Abstract
In most sexually reproducing animals, replication and maintenance of telomeres occurs in the germ line and during early development in embryogenesis through the use of telomerase. Somatic cells generally do not maintain telomere sequences, and these cells become senescent in adults as telomeres shorten to a critical length. Some animals reproduce clonally and must therefore require adult somatic mechanisms for maintaining their chromosome ends. Here we study the telomere biology of planarian flatworms with apparently limitless regenerative capacity fueled by a population of highly proliferative adult stem cells. We show that somatic telomere maintenance is different in asexual and sexual animals. Asexual animals maintain telomere length somatically during reproduction by fission or when regeneration is induced by amputation, whereas sexual animals only achieve telomere elongation through sexual reproduction. We demonstrate that this difference is reflected in the expression and alternate splicing of the protein subunit of the telomerase enzyme. Asexual adult planarian stem cells appear to maintain telomere length over evolutionary timescales without passage through a germ-line stage. The adaptations we observe demonstrate indefinite somatic telomerase activity in proliferating stem cells during regeneration or reproduction by fission, and establish planarians as a pertinent model for studying telomere structure, function, and maintenance.
Some animals may be potentially immortal or at least very long-lived. Understanding the mechanisms that have evolved to allow some animals to be immortal may shed further light on the possibilities for alleviating aging and age-related phenotypes in human cells (1, 2). These animals must have the capacity to replace aged, damaged, or diseased tissues and cells and hence use a population(s) of proliferative stem cells able to do this (3–5).
To ensure heritability and genetic stability, dividing eukaryotic cells must overcome the end-replication problem to maintain linear chromosomes (6). In sexually reproducing animals such as humans, telomere elongation occurs mainly during embryogenesis and the development of the germ line (7, 8). Somatic cells become senescent in the adult when chromosome ends shorten to a critical length to avoid deleterious genome instability and the emergence of cancerous cells (9). This protective senescence mechanism appears to be a central part of the aging process (10), and animals that are potentially immortal must have somatic mechanisms for maintaining chromosome ends. We set out to test this idea in potentially immortal planarian flatworms. Planarians have been described as “immortal under the edge of the knife” (11), and may have an indefinite capacity to renew their differentiated tissues from a pool of potentially immortal planarian adult stem cells (pASCs) (12, 13). For long-term survival over evolutionary timescales, these cells need to overcome the end-replication problem (6).
The model planarian Schmidtea mediterranea has both asexual and sexual strains, both with apparently indefinite regenerative capacities (3, 12). The agametic asexual strain reproduces by fission behind the pharynx and has no functional gonads (12). Thus, we hypothesize that it has developed somatically active mechanisms for the maintenance of chromosome ends without sexual reproduction per se. The sexual strain of this species does not fission naturally, instead reproducing as a cross-fertilizing hermaphrodite (12). We find that asexual but not sexual animals have telomere maintenance mechanisms that allow telomere maintenance somatically. This mechanism uses alternative splicing of active telomerase splice forms such that higher levels of active telomerase transcript can be somatically up-regulated in asexual, but not sexual, pASCs.
Results
Asexual but Not Sexual Animals Maintain Telomere Length Through Regeneration.
Other platyhelminthes (flatworms) have been previously described as having the same repeat unit as human telomeres at their chromosome ends (14, 15). We performed Bal31 nuclease digestion of genomic DNA that only digests the ends of DNA molecules, followed by terminal restriction fragment (TRF) length analysis and confirmed this was the case for S. mediterranea (Fig. S1) (16).
We investigated telomere lengths in individual asexual and sexual planarians of known age since their last reproductive event. For sexual animals, this is the period since hatching from cocoons in culture, and for asexual animals this is time since their last fission event. Telomeres of asexual animals that had undergone recent fission (7 d previously) had a longer average length (Fig. 1A; mean 28.0 kb, SD 0.7 kb, n = 10) in both the anterior and posterior pieces than those of newly born sexual animals (Fig. 1B; 21.2 kb, SD 1.67 kb, n = 7).
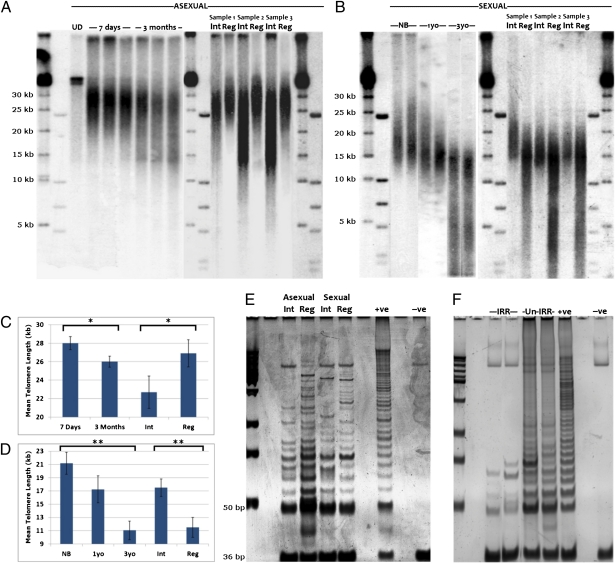
Fig. 1.
