Background: KEAP1 is a ubiquitin ligase adaptor that promotes the ubiquitination and degradation of NRF2, a transcription factor that drives the antioxidant response.
Results: Wilms tumor gene on the X chromosome (WTX) stabilizes NRF2 by competing with NRF2 for binding to KEAP1.
Conclusion: WTX regulates the antioxidant response.
Significance: This study reveals a novel regulatory mechanism governing the antioxidant response.
Keywords: Antioxidants, Nrf2, Ubiquitin, Ubiquitination, BTRC, KEAP1, WTX
Abstract
WTX is a tumor suppressor protein that is lost or mutated in up to 30% of cases of Wilms tumor. Among its known functions, WTX interacts with the β-transducin repeat containing family of ubiquitin ligase adaptors and promotes the ubiquitination and degradation of the transcription factor β-catenin, a key control point in the WNT/β-catenin signaling pathway. Here, we report that WTX interacts with a second ubiquitin ligase adaptor, KEAP1, which functions to regulate the ubiquitination of the transcription factor NRF2, a key control point in the antioxidant response. Surprisingly, we find that unlike its ability to promote the ubiquitination of β-catenin, WTX inhibits the ubiquitination of NRF2. WTX and NRF2 compete for binding to KEAP1, and thus loss of WTX leads to rapid ubiquitination and degradation of NRF2 and a reduced response to cytotoxic insult. These results expand our understanding of the molecular mechanisms of WTX and reveal a novel regulatory mechanism governing the antioxidant response.
Introduction
FAM123B/WTX/AMER1 (hereafter referred to as WTX) is located on the X chromosome and encodes a tumor suppressor protein that is lost or mutated in up to 30% of the cases of Wilms tumor, the most common pediatric kidney cancer (1–3). Recently, germ line mutations in WTX were also discovered in families suffering from osteopathia striata congenital with cranial sclerosis (OSCS),4 a debilitating skeletal dysplasia that is often accompanied by developmental abnormalities and fatality in males (4, 5).
We previously reported that WTX regulates the stability of β-catenin (6). The regulation of the stability of β-catenin is a key control point of the WNT/β-catenin signaling pathway and the broader protein networks with which it interacts (7). In the absence of WNT ligand, β-catenin is phosphorylated by a multiprotein complex often called the “destruction complex” and is subsequently recognized by the SCFBTRC (Skp, Cullin, F-box) ubiquitin ligase complex where it is ubiquitinated and targeted for proteasomal degradation (8–10). In the presence of WNT ligand, phosphorylation of β-catenin is attenuated, resulting in the nuclear accumulation of β-catenin and the regulation of transcription. Through proteomic and functional dissection of the WNT/β-catenin signaling pathway, we discovered that WTX associates with β-catenin as well as proteins in the destruction complex, including adenomatous polyposis coli (APC), AXIN1, BTRC (commonly referred to as β-TrCP), and FBXW11 (commonly referred to as β-TrCP2) (6). Although the precise mechanism(s) is unknown, WTX promotes the ubiquitination and degradation of β-catenin.
In addition to regulating the stability of β-catenin, WTX has also been shown to play a role in regulating WNT signal transduction at the membrane (11). Furthermore, WTX controls cell-cell adhesion through interactions with APC at the plasma membrane (12) and modulates the activity of the WT1 (Wilms tumor 1) transcription factor in the nucleus (13). Thus, it is possible that the loss of WTX contributes to disease through distinct mechanisms in specific subcellular compartments.
Our previous analysis of the WTX protein interaction network identified an association between WTX and KEAP1 (Kelch-like ECH-associated protein 1) (6). KEAP1 is a substrate recognition module for the CUL3-based E3 ubiquitin ligase that constitutively ubiquitinates the transcription factor NRF2 (NF-E2-related factor-2; NFE2L2) (14–16). In the presence of cytotoxic stress such as xenobiotics and antioxidants, KEAP1 is inhibited, and NRF2 is no longer targeted for ubiquitination and degradation. NRF2 then accumulates in the nucleus where it regulates transcription of genes involved in the “phase II” antioxidant response (17–19).
Although many studies have reported that small molecules and stressors regulate NRF2 stability and function, relatively little is known about regulation of KEAP1 and NRF2 through protein-protein interactions. Here, we report that WTX directly binds to KEAP1. Using gain-of-function and loss-of-function approaches, we found that WTX stabilizes NRF2 and positively regulates its transcriptional activity. We identified an ETGE motif within the KEAP1 interacting domain of WTX that is nearly identical to that of NRF2. We show that this motif is not only required for the interaction of WTX with KEAP1 but is also required for the ability of WTX to regulate NRF2 stability and activity. Our observations support a model whereby WTX competes with NRF2 for binding to KEAP1, thereby promoting the NRF2-mediated antioxidant response.
EXPERIMENTAL PROCEDURES
Tissue Culture, Transfections, and Small Interfering RNAs
All cell lines were grown in DMEM supplemented with 10% fetal bovine serum in a 37 °C humidified incubator with 5% CO2. Selection and passage of stable cell lines were performed with 1.5 μg/ml puromycin until cell death was no long apparent. Expression constructs were transiently transfected in HEK293T cells with Lipofectamine 2000 as directed by the manufacturer (Invitrogen). Transient transfection of siRNA was performed with Lipofectamine RNAiMAX, as directed by the manufacturer (Invitrogen). Sequences of the siRNA sense strands are as follows: WTX-A (CCU GGA GAU GAC UGC CUU U dTdT), WTX-B (UAU GCC AGG GAG GCC CAC A dTdT), NRF2 (GUA AGA AGC CAG AUG UUA A dTdT), and KEAP1 (GGG CGU GGC UGU CCU CAA U dTdT). Control siRNA was acquired from Ambion.
Plasmids and Expression Vectors
NRF2, WTX, KEAP1, CUL3, and GFP cDNAs were created with standard PCR-based cloning strategies. The reporter gene fusion construct for human NQO1-ARE (hNQO1-ARE-luciferase) was a kind gift from Jeffrey Johnson.
WTX Antibody Production
Amino acids 212–438 of WTX were expressed as a GST fusion protein in Escherichia coli. Purified GST-tagged WTX fragment was used as an immunogen in rabbits, following established protocols by Cocalico Biologicals, Inc. Prior to use in Western blot analysis or immunostaining, the antibody was affinity-purified over protein G beads.
Affinity Pulldowns and Western Blotting
For streptavidin affinity purification, cells were lysed in radioimmunoprecipitation buffer (RIPA: 25 mm Tris-HCl, pH 8.0, 150 mm NaCl, 10% glycerol, 1% Triton X-100, 0.25% deoxycholic acid, 2 mm EDTA) containing protease inhibitor mixture (Roche Applied Science) and phosphatase inhibitor mixture (Calbiochem). Cell lysates were cleared by centrifugation and incubated with streptavidin resin (Amersham Biosciences) before washing with lysis buffer and eluting in NuPAGE loading buffer (Invitrogen). Detection of proteins by Western blot was performed using the following antibodies: anti-NRF2 (H-300) polyclonal (sc-13032, Santa Cruz Biotechnology), anti-KEAP1 polyclonal (10503–2-AP, Proteintech), anti-HMOX1 monoclonal (ab13248; Abcam), anti-FLAG M2 monoclonal (Sigma), anti-HA polyclonal (1867423; Roche Applied Science), anti-CTNNB1 polyclonal (9562; Cell Signaling Technology), anti-GFP polyclonal (ab290, Abcam), and anti-TUBB1 monoclonal (T7816; Sigma).
In Vitro Binding Experiments
Human GST-VSV-WTX was purified from E. coli and mixed with purified CUL3 or KEAP1 in buffered 150 mm NaCl. Following incubation for 30 min at 4 °C, complexes were washed with five bed volumes of 350 mm buffered NaCl before elution and Western blot.
