Abstract
Protein kinases are attractive therapeutic targets, but their high sequence and structural conservation complicates the development of specific inhibitors. We recently discovered from a DNA-templated macrocycle library inhibitors with unusually high selectivity among Src-family kinases. Starting from these compounds, we developed and characterized in molecular detail potent macrocyclic inhibitors of Src kinase and its cancer-associated gatekeeper mutant. We solved two co-crystal structures of macrocycles bound to Src kinase. These structures reveal the molecular basis of the combined ATP- and substrate peptide-competitive inhibitory mechanism and the remarkable kinase specificity of the compounds. The most potent compounds inhibit Src activity in cultured mammalian cells. Our work establishes that macrocycles can inhibit protein kinases through a bi-substrate competitive mechanism with high potency and exceptional specificity, reveals the precise molecular basis for their desirable properties, and provides new insights into the development of Src-specific inhibitors with potential therapeutic relevance.
Keywords: kinase inhibition, Src, macrocycle, mixed inhibition, gatekeeper mutant, DNA-templated synthesis
Introduction
Since protein kinases play a central role in cell signaling, the discovery and development of protein kinase inhibitors have been the focus of intensive research over the past two decades.1 Currently, all FDA-approved small-molecule inhibitors of protein kinases target the binding site for ATP, the common substrate in the enzymatic reaction of all 518 human kinases.2 The high sequence conservation within the kinase ATP binding pocket, however, makes challenging the development of specific inhibitors of protein kinases. When such specificity is achieved, kinase inhibitors can become effective drugs such as the c-Abl kinase inhibitor imatinib, which treats chronic myelogenic leukemia (CML).3–5
Non-ATP competitive inhibitors have the potential for exhibiting excellent specificity due to the more varied nature of their binding sites among kinases. Allosteric inhibitors such as GNF-2 (targeting Abl kinase) and CI-1040 (targeting MEK kinases) have been shown to be potent and highly specific.6–8 Recently we developed computational approaches for identifying allosteric binding sites of Src kinase.9 Likewise, substrate peptide-competitive inhibitors such as tyrphostins (targeting EGFR kinase) have the potential to be highly specific because of the sequence variation in the substrate-peptide binding sites of protein kinases.10,11
In addition to inadequate specificity, vulnerability to drug-resistance mutations represents another major problem facing the use of kinase inhibitors as therapeutics. 12 One of the most common imatinib resistance mutations in Abl kinase, Thr315Ile, exhibits resistance to other available kinase inhibitor therapeutics.13 This threonine residue (Thr338 in chicken c-Src) regulates access to a hydrophobic pocket adjacent to the ATP binding pocket and is therefore often referred to as the “gatekeeper.” Replacement of the gatekeeper residue in Src, Abl, PDGFR, and EGFR kinases with hydrophobic residues increases kinase activity and leads to the transformation of Ba/F3 cells.14
Most small-molecule kinase inhibitor discovery efforts rely on combinatorial or diversity-oriented synthesis and high-throughput screening (HTS). In contrast, we developed DNA-templated synthesis as a method of translating DNA sequences into small-molecule libraries that can be directly subjected to in vitro selections for desired properties including target affinity.15–18 Selection-based approaches are typically much more efficient than screening because they enable the simultaneous evaluation of all library members in one experiment regardless of library size, obviating the time and infrastructure demands of screening.19 Recently, we reported the synthesis20 and selection21 of a 13,824-membered DNA-templated macrocycle library. We identified from this library a series of macrocycles that inhibit Src with IC50 values as potent as 680 nM. Two of these macrocyclic compounds (2 and 9, Fig. 1, Table 1) displayed a remarkable level of specificity, inhibiting Src kinase but not Abl kinase or closely related Src-family kinases including Hck.
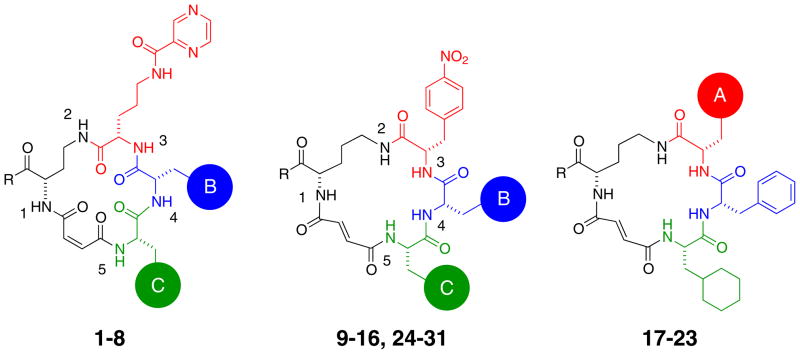
Figure 1.
Chemical structures of macrocycles described in this work. The compounds fall into two families: 1–8, which contain a diaminobutyric acid scaffold, cis-olefin, and N-pyrazinylcarbonyl-ornithine building block at the A-position, and 9–31, which contain an ornithine scaffold, trans-olefin, and para-substituted phenylalanine building block at the A-position.
Table 1.
Chemical composition and Src kinase inhibition activities of macrocycles. Macrocycles 1, 2, and 9 were previously referred to as cis-All-B8-C10-D7, cis-A11-B1-C5-D7, and trans-A10-B1-C5-D6 respectively. The concentration of compound that inhibited 50% of Src83-533 kinase activity (IC50) was determined using the Z′-LYTE kinase assay (see Methods). All experiments were performed in triplicate, and data represent mean values ± standard deviation.