Planarian telomere length dynamics. (A) Telomere length in asexual animals increases after both fission and regeneration induced by amputation.UD, undigested genomic DNA; 7 d, animals that underwent fission 7 d previously (mean 28 kb, SD 0.7 kb, n = 10); 3 mo, animals that underwent fission 85–95 d previously (mean 26 kb, SD 0.6 kb, n = 10, P < 0.02, two-tailed t test); Int, intact asexual animals that have not undergone fission for between 85 and 195 d (mean 22.6 kb, SD 1.75 kb, n = 10); Reg, animals that have undergone three rounds of regeneration (mean 26.9 kb, SD 1.48 kb, n = 10, P < 0.04, two-tailed t test). (B) Telomere length in sexual animals decreases with age, with newly born (NB, mean 21.2 kb, SD 1.67 kb, n = 7) animals from 3-y-old parents (3yo, mean 11.1 kb, SD 1.4 kb, n = 6, P < 0.002, two-tailed t test) showing rejuvenated lengths. Serial regeneration of 6- to 12-mo-old animals significantly decreases telomere lengths. Shown are representative animals before and after three rounds of regeneration. Int, sexual animals between 180 and 360 d old (mean 17.5 kb, SD 1.33 kb, n = 10); Reg, animals that have undergone three rounds of regeneration (mean 11.6 kb, SD 1.50 kb, n = 10, P < 0.007, two-tailed t test). (C and D) Graphical representation of asexual and sexual TRF data showing statistically significant comparisons (*P < 0.05, **P< 0.01). (E) TRAP assay indicates that telomerase activity increases at 72 h after regeneration (Reg) in both asexual and sexual animals, with a greater increase visible in asexuals during regeneration compared with intact animals (Int). Hela cells extract (+ve), and heat treated (-ve). (F) γ-Irradiation to remove proliferating pASCs and their recent progeny leads to loss of telomerase activity. IRR, asexual animals 7 d after irradiation; Un-IRR, mock-irradiated animals.
Telomere lengths in asexual animals 7 d postfission had significantly increased (Fig. 1A; mean 28 kb, SD 0.7 kb, n = 10, t test, two-tailed, P < 0.02) compared with animals that had not undergone fission for 3 mo (Fig. 1A; mean 26 kb, SD 0.6 kb, n = 10). Older sexual animals showed a clear age-correlated decline in telomere lengths compared with hatchlings and younger animals (Fig. 1B). Both asexual and sexual animals display age-related decline in telomere length; however, asexual animals are able to maintain telomere lengths somatically, whereas sexual animals restore telomeres by extension during sexual reproduction or during embryogenesis like other sexual species (8).
The differing dynamics in asexual and sexual telomere length could be attributable to differences in reproductive strategy. The act of fission in asexuals requires the regeneration of missing structures. As sexual animals are capable of the same regenerative feats (12, 13), they may also maintain telomere length during this process. Repeated rounds of regeneration in sexual individuals led to a profound erosion of telomere length after just three consecutive rounds of regeneration (Fig. 1B; intact mean 17.5 kb, SD 1.33 kb, three rounds regeneration mean 11.6 kb, SD 1.50 kb, t test, two-tailed, P < 0.01, n = 10 starting animals). Conversely, serial regeneration of asexual animals that had not undergone fission for between 3 and 6 mo led to an increase in mean telomere length (Fig. 1A; intact 22.6 kb, SD 1.75 kb, three rounds regeneration mean 26.9 kb, SD 1.48 kb, t test, two-tailed, P < 0.04, n = 10). Our data led us to conclude that asexual, but not sexual, animals are capable of somatic telomere maintenance.
Telomerase Activity in pASCs Maintains Telomere Length in Asexual Planarians During Regeneration.
Observed differences between sexual and asexual telomere maintenance could be related to telomerase activity. To measure hypothetical telomerase activity, we used the telomere repeat amplification protocol (TRAP) assay. We found that telomerase activity as measured in vitro by the TRAP assay was up-regulated during asexual regeneration (Fig. 1E). In irradiated worms, where proliferative pASCs and their progeny are removed (17), we observed loss of telomerase activity, suggesting that telomerase activity is mainly confined to proliferating pASCs and/or their recent progeny (Fig. 1F). Sexual animals also had detectable telomerase activity, which also showed a slight increase during regeneration, but not as much as that observed for asexual worms (Fig. 1E). In any case, this activity is not sufficient to maintain telomere length during regeneration in the sexual strain (Fig. 1B). We conclude that homeostatic telomerase activity observed in both asexual and sexual animals is not sufficient to maintain telomere length, whereas the increased activity in regenerating asexuals is sufficient to renew telomere length.
Smed-Tert Is Required for Somatic Telomere Length Maintenance.