ARE-Luciferase Quantification
For DNA, HEK293T cells were transfected with expression constructs, ARE-luciferase (firefly), and a control plasmid containing Renilla luciferase driven by a constitutive cytomegalovirus (CMV) promoter for normalization. Approximately 6 h post-transfection, cells were treated with 100 μm tBHQ and incubated for an additional 12–16 h. Activation was measured as the ratio of firefly to Renilla luciferase activity. For siRNA, HEK293T cells stably expressing the ARE-luciferase and Renilla control reporters were transfected with siRNA. Approximately 48 h later, 100 μm tBHQ was added, and cells were incubated for an additional 16 h. Activation was then measured as described above.
NQO1-GFP Quantification
H1299 cells containing a YFP fragment retrovirally inserted into intron 1 of the NQO1 gene (170407PL1A2-NQO1) were a gift from Uri Alon and the Kahn Protein Dynamics group. Ten thousand cells were seeded into each well of a 96-well clear bottom white plate. Following an 18-h incubation period, cells were transfected with siRNA. After an additional 24 h, 100 μm tBHQ was added, and cells were incubated for an additional 24 h. Cells were lysed in RIPA buffer, and fluorescence at λex = 485 nm was determined.
Cell Viability
HEK293T cells were transfected with siRNA as described above in 6-well tissue-culture plates. Twenty hours post-transfection, 30,000 cells were seeded into each well of a 96-well plate. Approximately 8 h later, cells were treated with either DMSO (vehicle) or etoposide. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell proliferation assay was performed 36 h post-treatment as described by the manufacturer (ATCC, catalog no. 30-1010K-A).
RNA Isolation, Reverse Transcription, and Semi-quantitative Real Time-PCR
Total RNA from cells was harvested in TRIzol (Invitrogen) reagent according to the manufacturer's instructions. RNA was quantified by UV spectrophotometry, and cDNA was created using the RevertAid First Strand cDNA synthesis kit (Fermentas). PCR was performed in duplicate with the LightCycler FastStart DNA SYBR Green kit (Roche Applied Science) using the LightCycler 480 instrument (Roche Applied Science). The PCR conditions are as follows: 35 cycles of amplification with 1 s denaturation at 95 °C and 5 s annealing at 58 °C. A template free negative control was included in each experiment. Quantitative light cycler PCR primers used are as follows: WTX (GAC CCA AAA GGA TGA AGC T and reverse CCC CTC CAA AGA AAC TAG GC) and β-actin (AGA GCA AGA GAG GCA TCC TC and reverse CTC AAA CAT GAT CTG GGT CA).
Cellular Extract Degradation
HEK293T cells were transfected with either WTX-A or control siRNA. Forty eight hours post-transfection, cells were resuspended in swelling buffer (20 mm HEPES, pH 7.7, 5 mm MgCl2, 5 mm KCl, 1 mm DTT, 0.2 mm ATP, and protease inhibitor) at a ratio of 4:3. Resuspended pellets were subjected to two rounds of freeze/thaw and passed through a chilled 27-gauge needle two times. Lysates were then centrifuged at 2,300 × g for 5 min, and supernatant was transferred to a new tube and centrifuged for 30 min at 16,000 × g. The middle layer containing the cytosolic fraction was isolated and used in subsequent NRF2 degradation experiments. In vitro transcription and translation of 35S-labeled NRF2 was performed using the coupled transcription-translation T7 system (Promega). Reactions consisted of 0.25 μl of 35S-labeled NRF2, 15 μl of cell extract, and 1 μl of NRG mixture containing a 1:1:1 ratio of energy mix (150 mm creatine phosphate, 20 mm ATP, 2 mm EGTA, and 20 mm MgCl2), cycloheximide (0.1 μg/ml), and 0.1 μg/ml recombinant ubiquitin. One μl of each reaction was taken at 0, 30, 120, and 240 min and snap-frozen in loading buffer. Samples were then run on a 3–12% gradient gel (Invitrogen), dried onto filter paper, and exposed to a phosphor screen overnight. Bands were quantified, and percent of NRF2 remaining was determined as the ratio of each time point compared with the zero time point.
In Vitro Ubiquitination
In vitro transcription/translation of CUL3, NRF2, KEAP1, WTX, and GFP was performed with the TnT® Quick coupled Transcription/Translation System (Promega). Five μl of each product was incubated in various combinations with recombinant ubiquitin (1.25 μg), UBE1 (12.5 μg), and UBCH5B (20 μg) in the presence of 2 mm ATP, 5 mm MgCl2. The mixtures were incubated at room temperature for 45 min. NRF2 was immunoprecipitated, and complexes were analyzed by Western blot for ubiquitin.
Immobilized Metal Affinity Chromatography and Mass Spectrometry
Affinity purification of GLUE-WTX and FLAG-KEAP1 was performed as described previously (6). The precipitated proteins were trypsinized directly off beads following reduction with 5 mm DTT and alkylation with 15 mm chloroacetamide. Following “stage-tip” desalting (20), peptides were resuspended in binding buffer (250 mm acetic acid and 30% acetonitrile) and incubated for 30 min with immobilized metal affinity chromatography gel (I1408, Sigma). The phosphorylated peptides were washed three times with binding buffer, eluted with 200 mm ammonium phosphate buffer, and processed for mass spectrometry. Raw mass spectrometry data were searched with SEQUEST (ThermoFisher), and phosphorylation was queried via specification of a differential modification of 79.6 atomic mass units on serine, threonine, or tyrosine. Proteins were scored using the Institute for Systems Biology trans-proteomic pipeline (21), and phosphosite accuracy was evaluated using Ascore (22).
AlphaScreen
Recombinant GST-NRF2 and HIS6-hKELCH were purified using standard techniques. Seventy ng of AlphaScreen GSH-Donor and AlphaLISA nickel-acceptor beads (PerkinElmer Life Sciences) were mixed with 100 nm HIS6-hKELCH, 20 nm GST-NRF2, and respective peptides in binding buffer containing 20 mm Tris, pH 8, 200 mm NaCl, 1 mm DTT, and 0.05% Tween 20. Reactions were incubated at room temperature for 30 min, and AlphaScreen Signal was determined with an Envision plate-reader (PerkinElmer Life Sciences). Peptide sequences are as follows: NRF2 16-mer, AFFAQLQLDEETGEFL; WTX 16-mer, ASSLEEPHSPETGEKV; and β-catenin 26-mer, KAAVSHWQQQSYLDSGIHSGATTTAP.
Statistical Analysis
Student's t test was used to assess the statistical significance of the differences between the different groups; a p value of <0.05 was considered significant.
RESULTS
WTX Directly Interacts with KEAP1
The WTX gene encodes an 1135-amino acid protein (WTX-WT) characterized by multiple protein-protein interaction domains and an N-terminal phosphatidylinositol 4,5-bisphosphate binding domain that mediates its localization to the plasma membrane (12). Through in-frame alternative splicing, a shorter isoform (WTX-S) of 858 amino acids is produced that lacks residues 50–326 and does not localize to the plasma membrane (13). Here, we employ an 804-amino acid variant of WTX, previously reported by us and by Grohmann et al. (12) that shares perfect identity with WTX-WT through residue 785. This isoform contains the N-terminal phosphatidylinositol 4,5-bisphosphate binding domain, as well as the binding domains for KEAP1, β-catenin, APC, AXIN1, BTRC, and FBXW11.