| Compound | A-building block | B-building block | C-building block | R-group | N-methyl amide | Src IC50 (μM) |
|---|---|---|---|---|---|---|
| 1 | N-pyrazinylcarbonyl-ornithine | cyclohexylalanine | styrylalanine | -NH2 | - | 25 ± 3.0 |
| 2 | N-pyrazinylcarbonyl-ornithine | furylalanine | cyclopropylalanine | -NH2 | - | 0.60 ± 0.05 |
| 3 | N-pyrazinylcarbonyl-ornithine | phenylalanine | cyclopropylalanine | -NH-CH2CH3 | - | 0.19 ± 0.01 |
| 4a | N-pyrazinylcarbonyl-ornithine | phenylalanine | cyclohexylalanine | -NHCH2CH3 | - | ≤ 0.004 |
| 4b | N-pyrazinylcarbonyl-ornithine | phenylalanine | cyclohexylalanine | -OH | - | ≤ 0.004 |
| 5 | N-pyrazinylcarbonyl-ornithine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 2 | ≤ 0.004 |
| 6 | N-pyrazinylcarbonyl-ornithine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 3 | 3.0 ± 0.51 |
| 7 | N-pyrazinylcarbonyl-ornithine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 4 | > 5.0 |
| 8 | N-pyrazinylcarbonyl-ornithine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 5 | 0.90 ± 0.10 |
| 9 | p-nitrophenylalanine | furylalanine | cyclopropylalanine | -NH2 | - | 0.25 ± 0.03 |
| 10 | p-nitrophenylalanine | phenylalanine | cyclopropylalanine | -NH2 | - | 0.08 ± 0.008 |
| 11 | p-nitrophenylalanine | cyclohexylalanine | cyclopropylalanine | -NH2 | - | 2.0 ± 0.10 |
| 12 | p-nitrophenylalanine | pentafluorophenylalanine | cyclopropylalanine | -NH2 | - | > 10 |
| 13 | p-nitrophenylalanine | phenylalanine | phenylalanine | -NH2 | - | 0.22 ± 0.005 |
| 14 | p-nitrophenylalanine | phenylalanine | diphenylalanine | -NH2 | - | 1.3 ± 0.20 |
| 15 | p-nitrophenylalanine | phenylalanine | 1-naphthylalanine | -NH2 | - | 0.37 ± 0.03 |
| 16 | p-nitrophenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.006 ± 0.002 |
| 17 | p-methylphenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.27 ± 0.04 |
| 18 | p-chlorophenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.047 ± 0.007 |
| 19 | p-bromophenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.030 ± 0.002 |
| 20 | p-trifluoromethylphenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.18 ± 0.04 |
| 21 | p-cyanophenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | ≤ 0.004 |
| 22 | p-carbamoylphenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | 0.036 ± 0.004 |
| 23 | p-tert-butyl-phenylalanine | phenylalanine | cyclohexylalanine | -NH2 | - | > 10 |
| 24 | p-nitrophenylalanine | tyrosine | cyclohexylalanine | -NH2 | - | 0.79 ± 0.005 |
| 25a | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -NH2 | - | ≤ 0.004 |
| 25b | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | - | ≤ 0.004 |
| 26 | p-nitrophenylalanine | p-methylphenylalanine | cyclohexylalanine | -NH2 | - | 0.37 ± 0.04 |
| 27 | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 1 | ≤ 0.004 |
| 28 | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 2 | 0.036 ± 0.001 |
| 29 | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 3 | 0.16 ± 0.006 |
| 30 | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 4 | 5.0 ± 0.30 |
| 31 | p-nitrophenylalanine | p-fluorophenylalanine | cyclohexylalanine | -OH | position 5 | 0.57 ± 0.02 |
In this study we have developed second-generation macrocycles based on 2 and 9 with potencies as high as IC50 ≤ 4 nM. The most potent macrocycles inhibit Src kinase activity in mammalian cells. We determined the inhibition mechanism of these inhibitors and the structural basis of their unusual specificity by solving the X-ray co-crystal structure of two macrocycles bound to Src kinase domain. Our studies reveal that the macrocycles force the kinase to adopt the Src/CDK-like inactive conformation. In addition, the inhibitors occupy the ATP binding pocket of the kinase and simultaneously disrupt the substrate-peptide binding patch. We identified three amino acid substitutions between Src and Hck that determine the specificity of macrocycles for Src over Hck. Finally, we discovered that macrocycles derived from 9 potently inhibit the Thr338Ile Src gatekeeper mutant. Taken together, these findings establish that macrocycles can serve as Src kinase inhibitors with exceptional kinase selectivity, potency against common drug-resistant forms of tyrosine kinases, and activity in cultured mammalian cells. In addition, our results reveal that the macrocycles studied here achieve these properties through a bi-substrate mode of kinase inhibition that locks the kinase in an inactive conformation.
Results
Improvement of Src-inhibiting macrocycle potency
Since ATP-competitive kinase inhibitors must compete with millimolar ATP concentrations in the cell, nanomolar in vitro potency (typically measured in the presence of ATP concentrations near KM, ATP) is often required for a kinase inhibitor to demonstrate cellular activity at micromolar concentrations.22 We therefore sought to improve the potency of pyrazine-containing 2 and p-nitrophenylalanine-containing 9 by the systematic optimization of macrocycle building blocks (Fig. 1, Table 1). Derivatives of 9 were synthesized by Fmoc solid-phase peptide synthesis as previously described21 and assayed against Src83-533. Since we previously observed the importance of the nitro-Phe group at position A in 9 for Src kinase inhibition21, we first replaced building blocks at the B and C positions (Fig. 1, Table 1). Holding the amino acids at the A and C positions constant, we synthesized and assayed variants of 9 containing phenylalanine (10), cyclohexylalanine (11), and pentafluorophenylalanine (12) at the B position in place of furylalanine (Fig. 1, Table 1). While substituting pentafluorophenylalanine at this position abolished activity (IC50 > 10 μM) and cyclohexylalanine resulted in a ~10-fold decrease in potency, the phenylalanine derivative (10) resulted in a 3-fold increase in Src inhibition potency to IC50 = 80 nM (Fig. 1, Table 1).
We next varied building blocks at the C position. Holding the A and B positions constant as p-nitrophenylalanine and phenylalanine, respectively, we substituted phenylalanine (13), diphenylalanine (14), 1-naphthylalanine (15), and cyclohexylalanine (16) at the C position in place of cyclopropylalanine (Fig. 1, Table 1). Phenylalanine and naphthylalanine decreased Src inhibition potency by 3- to 5-fold, while diphenylalanine resulted in a 16-fold decrease in potency. Cyclohexylalanine, however, improved inhibition potency by more than 10-fold, resulting in macrocycle 16 with Src IC50 = 6 nM (Fig. 1, Table 1). These results collectively demonstrate that modest modifications in the size and shape of the macrocycle side chains can result in significant gains in potency. Notably, none of the amino acids that increased Src inhibition potency were present at the appropriate position in the DNA-templated library from which the initial macrocycles were discovered, consistent with the accuracy of the structure-activity relationships resulting from the in vitro selection for Src binding.20
We next installed more subtly altered building blocks into the partially optimized macrocycle 16. We probed the importance of p-nitrophenylalanine at the A position by replacing the nitro group at the para position with methyl (17), chloro (18), bromo (19), trifluoromethyl (20), cyano (21), carbamoyl (22), or tert-butyl (23) substituents (Fig. 1, Table 1). Remarkably, all analogs except the electronically similar p-cyanophenylalanine exhibited substantial reductions in potency. We implemented similar changes to the optimized B position building block, phenylalanine, by replacing it with tyrosine (24), p-fluorophenylalanine (25a) or p-methylphenylalanine (26) (Fig. 1, Table 1). Introduction of a methyl or hydroxyl group onto the phenyl ring resulted in a substantial decrease in Src inhibition. In contrast, the p-fluorophenylalanine derivative 25a, a very conservative change, retained the activity of 16 (Fig. 1, Table 1).
The analogous building block substitutions in the pyrazine-containing macrocycle structures (starting from 2), changing furylalanine to phenylalanine and cyclopropylalanine to cyclohexylalanine, resulted in macrocycle 4a with >150-fold greater potency against Src kinase than the parent compound 2 (Fig. 1, Table 1). Indeed, we could not accurately measure the potency of 4a since its IC50 of ≤ 4 nM was equivalent to ≥50% of the enzyme concentration required in the in vitro kinase assay. These findings support a similar mode of binding for the B and C building blocks in 2- and 9-derived macrocycles.
