Abstract
Background
Nucleosome translocation along DNA is catalyzed by eukaryotic SNF2-type ATPases. One class of SNF2-ATPases is distinguished by the presence of a C-terminal bromodomain and is conserved from yeast to man and plants. This class of SNF2 enzymes forms rather large protein complexes that are collectively called SWI/SNF complexes. They are involved in transcription and DNA repair. Two broad types of SWI/SNF complexes have been reported in the literature; PBAF and BAF. These are distinguished by the inclusion or not of polybromo and several ARID subunits. Here we investigated human SS18, a protein that is conserved in plants and animals. SS18 is a putative SWI/SNF subunit which has been implicated in the etiology of synovial sarcomas by virtue of being a target for oncogenic chromosomal translocations that underlie synovial sarcomas.
Methodology/Principal Findings
We pursued a proteomic approach whereby the SS18 open reading frame was fused to a tandem affinity purification tag and expressed in amenable human cells. The fusion permitted efficient and exclusive purification of so-called BAF-type SWI/SNF complexes which bear ARID1A/BAF250a or ARID1B/BAF250b subunits. This demonstrates that SS18 is a BAF subtype-specific SWI/SNF complex subunit. The same result was obtained when using the SS18-SSX1 oncogenic translocation product. Furthermore, SS18L1, DPF1, DPF2, DPF3, BRD9, BCL7A, BCL7B and BCL7C were identified. ‘Complex walking’ showed that they all co-purify with each other, defining human BAF-type complexes. By contrast,we demonstrate that human PHF10 is part of the PBAF complex, which harbors both ARID2/BAF200 and polybromo/BAF180 subunits, but not SS18 and nor the above BAF-specific subunits.
Conclusions/Significance
SWI/SNF complexes are found in most eukaryotes and in the course of evolution new SWI/SNF subunits appeared. SS18 is found in plants as well as animals. Our results suggest that in both protostome and deuterostome animals, a class of BAF-type SWI/SNF complexes will be found that harbor SS18 or its paralogs, along with ARID1, DPF and BCL7 paralogs. Those BAF complexes are proteomically distinct from the eukaryote-wide PBAF-type SWI/SNF complexes. Finally, our results suggests that the human bromodomain factors BRD7 and BRD9 associate with PBAF and BAF, respectively.
Introduction
Gene expression programs determine cell identity and response to endocrine stimuli, as has been demonstrated most dramatically by the generation of induced pluripotent stem cells with the Oct4, Sox2, Klf4 and c-Myc transcription factors [1]. Such epigenetic programming involves many nucleosome remodeling activities [2]. Besides covalent nucleosome modifications such as acetylation and methylation, a second type of nucleosome remodelling involves translocation of nucleosomes along chromosomal DNA [3]–[5] as well as the catalysis of alternative nucleosome conformations, and even nucleosome eviction [6]–[8]. These nucleosome transactions are catalyzed by SNF2 type enzymes, a group of ATPases that belongs to the SFII ATPase superfamily that includes many helicases [9]. The present paper is concerned with a subtype of the nucleosome remodeling SNF2 enzymes that are uniquely characterized by a C-terminal bromodomain, represented in yeast by Snf2 and Sth1, in Drosophila by brahma and in humans by BRM and BRG1.
The C-terminal bromo domain-bearing SNF2 enzymes are found in so-called SWI/SNF multiprotein complexes and are conserved in most eukaryotes. They are implicated in transcriptional regulation and multiple DNA repair pathways [10]–[25]. These large multi-protein complexes consist of at least 4 evolutionarily conserved core subunits represented in man by SMARCB1 and the SMARCA2/A4, SMARCC1/C2 and SMARCD1/D2/D3 paralogs [26], and a large number of ancillary subunits, some of which define SWI/SNF complex subtypes. Interestingly, SWI/SNF complexes were identified as biochemical factors that dramatically reduce the amount of time required to reprogram mouse embryonic fibroblasts into iPS at the hand of recombinant transcription factors [27], underscoring the importance of SWI/SNF in epigenetic programming processes [28]. Indeed, SWI/SNF has been mapped to some 50,000 human chromosomal sites in one cultured human cell line, demonstrating that this protein complex is a feature of many cis-acting regulatory elements, including DNA replication origins [29].
In mice and humans, at least 20 different SWI/SNF complex subunits have been reported (Table 1) [10], [30]–[43]. ‘Core’ subunits are found in virtually all cellular SWI/SNF complexes, whilst others define SWI/SNF complex subtypes. There are two broad classes of SWI/SNF complexes known; BAF-type SWI/SNF complexes (BRG1/BRM-associated factors) bear either one of ARID1A/BAF250a or ARID1B/BAF250b, whilst PBAF (Polybromo-associated BAF) complexes harbor both ARID2/BAF200 and polybromo/BAF180 subunits [38], [39], [44], [45]. Functionally, ARID1B/BAF250b was shown to be required to maintain ES cell identity [46] whilst ARID1A/BAF250a is required to permit proper ES cell differentiation with retinoic acid [47]. Furthermore, the SMARCC variant BAF170 is expressed less upon ES cell differentiation [43], [48], [49]. Similarly, the switch from one actin related subunit, BAF53A, to its paralog BAF53B appears to play a key role in neuron progenitor differentiation [41], [50]. Tissue specific expression of paralogous subunits has also been reported for the BAF60 variants [32], [51]–[53], as well as for the minor SWI/SNF subunits DPF1 and 3 [41], [42], [50].
Table 1. Abundance of purified proteins‡ in each TAP-tag preparation.
| Protein | alternative names | polyA mRNA* | TAP | INI1 | SS18 | SS18SSX1 | BCL7A | BCL7C | DPF2 | BRD9 | PHF10 |
| BRG1 | SMARCA4 | 5.93 | 0 | 0.303 | 5.337 | 4.440 | 2.039 | 2.290 | 1.662 | 0.141 | 0.984 |
| BRM | SMARCA2 | 1.45 | 0 | 0 | 1.339 | 0.730 | 0.116 | 0.280 | 0.179 | 0 | 0.028 |
| BAF250A | SMARCF1, ARID1A | 1.63 | 0 | 0.027 | 5.692 | 2.079 | 0.379 | 1.766 | 1.485 | 0.027 | 0.027 |
| BAF250B | ARID1B, OSA1 | 2.04 | 0 | 0 | 2.728 | 1.540 | 0.179 | 0.638 | 0.315 | 0.028 | 0 |
| BAF200 | ARID2, zipzap | nd | 0 | 0.122 | 0 | 0.029 | 0.029 | 0.122 | 0 | 0 | 0.884 |
| BAF180 | Polybromo-1 | 4.36 | 0 | 0.457 | 0 | 0 | 0 | 0.248 | 0 | 0 | 1.769 |
| BAF170 | SMARCC2 | 9.25 | 0 | 1.532 | 2.793 | 2.360 | 0.438 | 1.432 | 1.154 | 0.084 | 1.745 |
| BAF155 | SMARCC1 | 6.94 | 0 | 2.981 | 5.813 | 3.467 | 1.239 | 2.043 | 2.831 | 0.080 | 0.468 |
| BAF60A | SMARCD1 | 4.01 | 0 | 0.15 | 3.037 | 3.037 | 0.784 | 2.054 | 1.477 | 0.072 | 0.630 |
| BAF60B | SMARCD2 | 7.69 | 0 | 0.719 | 7.161 | 4.080 | 1.762 | 2.875 | 2.384 | 0 | 0.607 |
| BAF60C | SMARCD3 | 3.02 | 0 | 0 | 1.102 | 0.346 | 0 | 0.346 | 0.16 | 0 | 0 |
| BAF57 | SMARCE1 | 9.97 | 0 | 0.957 | 11.115 | 5.190 | 1.154 | 3.642 | 3.217 | 0.957 | 1.61 |
| BAF53A | ACTL6A, ArpNß | 8.37 | 0.233 | 0.110 | 6.305 | 6.305 | 2.511 | 2.511 | 0.874 | 0.369 | 0.52 |
