Abstract
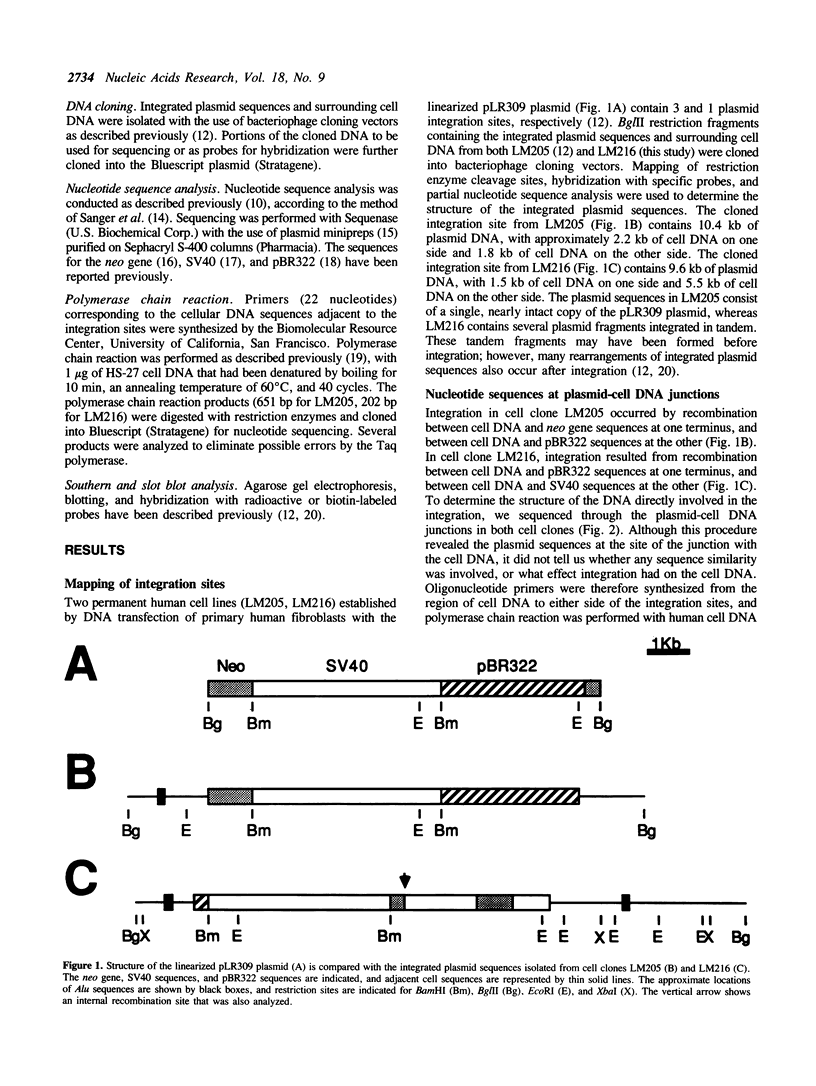
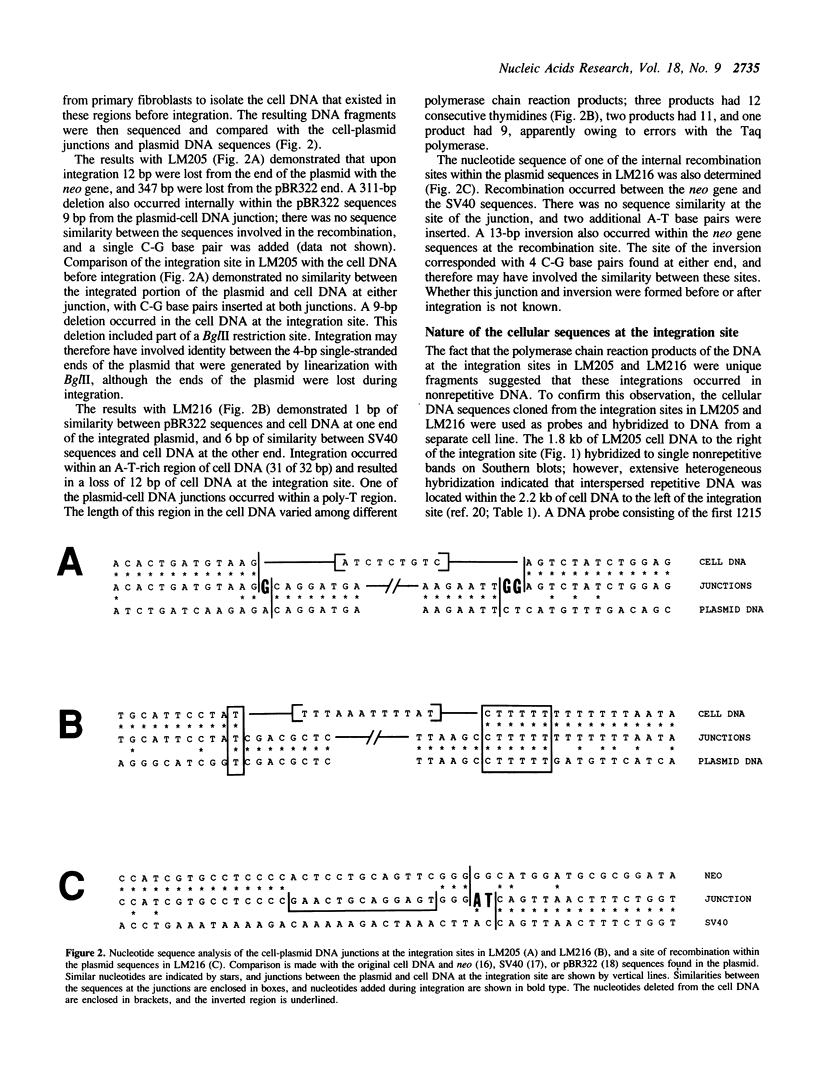
The mechanisms of recombination responsible for random integration of transfected DNA into the genome of normal human cells have been investigated by analysis of plasmid-cell DNA junctions. Cell clones containing integrated plasmid sequences were selected by morphological transformation of primary human fibroblasts after transfection with a plasmid containing simian virus 40 sequences. Nucleotide sequence analysis of the plasmid-cell DNA junctions was performed on cloned DNA fragments containing the integration sites from two of these cell clones. Polymerase chain reaction was then performed with human cell DNA from primary fibroblasts to isolate the cell DNA from the same sites before plasmid integration. Comparison of the sequences at the plasmid-cell DNA junctions with those of both the original plasmid and the cell DNA demonstrated short sequence similarities and additional nucleotides, typical of nonhomologous recombination. Evidence of short deletions in the cell DNA at the plasmid integration sites suggests that integration occurred by a mechanism similar to that used for repair of spontaneous or gamma ray-induced strand breaks. Plasmid integration occurred within nonrepetitive cell DNA with no major rearrangements, although rearrangements of the cell DNA at the integration site occurred in one of the clones after integration.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breimer L. H., Nalbantoglu J., Meuth M. Structure and sequence of mutations induced by ionizing radiation at selectable loci in Chinese hamster ovary cells. J Mol Biol. 1986 Dec 5;192(3):669–674. doi: 10.1016/0022-2836(86)90284-6. [DOI] [PubMed] [Google Scholar]
- Butner K. A., Lo C. W. High frequency DNA rearrangements associated with mouse centromeric satellite DNA. J Mol Biol. 1986 Feb 20;187(4):547–556. doi: 10.1016/0022-2836(86)90333-5. [DOI] [PubMed] [Google Scholar]
- Colbère-Garapin F., Ryhiner M. L., Stephany I., Kourilsky P., Garapin A. C. Patterns of integration of exogenous DNA sequences transfected into mammalian cells of primate and rodent origin. Gene. 1986;50(1-3):279–288. doi: 10.1016/0378-1119(86)90332-x. [DOI] [PubMed] [Google Scholar]