We also studied the effect of modifying the macrocycle peptide backbone on Src kinase inhibition. We systematically replaced each amide in the backbone of the improved p-nitrophenylalanine-containing macrocycle 25a with an N-methyl amide, generating 27–31 (Fig. 1, Table 1) and performed the same N-methyl amide backbone scan in the pyrazine-based macrocycle 4a (generating macrocycles 5–8, Fig. 1, Table 1) using Fukuyama-sulfonamide-alkylation chemistry on solid support.23 Nine of the ten possible N-methyl amide macrocycles were synthesized; methylation of the diaminobutyric acid α-nitrogen of 4a impaired macrocyclization. All N-methyl amide-containing macrocycles exhibited substantially lower activity against Src kinase, with the exception of compounds 5, 27 and 28, in which the methyl group resides on the scaffold diamino acid rather than on one of the three side-chain containing building blocks (Fig. 1, Table 1). The sensitivity of the macrocycle backbone to N-methylation suggests the importance of backbone hydrogen bonds or backbone conformation for kinase inhibition.
Specificity of macrocyclic Src kinase inhibitors
The development of small-molecule kinase inhibitors with specificity within the Src family has proven challenging. Notable successes in this area are a family of Lck-selective thienopyridine compounds24,25 and a catechol-based Src-selective inhibitor.26 Macrocycles 1, 2 and 9 inhibit Src kinase with exceptional specificity over the closely related Src family kinases Hck and Lck, or c-Abl kinase.21 To understand the origin of this unusual specificity we characterized in greater depth the activity and specificity of three previously reported macrocycles 1, 2, and 9 (Fig. 1, Table 1), as well as two second-generation compounds with greatly improved potency, 4b and 25b (the C-terminal carboxylate analogs of 4a and 25a, Fig. 1, Table 1). Since 4b and 25b are equally potent against Src kinase as carboxamides 4a and 25a, but offer higher aqueous solubility, we used these compounds for the biochemical and structural studies described below. Macrocycles 1, 2 and 4b share a diaminobutyric acid scaffold, cis-olefin stereochemistry, and a pyrazine group in position A, whereas macrocycles 9, and 25b contain an ornithine scaffold, trans-olefin stereochemistry, and nitrophenylalanine in position A (Fig. 1, Table 1).
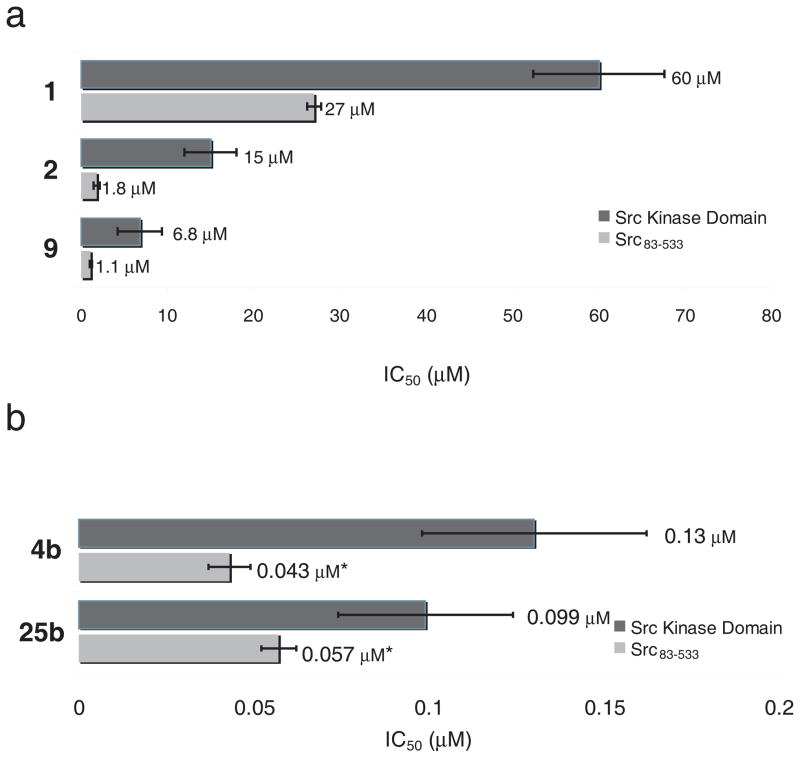
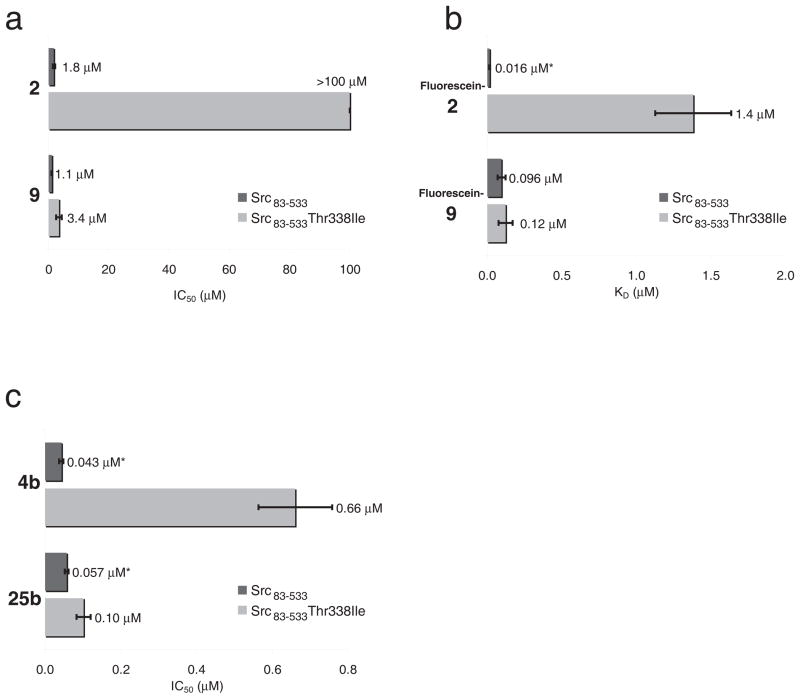
The original characterization of kinase inhibition was performed with a commercial kinase assay that relies on a fluorescently labeled peptide kinase substrate (Z′-LYTE, Invitrogen). For further characterization, we used a continuous spectrophotometric kinase activity assay that is easily adapted to different substrate peptides as well as a wider range of peptide concentrations. Using this assay, we found that the original compounds 1, 2, and 9 inhibited 50% of Src kinase domain activity (IC50) at 60 μM, 15 μM, and 6.8 μM respectively in the presence of 5 μM ATP and 100 μM Src-optimal substrate peptide (Supplementary Results, Supplementary Fig. 1). The IC50 values of these compounds against Hck, Lck, and Abl kinase were over 100 μM. Consistent with their improved potency in the Z′-LYTE assay, the second-generation compounds 4b and 25b exhibited more than 100-fold higher potency than the first generation compounds. Because the potency of the second-generation compounds approached the concentration of enzyme used in the assay, we increased the ATP concentration to 250 μM and the peptide concentration to 300 μM to determine their IC50 values. Under these more stringent conditions, 4b and 25b inhibited Src kinase domain with IC50 values of 0.13 μM and 0.099 μM, respectively, and exhibited only modest potency against the Src-family kinases Hck (IC50 = 0.86 μM and 8.4 μM, respectively) and Lck (IC50 = 2.4 μM and 6.1 μM, respectively). We noted that all compounds tested (1, 2, 4b, 9, and 25b) inhibit Src kinase constructs that include the regulatory SH3 and SH2 domains (chicken c-Src 83-533) approximately 2- to 10-fold more potently than the isolated kinase domain (chicken c-Src 251-533) (Fig. 2). The selectivity of the most potent second-generation compound 25b for Src over Lck, Hck and Abl kinase was maintained when the corresponding three-domain constructs were used (Supplementary Fig. 2).