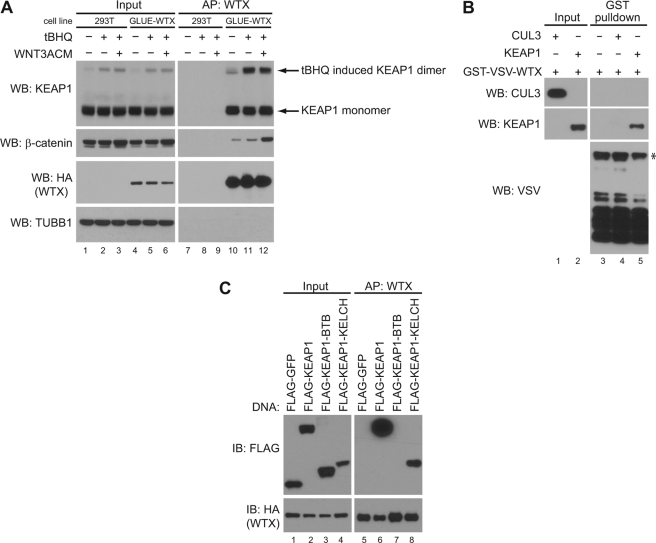
We previously observed that KEAP1 co-purifies with WTX protein complexes (6) as determined by mass spectrometry-based proteomics. To validate this interaction, we engineered HEK293T cells to express a fusion protein containing an N-terminal streptavidin-binding protein, calmodulin-binding protein, the hemagglutinin epitope (HA), and WTX (GLUE-WTX). Affinity purification of GLUE-WTX and Western blotting confirmed that endogenous KEAP1 forms a complex with GLUE-WTX (Fig. 1A, compare lane 10 with control lane 7). This interaction was independent of the activity of either the KEAP1/NRF2 or the WNT/β-catenin signaling pathways as neither the KEAP1 antagonist tBHQ (tert-butylhydroquinone) nor WNT3A conditioned media affected the interaction of WTX with KEAP1 (Fig. 1A, compare lanes 10, 11, and 12).
FIGURE 1.
WTX directly interacts with KEAP1. A, GLUE-WTX interacts with endogenous KEAP1. HEK293T parental or HEK293T cells stably expressing GLUE-WTX were treated with DMSO, 100 μm tBHQ, or 100 μm tBHQ and WNT3A conditioned media (WNT3ACM) for 2 h. Lysates were subjected to streptavidin affinity pulldown (AP) followed by Western blot (WB) analysis. B, WTX directly binds KEAP1. Recombinant GST-VSV-WTX was incubated with recombinant CUL3 or KEAP1. After GST affinity purification, complexes were washed, and associated proteins were resolved by Western blot. * denotes full-length GST-VSV-WTX. C, WTX interacts with the KELCH repeats of KEAP1. HEK293T cells stably expressing GLUE-WTX were transfected with FLAG-tagged KEAP1 constructs. Lysates were subjected to streptavidin affinity pulldown followed by Western blot analysis. IB, immunoblot.
To determine whether WTX directly interacts with KEAP1, we tested whether recombinant GST-WTX was able to pull down recombinant KEAP1 in vitro. Using this method, we determined that WTX directly interacts with KEAP1 but not with the KEAP1-associated CUL3 protein (Fig. 1B, compare lanes 4 and 5). Together with our previous work (6), these data demonstrate that WTX directly binds the substrate recognition modules of two different E3 ubiquitin ligases, namely BTRC and KEAP1.
KEAP1 has three well defined domains, an N-terminal BTB domain that binds CUL3 (14), an intervening region, and the C-terminal KELCH repeats that form a β-propeller fold that binds NRF2 (23). To determine the domain in KEAP1 that binds WTX, HEK293T cells expressing GLUE-WTX were transfected with FLAG-KEAP1, FLAG-BTB, or FLAG-KELCH constructs. Affinity purification of GLUE-WTX revealed an association with full-length KEAP1 and the KELCH repeats but not with the KEAP1 BTB domain (Fig. 1C, compare lanes 6, 7, and 8). Taken together, our data suggest that WTX directly associates with the KELCH repeats of KEAP1. Despite its association with the substrate-recognition domain of KEAP1, the steady-state levels of WTX are not regulated by KEAP1 or proteasome-mediated degradation (supplemental Fig. S1). Thus we conclude that WTX is not a substrate of KEAP1.
WTX Promotes NRF2-dependent Transcription
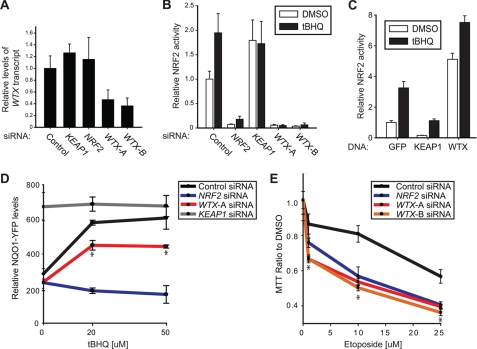
Based on the physical interaction between WTX and KEAP1, we next tested if WTX regulates NRF2 activity. First, we employed a luciferase reporter containing the ARE of NQO1, an NRF2 target gene (24). Two nonoverlapping WTX-specific siRNAs decreased both basal and tBHQ-induced activation of this ARE-luciferase reporter in HEK293T cells, similar to the effect of depleting NRF2 with siRNA (Fig. 2, A and B). Conversely, overexpression of WTX activated the luciferase reporter compared with control GFP (Fig. 2C). Second, because the engineered luciferase reporter may not accurately represent endogenous transcription, we tested whether WTX regulates the expression of endogenous NQO1. We utilized the H1299 non-small cell lung cancer cell line containing a YFP fragment retrovirally inserted into intron 1 of the endogenous NQO1 genomic locus resulting in an NQO1-YFP product (25). Induction of YFP in the presence of tBHQ was significantly lower in cells transfected with WTX siRNA compared with control (Fig. 2D). Taken together, these observations suggest that WTX is a positive regulator of NRF2-dependent transcription, unlike its role as a negative regulator of β-catenin-dependent transcription (6).
FIGURE 2.
WTX promotes NRF2-dependent transcription. A, two nonoverlapping siRNAs reduce WTX transcript levels. HEK293T cells were transfected with the indicated siRNAs. Total RNA was isolated 48 h after transfection, and relative transcript levels were quantified as the ratio compared with β-actin transcripts. B, WTX is required for activation of an ARE-luciferase reporter. HEK293T cells stably expressing an ARE-driven firefly-luciferase reporter as well as a control CMV-driven Renilla luciferase reporter were transfected with siRNAs and either DMSO or tBHQ. Relative levels of ARE-driven firefly luciferase compared with control CMV-driven Renilla luciferase are plotted. Error bars represent means ± S.D. of three technical replicates. Data are representative of three biological replicates. C, overexpression of WTX activates an ARE-luciferase reporter. HEK293T cells were transfected with the ARE-firefly luciferase reporter, a control CMV Renilla luciferase control reporter, and the indicated FLAG-tagged DNA constructs. Cells were treated with tBHQ, and luciferase levels were quantified as in B. D, loss of WTX inhibits transcription of endogenous NQO1. H1299 cells expressing an NQO1-YFP fusion protein under control of the endogenous promoter were transfected with the indicated siRNAs and treated with either DMSO or tBHQ. Relative YFP fluorescence is plotted. Error bars represent means ± S.D. of four technical replicates. Data are representative of three biological replicates. *, p < 0.02 when comparing WTX siRNA with control. E, loss of WTX increases the cellular sensitivity to etoposide. HEK293T cells were transfected with the indicated siRNAs and treated with etoposide. Cell viability was determined by measuring the metabolic activity of mitochondria (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay). *, p < 0.03 when comparing NRF2 and WTX siRNAs with control.
KEAP1 and NRF2 are known to play opposing roles in the cellular response to oxidative stress and chemotherapeutics, most notably in non-small cell lung cancer where constitutive activation of NRF2 promotes resistance to chemotherapeutics (26). To determine whether WTX plays a similar role, we asked if loss of WTX sensitizes cells to the chemotherapeutic reagent etoposide. Compared with control, cells transfected with siRNA targeting WTX were more sensitive to cell death in the presence of etoposide as determined by mitochondrial activity (Fig. 2E). We conclude from these experiments that WTX and NRF2 promote resistance to apoptotic inducing reagents.