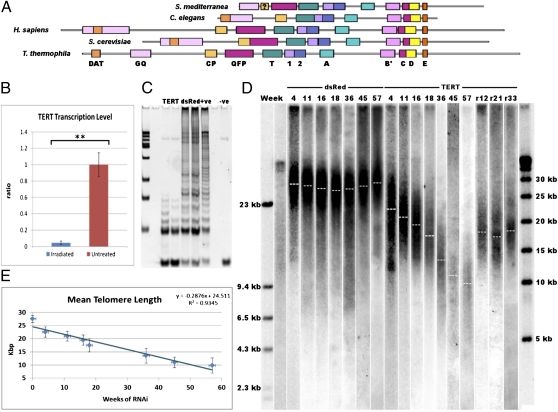
We identified and cloned a single gene with similarity to telomerase reverse transcriptase (TERT), the catalytic protein subunit of the telomerase enzyme (Fig. S2). Smed-Tert has the conserved sequence motifs common to TERT proteins across eukaryotes (Fig. 2A and Fig. S2). This includes a candidate QFP domain that has not previously been identified in other protostome TERT proteins (18). We compared the levels of Smed-tert mRNA in normal and irradiated animals and observed that irradiation led to almost complete loss of Smed-tert transcript (Fig. 2B; P < 0.01, two-tailed test). This correlates with the finding that telomerase activity as measured by the TRAP assay is also removed by irradiation (Fig. 1F).
Fig. 2.
Characterization and RNAi of Smed-TERT, the catalytic subunit of telomerase in S. mediterranea. (A) Smed-tert translation and multispecies alignment reveal that Smed-TERT has the conserved motifs of TERT genes across eukaryotes (Fig. S2). (B) RT-qPCR of Smed-tert in irradiated and mock-irradiated worms. Smed-tert is removed by irradiation (three replicate experiments on batches of five worms, **P < 0.01, two-tailed t test). (C) Smed-tert(RNAi) greatly reduces telomerase activity whereas control DsRed(RNAi) does not, as measured by TRAP assay (three replicate experiments on batches of five worms gave equivalent results). (D) Prolonged Smed-tert(RNAi) (TERT) leads to a reduction of telomere length over time as asexual animals fail to maintain their telomeres; removal of Smed-tert(RNAi) after 18 wk results in the stabilization of telomere length for times 12–33 wk later (r12, r21, r33). (E) Smed-tert(RNAi) leads to telomere erosion of 290 bp/wk in asexual animals. (Error bars are standard deviation of the mean telomere length.)
To confirm that Smed-tert was responsible for planarian telomere maintenance, we performed Smed-tert(RNAi). Smed-tert(RNAi) with two different regions of Smed-tert reduced telomerase activity as measured by the TRAP assay (Fig. 2C and Fig. S3). Control DsRed(RNAi) animals did not have reduced telomerase activity, and in fact wounding by injection raised telomerase activity (Fig. 2 C and D and Fig. S3). Control asexual DsRed(RNAi) animals also maintained their telomere length over 57 wk, suggesting a link between telomerase activity and raised levels of pASC proliferation caused by wounding (Fig. 2D) (19). Prolonged Smed-tert(RNAi) led to consistent and steady erosion of average telomere length of ∼290 bp per wk in asexual animals (Fig. 2 D and E). Neither Smed-tert(RNAi) nor reducing telomere length affected the proliferative rate of pASCs (Fig. S4). This suggests that telomerase activity and length are not controlling pASC division. Together, our data show that canonical telomerase activity is responsible for somatic telomere length maintenance in asexual planarians. The finding that telomerase activity and expression are acutely irradiation-sensitive suggests that it is confined to radiation-sensitive pASCs.
We confirmed Smed-tert(RNAi) efficacy in sexual and asexual animals by in situ hybridization (Fig. S5; see Fig. 3 for more details on normal expression). Smed-tert(RNAi) in sexual animals also led to a loss of TRAP activity in these animals compared with injected controls (Fig. S6). Whereas prolonged injection and wounding of sexual animals results in a drop in telomere length akin to regeneration, Smed-tert(RNAi) animals reduced telomere length more rapidly compared with controls as homeostatic telomerase activity was removed (Fig. S7). Together, our data confirm that Smed-tert is responsible for telomerase activity and adult telomere length maintenance in both strains of S. mediterranea, and telomere lengthening induced by regeneration in asexuals.
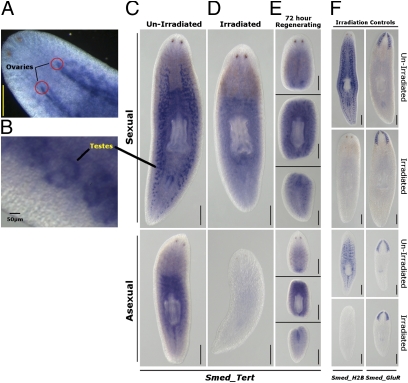
Fig. 3.