Figure 2.
Potency of macrocycle compounds against Src kinase domain constructs. (a) Inhibition of Src kinase domain (chicken c-Src251-533) and Src containing the N-terminal SH3 and SH2 domains in addition to the kinase domain (chicken c-Src83-533). IC50 values of 1, 2 and 9 were determined in the presence of 5 μM ATP and 100 μM Src-optimal substrate peptide. (b) Inhibition of Src kinase domain and Src83-533 by 4b, and 25b was determined in the presence of 250 μM ATP and 300 μM Src-optimal substrate peptide. *Values represent an upper estimate of the IC50 because the concentration of kinase used in the assay is higher than the measured apparent IC50. All experiments were performed in triplicate, and data represent mean values ± standard deviation.
The high specificity of the macrocycles for Src kinase over Lck and Hck is unusual because Src, Lck, and Hck typically behave very similarly towards known kinase inhibitors.27 In a recent study addressing the selectivity of kinase inhibitors, only two out of 38 compounds could discriminate Src from Hck with the 5-fold selectivity exhibited by 4b, and none with the 100-fold selectivity exhibited by 25b. In the same study, only two out of 38 compounds showed the 10-fold selectivity of 4b between Src and Lck, and none exhibited the 60-fold selectivity of 25b28. The molecular basis of the remarkable specificity of these macrocyclic kinase inhibitors is revealed in the experiments below.
ATP- and substrate peptide-competitive inhibition
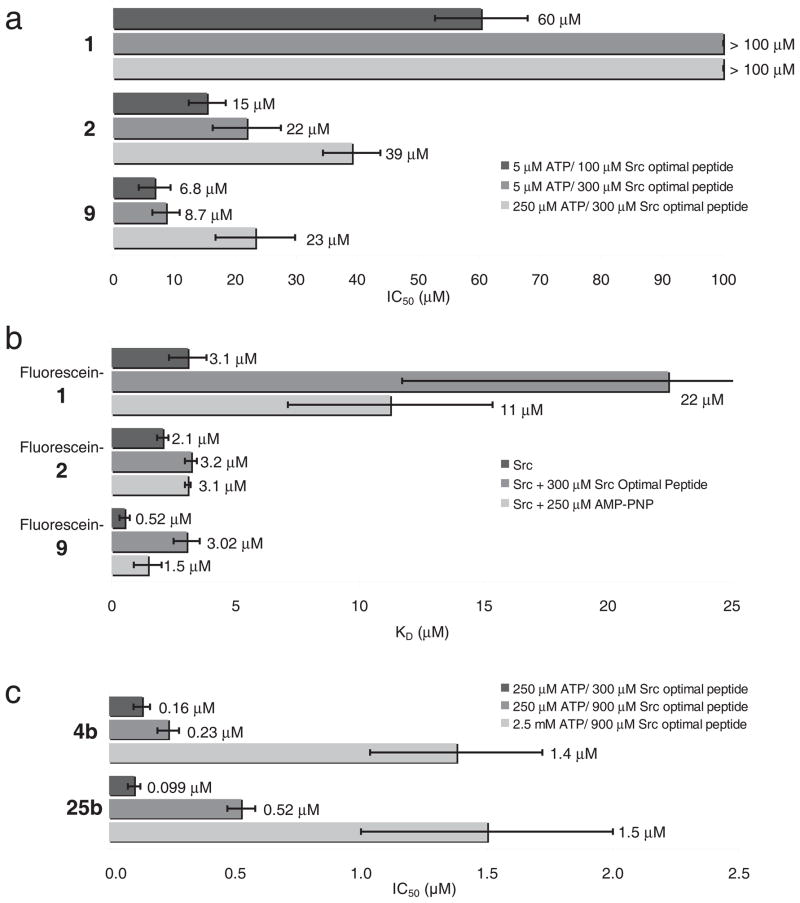
The large size and unusual specificity of the macrocyclic Src inhibitors led us to speculate that they may interfere with the binding of not only ATP, but the substrate peptide as well. To probe this possibility, we tripled the concentration of the Src-optimal substrate peptide in the kinase assay, and observed a 25% to 50% increase in the apparent IC50 for 1, 2, and 9 (Fig. 3). These results suggest that the macrocycle compounds could be substrate peptide-competitive. For comparison, we also raised the ATP concentration 50-fold and measured for 1, 2, 9 a 2-fold increase in IC50 consistent with the known ATP-competitive behavior of the compounds21 (Fig. 3). To clarify the inhibitory mechanism of the compounds further we synthesized fluorescein-labeled derivatives of 1, 2, and 9 (carboxyfluorescein-1, -2 and -9, respectively) and measured their binding affinity to Src kinase domain in a fluorescence anisotropy assay (Fig. 3). We found that Src kinase domain binds the fluorescently labeled macrocycles 1, 2 and 9 with dissociation constants (KD) of 3.1 μM, 2.1 μM, and 0.52 μM, respectively. The binding affinity of the compounds decreased in the presence of 250 μM of the non-hydrolyzable ATP analog AMP-PNP to 11 μM, 3.1 μM, and 1.5 μM, respectively (Fig. 3), as would be expected for an ATP-competitive inhibitor. Consistent with the increase in IC50 observed in the kinase assay, the presence of 300 μM Src-optimal substrate peptide decreased the apparent affinities of the fluorescent-macrocycle conjugates 7-fold for 1, 1.5-fold for 2, and 6-fold for 9 (Fig. 3). Because of their increased potency, we tested the substrate competitive behavior of the improved compounds 4b and 25b by varying the ATP concentration from 250 μM to 2.5 mM and the substrate peptide concentration from 300 μM to 900 μM. The ATP- and substrate peptide-competitive nature of both compounds is maintained under these conditions (Fig. 3). These results collectively indicate that the macrocycles are both ATP- and substrate peptide-competitive inhibitors.
Figure 3.
Macrocycle compounds are ATP- and peptide-competitive inhibitors. (a) IC50 values of 1, 2, and 9 for Src kinase domain in response to a 50-fold increase in ATP concentration (5 μM to 250 μM), and a 3-fold change in Src-optimal peptide concentration (100 μM to 300 μM). (b) Dissociation constant (KD) of fluorescein-labeled 1, 2, and 9 for Src kinase domain in the presence of 250 μM AMP-PNP or 300 μM Src-optimal peptide. (c) IC50 values of 4b, and 25b for Src kinase domain in response to a 10-fold increase in ATP (250 μM to 2.5 mM), and a 3-fold change in Src-optimal peptide (300 μM to 900 μM). All experiments were performed in triplicate, and data represent mean values ± standard deviation.
