Abstract
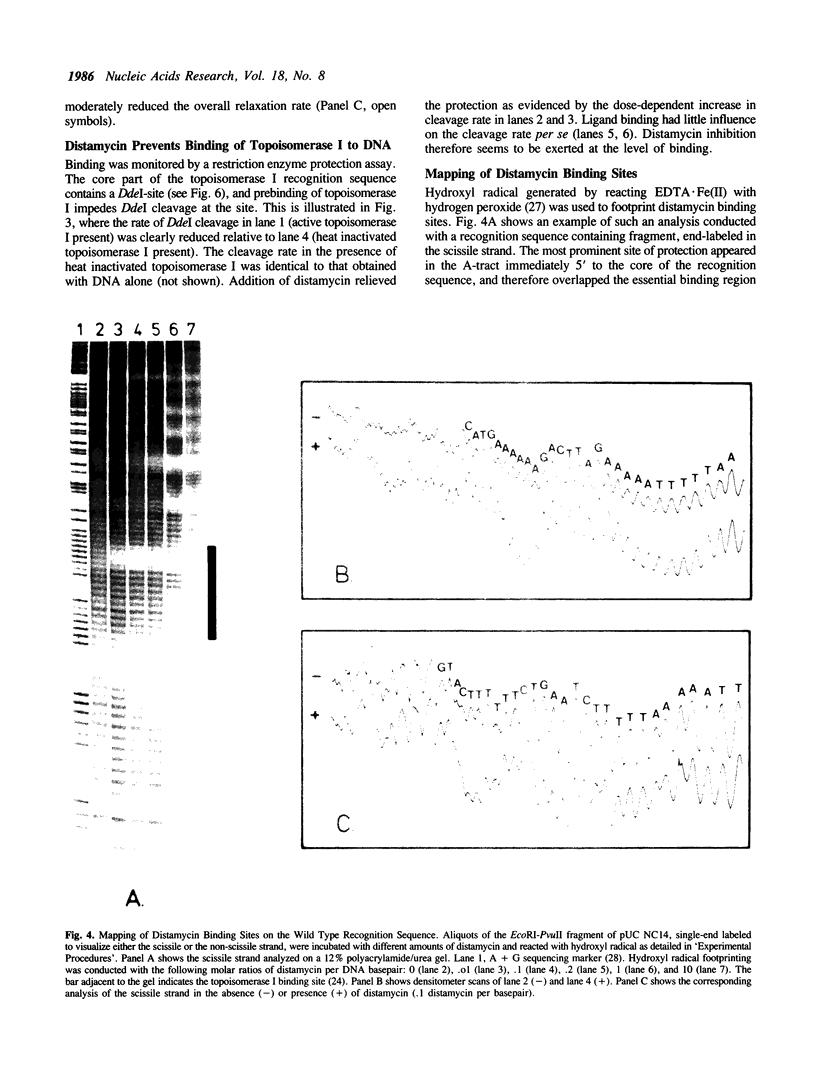
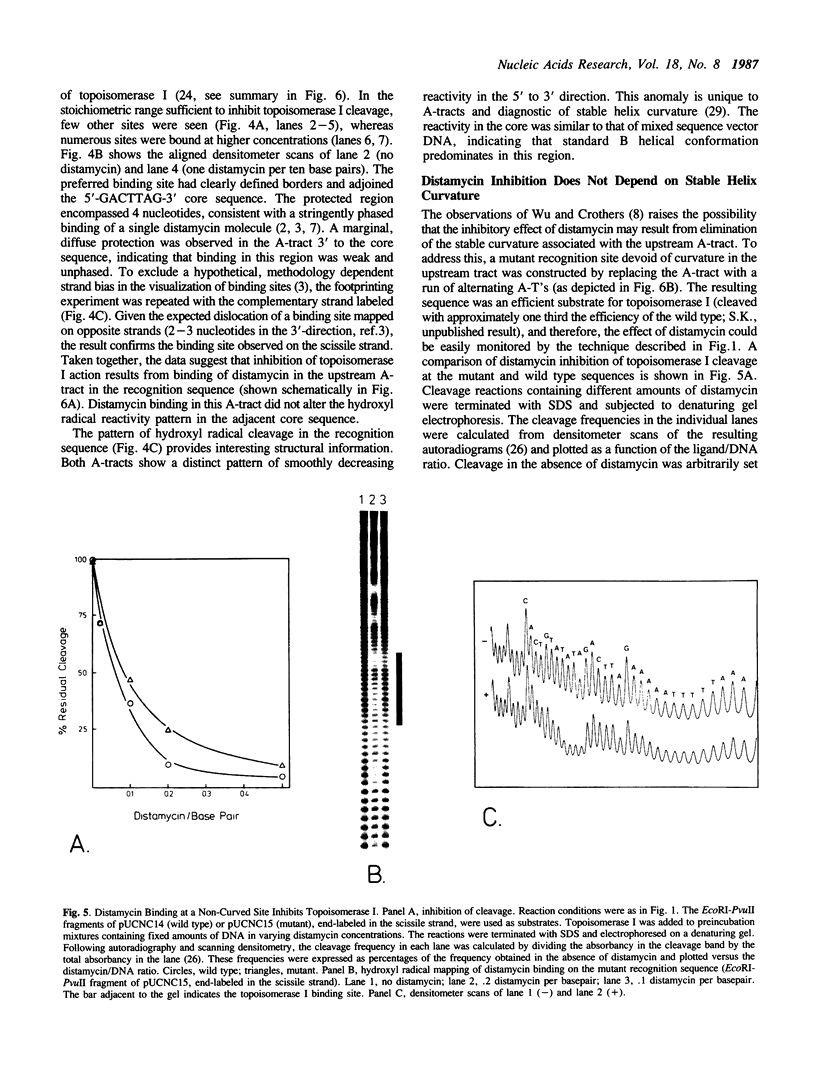
Inhibition of eukaryotic DNA topoisomerase I by the minor groove binding ligand, distamycin A, was investigated. Low concentrations of the ligand selectively prevented catalytic action at a high affinity topoisomerase I binding sequence. A restriction enzyme protection assay indicated that the catalytic cycle was blocked at the binding step. Distamycin binding sites on DNA were localized by hydroxyl radical footprinting. A strongly preferred site mapped to a homopolymeric (dA).(dT)-tract partially included in the essential topoisomerase I binding region. Mutational elimination of the stable helix curvature associated with this ligand binding site demonstrated that (i) the intrinsic bend was unessential for efficient binding of topoisomerase I, and (ii) distamycin inhibition did not occur by deformation of a stable band. Alternative modes of inhibition are discussed.
Full text
PDF

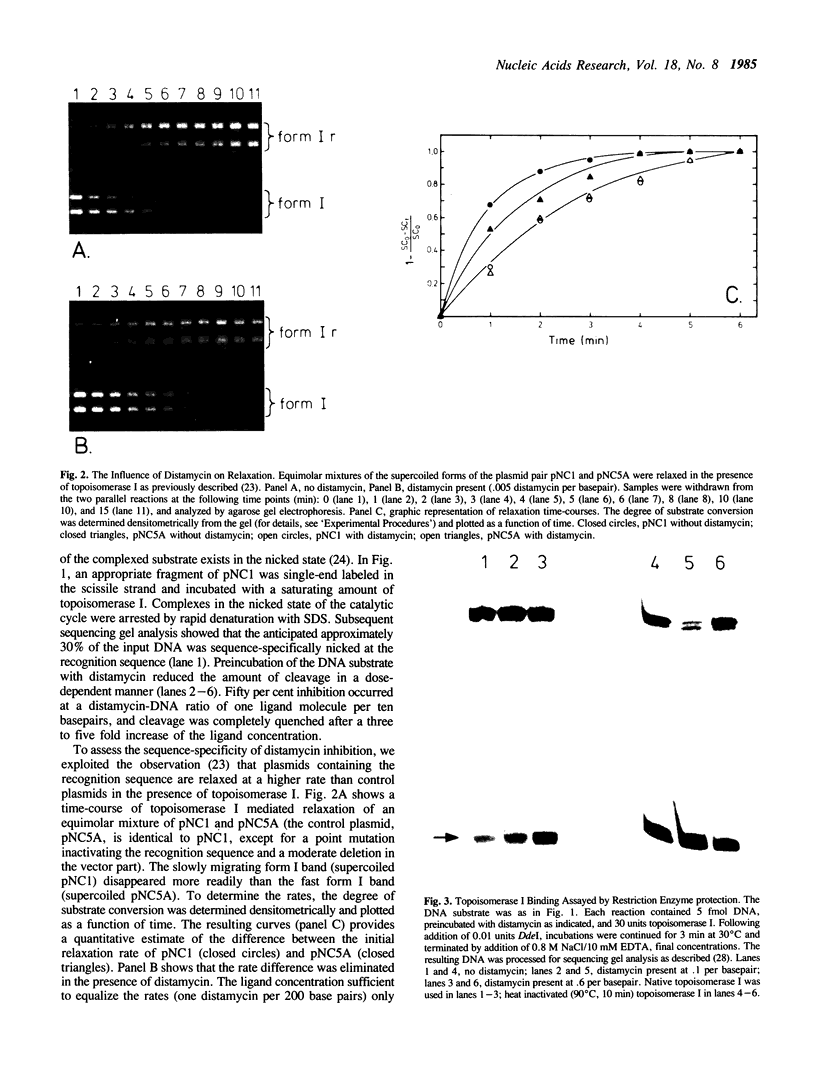


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen A. H., Gocke E., Bonven B. J., Nielsen O. F., Westergaard O. Topoisomerase I has a strong binding preference for a conserved hexadecameric sequence in the promoter region of the rRNA gene from Tetrahymena pyriformis. Nucleic Acids Res. 1985 Mar 11;13(5):1543–1557. doi: 10.1093/nar/13.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Been M. D., Burgess R. R., Champoux J. J. Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res. 1984 Apr 11;12(7):3097–3114. doi: 10.1093/nar/12.7.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Broggini M., Ponti M., Ottolenghi S., D'Incalci M., Mongelli N., Mantovani R. Distamycins inhibit the binding of OTF-1 and NFE-1 transfactors to their conserved DNA elements. Nucleic Acids Res. 1989 Feb 11;17(3):1051–1059. doi: 10.1093/nar/17.3.