WTX Regulates the Steady-state Levels of NRF2 by Inhibiting Its Ubiquitination
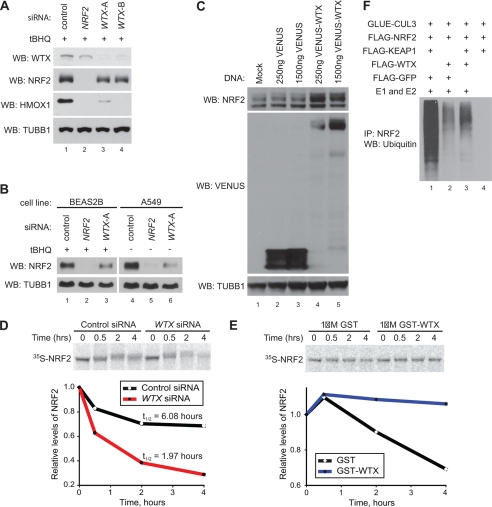
As WTX regulates the stability of β-catenin (6), we hypothesized that WTX also regulates the stability of NRF2. To test this, we transfected HEK293T cells with control, NRF2, or WTX siRNAs and monitored the levels of NRF2 and its target HMOX1 following treatment with tBHQ. siRNA-mediated silencing of WTX in HEK293T cells resulted in lower NRF2 and HMOX1 protein levels compared with control (Fig. 3A, compare lanes 3 and 4 with control lane 1). Similar results were also observed in cell lines derived from normal lung (BEAS2B) and lung carcinoma (A549) (Fig. 3B, compare lane 3 with control lane 1 and lane 6 with control lane 4). Of note, whereas HEK293T and BEAS2B cells have wild-type KEAP1, A549 cells harbor a G333C mutation in the KELCH repeats of KEAP1 that partially impairs the ubiquitination of NRF2, resulting in relatively high basal levels of NRF2. As a complement to the siRNA-based loss-of-function approach, we tested whether overexpression of WTX results in increased levels of NRF2 protein. In HEK293T cells transiently transfected with a VENUS-WTX expression construct, we observed increased levels of endogenous NRF2 (Fig. 3C, compare lane 4 with control lane 2, and lane 5 with control lane 3). Combined, these results suggest that WTX regulates the steady-state levels of NRF2.
FIGURE 3.
WTX regulates the steady-state levels of NRF2 by inhibiting its ubiquitination. A and B, knockdown of WTX inhibits stabilization of NRF2. A, HEK293T cells were transfected with the indicated siRNAs and treated with tBHQ (100 μm, 4 h) preceding lysis and Western blot (WB) analysis. B, BEAS2B and A549 cell lines were transfected with the indicated siRNAs. BEAS2B cells were treated with tBHQ preceding lysis and Western blot analysis. C, overexpression of WTX stabilizes NRF2. HEK293T cells were transfected with the indicated constructs and analyzed as in A and B. D and E, WTX regulates NRF2 steady-state levels. D, HEK293T cells were transfected with the indicated siRNAs and cellular extracts were purified. 35S-Radiolabeled NRF2 was added to cellular extracts, and degradation was monitored over time by autoradiography. A representative autoradiograph and quantification from three independent experiments is shown. E, 35S-NRF2 and recombinant GST or GST-WTX were added to HEK293T cellular extracts. 35S-NRF2 degradation was monitored and quantified as in E. F, in vitro transcribed and translated CUL3, NRF2, KEAP1, WTX, and GFP were mixed as indicated. NRF2 was immunoprecipitated (IP) from each reaction, and ubiquitination was observed by Western blot analysis.
We next asked if WTX regulates the steady-state levels of NRF2 by inhibiting the degradation of NRF2 protein. To test this, we measured the degradation of NRF2 in cell-free extracts generated from HEK293T cells. 35S-Labeled NRF2 was less stable in extracts purified from cells transfected with WTX siRNA compared with control extracts (Fig. 3D). Conversely, when extracts from HEK293T cells were supplemented with recombinant WTX, levels of 35S-labeled NRF2 were relatively stable compared with control, suggesting that WTX inhibits the degradation of NRF2 (Fig. 3E). Of note, although we observed that the half-life of exogenous NRF2 in cell extracts is ∼6 h, previous reports have demonstrated that exogenous and endogenous NRF2 has a half-life of ∼20 min in intact cells (15, 27). We also observed that NRF2 has a half-life of minutes in intact cells (supplemental Fig. S1B), and we attribute the differences in extracts to differences in experimental systems.
As WTX directly binds to KEAP1 and inhibits the degradation of NRF2, we hypothesized that WTX inhibits the ubiquitination of NRF2 by KEAP1. To test this hypothesis, we utilized an in vitro assay to monitor the ubiquitination of NRF2. In vitro transcribed/translated CUL3, KEAP1, and NRF2 were combined with either mock, GFP, or WTX and incubated with ubiquitination reagents. NRF2 was then immunoprecipitated, and ubiquitination was monitored by Western blot. Compared with control, WTX inhibited the ubiquitination of NRF2 (Fig. 3F, compare lane 3 with control lane 1). Taken together, these results indicate that WTX stabilizes NRF2 by inhibiting KEAP1-dependent ubiquitination.
WTX Interacts with KEAP1 through an ETGE Motif
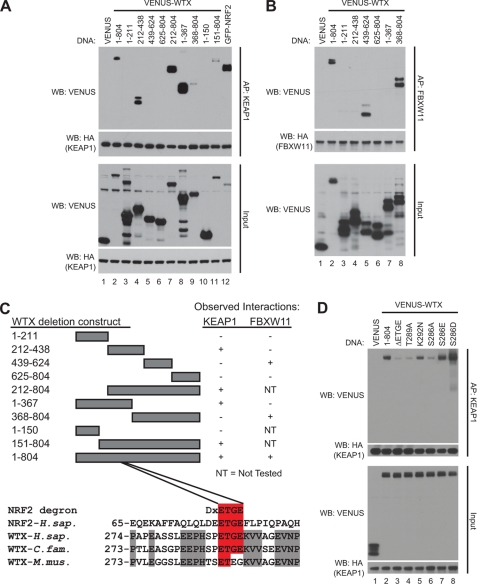
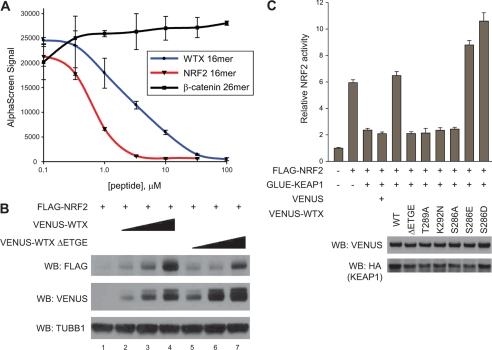
KEAP1 and members of the BTRC family (BTRC and FBXW11) are substrate-adaptor proteins that bind to CUL3- and CUL1-based E3 ubiquitin ligase complexes, respectively (9, 16). To gain insight into the mechanism(s) by which WTX regulates these complexes, we wanted to define the domain(s) within WTX that bind KEAP1 and the BTRC family (BTRC and FBXW11). Numerous fragments of the WTX protein were expressed as VENUS fusions in HEK293T cells engineered to express GLUE-KEAP1 or GLUE-FBXW11. Interestingly, affinity purification and Western blot revealed that WTX binds KEAP1 and FBXW11 through separable domains. We observed that WTX fragments containing amino acids 1–367 and 212–438 interacted with KEAP1 suggesting that amino acids 212–367 are required to bind KEAP1, whereas amino acids 439–624 are required to bind FBXW11 (Fig. 4, A and B, respectively; results are summarized in Fig. 4C).
FIGURE 4.
WTX interacts with KEAP1 through an ETGE motif. A and B, WTX associates with KEAP1 and FBXW11 through separable domains. A, HEK293T cells stably expressing GLUE-KEAP1 were transfected with the indicated VENUS-WTX deletion constructs. Lysates were subjected to streptavidin affinity pulldown (AP) followed by Western blot (WB) analysis. B, HEK293T cells stably expressing GLUE-FBXW11 were transfected with the indicated VENUS-WTX deletion constructs. Lysates were analyzed as in B. C, schematic representation of the observed interactions in Fig. 5, A and B. Below the schematic, conserved residues of WTX are indicated by gray shading. Conserved residues that are identical to the NRF2 degron are indicated by red shading. D, interaction between WTX and KEAP1 is mediated through an ETGE motif. The indicated VENUS-WTX mutation constructs were transfected into GLUE-KEAP1 cells. Complexes were purified and analyzed as in A and B.