Structural basis for Src kinase inhibition by macrocycles
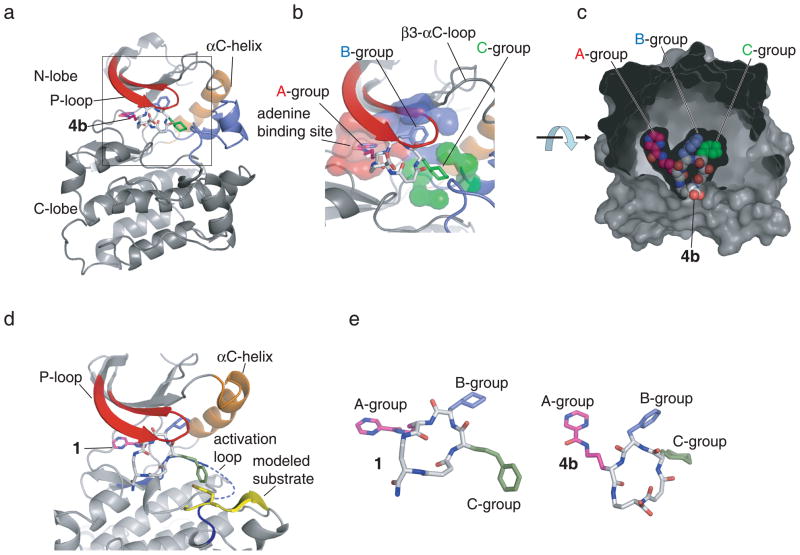
To determine the structural basis of Src inhibition, we solved the X-ray crystal structure of 4b in complex with c-Src kinase domain at a resolution of 1.9 Å (Fig. 4 and Supplementary Fig. 3). Surprisingly, the kinase domain adopts the Src/CDK-like inactive conformation; to our knowledge, this structure is the first in which the isolated kinase domain of Src has been observed in this conformation. Macrocycle 4b binds to the active site of the Src kinase domain (Fig. 4). The macrocycle forms five direct hydrogen bonds and four water-mediated hydrogen bonds with the kinase. Interestingly, the backbone of the macrocycle forms seven of these nine hydrogen bonds, explaining the loss of macrocycle potency upon backbone amide methylation (Fig. 1, Table 1). A total of 1392 Å2 of molecular surface is buried from solvent upon binding of 4b to the kinase.
Figure 4.
The three-dimensional structure of Src kinase domain bound to macrocyclic inhibitors. (a) 4b binds to the active site of the kinase underneath the phosphate-binding P-loop (shown in red). The kinase adopts the Src/CDK-like inactive conformation characterized by the outward orientation of helix αC (orange) and the disruption of the peptide-binding patch by the activation loop (blue). (b) The three building block positions of the macrocycle compounds occupy three distinct binding pockets: the A-building block binds to the adenine binding pocket (red), the B-building block binds to a hydrophobic pocket underneath the β3 -αC loop, and the C-building block occupies a binding pocket (green) facing the Asp-Phe-Gly motif of the activation loop. (c) 4b binding pocket in detail. The surface created by the protein and ordered water molecules is shown in grey. The carboxylate group, from which DNA was attached during in vitro selection of the DNA-templated library and from which the fluorescein group was attached during binding affinity measurements, is exposed to solvent. (d) Superposition of the experimental X-ray crystal structure of Src•1 with the structure of the substrate peptide (yellow) from the complex with IRK (pdb-entry: 1IR3).32 (e) Comparison of the structures of 1 and 4b when complexed with Src kinase domain. The macrocycle structures are shown from a perspective that fixes the kinase domains (not shown) in the same orientation.
Consistent with the macrocycle structure-activity relationships described above, 4b occupies three distinct binding sites (Fig. 4). The pyrazine group from building block A binds to the ATP binding pocket and forms a hydrogen bond with the backbone of the kinase, similar to the binding mode of adenine (Fig. 4).14 The phenylalanine side chain of building block B occupies a hydrophobic pocket between the β3-αC loop in the N-lobe of the kinase and the Asp-Phe-Gly (DFG) motif at the beginning of the activation loop (Fig. 4). The outward rotation of helix αC and the disruption of the salt bridge between Lys295 and Glu310 open up this hydrophobic pocket lined by Val281, Lys295, Leu297, Ile336, and Leu407. In the active conformation of the kinase, cyclohexylalanyl or phenylalanyl side chains at position B would clash with the side chain of Lys295 and Phe307, explaining the incompatibility of the bound macrocycles with the active conformation of the enzyme. The cyclohexylalanyl side chain of building block C faces into an amphipathic binding pocket around residues Phe278, Leu407, Ile411, Tyr416, Asp386, Arg388, and Asn391 (Fig. 4). The C-terminal carboxyl group of 4b, which represents the site of attachment of DNA in the library, faces the solvent and does not interact with the kinase, showing how the DNA-linked macrocycle could bind to Src during in vitro selection (Fig. 4).21
Structural basis of substrate peptide-competitive behavior
We were interested in the binding mode of 1 because it has the most pronounced substrate peptide-competitive behavior of the compounds tested (Fig 3). We solved the structure of Src kinase domain bound to 1 at a resolution of 2.2 Å (Fig. 4 and Supplementary Fig. 3). 1 and 4b belong to the same family of macrocycles, sharing a diaminobutyric acid backbone as well as a pyrazine group in position A. They differ in position B, where 1 contains cyclohexylalanine compared to phenylalanine in 4b, and in position C where 1 contains styrylalanine instead of cyclohexylalanine in 4b (Fig. 1, Table 1). The overall binding mode of 4b and 1 is similar, but 1 binds deeper into the active site of the kinase and the Cα-atoms of groups A, B and C in 1 are shifted 1.6 Å to 2.6 Å towards helix αC compared to 4b. While the three side chains of 1 and 4b occupy similar binding pockets of the kinase, the conformation of the macrocycle peptide backbone differs substantially between the two structures (Fig. 4). The intramolecular distances within the macrocycle backbone change on average by 0.74 Å and maximally by 4.19 Å between the structures of Src in complex with 1 and 4b (Fig. 4). These observations suggest that macrocycles of the 1 family are sufficiently rigid to bind with high affinity, but are flexible enough to accommodate subtle changes in binding conformation that optimize interactions with kinase residues.
Similar to the Src•4b structure, the side chain groups of macrocycle 1 occupy three distinct pockets in the kinase active site. The pyrazine group at the A-position binds to the ATP binding pocket in both structures. At the B-position, the cyclohexylalanine of 1 and the phenylalanine of 4b bind to the hydrophobic pocket towards the outwardly rotated helix αC. The styryl group at the C-position of 1 faces the C-terminal end of the activation loop of the kinase (residues 404–425), which is disordered between residues 405 and 423 (Fig. 4). The symmetry of the crystal would allow the activation loop in the Src•1 complex to access the same conformation as that observed in the Src•4b complex. Upon binding of 1 to the kinase, 1459 Å2 of surface area are buried, comparable to the amount of buried surface in the Src•4b complex. However, 1 forms only three hydrogen bonds with the backbone of the kinase and one hydrogen bond between its carboxamide terminus and the side-chain carboxylate of Asp348. The loss of five hydrogen bonds in the Src•1 complex compared to the Src•4b complex could explain the large difference in potency between these two inhibitors. This is largely the result of a 3.4 Å shift of the backbone Cα-atoms of 1 towards helix αC.
