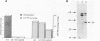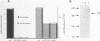Abstract
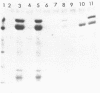
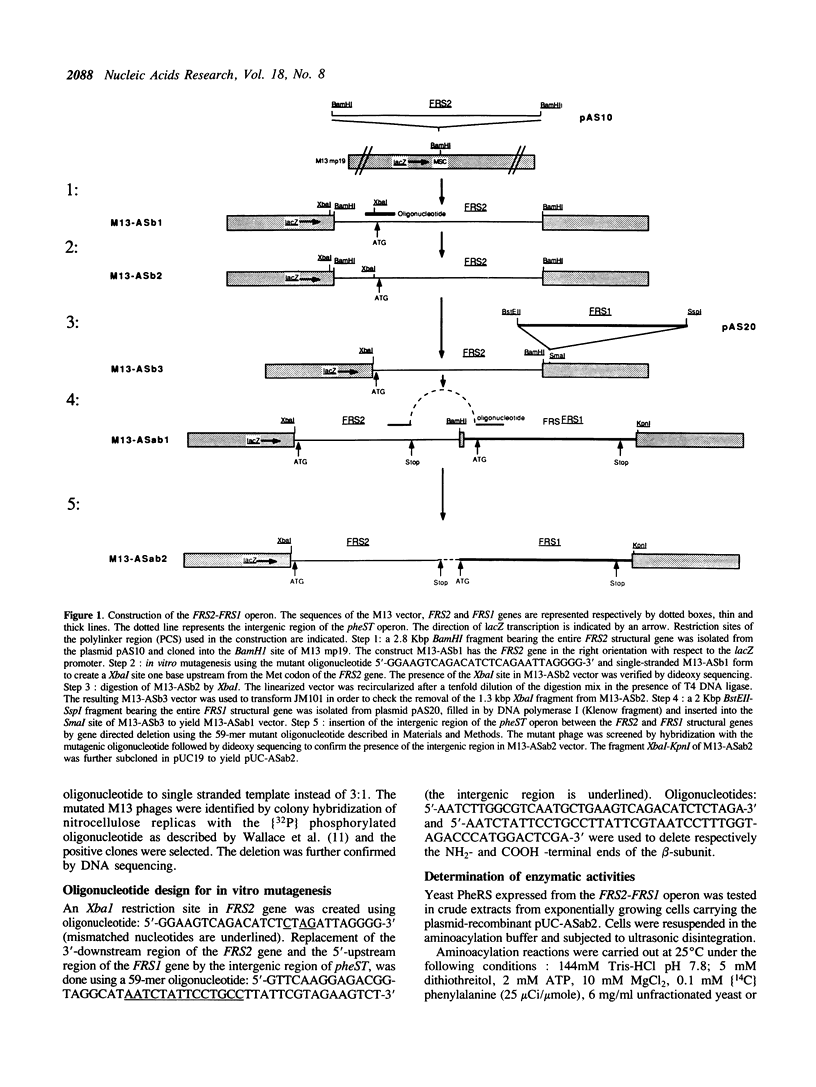
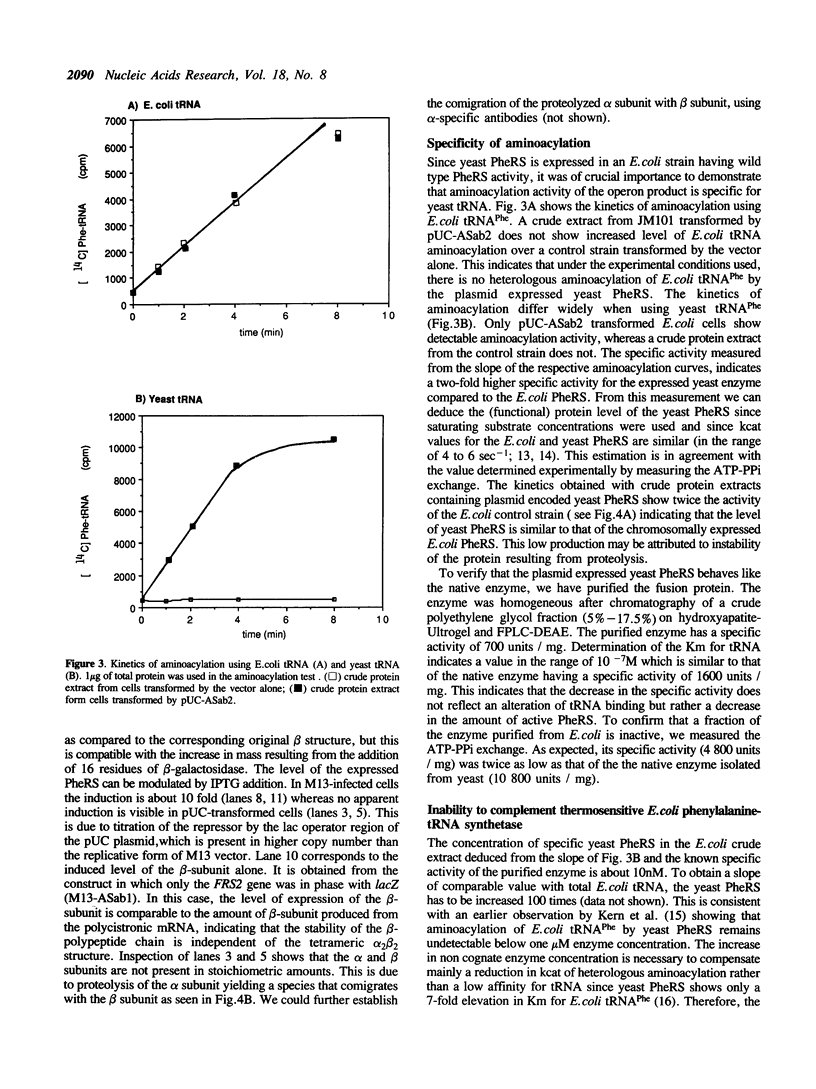
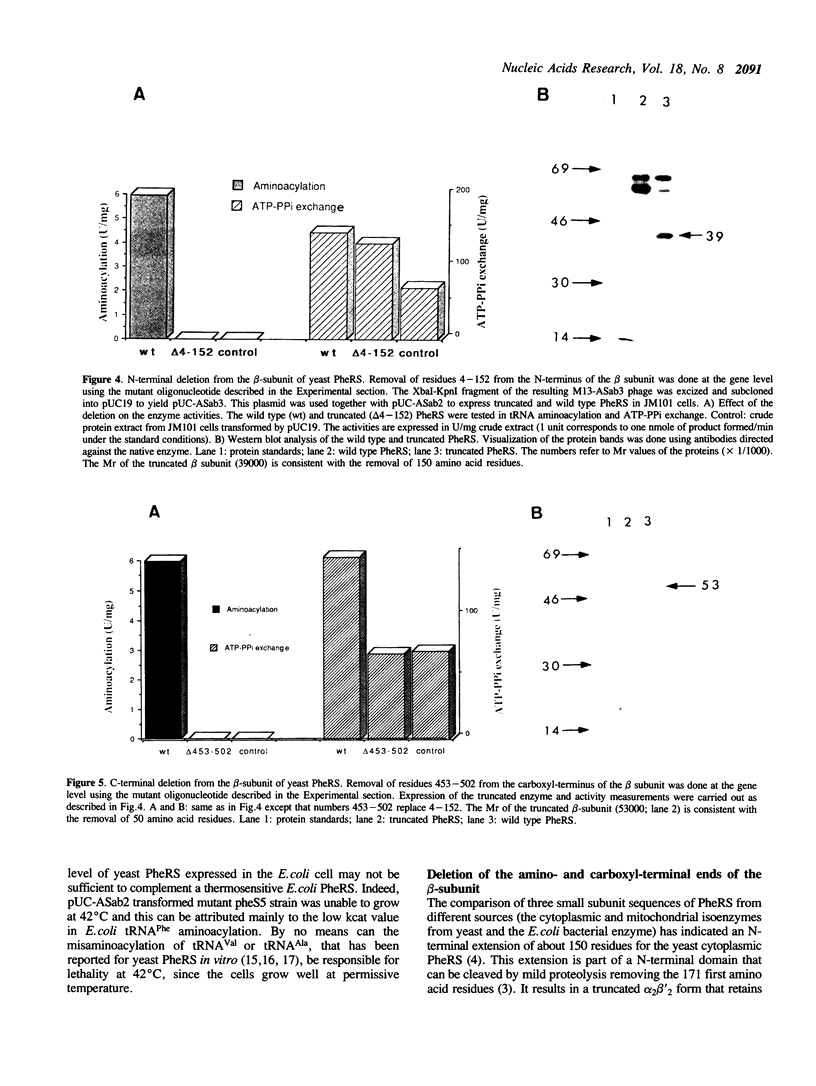
FRS1 and FRS2, the structural genes encoding the large (alpha) and small (beta) subunits of yeast phenylalanyl-tRNA synthetase (PheRS) were placed under the control of the lacZ promoter by creating an artificial operon. The FRS2 gene was fused next to the promoter, followed by a 14 base pair intergenic sequence containing a translation reinitiation site in front of the FRS1 coding sequences. The engineered PheRS has 16 N-terminal amino acids from beta-galactosidase fused to the beta subunit. However, the purified protein shows a Km value for tRNA(Phe) that is indistinguishable from that of the the native enzyme. The product of the FRS2-FRS1 operon is not able to complement thermosensitive E. coli PheRS, indicating the lack of heterologous aminoacylation in vivo. We made a deletion in the FRS2 gene that removed about 150 amino terminal residues of the beta subunit. The truncated protein showed intact ATP-PPi exchange, whereas tRNA aminoacylation was lost. This result is similar to that of limited proteolysis performed on the native enzyme that yielded a tetrameric alpha 2 beta'2 structure, able to form aminoacyladenylate but unable to bind tRNA(Phe). A deletion of 50 amino acids from the carboxyl terminus of the beta chain resulted in the loss of both enzyme activities; this suggests the participation of the C-terminal end of the beta subunit in the active site or in subunit assembly to yield a tetrameric functional enzyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltzinger M., Fasiolo F., Remy P. Yeast phenylalanyl-tRNA synthetase. Affinity and photoaffinity labelling of the stereospecific binding sites. Eur J Biochem. 1979 Jul;97(2):481–494. doi: 10.1111/j.1432-1033.1979.tb13136.x. [DOI] [PubMed] [Google Scholar]