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Busk H., Thomsen B., Bonven B. J., Kjeldsen E., Nielsen O. F., Westergaard O. Preferential relaxation of supercoiled DNA containing a hexadecameric recognition sequence for topoisomerase I. Nature. 1987 Jun 18;327(6123):638–640. doi: 10.1038/327638a0. [DOI] [PubMed] [Google Scholar]
- Caserta M., Amadei A., Di Mauro E., Camilloni G. In vitro preferential topoisomerization of bent DNA. Nucleic Acids Res. 1989 Nov 11;17(21):8463–8474. doi: 10.1093/nar/17.21.8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiansen K., Bonven B. J., Westergaard O. Mapping of sequence-specific chromatin proteins by a novel method: topoisomerase I on Tetrahymena ribosomal chromatin. J Mol Biol. 1987 Feb 5;193(3):517–525. doi: 10.1016/0022-2836(87)90264-6. [DOI] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesen M., Pommier Y. Mammalian topoisomerase II activity is modulated by the DNA minor groove binder distamycin in simian virus 40 DNA. J Biol Chem. 1989 Jul 5;264(19):11354–11359. [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transmission of allosteric effects in DNA. Nature. 1979 Apr 5;278(5704):521–524. doi: 10.1038/278521a0. [DOI] [PubMed] [Google Scholar]
- Ishii K., Hasegawa T., Fujisawa K., Andoh T. Rapid purification and characterization of DNA topoisomerase I from cultured mouse mammary carcinoma FM3A cells. J Biol Chem. 1983 Oct 25;258(20):12728–12732. [PubMed] [Google Scholar]
- Klevit R. E., Wemmer D. E., Reid B. R. 1H NMR studies on the interaction between distamycin A and a symmetrical DNA dodecamer. Biochemistry. 1986 Jun 3;25(11):3296–3303. doi: 10.1021/bi00359a032. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. The molecular origin of DNA-drug specificity in netropsin and distamycin. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1376–1380. doi: 10.1073/pnas.82.5.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupin W., Chazin W. J., Hyberts S., Denny W. A., Wüthrich K. NMR studies of the complex between the decadeoxynucleotide d-(GCATTAATGC)2 and a minor-groove-binding drug. Biochemistry. 1986 Oct 7;25(20):5902–5910. doi: 10.1021/bi00368a010. [DOI] [PubMed] [Google Scholar]