| BAF53B | ACTL6B, ArpNα | 1.68 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| BAF47 | SMARCB1, INI1, SNF5 | 5.81 | 0 | 0.931 | 5.449 | 3.160 | 1.154 | 3.160 | 0.551 | 0.245 | 0.823 |
| BAF45A | PHF10 | 3.08 | 0 | 0.086 | 0 | 0 | 0 | 0 | 0 | 0 | 2.728 |
| BAF45B | DPF1 | 0.50 | 0 | 0 | 0 | 0.110 | 0 | 0.110 | 0 | 0 | 0 |
| BAF45C | DPF3, CERD4 | 0.26 | 0 | 0 | 0 | 0 | 0 | 0.105 | 0 | 0 | 0 |
| BAF45D | DPF2, REQ, UBID4 | 2.80 | 0 | 0 | 4.623 | 1.610 | 0.101 | 1.371 | 1.873 | 0 | 0 |
| SS18 | SYT, SSXT | 3.73 | 0 | 0 | 9.00 | 9.00 | 0.78 | 0 | 2.16 | 1.000 | 0 |
| SS18L1 | CREST | 9.92 | 0 | 0 | 0 | 0 | 0 | 0.585 | 0.585 | 0 | 0 |
| BCL7A | - | nd | 0 | 0 | 0.874 | 1.310 | 3.329 | 0 | 0.585 | 0.233 | 0 |
| BCL7B | Hom s 3 | 4.77 | 0 | 0 | 0 | 0.292 | 0 | 0 | 0 | 0 | 0 |
| BCL7C | - | 4.18 | 0 | 0 | 0 | 1.783 | 0 | 11.915 | 0 | 0 | 0 |
| BRD7 | CELTIX-1 | nd | 0 | 0.064 | 0 | 0 | 0 | 0 | 0 | 0 | 0.645 |
| BRD9 | MU-RMS-40.8 | 0.18 | 0 | 0 | 0.186 | 0.668 | 0 | 0.089 | 0 | 4.505 | 0 |
| SSX1 | - | nd | 0 | 0 | 0 | 1.154 | 0 | 0 | 0 | 0 | 0 |
| GLTSCR1 | GSCR1 | 1.20 | 0 | 0 | 0.619 | 1.116 | 0.055 | 0.708 | 0 | 0 | 0 |
| SRRM2 | - | 8.15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| MYBBP1A | p160 | 3.72 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| NONO | NMT55, p54(nrb) | nd | 0.066 | 0 | 0 | 0.066 | 0 | 0 | 0 | 1.966 | 0.292 |
| NUMA1 | - | 3.23 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.167 | 0 |
| SFPQ | PSF | 11.42 | 0.199 | 0 | 0 | 0 | 0 | 0 | 0 | 0.624 | 0.528 |
| DDX3X | HLP2 | 12.18 | 0.116 | 0 | 0 | 0 | 0 | 0 | 0 | 10.159 | 0.315 |
| DDX17 | p72 | 11.85 | 0.058 | 0 | 0 | 0 | 0 | 0 | 0.058 | 9.578 | 0.136 |
| RBM14 | COAA | 7.10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.736 | 0 |
| DDX5 | p68 | 17.37 | 0 | 0 | 0 | 0 | 0 | 0 | 0.061 | 4.223 | 0.805 |
| actin | actg1 | 30.37 | 4.109 | 1.371 | 4.109 | 6.499 | 2.481 | 2.831 | 0.778 | 7.254 | 3.437 |
Strikingly, multiple SWI/SNF subunits function as tumor suppressors in man and mouse, adding a key medical dimension to SWI/SNF research [24], [54]–[66]. For instance: the INI1flox/flox mouse is the most lethal tumor suppressor mouse model reported to date [55], suggesting a decisive role for SWI/SNF in cell proliferation control. Cell cycle roles for SWI/SNF-type complexes have indeed been documented in human and in model organisms [67]–[78].
Another link to cancer is provided by the SS18-SSX oncofusion proteins [79]. Synovial sarcomas are aggressive soft-tissue tumors accounting for about ten percent of all human soft-tissue sarcomas [80]. Characteristic for synovial sarcomas is the t(X;18)(p11.2;q11.2) translocation which is found in over 95% of all synovial sarcoma cases and results in the fusion of the SS18 (also called Syt) gene on chromosome 18 with one of the highly homologous SSX genes, SSX1, SSX2 or SSX4, on the X chromosome and consequently the expression of SS18-SSX fusion proteins [81]–[84]. These translocation events are believed to be the main molecular basis of this disease [79]. Orthologs of the SS18 protein also exists in plants. They are positive regulator of cell proliferation in lateral organs, such as leaves and flowers and appear to control aspects of cell proliferation together with DNA sequence-specific GRF transcription factors [85], [86]. Mammalian SS18 has been reported to associate with SWI/SNF chromatin remodeling complexes and to interact with BRG1 and BRM proteins [87]–[90]. In order to identify protein interactors of the SS18 and SS18-SSX proteins and to characterize the SS18 and SS18-SSX complexes we exploited a Tandem Affinity Purification (TAP) tagging approach combined with mass spectrometric analysis [91].
We found SS18 to be present in BAF-class human SWI/SNF chromatin remodeling complexes. Purification of SS18-SSX1 revealed that this oncofusion protein resides in the same complexes. Interestingly, we detected several additional putative SWI/SNF interactors [92]–[95]. Complex walking revealed the presence of these proteins in the same BAF SWI/SNF complexes as SS18, refining observations made by others [41], [43]. Overall, we conclude that human SS18 and its paralogue SS18l1/CREST together with; double PHD finger factors (DPF1,-2,-3), the B-cell CLL/lymphoma 7 protein family members (BCL7A, -B, -C) and BRD9 are specific to BAF-class SWI/SNF complexes, whilst BRD7 and PHF10 characterize PBAF complexes. Furthermore, with the exception of BRD7 and BRD9, quantitative mass spectrometry analysis demonstrates that the major proteomic interaction partners of all these factors are SWI/SNF subunits, indicating that they are bona fide BAF-type SWI/SNF complex subunits.
Results
TAP-tag purification of SWI/SNF complexes
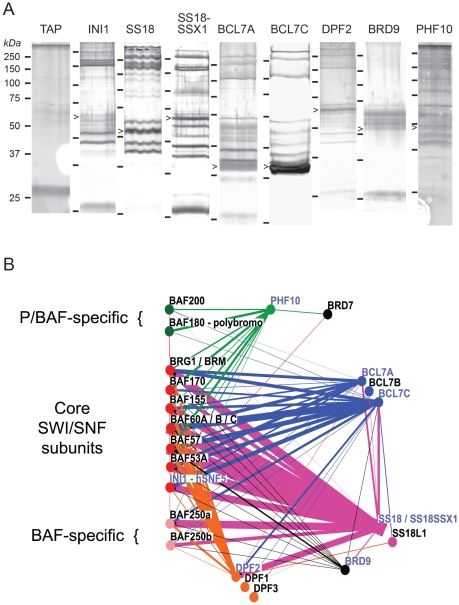
In order to define the protein complexes harboring known and suspected human SWI/SNF subunits we generated stable human embryonic kidney cell (Hek293) clones transduced with retroviral TAP-tag fusion expression constructs [91]. The following eight TAP-fusions were purified and analyzed by mass spectrometric analysis; INI1, SS18 and its oncogenic fusion product SS18-SSX1, BCL7A and BCL7C, DPF2, PHF10 and BRD9 (Figure 1A). The proteomic data we have collected (Table 1, Data S1) is schematized in Figure 1B, where the thickness of the edges reflect co-purification efficiency [96].
Figure 1. Complex walking.
(A) Silver-stained gels of the actual purified protein preparations that were analyzed by mass spectrometry. The respective TAP-tag fusion proteins are designated by black triangles. Size markers (Da) are indicated for every gel. Banding patterns differ because the gels were not all run under the same conditions. (B) Osprey interaction network [131] based on the mass spectrometry results obtained with the material shown in panel A. Blue name labels indicate the TAP-tag fusion employed here. The thickness of the lines reflect purification yield and are proportional to the emPAI values [96] shown on Table 1. The presence of orthologs in yeast and Drosophila genomes of the human factors that are displayed is indicated on Table 2.
INI1
INI1 is a core subunit of SWI/SNF complexes that is also known as hSNF5, SMARCB1 or BAF47. In our hands the yield of SWI/SNF complexes obtained with INI1TAP has consistently been comparatively low. For instance, in most INI1TAP preparations we detect BRG1 but not BRM, and ARID1A but not ARID1B (Table 1). This is consistent with BRG1 and ARID1A mRNA levels being 2–3 times that of their respective paralogs in Hek293 cells (Table 1, Data S2), a fact that is also reflected in the yields of these paralogous subunits in all the purifications (Table 1). Another indication that the INITAP construct is not amenable to very high yield SWI/SNF purifications is that of all the proteins we employed here to purify SWI/SNF complexes, only two, PHF10 and BRD7, are detected by INI1TAP, whilst INI1 was detected in all the reciprocal purifications (Table 1).