Within the KEAP1 binding domain of WTX, our attention was drawn to amino acids 286–291, containing the sequence SPETGE (Fig. 4C). KEAP1 directly interacts with a conserved DXETGE motif within NRF2 (28), raising the possibility that WTX binds KEAP1 through its ETGE motif. To test this, we expressed a WTX construct lacking amino acids 288–291 (VENUS-WTX ΔETGE) in GLUE-KEAP1 cells. Western blot analysis of KEAP1 affinity-purified complexes revealed substantially less WTX ΔETGE compared with wild-type WTX (Fig. 4D, compare lanes 2 and 3). Threonine 80 within the ETGE motif of NRF2 makes critical contacts with KEAP1 and is required for interacting with KEAP1 (29). Similarly, we observed that mutation of threonine 289 to alanine in the ETGE motif of WTX disrupts the interaction with KEAP1 (Fig. 4D, compare lanes 2 and 4). Interestingly, lysine 292, adjacent to the ETGE motif, is mutated to asparagine in several reported Wilms tumor cases and a single instance of acute myeloid leukemia (1, 3, 30). Compared with wild-type WTX, KEAP1 pulled down less WTX K292N, although more than WTX ΔETGE (Fig. 4D, compare lanes 2, 3, and 5). Combined, these results suggest that WTX binds to KEAP1 through a similar motif as NRF2.
One notable difference between the KEAP1 interaction domains of NRF2 and WTX is the two amino acids upstream of their respective ETGE motifs. The human NRF2 motif contains a conserved aspartic acid that interacts with two water molecules that in turn hydrogen bond with two arginine residues in KEAP1 (29). In place of the aspartic acid, WTX contains a serine (residue 286). Interestingly, mutation of serine 286 to alanine inhibited the ability of KEAP1 to pull down WTX (Fig. 4D, compare lanes 2 and 6). Based on the crystal structure of the KELCH repeats and an NRF2 peptide (23, 29), we reasoned that mutation of serine 286 to an acidic residue would enhance the interaction between WTX and KEAP1. Although we only observed a modest increase in the interaction between KEAP1 and WTX S286E, WTX S286D was significantly enriched relative to wild-type WTX in KEAP1 pulldowns (Fig. 4D, compare lanes 2, 7, and 8). We conclude from these experiments that in addition to the ETGE motif, serine 286 is also important in the KEAP1 interacting domain of WTX.
Serine 286 Is Phosphorylated in Vivo
Given that mutation of serine 286 to alanine and aspartic acid differentially affected the ability of KEAP1 to pull down WTX, we hypothesized that phosphorylation of WTX could affect its interaction with KEAP1. To directly test whether WTX is phosphorylated in vivo, we trypsinized GLUE-WTX complexes from HEK293T cells and enriched for phosphorylated peptide fragments by immobilized metal affinity chromatography. Peptides were then analyzed for the presence of phosphorylation by mass spectrometry. We identified 16 unique phosphorylation sites in 16 unique mono- or di-phosphorylated peptides (Table 1 and supplemental Fig. S2). Of the 16 unique peptides, two were phosphorylated at serine 286, including one with a second phosphorylated site at serine 280. The phosphorylation at serine 286 is considered definitive by the Ascore phosphorylation site algorithm, where any score above 19 has a greater than 99% chance of certainty (22).
TABLE 1.
Phosphorylated WTX peptides identified by mass spectrometry
Asterisk indicates phosphorylated residue. NA means not applicable. The particular peptides in this table that are important for our overall findings are shown in boldface type.
| Unique peptide sequence | Peptide probability | Index | Ascore1 | Ascore2 |
|---|---|---|---|---|
| KENANPQDAPGPKVS*PTPEPSPPATEK | 0.99 | 240 | 32.48 | NA |
| VSPT*PEPSPPATEK | 0.99 | 242 | 6.46 | NA |
| VSPTPEPS*PPATEK | 0.99 | 246 | 33.57 | NA |
| KENANPQDAPGPKVS*PTPEPS*PPATEK | 0.99 | 240, 246 | 32.48 | 33.57 |
| VSPT*PEPS*PPATEK | 0.99 | 242, 246 | 6.46 | 33.57 |
| PAPEASS*LEEPHSPETGEK | 0.89 | 280 | 9.7 | NA |
| PAPEASSLEEPHS*PETGEK | 0.99 | 286 | 46.85 | NA |
| PAPEASS*LEEPHS*PETGEK | 0.91 | 280, 286 | 9.7 | 46.85 |
| T*SLKSFDSLTGCGDIIAEQDMDSMTDSMASGGQR | 0.99 | 317 | 0 | NA |
| FDS*LT*GCGDIIAEQDMDSMTDSMASGGQR | 0.29 | 324, 326 | 11.63 | 12.28 |
| PNMNLGYHPTT*SPGHHGYMLLDPVR | 0.99 | 429 | 0 | NA |
| PNMNLGYHPTTS*PGHHGY | 0.57 | 430 | 0 | NA |
| DSYSGDALYEFYEPDDSLENS*PPGDDCLY*DLH | 0.85 | 518, 526 | 0 | 0 |
| SSEMFDPFLNFEPFLSS*RPPGAMET*EEE | 0.80 | 548, 556 | 2.99 | 5.45 |
| ITS*AFPTTASSEPDWR | 0.21 | 683 | 0 | NA |
| RQVTQACGT*W | 0.99 | 803a | 69.97 | NA |
a Threonine 803 is absent in the full-length isoform of WTX(1–1135).
Finally, we wanted to determine whether endogenous WTX is phosphorylated on serine 286 when associated with KEAP1. Immobilized metal affinity chromatography-enriched phosphopeptides from FLAG-KEAP1 affinity-purified protein complexes were identified by mass spectrometry. Within FLAG-KEAP1 complexes, we identified six phosphorylation sites on endogenous WTX, including serine 286 (Table 2 and supplemental Fig. S3). Although these data do not rule out the possibility that unphosphorylated WTX interacts with KEAP1, they confirm that endogenous WTX is phosphorylated at serine 286 and interacts with KEAP1.
TABLE 2.
Phosphorylated WTX peptides associated with KEAP1
Asterisk indicates phosphorylated residue. NA means not applicable. The particular peptides in this table that are important for our overall findings are shown in boldface type.
| Unique peptide sequence | Peptide probability | Index | Ascore1 | Ascore2 |
|---|---|---|---|---|
| KENANPQDAPGPKVS*PTPEPS*PPATEK | 0.81 | 240, 246 | 1 | 7.23 |
| VSPT*PEPS*PPATEK | 0.99 | 242, 246 | 13.7 | 38.5 |
| VSPTPEPS*PPATEK | 0.99 | 246 | 20.22 | NA |
| PAPEASSLEEPHS*PETGEK | 0.99 | 286 | 7.79 | NA |
| AYPTYS*PPEDPEEEEVEK | 0.99 | 749 | 19.85 | NA |
| FYQGLPWGVSS*LPR | 0.99 | 868 | 7.53 | NA |
WTX Competes with NRF2 for Binding to KEAP1
Our observation that WTX interacts with KEAP1 through a similar motif as NRF2 raises the possibility that WTX and NRF2 compete for the same binding site in KEAP1. To directly test this, we utilized an AlphaScreen approach to ask if a WTX peptide containing the SPETGE sequence could inhibit the interaction between NRF2 and KEAP1. We observed a strong interaction between GST-NRF2 and HIS6-hKELCH (KELCH repeats of KEAP1 that directly interact with NRF2) as indicated by a robust AlphaScreen signal. The specificity of the interaction was validated as a peptide containing the DXETGE motif of NRF2 inhibited the AlphaScreen signal in a dose-dependent manner (Fig. 5A). Whereas a control β-catenin peptide had no effect, a peptide containing the SPETGE motif of WTX (WTX 16-mer) inhibited the interaction between NRF2 and the KELCH repeats of KEAP1 in a dose-dependent manner (Fig. 5A). These results suggest that WTX and NRF2 compete for binding to KEAP1.