Whole-mount in situ hybridization of TERT in mature sexual and asexual worms. Expression can be observed in both (A) ovaries and (B) testes in mature sexual samples. (C) A transparenchyma expression concentrating around the pharynx can also be seen in asexual worms, suggesting expression in adult somatic stem cells. (D) Irradiation to remove proliferative stem cells removes Smed-tert expression in the germ line of sexual animals and the proliferative stem cells of asexual animals. (E) Major changes in the localization of Smed-tert expression during regeneration are not detected by in situ hybridization. (F) Controls in situ for irradiation indicating that Smed-H2B expression, a marker of proliferative stem cells and germ line, is removed by irradiation but expression of Smed-GluR, a marker of postmitotic differentiated brain cells, is not affected. All irradiated samples were treated with 85 Gy γ-irradiation. The Smed-tert samples were fixed at 7 d posttreatment, whereas the controls were fixed at 72 h postirradiation. (Scale bars, 1 mm.)
Expression of Smed-Tert in Intact Animals Has Changed from the Germ Line of Sexual Animals to the pASCs of Asexual Animals.
In sexual animals, we observed high transcript expression in both the testes and ovaries but only very low levels in the rest of the body (Fig. 3 A–C). Low expression in the rest of the body suggests that intact sexual animals do not express high levels of Smed-tert in pASCs. Germ-line expression was removed by irradiation, as the germ line is susceptible to irradiation (Fig. 3D). For example, expression of the pASC and germ-line marker Smed-H2B is abrogated by irradiation (Fig. 3F) (20). In asexual animals, Smed-tert expression, visualized by in situ hybridization, occurs in a pattern reminiscent of other genes enriched in pASCs (20, 21) and is also sensitive to irradiation, in agreement with our data suggesting telomerase activity and Smed-tert expression are confined to pASCs (Figs. 1F, 2B, and 3 C and D). Upon amputation, planarians reabsorb the germ line, and Smed-tert expression in regenerating sexual animals may reflect this as well as expression of new transcripts in pASCs during regeneration (Fig. 3E). New transcripts would be in agreement with TRAP assay data on regenerating sexual worms, suggesting that telomerase activity increases slightly (Fig. 1E).
Overall, our data reveal strong transcript expression in the germ line of sexual animals and irradiation-sensitive expression probably confined to the pASCs of asexual animals. This suggests a change in the regulatory control of Smed-tert in intact animals and that TRAP activity observed in intact sexual animals may be from the germ line.
Increased Transcript Levels and Alternate Splicing Activated During Regeneration Correlate with Telomere Maintenance in Asexual Animals.
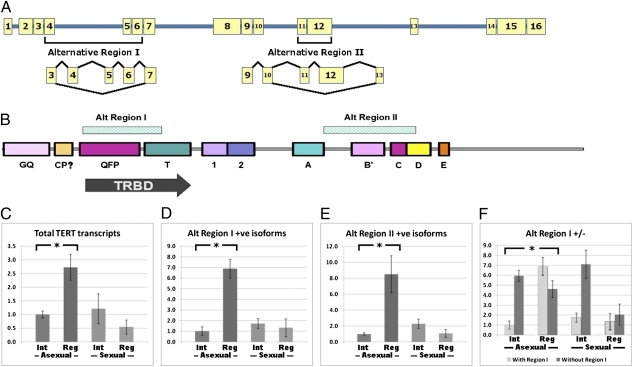
Analysis of transcriptome data from extensive next-generation short-read sequencing (22) and subsequent PCR analyses of possible exon–exon boundaries of full-length Smed-tert led to the identification of four different isoforms (Fig. 4A and Fig. S8). These four isoforms result from combinations of transcripts generated from two alternatively spliced regions (Fig. 4B). These exclude exon 4, 5, and 6 (alternate region 1) and exons 11 and 12 (alternate region 2) (Fig. 4B and Fig. S8). Exclusion of region 1 removes part of the telomere RNA-binding domain (TRBD), whereas exclusion of region 2 results in a premature stop codon at the end of motif B′ (Fig. 4B and Fig. S8).
Fig. 4.
Alternative splicing of Smed-tert. (A) Exonic structure of Smed-tert alternate splicing sites. All four possible transcripts can be detected by PCR. (B) Schematic showing that splicing events that skip exon numbers 4–6 remove the TRBD. (C–F) RT-qPCR analysis of (C) Smed-tert transcript levels; (D) alternate region 1-containing transcripts; (E) alternate region 2-containing transcripts; and (F) the ratio of alternate region 1-positive and -negative transcripts. Errors bars are ± 1 SD of the mean. In each case, transcript levels are significantly increased in regenerating asexual worms (three replicates of batches of five worms each, t test, two-tailed, *P < 0.03).
We performed quantitative (q)PCR to look at the levels of total Smed-tert transcript as well as the levels and relative ratios of the different isoforms. This approach allowed us to measure the relative amounts of total Smed-tert transcript, region 1-positive transcript, region 2-positive transcript, and the ratio of region 1-positive and -negative transcripts in intact and regenerating asexual and sexual animals.