In keeping with a role as a core SWI/SNF subunit, INI1TAP purifications harbored both PBAF and BAF-specific SWI/SNF subunits (Table 1, Table 2). The comparatively higher yield of the PBAF-specific subunits ARID2 and polybromo versus the BAF-specific ARID1A and ARID1B suggests that INI1-bearing PBAF complexes are more preponderant than INI1-bearing BAF complexes in Hek293 cells, in line with the higher expression level of the PBAF-specific polybromo subunit (Table 1).
Table 2. Orthologs of known SWI/SNF complex subunits in human, fly and yeast.
| Human Protein | Alternative human names | Human complex | Drosophila melanogaster | Fly complex | Saccharomyces cerevisiae SWI/SNF | Saccharomyces cerevisiae RSC [134] |
| BRG1 | SMARCA4 | Core | brahma/CG5942 | Core | SNF2 | STH1 |
| BRM | SMARCA2 | Core | brahma/CG5942 | SNF2 | STH1 | |
| BAF250A | SMARCF1, ARID1A | BAF | OSA/eyelid/CG7467 | BAP | SWI1 | - |
| BAF250B | ARID1B, OSA1 | BAF | OSA/eyelid/CG7467 | SWI1 | - | |
| BAF200 | ARID2, zipzap | PBAF | BAP170/CG3274 | PBAP | - | - |
| BAF180 | Polybromo-1 | PBAF | polybromo/BAP180/CG11375 | PBAP | - | RSC1, RSC2, RSC4 |
| BAF170 | SMARCC2 | Core | moira/BAP155/CG18740 | Core | SWI3 | RSC8 |
| BAF155 | SMARCC1 | Core | moira/BAP155/CG18740 | SWI3 | RSC8 | |
| BAF60A | SMARCD1 | Core | BAP60/CG4303 | Core | SWP73 | RSC6 |
| BAF60B | SMARCD2 | Core | BAP60/CG4303 | SWP73 | RSC6 | |
| BAF60C | SMARCD3 | Core | BAP60/CG4303 | SWP73 | RSC6 | |
| BAF57 | SMARCE1 | Core | dalao/BAP111/CG7055 | Core | - | - |
| BAF53A | ACTL6A, ArpNb | Core | BAP55/CG6546 | Core | ARP7 & ARP9 | ARP7 & ARP9 |
| BAF53B | ACTL6B, ArpNa | Core | ? | ARP7 & ARP9 | ARP7 & ARP9 | |
| BAF47 | SMARCB1, INI1, SNF5 | Core | SNR1/CG1064 | Core | SNF5 | SFH1 |
| BAF45A | PHF10 | PBAF | e(y)3/SAYP/CG12238 | PBAP | - | - |
| BAF45B | DPF1 | BAF | d4/CG2682 | ? | - | - |
| BAF45C | DPF3, CERD4 | BAF | d4/CG2682 | - | - | |
| BAF45D | DPF2, REQ, UBID4 | BAF | d4/CG2682 | - | - | |
| SS18 | SYT, SSXT | BAF | CG10555 | ? | - | - |
| SS18L1 | CREST | BAF | CG10555 | - | - | |
| BCL7A | - | BAF | BCL7-like/CG17252 | ? | - | - |
| BCL7B | Hom s 3 | BAF | BCL7-like/CG17252 | - | - | |
| BCL7C | - | BAF | BCL7-like/CG17252 | - | - | |
| BRD7 | CELTIX-1 | PBAF | CG7154 | ? | - | - |
| BRD9 | MU-RMS-40.8 | BAF | CG7154 | - | - | |
| actin | actg1 | actin | actin | actin | ||
| RTT102 | RTT102 | |||||
| SWP82 | NPL6 | |||||
| HTL1 | ||||||
| LDB7 | ||||||
| RSC3 | ||||||
| RSC30 | ||||||
| RSC58 | ||||||
| RSC9 | ||||||
| SNF6 |
SS18 and the oncogenic SS18-SSX fusions are BAF subunits
SS18TAP purifications yielded high levels of SWI/SNF (Table 1, Figure 1). All known core subunits were found, consistent with previous work [87], [97]. Since both ARID1A and ARID1B but no ARID2 nor polybromo peptides were found, SS18 appears to be specific to both the ARID1A and ARID1B-bearing BAF-class variants of SWI/SNF (Table 1). Furthermore, several other potential SS18 interactors were identified, including GLioma Tumor Suppressor Candidate Region gene 1 protein (GLTSCR1), zinc finger protein ubi-d4 (DPF2), B-cell CLL/lymphoma 7A (BCL7A) and bromodomain containing protein 9 (BRD9) (Table 1, Figure 1B).
Because the chromosomal translocation t(X;18)(p11.2;q11.2) results in production of the oncogenic SS18-SSX1 protein fusion it was of interest to compare the proteomic environments of SS18 and the SS18 oncofusions. Essentially, purification of SS18-SSX1TAP resulted in the same set of interactors as purification of SS18TAP, with the exception of peptides originating from the SSX1 moiety of the oncofusion protein (Table 1). All subunits of the SWI/SNF BAF variant complex were identified, as well as the novel interactors GLTSCR1, DPF2 and its paralog DPF1, BRD9, and BCL7A and its paralogs BCL7B and C (Table 1, Figure 1B). We conclude that, similarly to SS18, the SS18-SSX1 oncofusion protein also resides in both the ARID1A and ARID1B-bearing BAF variants of human SWI/SNF.
DPF2 resides in BAF
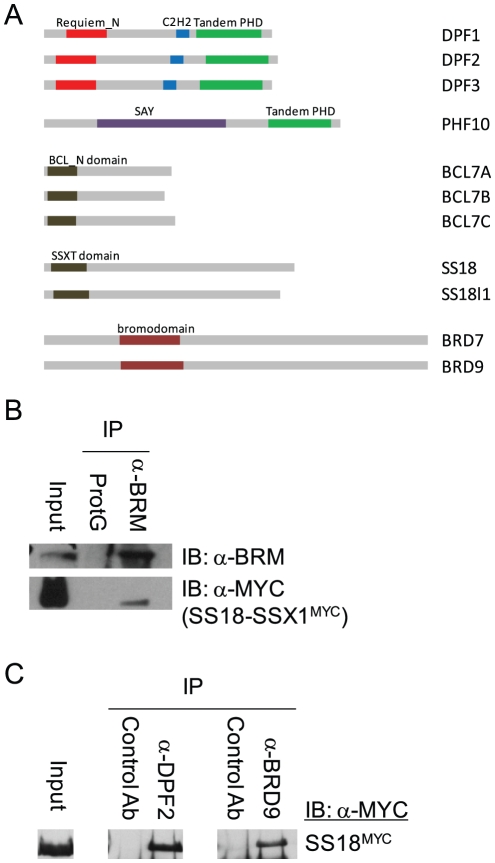
DPF2, also known as ubi-d4 or Requiem, is ubiquitously expressed and implicated in apoptosis [92]. It belongs to the d4 family which in humans consists of three paralogous genes: neuro-d4 (DPF1), ubi-d4 (DPF2) and cer-d4 (DPF3) [98], [99]. This gene family is not present in any of the currently sequenced plant genomes. Figure 2A shows that DPF factors harbor a conserved N-terminal domain (Pfam14051, [100]), a central C2H2-type Krüppel zinc finger motif with potential nucleic acid binding activity and C-terminal double paired finger PHD domains that have been shown to mediate conditional protein-protein interactions [42], [101], [102]. DPF1, 2, 3 and PHF10 were named BAF45A-D [41] because they were found in biochemical SWI/SNF preparations.
Figure 2. SS18 and the animal-specific SWI/SNF subunits.
(A) Domain organization of the human proteins DPF1,-2,-3; PHF10; SS18 and its paralog SS18l1/crest; and BCL7A,-B,-C. We note that while CG2682, the Drosophila melanogaster ortholog of DPF2 (Table 2), lacks the C2H2 domain, this domain is present in the Tribolium ortholog D6WFQ9_TRICA [125], [132], [133], suggesting conservation of this domain in protostome and in deuterostome animals (B) Co-immunoprecipitation of SS18-SSX1MYC by antibodies directed against human BRM. (C) Co-immunoprecipitation of SS18MYC by antibodies directed against DPF2 and BRD9.
The DPF2TAP purification results indicate that DPF2 resides mainly in ARID1-bearing BAF complexes, since no polybromo or ARID2 peptides were identified, whilst high confidence ARID1A and ARID1B peptides were detected (Table 1). Furthermore, like SS18TAP, DPF2TAP co-purified BCL7A and BRD9 as well as the SS18 paralog, SS18l1. Association of DPF2 with SS18 was further confirmed by co-immunoprecipitation (Figure 2C).