- Gahlmann R., Doerfler W. Integration of viral DNA into the genome of the adenovirus type 2-transformed hamster cell line HE5 without loss or alteration of cellular nucleotides. Nucleic Acids Res. 1983 Nov 11;11(21):7347–7361. doi: 10.1093/nar/11.21.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heartlein M. W., Knoll J. H., Latt S. A. Chromosome instability associated with human alphoid DNA transfected into the Chinese hamster genome. Mol Cell Biol. 1988 Sep;8(9):3611–3618. doi: 10.1128/mcb.8.9.3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeijmakers J. H., Odijk H., Westerveld A. Differences between rodent and human cell lines in the amount of integrated DNA after transfection. Exp Cell Res. 1987 Mar;169(1):111–119. doi: 10.1016/0014-4827(87)90230-8. [DOI] [PubMed] [Google Scholar]
- Hsiung N., Warrick H., deRiel J. K., Tuan D., Forget B. G., Skoultchi A., Kucherlapati R. Cotransfer of circular and linear prokaryotic and eukaryotic DNA sequences into mouse cells. Proc Natl Acad Sci U S A. 1980 Aug;77(8):4852–4856. doi: 10.1073/pnas.77.8.4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M., Berg P. Homologous integration in mammalian cells without target gene selection. Genes Dev. 1988 Nov;2(11):1353–1363. doi: 10.1101/gad.2.11.1353. [DOI] [PubMed] [Google Scholar]
- Kato S., Anderson R. A., Camerini-Otero R. D. Foreign DNA introduced by calcium phosphate is integrated into repetitive DNA elements of the mouse L cell genome. Mol Cell Biol. 1986 May;6(5):1787–1795. doi: 10.1128/mcb.6.5.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour S. L., Thomas K. R., Capecchi M. R. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988 Nov 24;336(6197):348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Murnane J. P. Inducible gene expression by DNA rearrangements in human cells. Mol Cell Biol. 1986 Feb;6(2):549–558. doi: 10.1128/mcb.6.2.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnane J. P., Yezzi M. J. Association of high rate of recombination with amplification of dominant selectable gene in human cells. Somat Cell Mol Genet. 1988 May;14(3):273–286. doi: 10.1007/BF01534588. [DOI] [PubMed] [Google Scholar]
- Murnane J. P., Young B. R. Nucleotide sequence analysis of novel junctions near an unstable integrated plasmid in human cells. Gene. 1989 Dec 7;84(1):201–205. doi: 10.1016/0378-1119(89)90157-1. [DOI] [PubMed] [Google Scholar]
- Nalbantoglu J., Hartley D., Phear G., Tear G., Meuth M. Spontaneous deletion formation at the aprt locus of hamster cells: the presence of short sequence homologies and dyad symmetries at deletion termini. EMBO J. 1986 Jun;5(6):1199–1204. doi: 10.1002/j.1460-2075.1986.tb04347.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbantoglu J., Miles C., Meuth M. Insertion of unique and repetitive DNA fragments into the aprt locus of hamster cells. J Mol Biol. 1988 Apr 5;200(3):449–459. doi: 10.1016/0022-2836(88)90535-9. [DOI] [PubMed] [Google Scholar]