We found that total Smed-tert transcripts, monitored by primers at the 3′ end of the transcript common to all isoforms, increased two- to threefold in regenerating asexual animals but decreased in sexual animals (Fig. 4C). This increase in total transcript included a seven- to eightfold increase in levels of transcript that include alternate regions 1 and 2, that are likely to encode active telomerase in asexual animals (Fig. 4 D and E). In sexually mature worms, we observed a twofold decrease in the levels of total transcript that were accompanied by smaller decreases in region 1- and 2-positive transcripts (Fig. 4 C–E). These data suggest that increases in transcript levels and regulation of alternative splicing events during regeneration lead to increased amounts of active Smed-tert isoforms, correlating with the ability of asexual animals, but not sexual animals, to maintain their telomeres during regeneration.
We also found that only regenerating asexual animals have more region 1-positive transcripts than region 1-negative transcripts. The overall increase in total transcript levels consists of a sevenfold increase in region 1-containing transcripts and a 1.5-fold decrease in transcripts without region 1. Given that this region would be absolutely necessary for telomerase activity, this shift in isoform expression may make the largest contribution to increased telomerase activity during regeneration (Fig. 4F). We also observed a decrease in the ratio of region 1-negative to -positive transcripts in regenerating sexual animals (Fig. 4F). The change in this ratio of transcripts is likely to account for the slight increase in sexual telomerase activity during regeneration observed by TRAP assay (Fig. 1E), despite a decrease in total Smed-tert transcript levels. Our data show that changes in the regulation of both Smed-tert transcript levels and alternate splicing produce active Smed-tert isoforms in asexual pASCs during regeneration. These changes explain our observations regarding differences in telomere length maintenance and telomerase activity in asexual and sexual animals.
Discussion
We find that in the model species S. mediterranea, asexual animals demonstrate the potential to maintain telomere length during regeneration. Sexual animals appear to only lengthen their telomeres through the sexual reproduction process. This finding suggests that asexual individuals will be able to avoid senescence over evolutionary timescales using telomerase, a prerequisite for the formation of an evolutionarily stable fissionating asexual lineage. We did not observe any adverse affects of telomere shortening through Smed-tert(RNAi) or serial regeneration. The difference we observe between asexual and sexual animals is surprising, given that sexual animals also appear to have an indefinite regenerative capacity. We conclude that either they would eventually show effects of telomere shortening or that they are able to use another chromosome end-maintenance mechanism not involving telomerase.
In most species, telomeres erode in the absence of telomerase until a senescent phenotype is seen. In mice, effects of telomere loss are observed by generation 4 and certainly by generation 6 (23, 24). In Arabidopsis, cell cultures can be maintained for a number of population doublings before senescence, whereas sexual generations exhibit effects after six or seven generations (25, 26). In yeast grown asexually, cells senesce after ∼70 generations (27). Trypanosoma brucei appears to escape senescence entirely, with telomere length stabilizing at a shorter length, before senescence, by an unknown mechanism (28).
It is possible that the erosion of telomeres we observe or induce by RNAi is counteracted by mechanisms that give rise to alternative lengthening of telomeres (ALT) not requiring TERT (29, 30). ALT is characterized by an abrupt change in telomere length (29, 30). An ALT mechanism is responsible for telomere elongation in blastomeres of the early mouse embryo (31). This abrupt change in telomere length has not been observed in adult Smed-tert(RNAi) animals or adult sexual animals with declining telomere length. This suggests that the shortened telomeres achieved by RNAi are not short enough to trigger any ALT mechanism, or that any ALT mechanism does not use the telomere repeat for elongation.
Telomere maintenance mechanisms show adaptation in asexuals is achieved at the level of Smed-tert expression. First, PCR and in situ hybridization expression data in the context of irradiated animals suggest that Smed-tert is expressed in irradiated pASCs, and this is also supported by TRAP assay data from intact and irradiated worms (Figs. 1F, 2B, and 3). However, the majority of Smed-tert transcripts present in the pASCs of intact asexual and germ-line cells of sexual animals are missing the TRBD required for binding the telomerase RNA component (Fig. 4). By analogy with all other species, these would not be able to engage in telomere extension. Alternate splicing has also been implicated in the generation of dominant-negative and inactive forms of TERT in vertebrate cells, and may be a common mechanism for differential control of telomerase activity in different animal cell types (32–34). Furthermore, we are able to show that during normal homeostatic turnover of planarian tissues telomerase activity is not sufficient to maintain telomere lengths. During regeneration and fission, asexual animals are able to increase telomere length by producing seven- to eightfold more TRBD-containing transcripts. Direct testing of the roles of the alternate transcripts awaits the development of transgenic techniques in S. mediterranea and/or the ability to use siRNA technology to target splice junctions and stretches of small sequences that are isoform-specific.