PHF10 resides in PBAF
PHF10 harbors two PHD domains but it is not a member of the DPF paralog group as it lacks a central Krüppel zinc finger motif, and harbors a SAY domain that is conserved in animals but not plants [103] (Figure 2A). In contrast to DPF2TAP, PHF10TAP was second only to INI1TAP in its yield of the PBAF-specific subunits ARID2 and polybromo (Table 1), demonstrating a strong association with PBAF-class SWI/SNF complexes. None of the BAF-associated SS18, DPF or BCL7 factors were detected in PHF10TAP preparations, suggesting that PHF10 indeed resides in a distinct subset of SWI/SNF complexes. Complete exclusion of PHF10 from BAF complexes may not be the case however, since one high confidence ARID1A-derived peptide was identified. Whether this reflects physiological subunit exchange between subtypes of SWI/SNF complexes or mal-assembled complexes remains an open question.
Notably, high confidence BRD7 peptides were detected, similar to the INI1TAP purification (Table 1), suggesting that PHF10 forms PBAF-type SWI/SNF complexes that can harbor BRD7 but not BRD9 since, PHF10TAP did not pull down BRD9, in contrast to SS18TAP, SS18-SSX1TAP and BCL7CTAP which did pull down BRD9 (Table 1, Figure 1B).
BCL7 proteins reside mainly in BAF
Similar to multiple SWI/SNF subunits, BCL7 family members have been implicated in carcinogenesis [93], [94]. The presence of BCL7 family members in SWI/SNF complexes has been reported before [43]. We succeeded in purifying BCL7ATAP and BCL7CTAP-associated proteins (Table 1, Figure 1B). In both cases and in contrast to INI1TAP and PHF10TAP, next to the core SWI/SNF subunits we also recovered more BAF-specific ARID1A and ARID1B subunit peptides (Table 1, Figure 1B), suggesting that BCL7 factors are mainly subunits of BAF complexes. The comparatively low levels of the PBAF-specific subunits in the BCL7CTAP preparations may be due to the high levels of BCL7CTAP in the cell line that was employed (Figure 1A) and indicate that the distinction between BAF and PBAF complexes by BAF subunits can be blurred operationally (Figure 1B). The absence of the PBAF-specific SWI/SNF subunit PHF10 from both BCL7TAP purifications strengthens the notion that BCL7 factors mainly associate with the BAF variants of SWI/SNF, however. Furthermore, the fact that BCL7CTAP co-purified DPF2, like BCL7ATAP, as well as the DPF2 paralogs DPF1 and DPF3 suggest that these two DPF2 paralogs also associate with BAF complexes. Indeed, co-purification of DPF1 and DPF3 parallels the purification results obtained with SS18-SSX1TAP (see above) again pointing towards the BAF variants of SWI/SNF. Finally, like DPF2TAP, BCL7CTAP also pulled down SS18l1 (Table 1, Figure 1B), a paralog of SS18 also known as CREST which has previously been linked physically to ARID1B [104], again strengthening the conclusion that BCL7A and BCL7C are mainly subunits of the BAF variants of SWI/SNF complexes. Interestingly, orthologs of BCL7 can only be found in sequenced animal genomes.
BRD9 associates with BAF
A well established function of bromodomains is to recognize specific acetylated lysines. The paralogous catalytic subunits of SWI/SNF, BRG1 and BRM harbor one C-terminal bromodomain that is closely related to the six bromodomains of polybromo, but quite distinct from the bromodomains of BRD7 and BRD9 [105]–[107].
A FLAG-BRD7 fusion has been reported to purify PBAF complexes [43]. We were not able to successfully perform BRD7TAP purifications (data not shown). However, BRD9TAP did yield significant mass spectrometry results (Table 1, Figure 1). BRD9TAP yielded peptide hits for at least one paralog of each core SWI/SNF subunit and, contrary to what was reported for BRD7 [43], [108], the presence of high confidence ARID1A and ARID1B peptides indicates inclusion of BRD9 in BAF complexes. This notion is buttressed by the presence of SS18 and BCL7C amongst the proteins co-purifying with BRD9TAP (Table 1, Figure 1, Data S1). Association of BRD9 with SS18TAP was further confirmed by co-immunoprecipitation (Figure 2C).
We quantified our mass spectrometry data on the basis of the exponentially modified protein abundance index (emPAI, Table 1, Figure 1) and this revealed that BRD9TAP did not efficiently purify SWI/SNF (Table 1), in keeping with our gel electrophoresis analysis (Figure 1A). The major factors we identified in our BRD9 preparation are the DEAD box ATP-dependent RNA helicases DDX3X, DDX5 and DDX17 and the RNA binding factor RBM14/COAA (Table 1). Since Emerson and co-workers reported substantially higher ATPase activity in their BRD7 preparations than predicted by BRG1/BRM content, it may perhaps be that BRD7 also co-purifies the DEAD box RNA helicases DDX3X, DDX5 and/or DDX17 [43], [109], [110]. Similarly, RBM14/COAA is a nuclear receptor co-activator [111]. Furthermore, RBM14/COAA has previously been reported to associate with SS18 in yeast two hybrid assays [112], [113]. However, arguing against a direct interaction between SS18 and RBM14, we did not detect RBM14/COAA when SS18TAP or SS18-SSX1TAP associated factors were purified (Table 1, Figure 1B).
Putative BAF associated proteins
Crabtree and colleagues [49] published a list of putative novel BAF-associated proteins which we have monitored in this data set. Hence, we also detected GLTSCR1 in our SWI/SNF complex preparations (Table 1). GLTSCR1 is a candidate tumor suppressor gene for gliomas [114]. As we detected GLTSCR1 in four of five BAF purifications (Table 1), our results support the notion that GLTSCR1 is a BAF-associated factor, but this will need to be confirmed directly.
Of the other putative novel BAF-associated proteins, we could detect NONO and its binding partner SFPQ [115], [116], however, at levels that were not much higher than in control purifications (Table 1). Thus, although our data do not exclude an interaction with SWI/SNF, more experimental evidence is needed on this front. Finally, the proposed putative BAF-associated factors NUMA1, SRRM2 and MYBBP1A [49] were not detected in any of our SWI/SNF purifications (Table 1), suggesting weak biochemical association with SWI/SNF in the ‘293’ human embryonic kidney cell line, if any.
Discussion
Paralogous human SWI/SNF subunits are known to be expressed in tissue and signal specific fashion, generating alternative SWI/SNF complex configurations that can cooperate with transcription factor networks to coordinate cell proliferation and differentiation. Here, we focus on SWI/SNF subunits that are absent from yeast but conserved in animals and plants (SS18) or only in animals (DPF, BCL7 and PHF10) (Table 2).
Essentially there are two types of human SWI/SNF complexes [32], [44]; those that harbor the polybromo/BAF180 and ARID2/BAF200 subunits (PBAF-class) and those that harbor either ARID1A/BAF250a or ARID1B/BAF250b (BAF-class) [117]. A similar bi-partition exists in Drosophila melanogaster except that there is only one ARID1 ortholog, namely OSA [118], [119]. Similarly, the fly genome only encodes one ortholog of the mammalian SMARCC (CG18740/moira) and of BRD7/9 (CG7154), SS18 (CG10555), DPF2 (CG2682/d4) and BCL7 (CG17252/BCL7-like) protein coding genes (Table 2).
Our mass spectrometry analysis of affinity tag-mediated protein complex purifications confirms bipartition of SWI/SNF complexes in BAF and PBAF-class complexes. We demonstrate here that the paralogous cancer-related minor SWI/SNF subunits DPF1, -2, -3; BCL7A, -B, -C; and SS18 and SS18L1 reside in BAF-class human SWI/SNF complexes and, that PHF10 marks PBAF SWI/SNF complexes. Moreover, because quantitative analysis indicates that the chief interaction partners of PHF10, DPF2, SS18, SS18-SSX1, BCL7A and BCL7C are the other SWI/SNF subunits, we speculate that they exert their molecular action through their respective SWI/SNF complexes. It remains to be seen indeed to what extent our results, which were obtained in one human cell line, can be extrapolated to other cell types and even other organisms. Considering the congruence between our data and a recent studies on Drosophila SAYP [120] and a large scale proteomic survey of nuclear receptor co-activators [121], we believe they can. Moreover, there may be more as yet ‘undiscovered’ human SWI/SNF subunits, such as the putative GLTSCR1 subunit [49], and these may also be present in our data sets (Data S1), which can be mined by interested investigators.
Since we could not detect notable differences between SS18 and its oncogenic fusion products at the proteomic level, the oncogenic activity of the SS18-SSX fusions may have to be sought either at the level of SWI/SNF (dis)assembly dynamics, post-translational modifications or an affinity for specific genomic loci [42], [106], [122], although it is formally possible that the SS18-SSX oncofusion proteins undergo different proteomic interactions in the elusive synovial sarcoma precursor cell type than those we detected in Hek293 cells [123].