- Baltzinger M., Holler E. Catalytic mechanism of phenylalanyl-tRNA synthetase of Escherichia coli K10. Conformational change and tRNAPhe phenylalanylation are concerted. Biochemistry. 1982 May 11;21(10):2467–2476. doi: 10.1021/bi00539a028. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Boulanger Y., Ebel J. Modification of phenylalanyl-tRNA synthetase from baker's yeast by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1975 May 6;53(2):487–492. doi: 10.1111/j.1432-1033.1975.tb04090.x. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Fersht A. R. The aminoacyladenylate mechanism in the aminoacylation reaction of yeast phenylalanyl-tRNA synthetase. Eur J Biochem. 1978 Apr;85(1):85–88. doi: 10.1111/j.1432-1033.1978.tb12214.x. [DOI] [PubMed] [Google Scholar]
- Fasiolo F., Sanni A., Potier S., Ebel J. P., Boulanger Y. Identification of the major tRNA(Phe) binding domain in the tetrameric structure of cytoplasmic phenylalanyl-tRNA synthetase from baker's yeast. FEBS Lett. 1989 Jan 2;242(2):351–356. doi: 10.1016/0014-5793(89)80500-9. [DOI] [PubMed] [Google Scholar]
- Kern D., Giegé R., Ebel J. P. Incorrect aminoacylatins catalysed by the phenylalanyl-and valyl-tRNA synthetases from yeast. Eur J Biochem. 1972 Nov 21;31(1):148–155. doi: 10.1111/j.1432-1033.1972.tb02513.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Fayat G., Blanquet S. Sequence of the Escherichia coli pheST operon and identification of the himA gene. J Bacteriol. 1985 Aug;163(2):787–791. doi: 10.1128/jb.163.2.787-791.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe B., Sirover M., Dudock B. Kinetics of homologous and heterologous aminoacylation with yeast phenylalanyl transfer ribonucleic acid synthetase. Biochemistry. 