Previous work on colonial ascidians (35) and oligochaete worms (36) that have asexual life-history phases and reproduce by fission suggests that passage through a sexual reproductive cycle is required to avoid telomere depletion (35) and senescence (35, 36). In both cases, these animals have the natural option of a sexually reproductive cycle. In the urochordate Botryllus schlosseri that also propagates sexually and asexually, telomerase activity appears to be up-regulated in asexually formed buds (37), although these animals also eventually undergo senescence (38). Longevity experiments to investigate senescence in Hydra that reproduced asexually suggest that they are immortal (4, 39), whereas a sexually reproducing species showed clear signs of degeneration and mortality (40). Although immortal Hydra also appears to share the (TTAGGG)n telomere repeat, there is as yet no data on how or whether they avoid chromosome end depletion (41). These data suggest the possibility that senescence or death of asexual individuals and colonies may in part result from a failure to maintain chromosome ends that are restored by going through a sexually reproductive cycle (35). In the case of “effectively immortal” and obligate asexual S. mediterranea, the end-replication problem in somatic stem cells has been solved by the simple evolutionary changes we have characterized. These allow increased telomerase activity in somatic pASCs, allowing them to be an effective cellular unit of inheritance.
Materials and Methods
Animal Culture and Amputation.
Planarian culture was performed as previously described (42).
Telomere Length Analysis.
Bal31 and TRF analysis was performed using 0.65 μg of genomic DNA with previously described protocols (16, 43). Scanned TRF images were analyzed with Quantity One software (Invitrogen).
TRAP Assay to Measure Telomerase Activity.
TRAP assays were performed with the TRAPeze Telomerase Detection Kit (Millipore). The amount of total extracted protein used for experiments comparing regeneration and intact worms was 0.5 μg per sample. For assessing the effects of irradiation and Smed-tert(RNAi), 1 μg per sample was used to allow detection of minimal residual activity.
Cloning of Smed-tert.
Full-length Smed-tert was cloned by RACE using the FirstChoice RLM-RACE Kit (Ambion). Isoforms were discovered by investigating transcriptome data and split reads using methods described in ref. 22 and further characterized by PCR, cloning, and sequencing of different-length products generated using the primers forward 5′-ATGGTTTTATGAAATTAGATCTTGG-3′ and reverse 5′-AATGGAGAATCCATTTCATTTGACC-3′ designed to amplify the full Smed-tert ORF.
Quantitative PCR.
RT-qPCR was performed with first-strand cDNA made from total RNA from TRIzol reagent (Invitrogen) with SuperScript III reverse transcriptase (Invitrogen) using Brilliant qPCR Master Mix (Agilent). Primers used are listed as follows with their coordinate position in relation to the full-length 1895-bp TERT isoform 1 (EMBL accession no. AEK12104): total TERT transcript (forward: 5′-TTATCGAGATTTGCAGGATT-3′ 1476–1495 bp; reverse: 5′-CACTACAGCAATTGTCATGG-3′ 1602–1583 bp); alternate region 1-positive (forward: 5′-TCTCGGCGATATTTTTCTAA-3′ 287–306 bp; reverse: 5′-TCTTCATTGACTTGCATACG-3′ 408–389 bp); alternate region 1-negative (forward: 5′-CAAAAACAAGTGTAGTGAAATTAAA-3′ 143–167 bp; reverse: 5′-CACCAGTGAAAATTTTGTTGA-3′ 520–504 bp); alternate region 2-positive (forward: 5′-CTGATTTGATTTCGAAGACTAAAG-3′ 1012–1035 bp; reverse: 5′-GAGGTATTCCGCATATTTGA-3′ 1149–1130 bp). Transcription levels were standardized with the internal control gene cystatin (forward: 5′-AACTCCATGGCTAGAACCGAA-3′; reverse: 5′-CCGTCGGGTAATCCAAGTACA-3′).
In Situ Hybridization and Irradiation.
Whole-mount in situ hybridization and irradiation were performed as previously described (44). A 601-bp probe generated with the forward primer 5′-CTTCAATATGTTGATGATGTTTTATTC-3′ and reverse primer 5′-AATGGAGAATCCATTTCATTTGACC-3′ was used for in situ hybridization.
Proliferation Assay.
Proliferation was assessed by counting total mitotic cells in regenerating animals, visualized with rabbit anti-phosphorylated histone H3 serine 10 (Upstate Biotechnology; 1:1,000) and goat anti-rabbit Alexa Fluor 568 (Molecular Probes; 1:1,000) with fluorescent microscopy.
RNAi Experiments.
RNAi experiments were performed as previously described with one round of three 33-nl injections of 2 μg/μl dsRNA every wk (42). Two different regions of the Smed-tert transcript were used in RNAi experiments: a 600-bp region generated with the forward primer 5′-CTTCAATATGTTGATGATGTTTTATTC-3′ and reverse primer 5′-AATGGAGAATCCATTTCATTTGACC-3′ and a 450-bp region amplified by the primers forward Smed_TERT_AltA_dsF 5′-TTGCATTTCTCAAGAGTCAA-3′ and reverse Smed_TERT_AltA_dsR 5′-TTCAAAATGGGAATAACAAAC-3′.