Interestingly, we detected the bromodomain proteins BRD7 and BRD9 in our SWI/SNF preparations. The fly protein CG7154 is that organism's sole BRD7/BRD9 ortholog. It will be interesting to determine whether it associates with the fly brahma SNF2 ATPase, and if so, whether it is specific to the BAP or PBAP fly equivalents of BAF and PBAF. In humans, BRD7 appears to promote cellular senescence [108], [124], and, in line with our results, it has convincingly been linked to the PBAF complex [43]. On the other hand, our data make a novel link between BRD9 and the BAF complex defined by SS18, DPF and BCL7, since we recovered it when BCL7CTAP, SS18TAP and the SS18-SSX1TAP were used as affinity bait and since in the reciprocal experiment, BRD9TAP purifications contained SS18 and BCL7A. We note however that BRD9TAP did not efficiently pull down BAF proteins in our experimental set-up (Figure 1A). Whether this reflects a status as a minor though BAF-specific subunit or a technical limitation is an open question.
Altogether, this work demonstrates that paralogs of SS18, BCL7 and DPF factors, which can be found in both protostome [125]) and deuterostome animals (Table 2), together define a novel class of BAF-type SWI/SNF complexes that is restricted to the animal lineage. Finally, our data indicate that PHF10 versus DPF1, -2, -3 respectively mark PBAF versus BAF-type SWI/SNF complexes in a mutually exclusive fashion.
Methods
Constructs
Tandem Affinity Purification (TAP) constructs were generated by PCR using the oligomers indicated in parentheses and cloned into the XhoI and EcoRI restriction sites in the retroviral expression vector pZXN, whereby the TAP-tag sequence was fused to the coding sequences at their N-terminus [91]. SS18 (isoform 2, cgtactGAATTCATGTCTGTGGCTTTCGCGG, tgacttCTCGAGTCACTGCTGGTAATTTCCATACT) and SS18-SSX1 (cgtactGAATTCATGTCTGTGGCTTTCGCGG, tgtcatCTCGAGTTACTCGTCATCTTCCTCAGGGT) coding sequence were amplified by PCR from pIRES2 vectors [126]. DPF2 (cgtatcGAATTCATGGCGGCTGTGGTGGAGAAT, tgtcttCTCGAGTCAAGAGGAGTTCTGGTTCTGGTA), BCL7C (cgtatcGAATTCATGGCCGGCCGGACTGTA, tgtcttCTCGAGTCAGGGGTCAGGGGCATTT), BRD9 (atacttGAATTCATGAAGGGATACCAAAGTCTTGTATTC, tactatCTCGAG TTAGGTCTTGGCAGAGGCCGCA) and PHF10 (gaattcGAATTCATGCTTCAAGAACAAGTCAGTG, aagcttAAGCTTTTATCCCTCTTTGCTGTTTTTCC, cloned into pBSIISK+ cut by the same enzymes and then released using SalI and EcoRI and further subcloned) coding sequences were amplified from cDNA clones (RZPD or OriGene). BCL7A (isoform 2, ttacttCAATTGATGTCGGGCAGGTCGGGT, tacttaGTCGACCTACATCTCTTCGGAGTTTTGTTG) was amplified from cDNA of Hek293 cells. INI1 (atacttGAATTCATGAAGGGATACCAAAGTCTTGTATTC, tactatCTCGAGTTAGGTCTTGGCAGAGGCCGCA) was amplified from pUHD-10-3-INI1 [127]. The retroviral vector pZXN is derived from the pZOME-1N vector (Euroscarf) and contains a TAP-tag consisting of one protein A domain followed by two tobacco etch virus cleavage sites (TEV) and then either a MYC epitope (GCCGGCAAGCCCCGGCATATGAATTTAATGGAGCAGAAGCT TATCAGCGAGGAGGACCTGGGCGGGGAATTC) or, in the case of PHF10, a TY1 epitope (GCCGGCGCCGATGCCGGCAAGCCCCGGCA TAGGACCGGTGAGGTGCACACCAACCAGGACCCCCTGGACGAATTC) 5′ of the EcoRI cloning site. Every clone was verified by DNA sequencing.
Cell culture and stable cell lines
Human Embryonic Kidney (Hek293, ATCC CRL-1573) and phoenix cells were grown in Dulbecco's modified Eagles medium (Invitrogen) supplemented with 10% FCS, penicillin 100 µg/ml and streptomycin 100 U/ml (Invitrogen) at 37°C in 5% CO2. Retroviral stable cell lines were generated as previously described [91]. Briefly, phoenix amphotropic packaging cells were transfected with 20 µg retroviral plasmid pZXN-SS18, pZXN- SS18-SSX1, pZXN- SS18-SSX2, pZXN-DPF2, pZXN-BRD9, pZXN-BCL7A, pZXN-BCL7C, pZXN-PHF10 or pZXN-INI1 after which Hek293 cells were transduced with virus containing supernatant in two infectious rounds of 24 hours in the presence of 8 µg/ml polybrene. Clones were selected with 1 µg/ml puromycin and tested for recombinant protein expression. Transduction of SS18 and SS18-SSX TAPtag fusions in the syo-1 synovial sarcoma cell line [128] were not successful (data not shown). Since Hek293 cells expressed the transduced transgenes efficiently and could be expanded as desired, we performed our study with this cell line.
Tandem Affinity Purification
Tandem Affinity Purification was performed as previously described in detail [91]. Shortly, whole cell extracts from cell lines expressing TAP-tagged proteins were incubated with IgG sepharose beads (Pharmacia). After TEV cleavage the TEV eluates were pre-cleared with protein A beads and used for immunoprecipitation with anti-MYC or anti TY1 epitope antibodies. Proteins were eluted from the beads by peptide elution, loaded on a SDS-PAGE gel and visualized by silver staining. The same protocol was employed for all the purifications reported here.
Co-immunoprecipitation
Co-immunoprecipitations were performed on TEV cleavage eluates obtained as described above, using antibodies directed against BRM (Abcam 15597), DPF2 (Aviva systems biology ARP33221_P050) or BRD9 (Aviva systems biology ARP34803_T200), under the same conditions as the anti-MYC or TY1 immunoprecipitations in the TAPtag purification protocol. The immunoprecipitated proteins were separated by SDS-PAGE, transferred onto nylon filters and probed with anti-MYC antibodies, which recognize the transduced SS18-SSX1 (Figure 2B) or SS18 (Figure 2C) proteins.
Mass spectrometry
The silver stained gel lanes were cut into small pieces. After reduction and alkylation the proteins were trypsin (Promega) digested and extracted from the gel using trifluoroacetic acid (TFA). Peptides were sequenced using a nano-high-pressure liquid chromatography Agilent 1100 nanoflow system connected online to a 7-Tesla linear quadrupole ion-trap Fourier transform (FT) mass spectrometer (Thermo Electron, Bremen, Germany) essentially as described previously [129]. MSquant software package (http://www.msquant.sourceforge.net) was used to parse the raw files and for generation of peak lists. The mascot algorithm was used to identify the proteins [130]. Exponentially Modified Protein Abundance Index (emPAI) factors were calculated as described previously [96], using high confidence peptides (Data S1, MASCOT score≥20, delta≥5, error≤5; 400–6000 Da).
Expression profiling
Expression profiling was performed on four Hek293 polyA mRNA samples by microarray analysis using Affymetrix human exon array 1.0 ST according to manufacturer instructions (Data S2).
Supporting Information
Mass spectrometry results, including; accession numbers, short protein descriptions, peptide sequences, associated Mascot score, peptide delta score and absolute calibrated mass relative error.
(XLS)
Quadruplate polyA mRNA expression profile of Hek293 cells determined with the Affymetrix human exon array 1.0 ST platform.
