Abstract
The arbuscular mycorrhizal (AM) symbiosis, formed by land plants and AM fungi, evolved an estimated 400 million years ago and has been maintained in angiosperms, gymnosperms, pteridophytes, and some bryophytes as a strategy for enhancing phosphate acquisition. During AM symbiosis, the AM fungus colonizes the root cortical cells where it forms branched hyphae called arbuscules that function in nutrient exchange with the plant. Each arbuscule is enveloped in a plant membrane, the periarbuscular membrane, that contains a unique set of proteins including phosphate transporters such as Medicago truncatula MtPT4 [Javot et al., (2007) Proc Natl Acad Sci USA 104:1720–1725], which are essential for symbiotic phosphate transport. The periarbuscular membrane is physically continuous with the plasma membrane of the cortical cell, but MtPT4 and other periarbuscular membrane-resident proteins are located only in the domain around the arbuscule branches. Establishing the distinct protein composition of the periarbuscular membrane is critical for AM symbiosis, but currently the mechanism by which this composition is achieved is unknown. Here we investigate the targeting of MtPT4 to the periarbuscular membrane. By expressing MtPT4 and other plasma membrane proteins from promoters active at different phases of the symbiosis, we show that polar targeting of MtPT4 is mediated by precise temporal expression coupled with a transient reorientation of secretion and alterations in the protein cargo entering the secretory system of the colonized root cell. In addition, analysis of phosphate transporter mutants implicates the trans-Golgi network in phosphate transporter secretion.
Keywords: biotrophic, endosymbiosis, legume, mutualistic, live imaging
The arbuscular mycorrhizal (AM) symbiosis, formed by plants and obligate biotrophic soil fungi of the phylum Glomeromycota, is a mutualistic endosymbiosis in which the fungal symbiont delivers phosphate to the plant and in return gains access to carbon (1, 2). Phosphorus is an essential mineral nutrient and often is limiting for plant growth; therefore additional phosphate acquired via the AM fungal symbiont has a significant impact on plant productivity and consequently on ecosystem function (3). In AM symbiosis, AM fungal hyphae colonize plant roots and penetrate the inner cortical cells, where they undergo terminal differentiation and branch extensively to form arbuscules through which phosphate is delivered to the root. Arbuscule development is accompanied by significant alterations in the subcellular architecture of the cortical cell, including the deposition of the periarbuscular membrane, which is thought to be synthesized de novo and surrounds the arbuscule, attaining a final area approximately eight times that of the plasma membrane of the cell (2, 4). Because arbuscules develop asynchronously deep inside the root cortex, the precise duration of their development is unknown; however, mature arbuscules are observed within 2 d of inoculation, suggesting rapid development (4–6). Arbuscules are short-lived structures; they degenerate approximately 1 wk after initiation, and the cortical cell resumes its original noncolonized configuration (2, 4).
Medicago truncatula phosphate transporter 4 (MtPT4) and its orthologs in other plants are expressed exclusively during AM symbiosis (7–9), and MtPT4 is essential for symbiotic phosphate transport and for maintenance of the symbiosis (5). Immunolocalization of native MtPT4 (7) and analysis of an MtPT4-GFP fusion expressed from the native promoter (10) reveal that this protein accumulates only in cortical cells with developing arbuscules. Furthermore, it localizes in the periarbuscular membrane surrounding arbuscule branches but not in the region of the membrane surrounding the arbuscule trunk and not in the plasma membrane around the cell periphery (Fig. 1A and Fig. S1A). The rice ortholog of MtPT4, OsPT11, displays similar localization (11). MtPT4 and OsPT11 are members of the PHT1 family of phosphate transporters, a large clade of major facilitator superfamily (MFS) proteins whose members are involved in phosphate transport in many cells throughout the plant (12). Where analyzed, other members of the PHT1 family have been found to localize in plasma membranes. For example in M. truncatula, MtPT1, which shares 61% amino acid identity with MtPT4, localizes in the plasma membrane of epidermal and root hair cells (Fig. 1B) (13) but is not expressed in cells with arbuscules (Fig. S1B). Likewise, MtPT1 orthologs show plasma membrane locations, and in Arabidopsis and rice these transporters contribute to phosphate acquisition from the soil (14, 15).
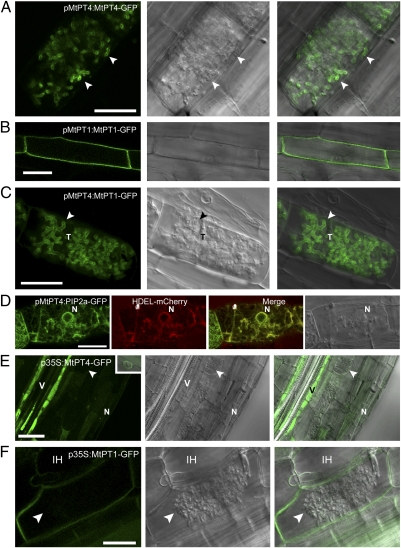
Fig. 1.
Protein localization in colonized cells is altered when genes are expressed from promoters that differ in activity relative to arbuscule development. Live-cell fluorescence and bright-field DIC images of longitudinal root sections of M. truncatula roots colonized with an AM fungus, G. versiforme. (A) pMtPT4:MtPT4-GFP localizes in the periarbuscular membrane (arrowhead) of colonized cortical cells. (B) pMtPT1:MtPT1-GFP localizes in the plasma membrane of root epidermal cells. (C) pMtPT4:MtPT1-GFP localizes in the periarbuscular membrane of colonized cortical cells and not in the plasma membrane. (D) pMtPT4:AtPIP2a-GFP does not localize in the periarbuscular or plasma membrane; instead it colocalizes with the HDEL-mCherry ER marker. (E) p35S:MtPT4-GFP accumulates mostly in the vacuole and ER, as shown by the presence of perinuclear signal (Inset), and not in the periarbuscular membrane (arrowhead). The signal is particularly strong in the vascular tissue. (F) p35S:MtPT1-GFP localizes in the plasma membrane and in the membrane that surrounds intracellular hyphae but not in the periarbuscular membrane. p35S:MtPT4-GFP includes the MtPT4 5′ UTR, whereas p35S:MtPT1-GFP contains the 35S 5′ UTR. For each construct, images are representative examples of at least 10 independent transgenic root systems; localization patterns were consistent among these independent experiments. Arrowheads indicate arbuscule branches. N, plant nucleus; T, arbuscule trunk; V, vascular tissue. (Scale bars: 20 μm.)