While a structure showing the interaction between Src and substrate peptide has not been reported, the closely related kinases Abl (PDB-entry: 2G2I31) and IRK (PDB-entry: 1IR332) have been crystallized in complex with substrate peptides. An analysis of these structures together with ours provides a plausible molecular basis for the observed substrate-competitive nature of our macrocycles. Substrate peptide binding is not compatible with the Src/CDK-like inactive conformation of the activation loop in the Src•1 complex likely because the peptide binding patch on the kinase is disrupted. When the substrate peptide from the complex with IRK is aligned onto the Src•1 complex, it clashes with the activation loop and the styryl moiety of 1 (Fig. 4). The cyclohexyl group of 4b does not clash with the substrate tyrosine in a model of a docked substrate peptide, but the inactive conformation of the activation loop in the Src•4b complex disrupts the substrate binding patch. Taken together, these observations reveal the molecular basis of the observed substrate peptide-competitive behavior of the macrocycles: the bulky groups in the B-position of the macrocycles (e.g. phenylalanyl or cyclohexylalanyl) bind to a pocket that is only present in the Src/CDK-like inactive conformation in which the salt bridge between Lys295 and Glu310 is disrupted. In the Src/CDK-like inactive conformation, the outward rotation of helix αC is coupled to a rearrangement of the activation loop, which subsequently disrupts the binding patch for substrate peptide.
Molecular basis of Src versus Hck inhibition specificity
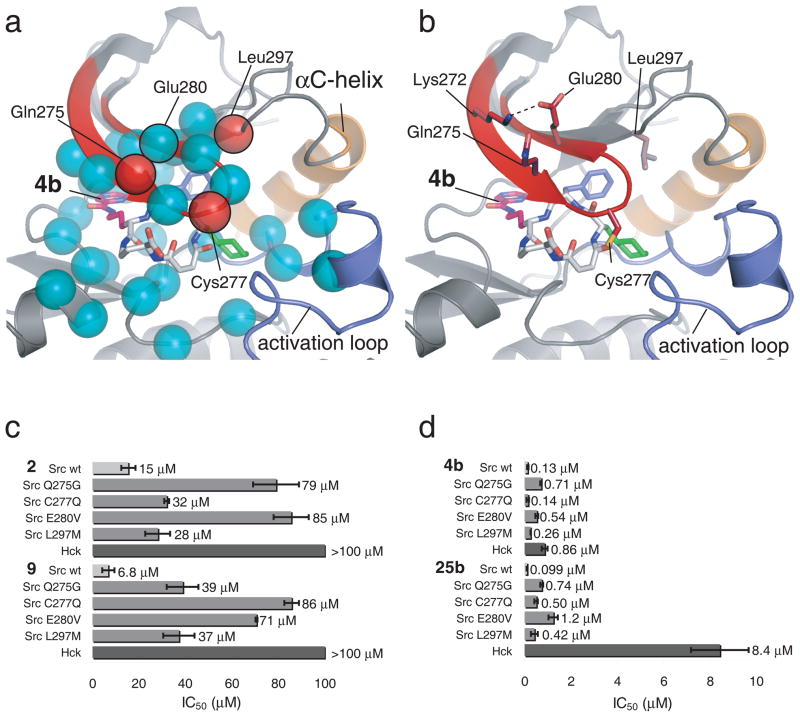
Based on the crystal structure of the Src•4b complex, we identified four Src amino acids that are within 5 Å of 4b and differ between Src and Hck (Fig. 5 Supplementary Fig. 4). Two of these Src residues, Gln275 and Cys277, are located in the phosphate-binding loop (P-loop) of the kinase. The third Src residue, Leu297, is in the β3 strand. The final difference, Src Tyr340 versus Hck Phe334, represents a modest change and either phenylalanine or tyrosine is found among kinases that are poorly inhibited by 4b (Lck Tyr318, Abl Phe317); therefore, this fourth residue was not studied further.
Figure 5.
Determinants of macrocycle specificity. (a) X-ray crystal structure of the Src 4b complex highlighting residues that differ between Src and Hck/Lck. Cα-atoms of amino acids within 5Å of 4b are depicted as spheres. Cyan spheres represent amino acids that are identical between Src and Hck/Lck, red spheres represent amino acids that differ between Src and Hck/Lck. (b) In the structural model of the Src 4b complex, the three amino acids within 5 Å of the macrocycle that differ between Src and Hck/Lck (Src Gln275, Cys277, and Leu297) are rendered as sticks, as well as the salt bridge forming Src residues Lys272 and Glu280. (c, d) IC50 of 2, 9, 4b and 25b for Src wt, Src Q275G, Src C277Q, Src E280V, Src L297M, and Hck kinase domains. IC50 values for 2 and 9 were determined in the presence of 5 μM ATP and 100 μM Src-optimal peptide. IC50 for 4b and 25b were determined in the presence of 250 μM ATP and 300 μM Src-optimal peptide. All experiments were performed in triplicate, and data represent mean values ± standard deviation.
Replacement of Cys277 in Src with the corresponding glutamine from Hck increased the IC50 of the mutant protein for the two ornithine-derived compounds 9 and 25b by 13-fold and 5-fold, respectively. In contrast, the inhibitory potency of the diaminobutyric acid-derived compounds 2 and 4b was hardly affected by the C277Q mutant (Fig. 5). We speculate that replacement of the small cysteine side chain with the larger glutamine side chain induces a steric clash between the kinase and the larger ornithine-containing macrocycle backbone. Substitutions at the C-building block, which is closest to the Cys277 side chain, do not correlate clearly with activity against this mutant. Since the C277Q mutation also removes a nucleophile that might react with the maleamide/fumaramide group found in all of the macrocycles, we performed competition binding experiments and time-dependent inhibition experiments (Supplementary Figs. 5 and 6). The results suggest that macrocycle inhibitors do not achieve Src selectivity by reacting with Cys277.
Gln275 in Src is replaced by alanine or glycine in Hck, Lck and Abl. The Q275G mutation in Src decreased the potencies of 2, 4b, 9 and 25b between 5 and 7-fold (Fig. 5). Interestingly, the side chain of Gln275 is facing away from 4b and hydrogen bonds to a salt bridge between Lys272 and Glu280 (Fig. 5). This salt bridge is thought to rigidify the otherwise flexible P-loop and its disruption decreases kinase activity.33 We speculate that the loss of this hydrogen bond in the Gln275Gly mutant destabilizes the P-loop and results in a reduction in potency for 4b. Likewise, disrupting the Lys272–Glu280 salt bridge by mutating Glu280 to valine in Src destabilizes the P-loop and results in a 5-fold loss in 4b potency (Fig. 5). Overall, the results of mutations to the P-loop of Src indicate that the binding of 4b to Src is likely dependent on the stability of the P-loop. In Hck, Lck, and Abl, which lack the glutamine in the salt-bridge, the P-loop may be more flexible and may adopt different conformations that make macrocycle binding more entropically unfavorable. Consistent with this model, some of the most common imatinib resistance mutations are in the P-loop of Abl kinase and include the residues corresponding to Src residues Gln275, Lys272 and Glu280.13
Leu297 is part of a cluster of hydrophobic residues that form the binding pocket for the B building blocks of 1 and 4b. Replacement of Leu297 in Src with the corresponding Hck residue (Met292) would likely decrease the size of the binding pocket and leads to a steric clash, explaining the lowered potency of 4b for Hck. All macrocycles tested contain a bulky 5- or 6-membered ring in the B-position and their potency against the Leu297Met mutant of Src decreased between 2- and 6-fold compared to their potencies against wild-type Src (Fig. 5). Because the two regions of sequence divergence between Src and Hck are independent, their effect on macrocycle potency could be at least partially additive.