Supplementary Material
Acknowledgments
We thank J. Jowett for maintaining the planarian colony, D. Kao for help with analysis of transcriptome splicing data, and Cristina Gonzalez-Estevez for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. JF693290–JF693293).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118885109/-/DCSupplemental.
References
- 1.Rando TA. Stem cells, ageing and the quest for immortality. Nature. 2006;441:1080–1086. doi: 10.1038/nature04958. [DOI] [PubMed] [Google Scholar]
- 2.Partridge L, Gems D. Benchmarks for ageing studies. Nature. 2007;450(7167):165–167. doi: 10.1038/450165a. [DOI] [PubMed] [Google Scholar]
- 3.Aboobaker AA. Planarian stem cells: A simple paradigm for regeneration. Trends Cell Biol. 2011;21:304–311. doi: 10.1016/j.tcb.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 4.Bosch TC. Hydra and the evolution of stem cells. Bioessays. 2009;31:478–486. doi: 10.1002/bies.200800183. [DOI] [PubMed] [Google Scholar]
- 5.Nilsson Sköld H, Obst M. Potential for clonal animals in longevity and ageing studies. Biogerontology. 2011;12:387–396. doi: 10.1007/s10522-011-9333-8. [DOI] [PubMed] [Google Scholar]
- 6.Olovnikov AM. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J Theor Biol. 1973;41(1):181–190. doi: 10.1016/0022-5193(73)90198-7. [DOI] [PubMed] [Google Scholar]
- 7.Gomes NM, Shay JW, Wright WE. Telomere biology in Metazoa. FEBS Lett. 2010;584:3741–3751. doi: 10.1016/j.febslet.2010.07.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schaetzlein S, et al. Telomere length is reset during early mammalian embryogenesis. Proc Natl Acad Sci USA. 2004;101:8034–8038. doi: 10.1073/pnas.0402400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Sullivan RJ, Karlseder J. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11(3):171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Flores I, Blasco MA. The role of telomeres and telomerase in stem cell aging. FEBS Lett. 2010;584:3826–3830. doi: 10.1016/j.febslet.2010.07.042. [DOI] [PubMed] [Google Scholar]
- 11.Dalyell JG. Observations on Some Interesting Phenomena in Animal Physiology, Exhibited by Several Species of Planariae. Edinburgh: Archibald Constable; 1814. [Google Scholar]
- 12.Saló E. The power of regeneration and the stem-cell kingdom: Freshwater planarians (Platyhelminthes) Bioessays. 2006;28:546–559. doi: 10.1002/bies.20416. [DOI] [PubMed] [Google Scholar]
- 13.Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–757. doi: 10.1146/annurev.cellbio.20.010403.095114. [DOI] [PubMed] [Google Scholar]
- 14.Joffee BI, Solovei IV, Macgregor HC. Ends of chromosomes in Polycelis tenuis (Platyhelminthes) have telomere repeat TTAGGG. Chromosome Res. 1996;4:323–324. doi: 10.1007/BF02263686. [DOI] [PubMed] [Google Scholar]
- 15.Bombarová M, Vítková M, Spakulová M, Koubková B. Telomere analysis of platyhelminths and acanthocephalans by FISH and Southern hybridization. Genome. 2009;52:897–903. doi: 10.1139/g09-063. [DOI] [PubMed] [Google Scholar]
- 16.Blackburn EH, Challoner PB. Identification of a telomeric DNA sequence in Trypanosoma brucei. Cell. 1984;36:447–457. doi: 10.1016/0092-8674(84)90238-1. [DOI] [PubMed] [Google Scholar]
- 17.Bardeen CR, Baetjer FH. The inhibitive action of the Roentgen rays on regeneration in planarians. J Exp Zool. 1904;1(1):191–195. [Google Scholar]
- 18.Meier B, et al. trt-1 is the Caenorhabditis elegans catalytic subunit of telomerase. PloS Genet. 2006;2:e18. doi: 10.1371/journal.pgen.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenemoser D, Reddien PW. Planarian regeneration involves distinct stem cell responses to wounds and tissue absence. Dev Biol. 2010;344:979–991. doi: 10.1016/j.ydbio.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo T, Peters AH, Newmark PA. A Bruno-like gene is required for stem cell maintenance in planarians. Dev Cell. 2006;11(2):159–169. doi: 10.1016/j.devcel.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 21.Reddien PW, Oviedo NJ, Jennings JR, Jenkin JC, Sánchez Alvarado A. SMEDWI-2 is a PIWI-like protein that regulates planarian stem cells. Science. 2005;310:1327–1330. doi: 10.1126/science.1116110. [DOI] [PubMed] [Google Scholar]
- 22.Blythe MJ, et al. A dual platform approach to transcript discovery for the planarian Schmidtea mediterranea to establish RNAseq for stem cell and regeneration biology. PLoS One. 2010;5:e15617. doi: 10.1371/journal.pone.0015617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco MA, et al. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997;91(1):25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 24.Lee HW, et al. Essential role of mouse telomerase in highly proliferative organs. Nature. 1998;392:569–574. doi: 10.1038/33345. [DOI] [PubMed] [Google Scholar]