A central role for MtPT4 in symbiotic phosphate transfer and maintenance of AM symbiosis has been demonstrated; however, the mechanism directing polar targeting of this phosphate transporter to the periarbuscular membrane has not been addressed. In plants, mechanisms conferring protein polarity in host–microbe interactions have not been reported. In contrast, polar localization of membrane proteins has been studied extensively for the PIN-formed (PIN) family of auxin efflux transporters. These proteins initially are secreted in a nonpolar manner, and polarity is generated by subsequent endocytosis and recycling (16). Here we reveal that polar targeting of MtPT4 is achieved by a mechanism that involves transient changes in the secretory system of the colonized root cell and requires expression synchronized with arbuscule development to achieve localization in the periarbuscular membrane.
Results and Discussion
To begin to dissect the mechanism conferring polarity to MtPT4, we tested the ability of mutated and truncated derivatives of MtPT4-GFP expressed from the MtPT4 promoter to localize in the periarbuscular membrane. We deleted the cytosolic amino- and carboxyl-terminal domains, which contain sorting motifs of importance in other MFS proteins (17) and mutated two putative tyrosine-based sorting/endocytic motifs (YXXΦ) in the large central loop. YXXΦ motifs are essential for the polar localization of many proteins, including the Arabidopsis boron transporter BOR1 (18, 19). All the mutant MtPT4 fusion proteins localized in the correct polar manner (Fig. S1 C and D).
Next, we examined localization of the plasma membrane- resident MtPT1 in cells with arbuscules by expressing an MtPT1-GFP fusion from the MtPT4 promoter. As expected, GFP signal from pMtPT4:MtPT1-GFP was detected only in cells with arbuscules, but, surprisingly, MtPT1-GFP localized in the periarbuscular membrane and not in the plasma membrane or in the membrane around the arbuscule trunk (Fig. 1C). This result was unexpected, so we examined the location of AtPIP2a, an aquaporin widely used as a plasma-membrane marker. Previously we showed that AtPIP2a localizes in the plasma membrane and in the membrane surrounding intracellular hyphae and arbuscule trunks when expressed from the CaMV 35S promoter (10). When expressed from the MtPT4 promoter, the same AtPIP2a marker did not localize to the plasma membrane or the periarbuscular membrane but rather was retained entirely in the endoplasmic reticulum (ER), as indicated by a reticulate pattern, perinuclear signal, and colocalization with a histidine-aspartic acid-glutamic acid-leucine (HDEL) ER marker (Fig. 1D).
Because AtPIP2a displayed different localizations when expressed from the 35S and MtPT4 promoters, we examined localization of MtPT4 and MtPT1 expressed from the 35S promoter. In roots expressing p35S:MtPT4-GFP, the GFP signal was weak in the cortex but strong in the vacuole of vascular tissue cells where 35S promoter activity is highest. Occasional cortical and vascular cells showed a weak perinuclear signal, suggesting ER retention (Fig. 1E, Inset). Similar observations were reported recently for OsPT11 (11). Surprisingly, when these roots were colonized by the AM fungus Glomus versiforme, MtPT4-GFP was not observed in the periarbuscular membrane (Fig. 1E). Roots expressing p35S:MtPT1-GFP displayed the anticipated plasma membrane signal in all cell types (Fig. S2 A and B). In colonized cortical cells, GFP signal was observed in the plasma membrane, and a weak signal was detected in the membrane surrounding arbuscule trunks (Fig. 1F). Additionally, signal was observed in the membrane surrounding intracellular hyphae. These patterns are fully consistent with the previous reports for the plasma-membrane marker p35S:AtPIP2a-mcherry (10). As seen with p35S:MtPT4-GFP, MtPT1-GFP signal was not observed in the periarbuscular membrane when expressed from the 35S promoter (Fig. 1F). In addition, the plasma membrane GFP signal appeared substantially weaker in cells harboring arbuscules than in other cortical cells (Fig. S2); together these results suggest that the 35S promoter may be strongly down-regulated in colonized cortical cells. Down-regulation of the 35S promoter has been observed in other plant–microbe interactions (20, 21). Thus, MtPT4 and MtPT1 both localize in the periarbuscular membrane when expressed from the MtPT4 promoter but not when expressed from the 35S promoter. The 5′ UTRs differ in these constructs, but UTR swaps indicated that the 5′ UTR does not influence localization of these proteins (Figs. S2 and S3).
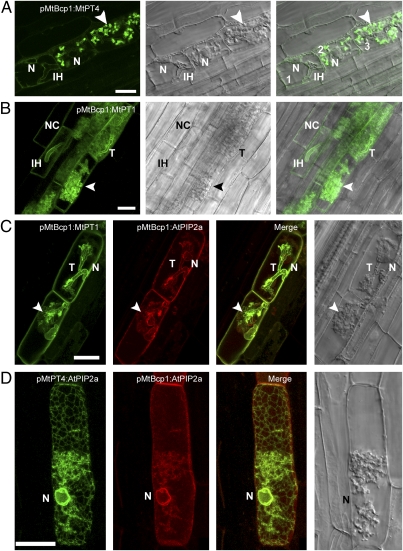
These results imply that, during arbuscule formation, secretion of newly synthesized phosphate transporters is directed predominantly toward the developing periarbuscular membrane and not toward the plasma membrane. To test this hypothesis, MtPT4- and MtPT1-GFP fusions were expressed from the MtBcp1 promoter, which is active in cortical cells both before and during arbuscule formation (10, 22) (Fig. S4) and therefore overlaps with phases of 35S and MtPT4 promoter activity. As predicted, expression from the MtBcp1 promoter produced a combination of the localization patterns obtained using the 35S and MtPT4 promoters: Roots expressing pMtBcp1:MtPT4-GFP displayed a relatively weak ER-localized signal in noncolonized cells, cells with intracellular hyphae, and cells with very young arbuscules, similar to the results of 35S promoter expression (Fig. 2A and Fig. S5). However, in cells harboring arbuscules with multiple branches, a strong GFP signal was observed in the periarbuscular membrane (Fig. 2A). Roots expressing pMtBcp1:MtPT1-GFP revealed GFP signal in the plasma membrane of noncolonized and colonized cells, in the membrane surrounding intracellular hyphae, and in the periarbuscular membrane surrounding arbuscule trunks and branches (Fig. 2B). pMtBcp1:AtPIP2a-mCherry displayed a pattern opposite to that of pMtBcp1:MtPT4-GFP, with signal in the plasma membrane of noncolonized and colonized cells and also surrounding arbuscule trunks and intracellular hyphae but in the ER of cells containing branched arbuscules (Fig. S5). Coexpression of pMtBcp1:MtPT1-GFP and pMtBcp1:AtPIP2a-mCherry revealed overlapping localization in the plasma membrane of cells with newly forming arbuscules but contrasting ER retention of AtPIP2a-mCherry and periarbuscular membrane localization of MtPT1-GFP (Fig. 2C). Similarly, coexpression of pMtBcp1:AtPIP2a-mCherry and pMtPT4:AtPIP2a-GFP highlighted the difference in cargo selection during arbuscule branching: In a single arbuscule-containing cell expressing AtPIP2a from two different promoters, AtPIP2a expressed from the MtBcp1 promoter was localized in the plasma membrane and ER, whereas AtPIP2a expressed from the MtPT4 promoter localized only in the ER. These localization patterns are consistent with the differential activities of these two promoters during arbuscule development (Fig. 2D).