- Linial M., Shlomai J. A unique endonuclease from Crithidia fasciculata which recognizes a bend in the DNA helix. Specificity of the cleavage reaction. J Biol Chem. 1988 Jan 5;263(1):290–297. [PubMed] [Google Scholar]
- Linial M., Shlomai J. Sequence-directed bent DNA helix is the specific binding site for Crithidia fasciculata nicking enzyme. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8205–8209. doi: 10.1073/pnas.84.23.8205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Maxwell A., Gellert M. Mechanistic aspects of DNA topoisomerases. Adv Protein Chem. 1986;38:69–107. doi: 10.1016/s0065-3233(08)60526-4. [DOI] [PubMed] [Google Scholar]
- McHugh M. M., Woynarowski J. M., Sigmund R. D., Beerman T. A. Effect of minor groove binding drugs on mammalian topoisomerase I activity. Biochem Pharmacol. 1989 Jul 15;38(14):2323–2328. doi: 10.1016/0006-2952(89)90472-3. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Patel I., Bastia D. Conformational changes in a replication origin induced by an initiator protein. Cell. 1985 Nov;43(1):189–197. doi: 10.1016/0092-8674(85)90023-6. [DOI] [PubMed] [Google Scholar]
- Perez-Stable C., Shen C. C., Shen C. K. Enrichment and depletion of Hela topoisomerase I recognition sites among specific types of DNA elements. Nucleic Acids Res. 1988 Aug 25;16(16):7975–7993. doi: 10.1093/nar/16.16.7975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portugal J., Waring M. J. Hydroxyl radical footprinting of the sequence-selective binding of netropsin and distamycin to DNA. FEBS Lett. 1987 Dec 10;225(1-2):195–200. doi: 10.1016/0014-5793(87)81156-0. [DOI] [PubMed] [Google Scholar]
