi: 10.1038/sj.bjc.6605141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akkiprik M, Feng Y, Wang H, Chen K, Hu L, Sahin A, et al. Multifunctional roles of insulin-like growth factor binding protein 5 in breast cancer. Breast Cancer Res. 2008;10(4):212. doi: 10.1186/bcr2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones JI, Gockerman A, Busby WH, Jr., Camacho-Hubner C, Clemmons DR. Extracellular matrix contains insulin-like growth factor binding protein-5: potentiation of the effects of IGF-I. J Cell Biol. 1993 May;121(3):679–87. doi: 10.1083/jcb.121.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van ’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002 Jan 31;415(6871):530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 28.Ahn BY, Elwi AN, Lee B, Trinh DL, Klimowicz AC, Yau A, et al. Genetic screen identifies insulin-like growth factor binding protein 5 as a modulator of tamoxifen resistance in breast cancer. Cancer Res. 2010 Apr 15;70(8):3013–9. doi: 10.1158/0008-5472.CAN-09-3108. [DOI] [PubMed] [Google Scholar]
- 29.Heidegger I, Pircher A, Klocker H, Massoner P. Targeting the insulin-like growth factor network in cancer therapy. Cancer Biol Ther. 2011 Apr 15;11(8) doi: 10.4161/cbt.11.8.14689. [DOI] [PubMed] [Google Scholar]
- 30.Quek R, Wang Q, Morgan JA, Shapiro GI, Butrynski JE, Ramaiya N, et al. Combination mTOR and IGF-1R inhibition: phase I trial of everolimus and figitumumab in patients with advanced sarcomas and other solid tumors. Clin Cancer Res. 2011 Feb 15;17(4):871–9. doi: 10.1158/1078-0432.CCR-10-2621. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.