Fig. 2.
The MtBcp1 promoter drives expression before and during arbuscule formation and confirms the importance of gene expression coordinated with arbuscule development for localization in the periarbuscular membrane. (A) In cells with intracellular hyphae (IH) but without arbuscule branches (cell 1), pMtBcp1:MtPT4-GFP localizes in the ER as indicated by the peripheral and perinuclear ER pattern. Upon arbuscule branching (cells 2 and 3), MtPT4 accumulates to high levels in the periarbuscular membrane (arrowhead). (B) pMtBcp1:MtPT1-GFP localizes in the plasma membrane of noncolonized cortical (NC) cells, in cells with intracellular hyphae, and in the periarbuscular membrane of cells with branched arbuscules. (C) Coexpression of pMtBcp1:MtPT1-GFP and pMtBcp1:AtPIP2a-mCherry in two cells with young arbuscules just beginning to branch illustrates plasma membrane localization of both proteins but contrasting perinuclear ER localization of AtPIP2a (N) and periarbuscular membrane localization of MtPT1 (arrowhead). (D) Coexpression of two AtPIP2a fusions reveals plasma membrane and ER localization for pMtBcp1:AtPIP2a-mCherry (red) but only ER localization for pMtPT4:AtPIP2a-GFP (green). (Scale bars: 20 μm.)
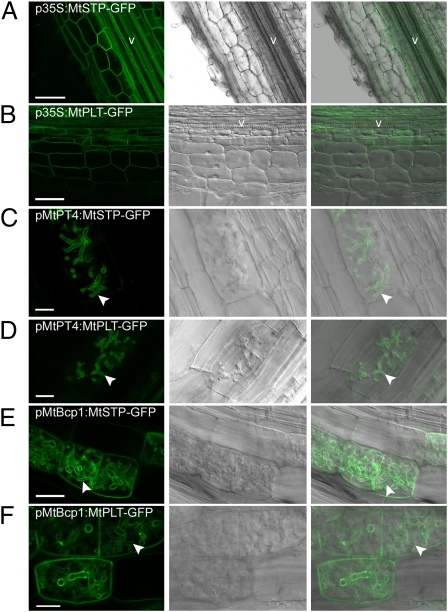
The data indicate that expression coincident with arbuscule formation is necessary and sufficient for localization of MtPT1 and MtPT4 in the periarbuscular membrane. To determine whether this principle extends to other transporters located in the plasma membrane, we evaluated the locations of two M. truncatula carbohydrate transporters when expressed from the p35S, pMtPT4, and MtBcp1 promoters. The transporters included an M. truncatula monosaccharide transporter (Medtr8g102860.1), referred to herein as “MtSTP,” that shares homology with the Arabidopsis galactose transporter AtSTP14, and an M. truncatula polyol transporter (TC183926), herein referred to as “MtPLT,” that shares homology with Arabidopsis AtPLT5. Both Arabidopsis AtSTP14 and AtPLT5 have demonstrated plasma membrane localization (23, 24). MtSTP and MtPLT are expressed in roots but are not differentially expressed during AM symbiosis (Medicago Gene Expression Atlas, http://mtgea.noble.org/v2/). As expected, expression of MtSTP and MtPLT from the 35S promoter resulted in location in the plasma membrane of noncolonized root cells (Fig. 3 A and B). In contrast, when expressed from the MtPT4 promoter, both proteins were located in the periarbuscular membrane and not in the plasma membrane of the colonized cell (Fig. 3 C and D). As observed previously for MtPT1, expression from the MtBcp1 promoter resulted in location in the plasma membrane of noncolonized cells and in the plasma membrane and periarbuscular membrane of cells containing arbuscules (Fig. 3 E and F). These patterns are consistent with those observed for MtPT1.
Fig. 3.
Localization of two carbohydrate transporters, MtSTP and MtPLT, when expressed from three promoters that differ in activity relative to arbuscule development. Live-cell fluorescence and bright-field DIC images of longitudinal root sections of M. truncatula roots (A and B) and M. truncatula roots colonized with the AM fungus, G. intraradices (C–F). (A and B) p35S:MtMST-GFP and p35S:MtPLT-GFP localize in the plasma membrane. (C and D) pMtPT4:MtMST-GFP and pMtPT4:MtPLT-GFP localize in the periarbuscular membrane around the branches of young arbuscules and not in the plasma membrane. (E and F) pMtBcp1:MtSTP-GFP and pMtBcp1:MtPLT-GFP are expressed before and during arbuscule formation and localize in the plasma membrane of noncolonized cortical cells and in the plasma membrane and periarbuscular membrane of cells containing arbuscules. Arrowheads indicate localization in the periarbuscular membrane. V, vascular tissue. (Scale bars: 75 μm in A, 50 μm in B, 10 μm in C and D, 25 μm in E, and 10 μm in F.)
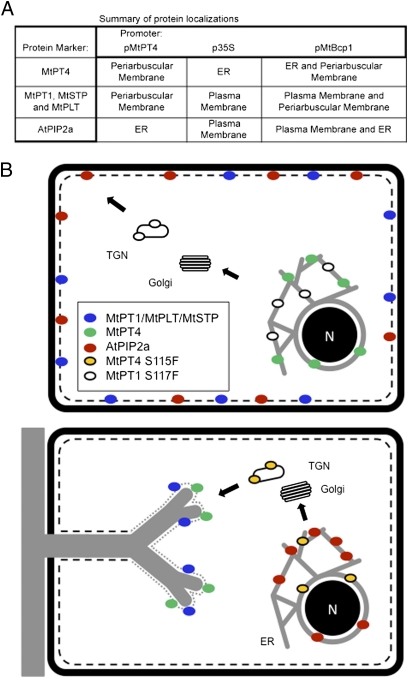
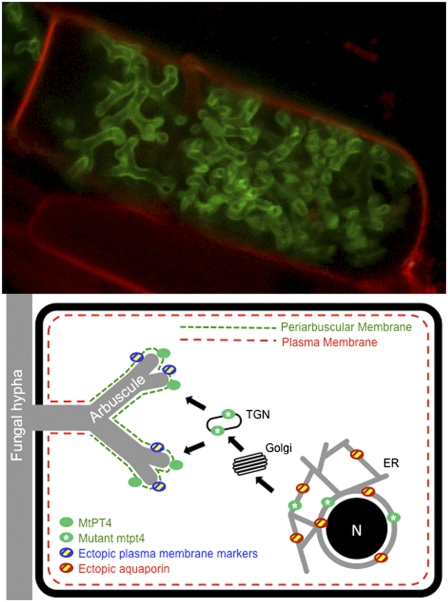
The localization results, summarized in Fig. 6A, indicate that expression coincident with arbuscule formation is necessary and sufficient not only to direct MtPT4 but also to direct the plasma membrane-resident proteins MtPT1, MtSTP, and MtPLT to the periarbuscular membrane. In addition, these results suggest that the protein cargo able to enter into the secretion pathway changes: In cells with very young arbuscules, ectopically expressed MtPT4 is retained in the ER, and AtPIP2a is secreted to the plasma membrane. After an arbuscule begins to branch, and the MtPT4 promoter is induced, MtPT4 traffics to the periarbuscular membrane, and AtPIP2a becomes retained in the ER (Fig. 6B).