(XLS)
Acknowledgments
We are thankful to Michiel Vermeulen for communicating unpublished data early on in this project and to the members of the molecular biology department for their generous help and support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the Dutch cancer research fund KWF KUN2006-3557 (CL) and KUN2006-3338 (HGS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell. 2011;145:835–850. doi: 10.1016/j.cell.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, et al. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 4.Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol Cell Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Vugt JJ, de Jager M, Murawska M, Brehm A, van Noort J, et al. Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. PLoS One. 2009;4:e6345. doi: 10.1371/journal.pone.0006345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, et al. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gkikopoulos T, Singh V, Tsui K, Awad S, Renshaw MJ, et al. The SWI/SNF complex acts to constrain distribution of the centromeric histone variant Cse4. Embo J. 2011;30:1919–1927. doi: 10.1038/emboj.2011.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Research. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. Embo J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiba H, Muramatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the estrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ichinose H, Garnier JM, Chambon P, Losson R. Ligand-dependent interaction between the estrogen receptor and the human homologues of SWI2/SNF2. Gene. 1997;188:95–100. doi: 10.1016/s0378-1119(96)00785-8. [DOI] [PubMed] [Google Scholar]
- 13.Kadam S, McAlpine GS, Phelan ML, Kingston RE, Jones KA, et al. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 2000;14:2441–2451. doi: 10.1101/gad.828000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiRenzo J, Shang Y, Phelan M, Sif S, Myers M, et al. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol Cell Biol. 2000;20:7541–7549. doi: 10.1128/mcb.20.20.7541-7549.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biggs JR, Yang J, Gullberg U, Muchardt C, Yaniv M, et al. The human brm protein is cleaved during apoptosis: the role of cathepsin G. Proc Natl Acad Sci U S A. 2001;98:3814–3819. doi: 10.1073/pnas.071057398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Belandia B, Orford RL, Hurst HC, Parker MG. Targeting of SWI/SNF chromatin remodelling complexes to estrogen-responsive genes. Embo J. 2002;21:4094–4103. doi: 10.1093/emboj/cdf412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Hankinson O. Functional involvement of the Brahma/SWI2-related gene 1 protein in cytochrome P4501A1 transcription mediated by the aryl hydrocarbon receptor complex. J Biol Chem. 2002;277:11821–11827. doi: 10.1074/jbc.M110122200. [DOI] [PubMed] [Google Scholar]
- 18.Lee D, Kim JW, Seo T, Hwang SG, Choi EJ, et al. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J Biol Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 19.Kitagawa H, Fujiki R, Yoshimura K, Mezaki Y, Uematsu Y, et al. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell. 2003;113:905–917. doi: 10.1016/s0092-8674(03)00436-7. [DOI] [PubMed] [Google Scholar]
- 20.Hsiao PW, Fryer CJ, Trotter KW, Wang W, Archer TK. BAF60a mediates critical interactions between nuclear receptors and the BRG1 chromatin-remodeling complex for transactivation. Mol Cell Biol. 2003;23:6210–6220. doi: 10.1128/MCB.23.17.6210-6220.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumann M, Mamais A, McBlane F, Xiao H, Boyes J. Regulation of V(D)J recombination by nucleosome positioning at recombination signal sequences. Embo J. 2003;22:5197–5207. doi: 10.1093/emboj/cdg487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X, Gao X, Diaz-Trelles R, Ruiz-Lozano P, Wang Z. Coronary development is regulated by ATP-dependent SWI/SNF chromatin remodeling component BAF180. Dev Biol. 2008;319:258–266. doi: 10.1016/j.ydbio.2008.04.020. [DOI] [PubMed] [Google Scholar]
- 23.Du H, Ishii H, Pazin MJ, Sen R. Activation of 12/23-RSS-dependent RAG cleavage by hSWI/SNF complex in the absence of transcription. Mol Cell. 2008;31:641–649. doi: 10.1016/j.molcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 25.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Molecular Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 27.Singhal N, Graumann J, Wu G, Arauzo-Bravo MJ, Han DW, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 28.Gresh L, Bourachot B, Reimann A, Guigas B, Fiette L, et al. The SWI/SNF chromatin-remodeling complex subunit SNF5 is essential for hepatocyte differentiation. Embo J. 2005;24:3313–3324. doi: 10.1038/sj.emboj.7600802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Euskirchen GM, Auerbach RK, Davidov E, Gianoulis TA, Zhong G, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 2011;7:e1002008. doi: 10.1371/journal.pgen.1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 31.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 32.Wang W, Xue Y, Zhou S, Kuo A, Cairns BR, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10:2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Chi T, Xue Y, Zhou S, Kuo A, et al. Architectural DNA binding by a high-mobility-group/kinesin-like subunit in mammalian SWI/SNF-related complexes. Proc Natl Acad Sci U S A. 1998;95:492–498. doi: 10.1073/pnas.95.2.492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ring HZ, Vameghi-Meyers V, Wang W, Crabtree GR, Francke U. Five SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARC) genes are dispersed in the human genome. Genomics. 1998;51:140–143. doi: 10.1006/geno.1998.5343. [DOI] [PubMed] [Google Scholar]
- 35.Harata M, Mochizuki R, Mizuno S. Two isoforms of a human actin-related protein show nuclear localization and mutually selective expression between brain and other tissues. Biosci Biotechnol Biochem. 1999;63:917–923. doi: 10.1271/bbb.63.917. [DOI] [PubMed] [Google Scholar]
- 36.Nie Z, Xue Y, Yang D, Zhou S, Deroo BJ, et al. A specificity and targeting subunit of a human SWI/SNF family-related chromatin-remodeling complex. Mol Cell Biol. 2000;20:8879–8888. doi: 10.1128/mcb.20.23.8879-8888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nie Z, Yan Z, Chen EH, Sechi S, Ling C, et al. Novel SWI/SNF chromatin-remodeling complexes contain a mixed-lineage leukemia chromosomal translocation partner. Mol Cell Biol. 2003;23:2942–2952. doi: 10.1128/MCB.23.8.2942-2952.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan Z, Cui K, Murray DM, Ling C, Xue Y, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19:1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xue Y, Canman JC, Lee CS, Nie Z, Yang D, et al. The human SWI/SNF-B chromatin-remodeling complex is related to yeast rsc and localizes at kinetochores of mitotic chromosomes. Proc Natl Acad Sci U S A. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, et al. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–636. doi: 10.1016/s0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 41.Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev. 2008;22:2370–2384. doi: 10.1101/gad.471408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaeser MD, Aslanian A, Dong MQ, Yates JR, 3rd, Emerson BM. BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J Biol Chem. 2008;283:32254–32263. doi: 10.1074/jbc.M806061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 45.Thompson M. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie. 2009;91:309–319. doi: 10.1016/j.biochi.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yan Z, Wang Z, Sharova L, Sharov AA, Ling C, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26:1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao X, Tate P, Hu P, Tjian R, Skarnes WC, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci U S A. 2008;105:6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schaniel C, Ang YS, Ratnakumar K, Cormier C, James T, et al. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–2991. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ho L, Ronan JL, Wu J, Staahl BT, Chen L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc Natl Acad Sci U S A. 2009;106:5181–5186. doi: 10.1073/pnas.0812889106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoo AS, Sun AX, Li L, Shcheglovitov A, Portmann T, et al. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature. 2011;476:228–231. doi: 10.1038/nature10323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lickert H, Takeuchi JK, Von Both I, Walls JR, McAuliffe F, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 52.Debril MB, Gelman L, Fayard E, Annicotte JS, Rocchi S, et al. Transcription factors and nuclear receptors interact with the SWI/SNF complex through the BAF60c subunit. J Biol Chem. 2004;279:16677–16686. doi: 10.1074/jbc.M312288200. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi JK, Lickert H, Bisgrove BW, Sun X, Yamamoto M, et al. Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry. Proc Natl Acad Sci U S A. 2007;104:846–851. doi: 10.1073/pnas.0608118104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Versteege I, Sevenet N, Lange J, Rousseau-Merck MF, Ambros P, et al. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature. 1998;394:203–206. doi: 10.1038/28212. [DOI] [PubMed] [Google Scholar]