Taken together, these mutational studies interpreted in light of the crystal structures suggest that specific interactions involving Src residues Gln275, Cys277, and Leu297 are the molecular basis for the unusual selectivity of these macrocycles for Src versus other Src-family kinases such as Hck.
Macrocycles are active against the Src gatekeeper mutant
Remarkably, p-nitrophenylalanine-containing macrocycles 9 and 25b inhibited the three-domain construct of the gatekeeper mutant Src Thr338Ile (residues 83-533) with potencies only 1.8- to 3.1-fold less than that of wild-type Src (Fig. 6). In contrast, the gatekeeper mutation had a much stronger impact on the potency of macrocycles containing the N-pyrazinylcarbonyl-ornithine building block at position A— 2 and 4b inhibit the gatekeeper mutant 100-fold and 15-fold less potently, respectively, than wild-type Src (Fig 6). Similarly, the gatekeeper mutation increases the dissociation constant almost 100-fold for fluorescein-labeled 2 but only 1.3-fold for fluorescein-labeled 9 (Fig. 6). This decrease in potency and affinity is consistent with the Src•4b structure (Supplementary Fig. 7), which suggests that substitution of Thr338 with isoleucine would create a steric clash with the N-pyrazinylcarbonyl-ornithine building block found in the A-position of 2 and 4b. In contrast, macrocycles 9 and 25b contain a smaller p-nitrophenylalanine building block at this position. We speculate that the smaller nitrophenyl group avoids steric repulsion with an isoleucine at Src residue 338, enabling compounds 9 and 25b to retain their potency against the Src Thr338Ile gatekeeper mutant.
Figure 6.
Activity of the macrocycles against the Thr338Ile gatekeeper mutant of Src kinase. (a) IC50 of 2, and 9 for Src83-533 and Src83-533 T338I. IC50 values for 2 and 9 were determined in the presence of 5 μM ATP, 100 μM Src-optimal peptide. (b) Measurement of the dissociation constant KD of fluorescein-labeled 2 and 9 for Src83-533 and Src83-533 T338I. *Value is an upper estimate of the KD because the KD is lower than the concentration of macrocycle compound used in the assay. (c) IC50 of 4b, and 25b for Src83-533, and Src83-533 T338I. IC50 values for 4b and 25b were determined in the presence of 250 μM ATP and 300 μM Src-optimal peptide. *Values are an upper estimate of the IC50 because the apparent IC50 is lower than the concentration of kinase used in the assay. All experiments were performed in triplicate, and data represent mean values ± standard deviation.
Src kinase inhibition in cell culture
The development of macrocyclic Src kinase inhibitors with low nanomolar in vitro potency raised the possibility that these compounds may inhibit Src kinase activity in living cells. We assayed our most potent carboxamide-containing macrocycles, 16, 25a, and 4a (Fig. 1, Table 1), against 3T3 (src−/−) cells transfected with a constitutively active form of murine c-Src (c-Src Y529F) 34. In these cells, Src kinase activity is responsible for nearly all the tyrosine phosphorylation detected by a phosphotyrosine antibody (Supplementary Fig. 8). Upon treatment with high micromolar concentrations of p-nitrophenylalanine-containing 25a, global phosphotyrosine levels were significantly reduced (Supplementary Fig. 9). Surprisingly, treating cells with 16, which differs in structure from 25a by the loss of a single fluorine atom, resulted in lower levels of tyrosine phosphorylation inhibition at comparable small molecule concentrations (Supplementary Fig. 9). Pyrazine-containing 4a at 100 μM did not reduce global tyrosine phosphorylation.
In addition to the abovementioned competition with high concentrations of intracellular ATP, we speculate that factors including modest cell-membrane permeability or differences in the inhibition of overexpressed and activated SrcY529F versus wild-type Src may explain the substantial differences between the in vitro potency and cell culture activity of these compounds.
Discussion
We systematically modified the building blocks of macrocyclic kinase inhibitors identified from the in vitro selection of a DNA-templated library and improved their potency by up to 240-fold while maintaining their unusually high specificity for Src kinase. Characterization of the inhibitory mechanism revealed that the compounds are both ATP-competitive and substrate peptide-competitive. Because the peptide binding patch on kinases is less conserved than the ATP binding pocket, compounds interacting with the former offer rich opportunities to develop kinase-specific inhibitors.
The three-dimensional structures of Src kinase domain complexed with a first-generation inhibitor and with an improved second-generation compound reveal that Src adopts the Src/CDK-like inactive conformation. These structures represent to our knowledge the first time that Src kinase domain alone has been observed to adopt this conformation, which has previously only been observed in the structures of larger Src constructs containing the N-terminal SH3/SH2 domains. This inactive kinase conformation is incompatible with substrate-peptide binding and provides the molecular basis for the substrate-peptide competitive behavior of the inhibitors. Macrocycle binding demands the Src/CDK-like inactive conformation for at least two reasons: first, the outward rotation of helix αC and the disruption of the salt bridge between Lys295 and Glu310 is required to form a binding pocket for building blocks in the B-position; second, the conformation of the activation loop of the kinase in its active form would clash with the backbone of the inhibitors as well as building blocks in the C-position.
The macrocycles studied here inhibit the larger Src constructs about 10-fold more potently than the Src kinase domain. Our structural data supports a model in which the ligands form no extra interactions with the larger constructs and therefore the observed differences in inhibitory potency are likely due to the relative stabilities of the conformational states between isolated kinase domain and the SH3-SH2-kinase domain constructs. This reasoning suggests that the SH3-SH2 domains stabilize the Src/CDK-like inactive conformation by approximately 1.3 kcal/mol. 35,36
Based on the structures and subsequent assays of mutant Src kinases, we identified two Src residues in the phosphate binding P-loop (Gln275 and Cys277) and one residue lining a hydrophobic pocket (Leu297) that largely explain the selectivity of the macrocycles for Src compared with Hck and, by inference, other Src-family kinases.
The two different families of Src-inhibiting macrocycles developed and characterized here differ mainly in their A-position building blocks and in the length and olefin stereochemistry of their backbones. The p-nitrophenylalanine-based macrocycles inhibit the gatekeeper mutant of Src kinase (Thr338Ile) with comparable potency as wild-type Src. In contrast, the pyrazine-containing macrocycles based on our structural data are predicted to clash with the Thr338Ile mutant, and indeed exhibit much lower ability to inhibit the gatekeeper mutant form of Src.