Fig. 6.
Summary and model for phosphate transporter secretion during arbuscule development. (A) Table summarizing membrane localizations and illustrating that proteins expressed from the MtBcp1 promoter show localization patterns that combine those obtained from MtPT4 and p35S promoters. (B) Proposed model derived from data obtained through analysis of protein-GFP fusions expressed either from their native promoters and/or expressed ectopically from promoters that show differential activity during arbuscule formation. The cartoon depicts an uninfected cell (Upper) and a cortical cell harboring a developing arbuscule (Lower). In an uninfected cell, MtPT1, MtSTP, MtPLT, and AtPIP2a are secreted to the plasma membrane (black dashed line), whereas MtPT4 is retained in the ER. The MtPT1S117F mutant protein is retained in the ER and likely in the TGN. During arbuscule development (Lower), newly synthesized MtPT4, MtPT1, MtSTP, and MtPLT, expressed from an arbuscule-specific promoter, are secreted to the periarbuscular membrane (gray dotted line), whereas AtPIP2a is retained in the ER. The mutant protein MtPT4S115F is retained in the ER and likely in the TGN.
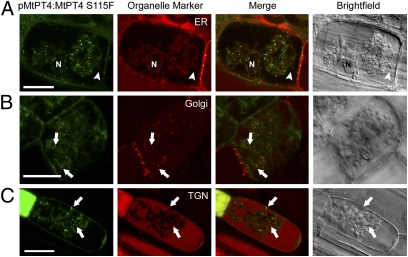
Although the various MtPT4 deletion mutants that we generated did not mislocalize, mtpt4-3, a mutant allele with a strong mtpt4 phenotype (Fig. S6) (25), led to the finding that mutation of a conserved serine, S115, to phenylalanine results in the loss of MtPT4 periarbuscular membrane localization. A pMtPT4:MtPT4S115F-GFP fusion did not localize in the periarbuscular membrane but accumulated in the endomembrane system, colocalizing with an HDEL ER marker and Syp61, a trans-Golgi network (TGN) marker, partially colocalizing with the VHA1 TGN marker but not with the GmMan1 Golgi marker (Fig. 4 and Fig. S7). In mtpt4-3 roots, immunolocalization with an MtPT4 antibody (7) detected native MtPT4S115F protein localized in a punctate pattern, whereas wild-type roots displayed periarbuscular membrane localization of MtPT4 (Fig. S8). Syp61 and VHA1 display partial colocalization in M. truncatula roots (Fig. S7), suggesting that MtPT4S115F is retained in a subset of TGN compartments. Site-directed mutagenesis of serine 115 to either alanine or a phosphoserine mimic, glutamic acid, did not support a role for phosphorylation of serine 115 and instead suggested that MtPT4S115F accumulates in the endomembrane system because of the addition of phenylalanine rather than because of a loss of serine (Fig. S9).
Fig. 4.
Mutant MtPT4S115F is retained in the endomembrane system and colocalizes with ER and TGN markers. Localization of MtPT4S115F-GFP expressed under pMtPT4 and coexpressed with HDEL-mCherry ER marker (A), GmMAN1-mCherry Golgi marker (B), and AtSyp61-mRFP TGN marker (C). MtPT4S115F signal overlaps with the ER and Syp61 signals. MtPT4S115F signals often occur adjacent to GmMAN1 signals but do not colocalize. (Left ) GFP fluorescence. (Center Left) mCherry/mRFP fluorescence. (Center Right) Overlay of both channels. (Right) DIC. Arrowheads indicate the arbuscule branch; arrows indicate endosomal localization of MtPT4S115F-GFP. In addition to TGN, AtSyp61-mRFP also produces signal in the vacuole (C), likely because of overexpression and turnover. ER, Golgi, and TGN markers are expressed from the 35S promoter. Localization of these markers has been verified and is consistent in colonized and noncolonized cells (10). (Scale bars: 20 μm.) N, nucleus.
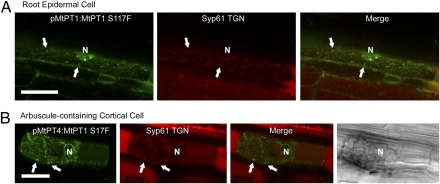
To determine whether mutation of the conserved serine impaired an MtPT4-specific secretion process or caused a general disruption of phosphate transporter secretion, the equivalent mutation (S117F) was introduced into the MtPT1-GFP constructs. When expressed from the MtPT4 promoter, MtPT1S117F protein failed to localize in the periarbuscular membrane and instead was retained in the ER and in TGN-labeled endomembrane compartments (Fig. 5B). Likewise, MtPT1S117F expressed from its native promoter localized in ER and TGN-labeled bodies and failed to accumulate in the plasma membrane of root epidermal cells (Fig. 5A). These results suggest that the TGN is an intermediate secretion compartment for PHT1-type phosphate transporters, as shown recently for a polar-localized boron transporter, BOR1 (19, 26) and also suggested for the Arabidopsis phosphate transporter PHT1;1 (27), and that the MtPT4S115F and MtPT1S117F mutations disrupt a TGN export mechanism.
Fig. 5.
MtPT1S117F is retained in endomembranes. AtSyp61-mRFP TGN marker coexpressed with pMtPT1:MtPT1S117F-GFP (A) and pMtPT4:MtPT1S117F-GFP (B). Colocalization between markers is observed in epidermal cells (A) and cortical cells harboring arbuscules (B). Retention in the ER is evident by reticulate and perinuclear signals. (Scale bars: 20 μm.)