- Pellicer A., Wigler M., Axel R., Silverstein S. The transfer and stable integration of the HSV thymidine kinase gene into mouse cells. Cell. 1978 May;14(1):133–141. doi: 10.1016/0092-8674(78)90308-2. [DOI] [PubMed] [Google Scholar]
- Perucho M., Hanahan D., Wigler M. Genetic and physical linkage of exogenous sequences in transformed cells. Cell. 1980 Nov;22(1 Pt 1):309–317. doi: 10.1016/0092-8674(80)90178-6. [DOI] [PubMed] [Google Scholar]
- Pfeiffer P., Vielmetter W. Joining of nonhomologous DNA double strand breaks in vitro. Nucleic Acids Res. 1988 Feb 11;16(3):907–924. doi: 10.1093/nar/16.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy V. B., Thimmappaya B., Dhar R., Subramanian K. N., Zain B. S., Pan J., Ghosh P. K., Celma M. L., Weissman S. M. The genome of simian virus 40. Science. 1978 May 5;200(4341):494–502. doi: 10.1126/science.205947. [DOI] [PubMed] [Google Scholar]
- Robins D. M., Ripley S., Henderson A. S., Axel R. Transforming DNA integrates into the host chromosome. Cell. 1981 Jan;23(1):29–39. doi: 10.1016/0092-8674(81)90267-1. [DOI] [PubMed] [Google Scholar]
- Rogers J. H. The origin and evolution of retroposons. Int Rev Cytol. 1985;93:187–279. doi: 10.1016/s0074-7696(08)61375-3. [DOI] [PubMed] [Google Scholar]
- Roth D. B., Chang X. B., Wilson J. H. Comparison of filler DNA at immune, nonimmune, and oncogenic rearrangements suggests multiple mechanisms of formation. Mol Cell Biol. 1989 Jul;9(7):3049–3057. doi: 10.1128/mcb.9.7.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth D. B., Wilson J. H. Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol. 1986 Dec;6(12):4295–4304. doi: 10.1128/mcb.6.12.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sager R., Gadi I. K., Stephens L., Grabowy C. T. Gene amplification: an example of accelerated evolution in tumorigenic cells. Proc Natl Acad Sci U S A. 1985 Oct;82(20):7015–7019. doi: 10.1073/pnas.82.20.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Greene R., Stringer J., Mitchison T., Hu S. L., Botchan M. Analysis of the sites of integration of viral DNA sequences in rat cells transformed by adenovirus 2 or SV40. Cold Spring Harb Symp Quant Biol. 1980;44(Pt 1):569–584. doi: 10.1101/sqb.1980.044.01.059. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small M. B., Gluzman Y., Ozer H. L. Enhanced transformation of human fibroblasts by origin-defective simian virus 40. Nature. 1982 Apr 15;296(5858):671–672. doi: 10.1038/296671a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]