FIGURE 5.
WTX competes with NRF2 for binding to KEAP1. A, WTX peptide containing the ETGE motif inhibits the interaction between KEAP1 and NRF2. A fixed concentration of GST-NRF2 and HIS6-hKELCH was mixed with increasing amounts of the indicated peptides in the presence of GSH donor and nickel acceptor AlphaScreen beads. The interaction between GST-NRF2 and HIS6-hKELCH is represented as AlphaScreen signal. Error bars represent means ± S.D. of three technical replicates. Data are representative of three biological replicates. B, ETGE motif is required to stabilize NRF2. HEK293T cells were transfected with the indicated expression constructs. Proteins were analyzed by Western blot (WB). C, ETGE motif is required to regulate NRF2 transcriptional activity. HEK293T cells in a 48-well tissue culture plate were transfected with 5 ng of an ARE-luciferase reporter, 10 ng of a control CMV Renilla luciferase control reporter, and the indicated DNA (2 ng of FLAG-NRF2, 10 ng of GLUE-KEAP1, and 10 ng of VENUS constructs). The relative levels of ARE-driven firefly luciferase compared with control CMV-driven Renilla luciferase are plotted. Error bars represent the means ± S.D. for three replicates. Data are representative of three independent experiments. Lysates were also analyzed by Western blot analysis for relative WTX and KEAP1 expression.
If WTX regulates NRF2 steady-state levels through direct competition with KEAP1, we would expect that mutations in WTX that inhibit its interaction with KEAP1 should impair its ability to regulate the steady-state levels of NRF2. To test this, we assessed the ability of VENUS-WTX ΔETGE to stabilize NRF2. Whereas overexpression of wild-type WTX robustly stabilized NRF2 in a dose-dependent manner, WTX ΔETGE only modestly stabilized NRF2 at significantly higher levels of expression (Fig. 5B, compare lane 4 with lanes 6 and 7). In addition to regulating the steady-state levels of NRF2, the interaction of WTX with KEAP1 should also regulate NRF2-dependent transcription. To test this, we examined whether the various WTX mutants could regulate NRF2-dependent transcription in the presence of exogenous KEAP1. Overexpression of FLAG-NRF2 activated an ARE-luciferase reporter and was partially inhibited by overexpression of KEAP1. Concomitant expression of the various WTX mutants rescued FLAG-NRF2 activity correlating with the ability of the WTX mutants to bind KEAP1 (Fig. 5C). We conclude from these experiments that the ability of WTX to regulate the steady-state levels of NRF2 and NRF2-dependent transcription directly correlates with its ability to bind KEAP1. Taken together, our data suggest that WTX regulates NRF2 steady-state levels and NRF2-dependent transcription by competing for binding to KEAP1.
DISCUSSION
The WTX gene encodes a tumor suppressor protein; it is located on the X chromosome and is somatically lost or mutated in 7–30% of cases of Wilms tumor (1–3). Germ line mutations in WTX give rise to OSCS, a debilitating and fatal disease that largely affects the skeletal system (4, 5). As the discovery of WTX and its mutations in human disease is relatively new, the molecular function(s) and developmental or homeostatic consequences of its loss is only beginning to be unraveled. Compelling data from numerous research groups have described functions for WTX in controlling WNT/β-catenin signaling, cell-cell adhesion, apoptosis, and transcription (1, 6, 11–13). Recently, characterization of Wtx deletion in mice revealed that Wtx regulates mesenchymal progenitor cell fate specification in part through β-catenin (31).
Here, we expand on the knowledge of the molecular mechanisms of action of WTX with the observation that WTX regulates the steady-state levels of NRF2 and NRF2-dependent transcription by competing with NRF2 for binding to KEAP1. KEAP1 forms a homodimer and interacts with CUL3 through its BTB domain (32, 33) and interacts with the N-terminal Neh2 domain of a single NRF2 molecule through its KELCH repeats. The Neh2 domain of NRF2 contains two highly conserved regions, one bearing an LXXQDXDLG (DLG) motif (34, 35) and the other bearing a DXETGE motif (28). These motifs interact with a separate KEAP1 molecule in the KEAP1 homodimer, although the DXETGE motif binds with ∼100-fold higher affinity (36). WTX contains a similar SPETGE motif that can directly inhibit the interaction between NRF2 and the KELCH repeats of KEAP1 in vitro. As WTX does not contain a DLG motif similar to NRF2, it is possible that NRF2 can still interact with a KEAP1 dimer through this low affinity interaction motif. We hypothesize that WTX enhances NRF2 steady-state levels by disrupting the conformation of the E3 ubiquitin ligase complex, resulting in lower ubiquitination of NRF2. In support of our findings, expression of the NRF2 target genes NQO1 and HMOX1 was significantly enhanced in HEK293T cells overexpressing a fragment of WTX containing the KEAP1 interaction domain (37).
In addition to the ETGE motif, we determined that serine 286 is also required for the ability of WTX to interact with KEAP1 and regulate NRF2. Interestingly, mutation of serine 286 to either glutamic or aspartic acid enhanced the functional effects of WTX. Although these mutations may only present conformational changes in WTX that make it more suitable for binding to KEAP1, both glutamic and aspartic acid resemble phosphorylated serine. Thus, phosphorylation of WTX at serine 286 may increase its affinity for KEAP1. Furthermore, we found that WTX is phosphorylated in vivo, and KEAP1 interacts with endogenous WTX that is phosphorylated at serine 286, raising the intriguing possibility that the interaction between WTX and KEAP1 is regulated by a yet to be identified kinase(s).
Under normal conditions, NRF2 is constitutively ubiquitinated through its association with KEAP1 (14–16). In the presence of oxidative stress, cysteine residues in KEAP1 are modified resulting in a conformational change that disrupts the ubiquitination of NRF2, and NRF2 accumulates in the nucleus where it regulates gene transcription (16, 38). In addition to our findings, several recent studies have also revealed that NRF2 signaling is regulated through protein-protein interactions. The cyclin-dependent kinase inhibitor p21, a p53-regulated gene with pro-survival properties, was recently shown to bind the DLG motif of NRF2 and inhibit its interaction with KEAP1, resulting in elevated NRF2 levels under both basal and chemically induced conditions (39). Another recent study identified p62 as a novel interactor of KEAP1 (40). During autophagy, p62 directs ubiquitinated proteins to degradation by the lysosome. By binding to KEAP1 through an ETGE-like motif similar to that of NRF2 and WTX, p62 targets KEAP1 for autophagic degradation, thus contributing to the stabilization of NRF2. Combined with WTX, these examples highlight the complex regulation of NRF2 degradation and its importance in both homeostasis and response to cytotoxic stress.
Whereas WTX inhibits the ubiquitination of NRF2, we previously reported that WTX promotes the ubiquitination of β-catenin by the SCFBTRC ubiquitin ligase complex. To our knowledge, WTX is the first described protein to interact with two E3 ubiquitin ligase adaptors and have opposite regulatory effects on their respective substrates. How is this possible? WTX interacts with the KELCH repeats that form the β-propeller fold of KEAP1 and likely inhibits the formation of a functional CUL3-KEAP1-NRF2 complex. Through a separable domain, WTX interacts with both BTRC and β-catenin, suggesting that it binds to an intact SCFBTRC-substrate complex (6). BTRC also employs a β-propeller fold for substrate capture (41), but it is unlikely that WTX interacts with the β-propeller fold of BTRC in a similar fashion as KEAP1 as this would inhibit the formation of a functional CUL1-SKP1-BTRC-β-catenin complex and result in elevated β-catenin levels. Interestingly, a recent study demonstrated that NRF2 is phosphorylated by GSK3β in the central Neh6 domain (DSGIS, residues 334–338), creating an SCFBTRC destruction motif similar to that of β-catenin (42). This phosphorylated form of NRF2 is recognized by BTRC. Additionally, KEAP1 and BTRC have been identified in the same complex by mass spectrometry (43). This raises the possibility that multiple E3 ubiquitin ligase complexes consisting of a KEAP1 homodimer, a BTRC homodimer, or a KEAP1/BTRC heterodimer regulate the ubiquitination of NRF2. As WTX interacts with BTRC and KEAP1 through separable domains, we hypothesize that WTX coordinates the adaptors in the E3 ubiquitin ligase complexes, resulting in variable ubiquitination of substrates.