We have observed two compartments in which phosphate transporters can be retained in the endomembrane system: the ER and TGN-labeled compartments (Fig. 6B). Ectopic expression of MtPT4 leads to ER retention, as does heterologous expression of OsPT11 (11). In Arabidopsis, PHT1-type phosphate transporters require a specific chaperone, PHF1, to exit the ER (28), whereas in yeast, PHO86, a protein unrelated to PHF1, serves this function (28, 29). Similarly, AXR4 and TWD1 are required specifically for ER exit of the transporter proteins AUX1 and ABCB, respectively (30, 31). The opposite ER retention patterns observed for MtPT4 and AtPIP2a suggest that arbuscule development elicits a symbiosis-specific modification in ER exit, possibly mediated by a change in chaperone functions. ER retention of aquaporins has been demonstrated to result from alterations in stoichiometry and posttranslational modifications (32, 33). In contrast, accumulation of both MtPT4S115F and MtPT1S117F in TGN-labeled endomembrane compartments suggests that mutation of this residue disrupts a conserved process in phosphate transporter trafficking. Because this mutation causes a similar effect in cells with arbuscules and in epidermal cells, it seems likely that periarbuscular membrane secretion employs some mechanisms used for basic plant function, as identified recently for secretion during a plant–pathogen interaction (34). In AM symbiosis this idea is supported further by recent data showing the localization of exocytosis markers at perifungal membranes (35).
Based on the outcome of these promoter/protein combinations, we propose that correct localization of MtPT4 is regulated by a polarization of the bulk secretory pathway favoring vesicle fusion with the developing periarbuscular membrane rather than with the plasma membrane, and that there is a coincident change in the newly synthesized protein cargo entering into the secretion system. The surprising results that three plasma membrane-resident proteins, MtPT1, MtSTP, and MtPLT, localize in the periarbuscular membrane when expressed during arbuscule development and that expression before arbuscule development is insufficient for localization in this domain argue strongly that the driving factor is a transient reorientation of secretion rather than a protein-sorting process (Fig. 6B).
In plants, polar localization of membrane proteins has been studied extensively for the PIN family of auxin efflux transporters, which localize to the basal or apical membrane of cells to direct polar auxin transport. PINs initially are secreted in a nonpolar manner to the plasma membrane, and polarity is generated subsequently by endocytosis and polar resecretion (16, 36). Under this model, MtPT1 expressed from the 35S promoter before arbuscule development should localize subsequently in the periarbuscular membrane through endocytosis and resecretion. This result was not observed, indicating a fundamentally different mechanism for polar targeting to the periarbuscular membrane. Recent work on the outer membrane-localized transporters BOR4, ABCG37, and PEN3 in Arabidopsis roots identified direct polar secretion rather than recycling as a mechanism (37). It is tempting to speculate that this secretion mechanism dominates during AM symbiosis; however, experiments to test overlapping mechanisms are not currently feasible, because polar-secretion mutants, such as gnom or van3 (37), are not available in M. truncatula, and pharmacological studies are complicated by negative effects on the AM fungus.
The PIN proteins function in relatively mature cells, and polarity is maintained and modified over the life of the cell (16). In contrast, MtPT4 functions in a cell that must rapidly secrete up to eight times the amount of its plasma membrane, and the membrane and its resident proteins function for only the few days before the arbuscule is degraded. Transient reorientation of secretion toward the developing membrane could facilitate its rapid development, and a significant change in secreted cargo is consistent with the large transcriptional changes that occur in these cells (22, 38–40). Whether this mechanism also directs the polar localization of other proteins that function in the AM symbiosis awaits further study.
Analogous mechanisms may function in other dynamic cell types as well. Ectopic expression from the Knolle promoter results in localization of plasma membrane-resident syntaxins to the developing cell plate during cytokinesis (41), suggesting, according to Völker et al. (42), a “general redirection of membrane flow,” which essentially matches our proposed mechanism. AM fungal arbuscules also share similarities with haustoria formed by some biotrophic pathogenic fungi and oomycetes. Haustoria are nutrient-extracting hyphae that are surrounded by a plant membrane called the “extrahaustorial membrane” (43). The Arabidopsis resistance protein RPW8.2 localizes specifically in this membrane, but the underlying mechanism is unknown (44). In parallel with our findings, expression of RPW8.2 from the 35S promoter does not result in correct localization or function (21). It will be of interest to determine whether a transient reorientation of secretion, coupled with alterations in protein cargo entering the secretory system, operates to enable polar localization of proteins during other host–microbe interactions.
Materials and Methods
Growth and Transformation.
M. truncatula cv. Jemalong line A17 was used for all experiments described except those involving the MtPT4 mutant, mtpt4-3. The mtpt4-3 allele has a C-to-T transition resulting in a substitution of phenylalanine for serine at position 115 (25). Transgenic roots were generated by Agrobacterium rhizogenes-mediated root transformation as described (45), using the strain ARquaI. Cotransformed composite roots were generated by mixing ARquaI strains harboring appropriate plasmids before seedling inoculation. M. truncatula seeds were surface-sterilized and germinated by a 24-h dark treatment at 30 °C; elongating hypocotyls were excised and inoculated with ARquaI strains. Seedlings were transferred to modified Fahraeus medium containing 25 mg/L kanamycin for selection of transgenic roots. After 20 d, plants were transferred to a sterilized surface and were grown for 10 d before inoculation with surface-sterilized G. versiforme or Glomus intraradices spores. Plants were grown in a growth chamber under 16-h light (25 °C)/8 h dark (22 °C) cycle and fertilized once per week with half-strength Hoagland's solution modified with full-strength nitrogen and 20 μM potassium phosphate. Mycorrhizal roots were analyzed 3–5 wk after inoculation. Wild-type plants were used for all experiments except where indicated.
Plasmid Construction.
All constructs created for this report were expressed in the pCAMBIA2301 vector (www.cambia.org) and fused to the S65T variant of GFP (46), except where indicated.