- Ryder K., Silver S., DeLucia A. L., Fanning E., Tegtmeyer P. An altered DNA conformation in origin region I is a determinant for the binding of SV40 large T antigen. Cell. 1986 Mar 14;44(5):719–725. doi: 10.1016/0092-8674(86)90838-x. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Rosenberg J. M., Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976 Mar;73(3):804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevnsner T., Mortensen U. H., Westergaard O., Bonven B. J. Interactions between eukaryotic DNA topoisomerase I and a specific binding sequence. J Biol Chem. 1989 Jun 15;264(17):10110–10113. [PubMed] [Google Scholar]
- Thomsen B., Mollerup S., Bonven B. J., Frank R., Blöcker H., Nielsen O. F., Westergaard O. Sequence specificity of DNA topoisomerase I in the presence and absence of camptothecin. EMBO J. 1987 Jun;6(6):1817–1823. doi: 10.1002/j.1460-2075.1987.tb02436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A. Iron(II) EDTA used to measure the helical twist along any DNA molecule. Science. 1985 Nov 8;230(4726):679–681. doi: 10.1126/science.2996145. [DOI] [PubMed] [Google Scholar]
- Van Dyke M. W., Hertzberg R. P., Dervan P. B. Map of distamycin, netropsin, and actinomycin binding sites on heterogeneous DNA: DNA cleavage-inhibition patterns with methidiumpropyl-EDTA.Fe(II). Proc Natl Acad Sci U S A. 1982 Sep;79(18):5470–5474. doi: 10.1073/pnas.79.18.5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosberg H. P. DNA topoisomerases: enzymes that control DNA conformation. Curr Top Microbiol Immunol. 1985;114:19–102. doi: 10.1007/978-3-642-70227-3_2. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zimmer C., Wähnert U. Nonintercalating DNA-binding ligands: specificity of the interaction and their use as tools in biophysical, biochemical and biological investigations of the genetic material. Prog Biophys Mol Biol. 1986;47(1):31–112. doi: 10.1016/0079-6107(86)90005-2. [DOI] [PubMed] [Google Scholar]