Although most commonly lost through gene-encompassing deletions, a small percentage of WTX mutations identified in Wilms tumor yield single amino acid substitutions and truncated proteins. Conversely, the majority of WTX mutations identified in OSCS yield truncated proteins. Aligning the location of these mutations with the WTX protein interaction domains suggests relationships of these diseases with specific binding interfaces. The KEAP1 interaction domain lies N-terminal to the domains that bind β-catenin/BTRC/APC and WT1 and remains intact in 9 out of the 20 reported mutation products. Of note, we determined that the K292N substitution inhibited the association of WTX and KEAP1. The β-catenin/BTRC/APC interacting domains encompass residues 280–839. Of the 20 reported mutations, 11 are predicted to alter these binding activities. As the C terminus of WTX binds WT1, this interaction is lost in all truncation products derived from mutations in WTX. These correlations suggest that whereas WT1 is likely central to WTX-associated diseases, the WTX-KEAP1 and WTX-β-catenin/BTRC/APC functional relationships may contribute to a subset of Wilms tumor and OSCS, perhaps accounting for variability in disease onset or progression.
The observation that WTX regulates the NRF2-mediated antioxidant response supports further study of its role in disease beyond Wilms tumor and OSCS. One of the best-studied diseases with altered KEAP1/NRF2 activity is non-small cell lung cancer, where mutations in KEAP1 lead to constitutive NRF2-mediated transcription. Consequently, cultured lung cancer cell lines with constitutive NRF2 activity are resistant to cell death induced by etoposide (26). Similarly, silencing NRF2 restores resistance to chemotherapeutics in KEAP1 mutant cells (44). We find that silencing WTX sensitizes HEK293T cells to death induced by etoposide, similar to loss of NRF2. Interestingly, although WTX expression is diminished in adult mouse brain and kidney compared with embryonic tissues, expression levels remain high in the lung, suggesting it may contribute to lung cell homeostasis in the adult (1). Coupled with the established roles of WTX in Wilms tumor and KEAP1/NRF2 in lung cancer, our data support future investigations into a functional role for WTX in lung cancer and other diseases associated with aberrant NRF2 activity.
Supplementary Material
Acknowledgments
We thank Priscila Siesser for unpublished data, Jeffrey Johnson and Seth Goldenberg for reagents, and other members of the Moon laboratory and Major laboratory for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant T32 GM07270 from USPHS and NRSA (to N. D. C.).

This article contains supplemental Figs. S1–S3.
- OSCS
- osteopathia striata congenital with cranial sclerosis
- APC
- adenomatous polyposis coli
- ARE
- antioxidant-response element
- BTRC
- β-transducin repeat containing
- tBHQ
- tert-butylhydroquinone.
REFERENCES
- 1. Rivera M. N., Kim W. J., Wells J., Driscoll D. R., Brannigan B. W., Han M., Kim J. C., Feinberg A. P., Gerald W. L., Vargas S. O., Chin L., Iafrate A. J., Bell D. W., Haber D. A. (2007) An X chromosome gene, WTX, is commonly inactivated in Wilms tumor. Science 315, 642–645 [DOI] [PubMed] [Google Scholar]
- 2. Perotti D., Gamba B., Sardella M., Spreafico F., Terenziani M., Collini P., Pession A., Nantron M., Fossati-Bellani F., Radice P. (2008) Functional inactivation of the WTX gene is not a frequent event in Wilms' tumors. Oncogene 27, 4625–4632 [DOI] [PubMed] [Google Scholar]
- 3. Ruteshouser E. C., Robinson S. M., Huff V. (2008) Wilms tumor genetics. Mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer 47, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Perdu B., de Freitas F., Frints S. G., Schouten M., Schrander-Stumpel C., Barbosa M., Pinto-Basto J., Reis-Lima M., de Vernejoul M. C., Becker K., Freckmann M. L., Keymolen K., Haan E., Savarirayan R., Koenig R., Zabel B., Vanhoenacker F. M., Van Hul W. (2009) Osteopathia striata with cranial sclerosis owing to WTX gene defect. J. Bone Miner. Res. 25, 82–90 [DOI] [PubMed] [Google Scholar]
- 5. Jenkins Z. A., van Kogelenberg M., Morgan T., Jeffs A., Fukuzawa R., Pearl E., Thaller C., Hing A. V., Porteous M. E., Garcia-Miñaur S., Bohring A., Lacombe D., Stewart F., Fiskerstrand T., Bindoff L., Berland S., Adès L. C., Tchan M., David A., Wilson L. C., Hennekam R. C., Donnai D., Mansour S., Cormier-Daire V., Robertson S. P. (2009) Germ line mutations in WTX cause a sclerosing skeletal dysplasia but do not predispose to tumorigenesis. Nat. Genet. 41, 95–100 [DOI] [PubMed] [Google Scholar]
- 6. Major M. B., Camp N. D., Berndt J. D., Yi X., Goldenberg S. J., Hubbert C., Biechele T. L., Gingras A. C., Zheng N., Maccoss M. J., Angers S., Moon R. T. (2007) Wilms tumor suppressor WTX negatively regulates WNT/β-catenin signaling. Science 316, 1043–1046 [DOI] [PubMed] [Google Scholar]
- 7. van Amerongen R., Mikels A., Nusse R. (2008) Alternative wnt signaling is initiated by distinct receptors. Sci. Signal. 1, re9. [DOI] [PubMed] [Google Scholar]
- 8. Yost C., Torres M., Miller J. R., Huang E., Kimelman D., Moon R. T. (1996) The axis-inducing activity, stability, and subcellular distribution of β-catenin is regulated in Xenopus embryos by glycogen synthase kinase 3. Genes Dev. 10, 1443–1454 [DOI] [PubMed] [Google Scholar]
- 9. Winston J. T., Strack P., Beer-Romero P., Chu C. Y., Elledge S. J., Harper J. W. (1999) The SCFβ-TRCP-ubiquitin ligase complex associates specifically with phosphorylated destruction motifs in IκBα and β-catenin and stimulates IκBα ubiquitination in vitro. Genes Dev. 13, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu C., Kato Y., Zhang Z., Do V. M., Yankner B. A., He X. (1999) β-Trcp couples β-catenin phosphorylation-degradation and regulates Xenopus axis formation. Proc. Natl. Acad. Sci. U.S.A. 96, 6273–6278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tanneberger K., Pfister A. S., Brauburger K., Schneikert J., Hadjihannas M. V., Kriz V., Schulte G., Bryja V., Behrens J. (2011) Amer1/WTX couples Wnt-induced formation of PtdIns(4,5)P2 to LRP6 phosphorylation. EMBO J. 30, 1433–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Grohmann A., Tanneberger K., Alzner A., Schneikert J., Behrens J. (2007) AMER1 regulates the distribution of the tumor suppressor APC between microtubules and the plasma membrane. J. Cell Sci. 120, 3738–3747 [DOI] [PubMed] [Google Scholar]
- 13. Rivera M. N., Kim W. J., Wells J., Stone A., Burger A., Coffman E. J., Zhang J., Haber D. A. (2009) The tumor suppressor WTX shuttles to the nucleus and modulates WT1 activity. Proc. Natl. Acad. Sci. U.S.A. 106, 8338–8343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cullinan S. B., Gordan J. D., Jin J., Harper J. W., Diehl J. A. (2004) The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase. Oxidative stress sensing by a Cul3-Keap1 ligase. Mol. Cell. Biol. 24, 8477–8486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kobayashi A., Kang M. I., Okawa H., Ohtsuji M., Zenke Y., Chiba T., Igarashi K., Yamamoto M. (2004) Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol. Cell. Biol. 24, 7130–7139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang D. D., Lo S. C., Cross J. V., Templeton D. J., Hannink M. (2004) Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol. Cell. Biol. 24, 10941–10953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kensler T. W., Wakabayashi N., Biswal S. (2007) Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu. Rev. Pharmacol. Toxicol. 47, 89–116 [DOI] [PubMed] [Google Scholar]