- 55.Roberts CW, Leroux MM, Fleming MD, Orkin SH. Highly penetrant, rapid tumorigenesis through conditional inversion of the tumor suppressor gene Snf5. Cancer Cell. 2002;2:415–425. doi: 10.1016/s1535-6108(02)00185-x. [DOI] [PubMed] [Google Scholar]
- 56.Reisman DN, Sciarrotta J, Wang W, Funkhouser WK, Weissman BE. Loss of BRG1/BRM in human lung cancer cell lines and primary lung cancers: correlation with poor prognosis. Cancer Res. 2003;63:560–566. [PubMed] [Google Scholar]
- 57.Wang X, Nagl NG, Jr, Flowers S, Zweitzig D, Dallas PB, et al. Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int J Cancer. 2004;112:636. doi: 10.1002/ijc.20450. [DOI] [PubMed] [Google Scholar]
- 58.Glaros S, Cirrincione GM, Muchardt C, Kleer CG, Michael CW, et al. The reversible epigenetic silencing of BRM: implications for clinical targeted therapy. Oncogene. 2007;26:7058–7066. doi: 10.1038/sj.onc.1210514. [DOI] [PubMed] [Google Scholar]
- 59.Xia W, Nagase S, Montia AG, Kalachikov SM, Keniry M, et al. BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 2008;68:1667–1674. doi: 10.1158/0008-5472.CAN-07-5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones S, Wang TL, Shih Ie M, Mao TL, Nakayama K, et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science. 2010;330:228–231. doi: 10.1126/science.1196333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wiegand KC, Shah SP, Al-Agha OM, Zhao Y, Tse K, et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N Engl J Med. 2010;363:1532–1543. doi: 10.1056/NEJMoa1008433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Varela I, Tarpey P, Raine K, Huang D, Ong CK, et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gui Y, Guo G, Huang Y, Hu X, Tang A, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mamo A, Cavallone L, Tuzmen S, Chabot C, Ferrario C, et al. An integrated genomic approach identifies ARID1A as a candidate tumor-suppressor gene in breast cancer. Oncogene. 2011 doi: 10.1038/onc.2011.386. [DOI] [PubMed] [Google Scholar]
- 65.Li M, Zhao H, Zhang X, Wood LD, Anders RA, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu G, Gramling S, Munoz D, Cheng D, Azad AK, et al. Two novel BRM insertion promoter sequence variants are associated with loss of BRM expression and lung cancer risk. Oncogene. 2011;30:3295–3304. doi: 10.1038/onc.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, et al. RSC, an essential, abundant chromatin-remodeling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 68.Trouche D, Le Chalony C, Muchardt C, Yaniv M, Kouzarides T. RB and hbrm cooperate to repress the activation functions of E2F1. Proc Natl Acad Sci U S A. 1997;94:11268–11273. doi: 10.1073/pnas.94.21.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, et al. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2alpha). Embo J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cosma MP, Tanaka T, Nasmyth K. Ordered recruitment of transcription and chromatin remodeling factors to a cell cycle- and developmentally regulated promoter. Cell. 1999;97:299–311. doi: 10.1016/s0092-8674(00)80740-0. [DOI] [PubMed] [Google Scholar]
- 71.Zhang HS, Gavin M, Dahiya A, Postigo AA, Ma D, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101:79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 72.Coisy M, Roure V, Ribot M, Philips A, Muchardt C, et al. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2alpha. Mol Cell. 2004;15:43–56. doi: 10.1016/j.molcel.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 73.Nagl NG, Jr, Patsialou A, Haines DS, Dallas PB, Beck GR, Jr, et al. The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 2005;65:9236–9244. doi: 10.1158/0008-5472.CAN-05-1225. [DOI] [PubMed] [Google Scholar]
- 74.Vries RG, Bezrookove V, Zuijderduijn LM, Kia SK, Houweling A, et al. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 2005;19:665–670. doi: 10.1101/gad.335805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sansam CG, Roberts CW. Epigenetics and cancer: altered chromatin remodeling via Snf5 loss leads to aberrant cell cycle regulation. Cell Cycle. 2006;5:621–624. doi: 10.4161/cc.5.6.2579. [DOI] [PubMed] [Google Scholar]
- 76.Campsteijn C, Wijnands-Collin AM, Logie C. Reverse genetic analysis of the yeast RSC chromatin remodeler reveals a role for RSC3 and SNF5 homolog 1 in ploidy maintenance. PLoS Genet. 2007;3:e92. doi: 10.1371/journal.pgen.0030092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Becker TM, Haferkamp S, Dijkstra MK, Scurr LL, Frausto M, et al. The chromatin remodelling factor BRG1 is a novel binding partner of the tumor suppressor p16INK4a. Mol Cancer. 2009;8:4. doi: 10.1186/1476-4598-8-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Inoue H, Giannakopoulos S, Parkhurst CN, Matsumura T, Kono EA, et al. Target genes of the largest human SWI/SNF complex subunit control cell growth. Biochem J. 2011;434:83–92. doi: 10.1042/BJ20101358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haldar M, Randall RL, Capecchi MR. Synovial sarcoma: from genetics to genetic-based animal modeling. Clin Orthop Relat Res. 2008;466:2156–2167. doi: 10.1007/s11999-008-0340-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ladanyi M, Antonescu CR, Leung DH, Woodruff JM, Kawai A, et al. Impact of SYT-SSX fusion type on the clinical behavior of synovial sarcoma: a multi-institutional retrospective study of 243 patients. Cancer Res. 2002;62:135–140. [PubMed] [Google Scholar]
- 81.Clark J, Rocques PJ, Crew AJ, Gill S, Shipley J, et al. Identification of novel genes, SYT and SSX, involved in the t(X;18)(p11.2;q11.2) translocation found in human synovial sarcoma. Nat Genet. 1994;7:502–508. doi: 10.1038/ng0894-502. [DOI] [PubMed] [Google Scholar]
- 82.de Leeuw B, Balemans M, Olde Weghuis D, Geurts van Kessel A. Identification of two alternative fusion genes, SYT-SSX1 and SYT-SSX2, in t(X;18)(p11.2;q11.2)-positive synovial sarcomas. Hum Mol Genet. 1995;4:1097–1099. doi: 10.1093/hmg/4.6.1097. [DOI] [PubMed] [Google Scholar]
- 83.Crew AJ, Clark J, Fisher C, Gill S, Grimer R, et al. Fusion of SYT to two genes, SSX1 and SSX2, encoding proteins with homology to the Kruppel-associated box in human synovial sarcoma. Embo J. 1995;14:2333–2340. doi: 10.1002/j.1460-2075.1995.tb07228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Skytting B, Nilsson G, Brodin B, Xie Y, Lundeberg J, et al. A novel fusion gene, SYT-SSX4, in synovial sarcoma. J Natl Cancer Inst. 1999;91:974–975. doi: 10.1093/jnci/91.11.974. [DOI] [PubMed] [Google Scholar]
- 85.Kim JH, Kende H. A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc Natl Acad Sci U S A. 2004;101:13374–13379. doi: 10.1073/pnas.0405450101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee BH, Ko JH, Lee S, Lee Y, Pak JH, et al. The Arabidopsis GRF-INTERACTING FACTOR gene family performs an overlapping function in determining organ size as well as multiple developmental properties. Plant Physiol. 2009;151:655–668. doi: 10.1104/pp.109.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kato H, Tjernberg A, Zhang W, Krutchinsky AN, An W, et al. SYT associates with human SNF/SWI complexes and the C-terminal region of its fusion partner SSX1 targets histones. J Biol Chem. 2002;277:5498–5505. doi: 10.1074/jbc.M108702200. [DOI] [PubMed] [Google Scholar]
- 88.Thaete C, Brett D, Monaghan P, Whitehouse S, Rennie G, et al. Functional domains of the SYT and SYT-SSX synovial sarcoma translocation proteins and co-localization with the SNF protein BRM in the nucleus. Hum Mol Genet. 1999;8:585–591. doi: 10.1093/hmg/8.4.585. [DOI] [PubMed] [Google Scholar]
- 89.Nagai M, Tanaka S, Tsuda M, Endo S, Kato H, et al. Analysis of transforming activity of human synovial sarcoma-associated chimeric protein SYT-SSX1 bound to chromatin remodeling factor hBRM/hSNF2 alpha. Proc Natl Acad Sci U S A. 2001;98:3843–3848. doi: 10.1073/pnas.061036798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ishida M, Tanaka S, Ohki M, Ohta T. Transcriptional co-activator activity of SYT is negatively regulated by BRM and Brg1. Genes Cells. 2004;9:419–428. doi: 10.1111/j.1356-9597.2004.00737.x. [DOI] [PubMed] [Google Scholar]