Creation of pMtPT4:MtPT4-GFP has been described (10). pMtPT4:MtPT4Δ506-GFP was created by amplifying the full-length MtPT4 plasmid with a reverse primer that truncated the gene 1,518 bp downstream of the ATG, corresponding to a protein truncation at amino acid 506. The resulting fragment was cloned into the XbaI and NcoI restriction sites of pMtPT4:MtPT4-GFP. pMtPT4:MtPT4S115F-GFP was created by replacing an Age1 fragment from the wild-type MtPT4 gene with that of the mtpt4-3 gene.
pMtPT1:MtPT1-GFP was created by amplifying a 3,164-bp genomic fragment corresponding to 1,545 bp of promoter and the 1,611 bp coding sequence (CDS) with primers that added a 5′ XbaI and 3′ NcoI restriction site. This fragment was digested and ligated into a modified pCAMBIA2301 containing the S65T GFP variant and nopaline synthase (Nos) terminator, resulting in an in-frame 3′ fusion to GFP. pMtPT4:MtPT1-GFP was created by amplifying the MtPT4 864-bp promoter fragment with primers adding 5′XbaI and 3′ KpnI. This fragment was cloned into the pGEM-T EZ vector. The MtPT1 ORF was amplified with primers adding 5′ KpnI and 3′ BamHI sites and was cloned into the MtPT4 promoter vector. This fusion was digested with XbaI and BamHI and was ligated into pCAMBIA2301.
pMtPT4:AtPIP2a-GFP was created by amplifying the 861-bp ORF of AtPIP2a from the 35S:AtPIP2a-mCherry vector (47) with primers adding 5′ KpnI and 3′ BamHI restriction sites. This fragment was digested and ligated into the similarly digested pMtPT4:MtPT1-GFP vector, effectively replacing the MtPT1 ORF for AtPIP2a.
p35S:MtPT4-GFP was created by amplifying the 1,581-bp MtPT4 cDNA with primers adding 5′ SalI and 3′ NcoI restriction sites. This fragment was digested and ligated into the 35S:sGFP (S65T)-Nos vector, creating an in-frame fusion of GFP to the 3′ end of MtPT4. This vector was digested with HindIII and EcoRI and was ligated into pCAMBIA2301. p35S:MtPT1-GFP was created in the same manner, using primers that added 5′ SalI and 3′ BamHI restriction sites.
pMtBcp1:MtPT4-GFP and pMtBcp1:MtPT1-GFP were created by amplifying the 1,179-bp MtBcp1 promoter with primers that added 5′ XbaI and 3′ SalI restriction sites. This fragment was digested with XbaI and SalI and ligated into similarly digested p35S:MtPT4-sGFP (S65T)-Nos or p35S:MtPT4-sGFP (S65T)-Nos vector, replacing the promoter with pMtBcp1. These plasmids then were digested with XbaI and EcoRI and ligated into pCAMBIA2301. pMtBcp1:AtPIP2a-mCherry was created by amplifying the AtPIP2a-mCherry fusion (47) with primers that added 5′ SalI and 3′ NotI restriction sites. This fragment was digested with SalI and NotI and was ligated into the pMtBcp1:MtPT1-GFP pUC vector digested with Sal1 and Not1. The resulting pMtBcp1:AtPIP2a-mCherry plasmid was digested with XbaI and EcoRI and ligated into pCAMBIA2301.
p35S:MtSTP-GFP was created by amplifying a 1,626-bp ORF of MtSTP (Medtr8g102860.1) with primers that add 5′ SalI and 3′ BamHI restriction sites. The digested fragment was ligated into a modified pCAMBIA2301 containing pMtPT4:AtPIP2a-GFP and Nos terminator, replacing pMtPT4:AtPIP2a with MtSTP, which results in an in-frame MtSTP-GFP with a Nos terminator. The MtSTP-GFP plasmid was digested with SalI and EcoRI and then was cloned into the pCAMBIA2300-35S:sGFP (S65T)-Nos vector, creating p35S:MtSTP-GFP. pMtBCP1:MtSTP-GFP was created by replacing the 35S promoter with MtBCP1 promoter digested with XbaI and SalI from pMtBcp1:MtPT4-GFP. pMtPT4:MtSTP-GFP was created in the same manner by replacing the 35S promoter with MtPT4 promoter digested from pMtPT4:MtPT1-GFP. Creation of p35S:MtPLT-GFP, pMtBCP1:MtPLT-GFP, and pMtPT4:MtPLT-GFP was carried out as described above. The 1,569-bp ORF of MtPLT was amplified with primers adding 5′ SalI and 3′ BamHI restriction sites.
p35S:MtPT4 5′ UTR-MtPT4-GFP and p35S:MtPT4 5′UTR-MtPT1-GFP UTR-swap constructs were created by fusion PCR. A primer was designed to anneal to the 5′ end of the 5′ UTR of MtPT4 and to contain an overhang corresponding to the 35S promoter region at the TATA box. This primer was used together with a reverse primer that amplified either MtPT4 or MtPT1 from the pMtPT4:MtPT1 vector and added a 3′ NcoI restriction site. A second fragment corresponding to the 35S promoter was amplified using a forward primer that added a 5′ XbaI site and a reverse primer that corresponded to the 35S TATA box and an overhang corresponding to the 5′ end of the MtPT4 5′ UTR. These two fragments then were purified and amplified in a combined PCR using the forward primer that added a 5′ XbaI and the appropriate reverse primer that added a 3′ NcoI to create the UTR swap. This fused fragment then was digested with XbaI and NcoI and cloned into the 35S:sGFP (S65T)-Nos vector. This plasmid subsequently was digested with XbaI and EcoRI, and the resulting fragment was ligated into pCAMBIA2301. pMtPT4:35S 5′UTR-MtPT4-GFP and pMtPT4:35S 5′UTR-MtPT1-GFP were created using the strategy and restriction sites described above.
pMtPT4:MtPT4S115A-GFP and pMtPT4:MtPT4S115E-GFP were created by site-directed mutagenesis using the Quikchange II Mutagenesis kit (Stratagene) starting with pMtPT4:MtPT4-GFP in the pUC vector as template and resulting in a mutated MtPT4 protein with serine 115 substituted for alanine or glutamic acid, respectively. After mutagenesis, the modified fusion construct was digested with XbaI and EcoRI and ligated into pCAMBIA2301. pMtPT4:MtPT4Y228/239A-GFP was created in similar manner, using primers that introduced both mutations, changing tyrosine 228 and 239 to alanine. pMtPT4:MtPT1S117F-GFP and pMtPT1:MtPT1S117F-GFP were created similarly by the Quikchange II Mutagenesis kit using pMtPT4:MtPT1-GFP and pMtPT1:MtPT1-GFP, respectively, in pUC vector as template. Fusions also were subcloned into pCAMBIA2301 for expression.
p35S:HDEL-mCherry (ER) and p35S:GmMAN1-mCherry (Golgi) markers in pBIN vectors are described in ref. 47 and are available from the Arabidopsis Biological Resource Center (http://abrc.osu.edu/). The p35S:AtSyp61-mRFP TGN marker and pUbq10:VHA1-GFP39 have been described previously (48, 49).
Live Imaging Using Confocal Microscopy.