- 25.Fitzgerald MS, et al. Disruption of the telomerase catalytic subunit gene from Arabidopsis inactivates telomerase and leads to a slow loss of telomeric DNA. Proc Natl Acad Sci USA. 1999;96:14813–14818. doi: 10.1073/pnas.96.26.14813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riha K, McKnight TD, Griffing LR, Shippen DE. Living with genome instability: Plant responses to telomere dysfunction. Science. 2001;291:1797–1800. doi: 10.1126/science.1057110. [DOI] [PubMed] [Google Scholar]
- 27.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dreesen O, Cross GA. Telomerase-independent stabilization of short telomeres in Trypanosoma brucei. Mol Cell Biol. 2006;26:4911–4919. doi: 10.1128/MCB.00212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McEachern MJ, Haber JE. Break-induced replication and recombinational telomere elongation in yeast. Annu Rev Biochem. 2006;75:111–135. doi: 10.1146/annurev.biochem.74.082803.133234. [DOI] [PubMed] [Google Scholar]
- 30.Henson JD, Neumann AA, Yeager TR, Reddel RR. Alternative lengthening of telomeres in mammalian cells. Oncogene. 2002;21:598–610. doi: 10.1038/sj.onc.1205058. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, et al. Telomere lengthening early in development. Nat Cell Biol. 2007;9:1436–1441. doi: 10.1038/ncb1664. [DOI] [PubMed] [Google Scholar]
- 32.Yi X, et al. An alternate splicing variant of the human telomerase catalytic subunit inhibits telomerase activity. Neoplasia. 2000;2:433–440. doi: 10.1038/sj.neo.7900113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ulaner GA, Hu JF, Vu TH, Giudice LC, Hoffman AR. Telomerase activity in human development is regulated by human telomerase reverse transcriptase (hTERT) transcription and by alternate splicing of hTERT transcripts. Cancer Res. 1998;58:4168–4172. [PubMed] [Google Scholar]
- 34.Kilian A, et al. Isolation of a candidate human telomerase catalytic subunit gene, which reveals complex splicing patterns in different cell types. Hum Mol Genet. 1997;6:2011–2019. doi: 10.1093/hmg/6.12.2011. [DOI] [PubMed] [Google Scholar]
- 35.Sköld HN, Asplund ME, Wood CA, Bishop JD. Telomerase deficiency in a colonial ascidian after prolonged asexual propagation. J Exp Zool B Mol Dev Evol. 2011;316:276–283. doi: 10.1002/jez.b.21399. [DOI] [PubMed] [Google Scholar]
- 36.Martínez DE, Levinton JS. Asexual metazoans undergo senescence. Proc Natl Acad Sci USA. 1992;89:9920–9923. doi: 10.1073/pnas.89.20.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laird DJ, Weissman IL. Telomerase maintained in self-renewing tissues during serial regeneration of the urochordate Botryllus schlosseri. Dev Biol. 2004;273(2):185–194. doi: 10.1016/j.ydbio.2004.05.029. [DOI] [PubMed] [Google Scholar]
- 38.Lauzon RJ, Rinkevich B, Patton CW, Weissman IL. A morphological study of nonrandom senescence in a colonial urochordate. Biol Bull. 2000;198:367–378. doi: 10.2307/1542692. [DOI] [PubMed] [Google Scholar]
- 39.Martínez DE. Mortality patterns suggest lack of senescence in Hydra. Exp Gerontol. 1998;33:217–225. doi: 10.1016/s0531-5565(97)00113-7. [DOI] [PubMed] [Google Scholar]
- 40.Watanabe H, Hoang VT, Mättner R, Holstein TW. Immortality and the base of multicellular life: Lessons from cnidarian stem cells. Semin Cell Dev Biol. 2009;20:1114–1125. doi: 10.1016/j.semcdb.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 41.Traut W, et al. The telomere repeat motif of basal Metazoa. Chromosome Res. 2007;15:371–382. doi: 10.1007/s10577-007-1132-3. [DOI] [PubMed] [Google Scholar]
- 42.Felix DA, Aboobaker AA. The TALE class homeobox gene Smed-prep defines the anterior compartment for head regeneration. PLoS Genet. 2010;6:e1000915. doi: 10.1371/journal.pgen.1000915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Harley CB, Futcher AB, Greider CW. Telomeres shorten during ageing of human fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 44.González-Estévez C, Arseni V, Thambyrajah RS, Felix DA, Aboobaker AA. Diverse miRNA spatial expression patterns suggest important roles in homeostasis and regeneration in planarians. Int J Dev Biol. 2009;53:493–505. doi: 10.1387/ijdb.082825cg. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.