- 18. Hayes J. D., McMahon M. (2009) NRF2 and KEAP1 mutations. Permanent activation of an adaptive response in cancer. Trends Biochem. Sci. 34, 176–188 [DOI] [PubMed] [Google Scholar]
- 19. Venugopal R., Jaiswal A. K. (1996) Nrf1 and Nrf2 positively and c-Fos and Fra1 negatively regulate the human antioxidant response element-mediated expression of NAD(P)H:quinone oxidoreductase1 gene. Proc. Natl. Acad. Sci. U.S.A. 93, 14960–14965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rappsilber J., Mann M., Ishihama Y. (2007) Protocol for micro-purification, enrichment, pre-fractionation, and storage of peptides for proteomics using StageTips. Nat. Protoc. 2, 1896–1906 [DOI] [PubMed] [Google Scholar]
- 21. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 22. Beausoleil S. A., Villén J., Gerber S. A., Rush J., Gygi S. P. (2006) A probability-based approach for high throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 24, 1285–1292 [DOI] [PubMed] [Google Scholar]
- 23. Padmanabhan B., Tong K. I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M. I., Kobayashi A., Yokoyama S., Yamamoto M. (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700 [DOI] [PubMed] [Google Scholar]
- 24. Lee J. M., Hanson J. M., Chu W. A., Johnson J. A. (2001) Phosphatidylinositol 3-kinase, not extracellular signal-regulated kinase, regulates activation of the antioxidant-responsive element in IMR-32 human neuroblastoma cells. J. Biol. Chem. 276, 20011–20016 [DOI] [PubMed] [Google Scholar]
- 25. Cohen A. A., Geva-Zatorsky N., Eden E., Frenkel-Morgenstern M., Issaeva I., Sigal A., Milo R., Cohen-Saidon C., Liron Y., Kam Z., Cohen L., Danon T., Perzov N., Alon U. (2008) Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 [DOI] [PubMed] [Google Scholar]
- 26. Singh A., Misra V., Thimmulappa R. K., Lee H., Ames S., Hoque M. O., Herman J. G., Baylin S. B., Sidransky D., Gabrielson E., Brock M. V., Biswal S. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small cell lung cancer. PLoS Med. 3, e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen T., Sherratt P. J., Huang H. C., Yang C. S., Pickett C. B. (2003) Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J. Biol. Chem. 278, 4536–4541 [DOI] [PubMed] [Google Scholar]
- 28. Kobayashi M., Itoh K., Suzuki T., Osanai H., Nishikawa K., Katoh Y., Takagi Y., Yamamoto M. (2002) Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells 7, 807–820 [DOI] [PubMed] [Google Scholar]
- 29. Lo S. C., Li X., Henzl M. T., Beamer L. J., Hannink M. (2006) Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J. 25, 3605–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Owen C., Virappane P., Alikian M., Stasevich I., Summers K., Lillington D., Bonnet D., Burnett A., Mills K., Lister T. A., Fitzgibbon J. (2008) WTX is rarely mutated in acute myeloid leukemia. Haematologica 93, 947–948 [DOI] [PubMed] [Google Scholar]
- 31. Moisan A., Rivera M. N., Lotinun S., Akhavanfard S., Coffman E. J., Cook E. B., Stoykova S., Mukherjee S., Schoonmaker J. A., Burger A., Kim W. J., Kronenberg H. M., Baron R., Haber D. A., Bardeesy N. (2011) The WTX tumor suppressor regulates mesenchymal progenitor cell fate specification. Dev. Cell 20, 583–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ogura T., Tong K. I., Mio K., Maruyama Y., Kurokawa H., Sato C., Yamamoto M. (2010) Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening double glycine repeat and C-terminal domains. Proc. Natl. Acad. Sci. U.S.A. 107, 2842–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zipper L. M., Mulcahy R. T. (2002) The Keap1 BTB/POZ dimerization function is required to sequester Nrf2 in cytoplasm. J. Biol. Chem. 277, 36544–36552 [DOI] [PubMed] [Google Scholar]
- 34. McMahon M., Thomas N., Itoh K., Yamamoto M., Hayes J. D. (2004) Redox-regulated turnover of Nrf2 is determined by at least two separate protein domains, the redox-sensitive Neh2 degron and the redox-insensitive Neh6 degron. J. Biol. Chem. 279, 31556–31567 [DOI] [PubMed] [Google Scholar]
- 35. Katoh Y., Iida K., Kang M. I., Kobayashi A., Mizukami M., Tong K. I., McMahon M., Hayes J. D., Itoh K., Yamamoto M. (2005) Evolutionary conserved N-terminal domain of Nrf2 is essential for the Keap1-mediated degradation of the protein by proteasome. Arch. Biochem. Biophys. 433, 342–350 [DOI] [PubMed] [Google Scholar]
- 36. Tong K. I., Katoh Y., Kusunoki H., Itoh K., Tanaka T., Yamamoto M. (2006) Keap1 recruits Neh2 through binding to ETGE and DLG motifs. Characterization of the two-site molecular recognition model. Mol. Cell. Biol. 26, 2887–2900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim M. K., Min D. J., Rabin M., Licht J. D. (2011) Functional characterization of Wilms tumor-suppressor WTX and tumor-associated mutants. Oncogene 30, 832–842 [DOI] [PubMed] [Google Scholar]
- 38. Yamamoto T., Suzuki T., Kobayashi A., Wakabayashi J., Maher J., Motohashi H., Yamamoto M. (2008) Physiological significance of reactive cysteine residues of Keap1 in determining Nrf2 activity. Mol. Cell. Biol. 28, 2758–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chen W., Sun Z., Wang X. J., Jiang T., Huang Z., Fang D., Zhang D. D. (2009) Direct interaction between Nrf2 and p21(Cip1/WAF1) up-regulates the Nrf2-mediated antioxidant response. Mol. Cell 34, 663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y. S., Ueno I., Sakamoto A., Tong K. I., Kim M., Nishito Y., Iemura S., Natsume T., Ueno T., Kominami E., Motohashi H., Tanaka K., Yamamoto M. (2010) The selective autophagy substrate p62 activates the stress-responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 12, 213–223 [DOI] [PubMed] [Google Scholar]
- 41. Wu G., Xu G., Schulman B. A., Jeffrey P. D., Harper J. W., Pavletich N. P. (2003) Structure of a β-TrCP1-Skp1-β-catenin complex. Destruction motif binding and lysine specificity of the SCF(β-TrCP1) ubiquitin ligase. Mol. Cell 11, 1445–1456 [DOI] [PubMed] [Google Scholar]
- 42. Rada P., Rojo A. I., Chowdhry S., McMahon M., Hayes J. D., Cuadrado A. (2011) SCF/β-TrCP promotes glycogen synthase kinase 3-dependent degradation of the Nrf2 transcription factor in a Keap1-independent manner. Mol. Cell. Biol. 31, 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Defining the human deubiquitinating enzyme interaction landscape. Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Homma S., Ishii Y., Morishima Y., Yamadori T., Matsuno Y., Haraguchi N., Kikuchi N., Satoh H., Sakamoto T., Hizawa N., Itoh K., Yamamoto M. (2009) Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin. Cancer Res. 15, 3423–3432 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.