Imaging of fluorescent protein fusions was performed as described by Pumplin and Harrison (10). Briefly, wild-type transgenic roots expressing appropriate fluorescence were excised under an Olympus SZX-12 stereo microscope, bisected longitudinally, and sealed under a coverslip with Vaseline:lanolin:paraffin (VALAP), 1:1:1. Samples then were imaged on a Leica TCS-SP5 confocal microscope (Leica Microsystems) using a 63×, NA 1.2 water-immersion objective. GFP was excited with a blue argon ion laser at 488 nm, and emitted fluorescence was collected from 505–545 nm; mCherry and mRFP were excited with a diode-pumped solid-state laser, 561 nm, and emitted fluorescence was collected from 590–640 nm for mCherry and from570–600 nm for mRFP. Bright-field differential interference contrast (DIC) images were collected with the transmitted light detector. Image data were processed with the Leica LAS-AF Version 1.7.0 and Adobe Photoshop CS Version 8 (Adobe Systems) software. Only undisrupted cells below the sectioning plane were imaged. Figures include representative images from at least three independent experiments for each construct.
Immunolocalization.
Immunolocalization of MtPT4 was performed according to Harrison and co-workers (7) with a modified fixation buffer. Colonized root segments from wild-type and mtpt4-3 plants, stably transformed with the symbiosis-specific pMtScp1:GFP reporter (38), were isolated under a dissecting microscope. Samples were fixed for 2 h on a rotary shaker in a PME buffer (50 mM Pipes, 5 mM MgSO4, and 10 mM ethylene glycol tetraacetic acid, pH 6.9) containing 4% (vol/vol) formaldehyde, 1% (vol/vol) glutaraldehyde, and 5% (vol/vol) dimethyl sulfoxide. Roots then were bisected longitudinally, placed on a coverslip, and overlaid with a thin film of 1% (wt/vol) agar according to the procedures in ref. 50, followed by a 10-min cell-wall digestion in PME buffer containing 0.1% (wt/vol) BSA, 1% (wt/vol) cellulase RS, and 0.01% (wt/vol) pectolyase Y23 (Karlan Research Products). This digestion was followed by three 5-min rinses with PME and 10-min incubation in PBS buffer (135 mM NaCl, 25 mM KCl, and 10 mM Na2HPO4, pH 7.4) containing 1% (wt/vol) BSA and overnight incubation in a 1/500 dilution of MtPT4 antibody in PBS buffer containing 0.5% (wt/vol) BSA (7). The next day, samples were rinsed three times with PBS and were incubated for 2 h with a 1/100 dilution in PBS of goat anti-rabbit IgG conjugated to Alexa Fluor 488 (Molecular Probes) followed by additional rinses and a 30-min incubation with 0.05 mg/mL wheat germ agglutinin conjugated to Alexa Fluor 594 (Molecular Probes) in PBS. After four rinses with PBS, samples were rinsed once with PBS (pH 8.5) and mounted onto microscope slides in PBS (pH 8.5) containing 13% (wt/vol) Mowiol 4–88 (Calbiochem).
Supplementary Material
Acknowledgments
We thank K. Schumacher (Center for Organismal Studies, Heidelberg) and T. Ueda (RIKEN, Japan) for markers, M. Farman (University of Kentucky, Lexington, KY) for generation of the MtPT4ΔN construct, and Dierdra Daniels for assistance with transgenic roots expressing carbohydrate transporter constructs. Financial support was provided by National Science Foundation Grants DBI-0421677 and IOS-0842720 and by the Atlantic Philanthropies Molecular and Chemical Ecology (MACE) program. A Cornell Presidential Life Sciences Fellowship provided a 2-y stipend for N.P.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. A.V. is a guest editor invited by the Editorial Board.
See Author Summary on page 4041 (volume 109, number 11).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110215109/-/DCSupplemental.
References
- 1.Parniske M. Arbuscular mycorrhiza: The mother of plant root endosymbioses. Nat Rev Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 2.Bonfante P, Genre A. (2010) Mechanisms underlying beneficial plant-fungus interactions in mycorrhizal symbiosis. Nature Communications, 10.1038/ncomms1046. [DOI] [PubMed]
- 3.van der Heijden MGA, et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature. 1998;396:69–72. [Google Scholar]
- 4.Alexander T, Toth R, Meier R, Weber HC. Dynamics of arbuscule development and degeneration in onion, bean and tomato with reference to vesicular-arbuscular mycorrhizae in grasses. Can J Bot. 1989;67:2505–2513. [Google Scholar]
- 5.Javot H, Penmetsa RV, Terzaghi N, Cook DR, Harrison MJ. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2007;104:1720–1725. doi: 10.1073/pnas.0608136104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Q, Blaylock LA, Harrison MJ. Two Medicago truncatula half-ABC transporters are essential for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Cell. 2010;22:1483–1497. doi: 10.1105/tpc.110.074955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harrison MJ, Dewbre GR, Liu J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell. 2002;14:2413–2429. doi: 10.1105/tpc.004861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paszkowski U, Kroken S, Roux C, Briggs SP. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc Natl Acad Sci USA. 2002;99:13324–13329. doi: 10.1073/pnas.202474599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy R, et al. The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 2005;42:236–250. doi: 10.1111/j.1365-313X.2005.02364.x. [DOI] [PubMed] [Google Scholar]
- 10.Pumplin N, Harrison MJ. Live-cell imaging reveals periarbuscular membrane domains and organelle location in Medicago truncatula roots during arbuscular mycorrhizal symbiosis. Plant Physiol. 2009;151:809–819. doi: 10.1104/pp.109.141879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobae Y, Hata S. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 2010;51:341–353. doi: 10.1093/pcp/pcq013. [DOI] [PubMed] [Google Scholar]
- 12.Bucher M. Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 2007;173:11–26. doi: 10.1111/j.1469-8137.2006.01935.x. [DOI] [PubMed] [Google Scholar]
- 13.Chiou TJ, Liu H, Harrison MJ. The spatial expression patterns of a phosphate transporter (MtPT1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 2001;25:1–15. doi: 10.1046/j.1365-313x.2001.00963.x. [DOI] [PubMed] [Google Scholar]
- 14.Shin H, Shin HS, Dewbre GR, Harrison MJ. Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004;39:629–642. doi: 10.1111/j.1365-313X.2004.02161.x. [DOI] [PubMed] [Google Scholar]
- 15.Ai PH, et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009;57:798–809. doi: 10.1111/j.1365-313X.2008.03726.x. [DOI] [PubMed] [Google Scholar]