1973 Oct 9;12(21):4146–4154. doi: 10.1021/bi00745a018. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanni A., Mirande M., Ebel J. P., Boulanger Y., Waller J. P., Fasiolo F. Structure and expression of the genes encoding the alpha and beta subunits of yeast phenylalanyl-tRNA synthetase. J Biol Chem. 1988 Oct 25;263(30):15407–15415. [PubMed] [Google Scholar]
- Sayers J. R., Schmidt W., Eckstein F. 5'-3' exonucleases in phosphorothioate-based oligonucleotide-directed mutagenesis. Nucleic Acids Res. 1988 Feb 11;16(3):791–802. doi: 10.1093/nar/16.3.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M., Mayaux J. F., Fayat G., Plumbridge J. A., Graffe M., Blanquet S., Grunberg-Manago M. Attenuation control of the Escherichia coli phenylalanyl-tRNA synthetase operon. J Mol Biol. 1985 Feb 20;181(4):467–478. doi: 10.1016/0022-2836(85)90420-6. [DOI] [PubMed] [Google Scholar]
- Taglang R., Waller J. P., Befort N., Fasiolo F. Amino-acylation du tRNA-1-Val de Escherichia coli par la phénylalanyl-tRNA synthétase de levure. Eur J Biochem. 1970 Feb;12(3):550–557. doi: 10.1111/j.1432-1033.1970.tb00886.x. [DOI] [PubMed] [Google Scholar]
- Wallace R. B., Shaffer J., Murphy R. F., Bonner J., Hirose T., Itakura K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: the effect of single base pair mismatch. Nucleic Acids Res. 1979 Aug 10;6(11):3543–3557. doi: 10.1093/nar/6.11.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfes H., Alves J., Fliess A., Geiger R., Pingoud A. Site directed mutagenesis experiments suggest that Glu 111, Glu 144 and Arg 145 are essential for endonucleolytic activity of EcoRI. Nucleic Acids Res. 1986 Nov 25;14(22):9063–9080. doi: 10.1093/nar/14.22.9063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]