- 91.Le Guezennec X, Vermeulen M, Brinkman AB, Hoeijmakers WA, Cohen A, et al. MBD2/NuRD and MBD3/NuRD, two distinct complexes with different biochemical and functional properties. Mol Cell Biol. 2006;26:843–851. doi: 10.1128/MCB.26.3.843-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gabig TG, Mantel PL, Rosli R, Crean CD. Requiem: a novel zinc finger gene essential for apoptosis in myeloid cells. J Biol Chem. 1994;269:29515–29519. [PubMed] [Google Scholar]
- 93.Zani VJ, Asou N, Jadayel D, Heward JM, Shipley J, et al. Molecular cloning of complex chromosomal translocation t(8;14;12)(q24.1;q32.3;q24.1) in a Burkitt lymphoma cell line defines a new gene (BCL7A) with homology to caldesmon. Blood. 1996;87:3124–3134. [PubMed] [Google Scholar]
- 94.Jadayel DM, Osborne LR, Coignet LJ, Zani VJ, Tsui LC, et al. The BCL7 gene family: deletion of BCL7B in Williams syndrome. Gene. 1998;224:35–44. doi: 10.1016/s0378-1119(98)00514-9. [DOI] [PubMed] [Google Scholar]
- 95.Wu M, Li X, Li X, Li G. Signaling Transduction Network Mediated by Tumor Suppressor/Susceptibility Genes in NPC. Curr Genomics. 2009;10:216–222. doi: 10.2174/138920209788488481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 97.Perani M, Ingram CJ, Cooper CS, Garrett MD, Goodwin GH. Conserved SNH domain of the proto-oncoprotein SYT interacts with components of the human chromatin remodelling complexes, while the QPGY repeat domain forms homo-oligomers. Oncogene. 2003;22:8156–8167. doi: 10.1038/sj.onc.1207031. [DOI] [PubMed] [Google Scholar]
- 98.Chestkov AV, Baka ID, Kost MV, Georgiev GP, Buchman VL. The d4 gene family in the human genome. Genomics. 1996;36:174–177. doi: 10.1006/geno.1996.0440. [DOI] [PubMed] [Google Scholar]
- 99.Ninkina NN, Mertsalov IB, Kulikova DA, Alimova-Kost MV, Simonova OB, et al. Cerd4, third member of the d4 gene family: expression and organization of genomic locus. Mamm Genome. 2001;12:862–866. doi: 10.1007/s00335-001-3039-1. [DOI] [PubMed] [Google Scholar]
- 100.Tando T, Ishizaka A, Watanabe H, Ito T, Iida S, et al. Requiem protein links RelB/p52 and the Brm-type SWI/SNF complex in a noncanonical NF-kappaB pathway. J Biol Chem. 2010;285:21951–21960. doi: 10.1074/jbc.M109.087783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- 102.Musselman CA, Kutateladze TG. PHD fingers: epigenetic effectors and potential drug targets. Mol Interv. 2009;9:314–323. doi: 10.1124/mi.9.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shidlovskii YV, Krasnov AN, Nikolenko JV, Lebedeva LA, Kopantseva M, et al. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. Embo J. 2005;24:97–107. doi: 10.1038/sj.emboj.7600508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiu Z, Ghosh A. A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron. 2008;60:775–787. doi: 10.1016/j.neuron.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sun H, Liu J, Zhang J, Shen W, Huang H, et al. Solution structure of BRD7 bromodomain and its interaction with acetylated peptides from histone H3 and H4. Biochem Biophys Res Commun. 2007;358:435–441. doi: 10.1016/j.bbrc.2007.04.139. [DOI] [PubMed] [Google Scholar]
- 106.Kupitz C, Chandrasekaran R, Thompson M. Kinetic analysis of acetylation-dependent Pb1 bromodomain-histone interactions. Biophys Chem. 2008;136:7–12. doi: 10.1016/j.bpc.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Burrows AE, Smogorzewska A, Elledge SJ. Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc Natl Acad Sci U S A. 2010;107:14280–14285. doi: 10.1073/pnas.1009559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schroder M. Viruses and the human DEAD-box helicase DDX3: inhibition or exploitation? Biochem Soc Trans. 2011;39:679–683. doi: 10.1042/BST0390679. [DOI] [PubMed] [Google Scholar]
- 110.Fuller-Pace FV, Ali S. The DEAD box RNA helicases p68 (Ddx5) and p72 (Ddx17): novel transcriptional co-regulators. Biochem Soc Trans. 2008;36:609–612. doi: 10.1042/BST0360609. [DOI] [PubMed] [Google Scholar]
- 111.Auboeuf D, Dowhan DH, Li X, Larkin K, Ko L, et al. CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol Cell Biol. 2004;24:442–453. doi: 10.1128/MCB.24.1.442-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Iwasaki T, Chin WW, Ko L. Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J Biol Chem. 2001;276:33375–33383. doi: 10.1074/jbc.M101517200. [DOI] [PubMed] [Google Scholar]
- 113.Perani M, Antonson P, Hamoudi R, Ingram CJ, Cooper CS, et al. The proto-oncoprotein SYT interacts with SYT-interacting protein/co-activator activator (SIP/CoAA), a human nuclear receptor co-activator with similarity to EWS and TLS/FUS family of proteins. J Biol Chem. 2005;280:42863–42876. doi: 10.1074/jbc.M502963200. [DOI] [PubMed] [Google Scholar]
- 114.Smith JS, Tachibana I, Pohl U, Lee HK, Thanarajasingam U, et al. A transcript map of the chromosome 19q-arm glioma tumor suppressor region. Genomics. 2000;64:44–50. doi: 10.1006/geno.1999.6101. [DOI] [PubMed] [Google Scholar]
- 115.Guillaumond F, Boyer B, Becquet D, Guillen S, Kuhn L, et al. Chromatin remodeling as a mechanism for circadian prolactin transcription: rhythmic NONO and SFPQ recruitment to HLTF. Faseb J. 2011;25:2740–2756. doi: 10.1096/fj.10-178616. [DOI] [PubMed] [Google Scholar]
- 116.Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2011;9:1568–1576. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- 117.Flores-Alcantar A, Gonzalez-Sandoval A, Escalante-Alcalde D, Lomeli H. Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res. 2011;345:137–148. doi: 10.1007/s00441-011-1182-x. [DOI] [PubMed] [Google Scholar]
- 118.Mohrmann L, Langenberg K, Krijgsveld J, Kal AJ, Heck AJ, et al. Differential targeting of two distinct SWI/SNF-related Drosophila chromatin-remodeling complexes. Mol Cell Biol. 2004;24:3077–3088. doi: 10.1128/MCB.24.8.3077-3088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moshkin YM, Mohrmann L, van Ijcken WF, Verrijzer CP. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol Cell Biol. 2007;27:651–661. doi: 10.1128/MCB.01257-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vorobyeva NE, Soshnikova NV, Nikolenko JV, Kuzmina JL, Nabirochkina EN, et al. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A. 2009;106:11049–11054. doi: 10.1073/pnas.0901801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, et al. Analysis of the human endogenous coregulator complexome. Cell. 2011;145:787–799. doi: 10.1016/j.cell.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142:967–980. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 123.Haldar M, Hancock JD, Coffin CM, Lessnick SL, Capecchi MR. A conditional mouse model of synovial sarcoma: insights into a myogenic origin. Cancer Cell. 2007;11:375–388. doi: 10.1016/j.ccr.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 124.Drost J, Mantovani F, Tocco F, Elkon R, Comel A, et al. BRD7 is a candidate tumour suppressor gene required for p53 function. Nat Cell Biol. 2010;12:380–389. doi: 10.1038/ncb2038. [DOI] [PubMed] [Google Scholar]
- 125.Richards S, Gibbs RA, Weinstock GM, Brown SJ, Denell R, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 126.de Bruijn DR, van Dijk AH, Willemse MP, van Kessel AG. The C terminus of the synovial sarcoma-associated SSX proteins interacts with the LIM homeobox protein LHX4. Oncogene. 2008;27:653–662. doi: 10.1038/sj.onc.1210688. [DOI] [PubMed] [Google Scholar]
- 127.Craig E, Zhang ZK, Davies KP, Kalpana GV. A masked NES in INI1/hSNF5 mediates hCRM1-dependent nuclear export: implications for tumorigenesis. Embo J. 2002;21:31–42. doi: 10.1093/emboj/21.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kawai A, Naito N, Yoshida A, Morimoto Y, Ouchida M, et al. Establishment and characterization of a biphasic synovial sarcoma cell line, SYO-1. Cancer Lett. 2004;204:105–113. doi: 10.1016/j.canlet.2003.09.031. [DOI] [PubMed] [Google Scholar]
- 129.Olsen JV, Ong SE, Mann M. Trypsin cleaves exclusively C-terminal to arginine and lysine residues. Mol Cell Proteomics. 2004;3:608–614. doi: 10.1074/mcp.T400003-MCP200. [DOI] [PubMed] [Google Scholar]
- 130.Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 131.Breitkreutz BJ, Stark C, Tyers M. Osprey: a network visualization system. Genome Biol. 2003;4:R22. doi: 10.1186/gb-2003-4-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hunter S, Jones P, Mitchell A, Apweiler R, Attwood TK, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Vugt JJ, Ranes M, Campsteijn C, Logie C. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim Biophys Acta. 2007;1769:153–71. doi: 10.1016/j.bbaexp.2007.01.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mass spectrometry results, including; accession numbers, short protein descriptions, peptide sequences, associated Mascot score, peptide delta score and absolute calibrated mass relative error.
(XLS)
Quadruplate polyA mRNA expression profile of Hek293 cells determined with the Affymetrix human exon array 1.0 ST platform.
(XLS)