- 16.Geldner N. Cell polarity in plants: A PARspective on PINs. Curr Opin Plant Biol. 2009;12:42–48. doi: 10.1016/j.pbi.2008.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Shewan AM, et al. GLUT4 recycles via a trans-Golgi network (TGN) subdomain enriched in Syntaxins 6 and 16 but not TGN38: Involvement of an acidic targeting motif. Mol Biol Cell. 2003;14:973–986. doi: 10.1091/mbc.E02-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurten RC. Sorting motifs in receptor trafficking. Adv Drug Deliv Rev. 2003;55:1405–1419. doi: 10.1016/j.addr.2003.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Takano J, et al. Polar localization and degradation of Arabidopsis boron transporters through distinct trafficking pathways. Proc Natl Acad Sci USA. 2010;107:5220–5225. doi: 10.1073/pnas.0910744107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Auriac MC, Timmers AC. Nodulation studies in the model legume Medicago truncatula: Advantages of using the constitutive EF1alpha promoter and limitations in detecting fluorescent reporter proteins in nodule tissues. Mol Plant Microbe Interact. 2007;20:1040–1047. doi: 10.1094/MPMI-20-9-1040. [DOI] [PubMed] [Google Scholar]
- 21.Wang W, Wen Y, Berkey R, Xiao S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell. 2009;21:2898–2913. doi: 10.1105/tpc.109.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hohnjec N, Vieweg MF, Pühler A, Becker A, Küster H. Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 2005;137:1283–1301. doi: 10.1104/pp.104.056572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klepek YS, et al. Arabidopsis thaliana POLYOL/MONOSACCHARIDE TRANSPORTERS 1 and 2: Fructose and xylitol/H+ symporters in pollen and young xylem cells. J Exp Bot. 2010;61:537–550. doi: 10.1093/jxb/erp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poschet G, Hannich B, Büttner M. Identification and characterization of AtSTP14, a novel galactose transporter from Arabidopsis. Plant Cell Physiol. 2010;51:1571–1580. doi: 10.1093/pcp/pcq100. [DOI] [PubMed] [Google Scholar]
- 25.Javot H, et al. Medicago truncatula mtpt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 2011;68:954–965. doi: 10.1111/j.1365-313X.2011.04746.x. [DOI] [PubMed] [Google Scholar]
- 26.Viotti C, et al. Endocytic and secretory traffic in Arabidopsis merge in the trans-Golgi network/early endosome, an independent and highly dynamic organelle. Plant Cell. 2010;22:1344–1357. doi: 10.1105/tpc.109.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bayle V, et al. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell. 2011;23:1523–1535. doi: 10.1105/tpc.110.081067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González E, Solano R, Rubio V, Leyva A, Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell. 2005;17:3500–3512. doi: 10.1105/tpc.105.036640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lau WT, Howson RW, Malkus P, Schekman R, O'Shea EK. Pho86p, an endoplasmic reticulum (ER) resident protein in Saccharomyces cerevisiae, is required for ER exit of the high-affinity phosphate transporter Pho84p. Proc Natl Acad Sci USA. 2000;97:1107–1112. doi: 10.1073/pnas.97.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dharmasiri S, et al. AXR4 is required for localization of the auxin influx facilitator AUX1. Science. 2006;312:1218–1220. doi: 10.1126/science.1122847. [DOI] [PubMed] [Google Scholar]
- 31.Wu GS, Otegui MS, Spalding EP. The ER-localized TWD1 immunophilin is necessary for localization of multidrug resistance-like proteins required for polar auxin transport in Arabidopsis roots. Plant Cell. 2010;22:3295–3304. doi: 10.1105/tpc.110.078360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F. An N-terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. Plant J. 2009;57:346–355. doi: 10.1111/j.1365-313X.2008.03691.x. [DOI] [PubMed] [Google Scholar]
- 33.Prak S, et al. Multiple phosphorylations in the C-terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of AtPIP2;1 in response to salt stress. Mol Cell Proteomics. 2008;7:1019–1030. doi: 10.1074/mcp.M700566-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Kwon C, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 35.Genre A, et al. Multiple exocytotic markers accumulate at the sites of perifungal membrane biogenesis in arbuscular mycorrhizas. Plant Cell Physiol. 2012;53:244–255. doi: 10.1093/pcp/pcr170. [DOI] [PubMed] [Google Scholar]
- 36.Dhonukshe P, et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature. 2008;456:962–966. doi: 10.1038/nature07409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Langowski L, Růzicka K, Naramoto S, Kleine-Vehn J, Friml J. Trafficking to the outer polar domain defines the root-soil interface. Curr Biol. 2010;20:904–908. doi: 10.1016/j.cub.2010.03.059. [DOI] [PubMed] [Google Scholar]
- 38.Liu JY, et al. Transcript profiling coupled with spatial expression analyses reveals genes involved in distinct developmental stages of an arbuscular mycorrhizal symbiosis. Plant Cell. 2003;15:2106–2123. doi: 10.1105/tpc.014183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomez SK, et al. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009;9:1–19. doi: 10.1186/1471-2229-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guether M, et al. Genome-wide reprogramming of regulatory networks, transport, cell wall and membrane biogenesis during arbuscular mycorrhizal symbiosis in Lotus japonicus. New Phytol. 2009;182:200–212. doi: 10.1111/j.1469-8137.2008.02725.x. [DOI] [PubMed] [Google Scholar]
- 41.Müller I, et al. Syntaxin specificity of cytokinesis in Arabidopsis. Nat Cell Biol. 2003;5:531–534. doi: 10.1038/ncb991. [DOI] [PubMed] [Google Scholar]
- 42.Völker A, Stierhof YD, Jürgens G. Cell cycle-independent expression of the Arabidopsis cytokinesis-specific syntaxin KNOLLE results in mistargeting to the plasma membrane and is not sufficient for cytokinesis. J Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- 43.Koh S, André A, Edwards H, Ehrhardt D, Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 2005;44:516–529. doi: 10.1111/j.1365-313X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, Berkey R, Wen Y, Xiao S. Accurate and adequate spatiotemporal expression and localization of RPW8.2 is key to activation of resistance at the host-pathogen interface. Plant Signal Behav. 2010;5:1002–1005. doi: 10.4161/psb.5.8.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 46.Chiu W, et al. Engineered GFP as a vital reporter in plants. Curr Biol. 1996;6:325–330. doi: 10.1016/s0960-9822(02)00483-9. [DOI] [PubMed] [Google Scholar]
- 47.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 48.Sanderfoot AA, Kovaleva V, Bassham DC, Raikhel NV. Interactions between syntaxins identify at least five SNARE complexes within the Golgi/prevacuolar system of the Arabidopsis cell. Mol Biol Cell. 2001;12:3733–3743. doi: 10.1091/mbc.12.12.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dettmer J, Hong-Hermesdorf A, Stierhof YD, Schumacher K. Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell. 2006;18:715–730. doi: 10.1105/tpc.105.037978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown RC, Lemmon BE. Methods in plant immunolight microscopy. Methods Cell Biol. 1995;49:85–107. doi: 10.1016/s0091-679x(08)61448-x. [DOI] [PubMed] [Google Scholar]