The dysregulation and activity of many kinases is associated with human disease. Highly specific kinase inhibitors such as the macrocyclic compounds presented here that compete not only with ATP binding but also with substrate-peptide binding could inspire the development of new inhibitors with two qualities unavailable to ATP-competitive inhibitors. First, we have shown that the level of peptide competition is tunable in these macrocycles. Second, a single peptide-competitive kinase inhibitor could in principle reshape signaling pathways downstream of the kinase by favoring the phosphorylation of strongly competing substrate peptides over weakly competing substrate peptides, rather than simply inhibiting the ability of the kinase to phosphorylate all downstream targets. Finally, the potency of some of the macrocycles characterized here, including those that inhibit the cancer-associated gatekeeper mutant, together with their observed activity in cell culture, are encouraging for the future development of macrocyclic kinase inhibitors with potential therapeutic relevance.
Methods
Macrocycle Synthesis
Carboxamide-containing macrocycles, which most closely resemble DNA-templated macrocycles, were synthesized on multi-milligram scale using Fmoc solid-phase peptide synthesis as previously described.21 Carboxylate-containing macrocycles, which in general exhibit comparable potency and increased solubility compared with the carboxamide-containing macrocycles, were synthesized using standard macrocycle synthesis protocols substituting 2-chlorotrityl resin (EMD Biosciences) in place of Rink amide resin. Fluorescein-macrocycle conjugates were synthesized using standard macrocycle synthesis protocols substituting 1,6-diaminohexane trityl resin (EMD Biosciences) in place of Rink amide resin. After HPLC purification, the 6-aminohexane-conjugate was reacted with 5 equivalents of 5-carboxyfluorescein N-succinimidyl ester (Sigma-Aldrich) and 10 equivalents of DIPEA in DMF. The fluorescein-macrocycle conjugate was then purified by reverse-phase HPLC using a C18 stationary phase and eluting with a gradient of water/acetonitrile. Proton NMR spectra and mass spectrometry data are provided in the Supplementary Information (Supplementary Table 2).
Protein Purification
Kinase domain constructs of human c-Abl (residues 229–512), chicken c-Src (residues 251-533 and residues 83-533), murine Lck (residues 227-509), and human Hck (residues 166-445) were expressed as previously described37,38. Mutations were introduced into chicken c-Src (residues 251-533) (Q275G, C277Q, E280V, L297M, and T338I) by site-directed mutagenesis and verified by DNA sequencing.
Crystallization
The complex between 1 and c-Src kinase domain (residues 251-533) was formed in a solution of 190 μM kinase domain, 476 μM 1, 50 mM Tris (pH 8.0), 125 mM NaCl, 5% DMSO, 2.5% Glycerol. Using the hanging drop vapor diffusion method (1 μL Src•1 complex + 1 μL mother liquor), crystals grew overnight at 24°C in a mother liquor of 0.1 M Bis-Tris (pH 6.5), 12% PEG 3350, 1% Tacsimate (pH 6.0). Crystals were cryoprotected in mother liquor plus 20% ethylene glycol, frozen, and stored in liquid nitrogen.
The complex of c-Src kinase domain and 4b was formed by concentrating 10-fold a mixture of 10 μM kinase domain and 12.5 μM 4b in 50 mM NaCl, 20 mM Tris (pH 8.0), and 5% glycerol. Using the hanging drop vapor diffusion method (1 μL Src•4b complex + 1 μL mother liquor), crystals grew in a mother liquor containing 200 mM ammonium sulfate, and 3% glycerol overnight at 24°C. Crystals were cryoprotected in mother liquor with 20% glycerol, frozen and stored in liquid nitrogen.
X-Ray Diffraction
X-ray diffraction data were collected at the National Synchrotron Light Source at Brookhaven National Laboratories beamline X29. Data for both complexes were collected at 100K and 1.075 Å wavelength.
Structure Determination of Src•1 complex
Data were processed in space group P21 (Supplementary Table 1) with DENZO and Scalepack via the HKL2000 suite.39 While the unit cell parameters are almost compatible with space group p222, processing of the data in p222 yielded poor statistics and molecular replacement with phaser40 failed. Analysis of the data for possible twinning in space group p21 with phenix.xtriage showed significant pseudo-merohedral twinning with the h, -k, -l operator. We therefore included the twin law in further refinement with Phenix, which improved refinement statistics and electron density maps. The structure was solved by molecular replacement using the kinase domain of human c-Src (PDB: 1Y57)41 (residues 260–520) without the αC-helix (residues 298–310) and the activation loop (residues 400–425) as a search model in Phaser.40 The structure was built in Coot,42 and refined with PHENIX.43 In the Ramachandran plot, 99.6% of the residues appear in the allowed regions, 96.4% of the residues appear in the favored regions and 0.4% of the residues appear in the outlier regions.
Structure Determination of Src•4b complex
Data were processed in space group P321 (Supplementary Table 1) using Mosflm and Scala in iMosflm.44 The structure was phased by molecular replacement using the kinase domain of inactive c-Src (PDB: 2SRC)45 (residues 250-533) without the αC-helix (residues 298-310) and activation loop (residues 400-425) as a search model in Phaser40 The model of the structure was built in Coot42 and refined in PHENIX.43 In the Ramachandran plot, 100% of the residues appear in the allowed regions, 98.5% of the residues appear in the favored regions, and 0% of the residues appear in the outlier regions.
Kinase Activity Assays
For the continuous spectrophotometric assay,46 100 μM Src-optimal substrate peptide (AEEEIYGEFAKKK)29,30 was combined with 5 μM ATP for 1, 2, and 9. Concentrations of kinase used for these assays were as follows: 0.125 μM for Src kinase domain, 0.33 μM for Src83-533, 0.33 μM for Src Q275G, 0.4 μM for Src C277Q, 0.8 μM for Src L297M, 0.4 μM for Src E280V, 0.042 μM for Hck, 0.25 μM for Lck, and 0.5 μM for Abl. For 4b, and 25b, the assay was performed using 250 μM ATP and 300 μM Src-optimal peptide (see the main text). The concentrations of kinase constructs used in these assays were as follows: 0.0125 μM for Src kinase domain, 0.1 μM for Src83-533, 0.033 μM for Src Q275G, 0.02 μM for Src C277Q, 0.04 μM for Src L297M, 0.02 μM for Src E280V, 0.0042 μM for Hck, 0.025 μM for Lck, and 0.05 μM for Abl. Titrations of 1, 2, 4b, 9, and 25b (ranging from 0 μM to 83.3 μM) were performed at 30 °C as described before for imatinib38 to determine the concentration at which 50% of the initial kinase activity is inhibited (IC50).
For the FRET-based endpoint assay, kinase inhibition was measured with the Z′-LYTE kinase assay (Invitrogen) using the Tyr 02 substrate peptide (based upon the optimal Abl substrate peptide EAIYAAPF) according to the manufacturers instructions. Kinase reactions were performed in the presence of 2 μM Tyr 02 substrate peptide, 50 μM ATP and 8 nM chicken c-Src83-533.
Anisotropy Binding Assay
Src kinase domain (residues 251-533) was titrated to 0.5 μM of the fluorescein-labeled macrocycle, in 100 mM Tris pH 8.0, 10 mM MgCl2 at 25°C. Src83-533 was titrated to 0.05 μM fluorescein-labeled macrocycle in 100 mM Tris pH 8.0, 10 mM MgCl2 at 25°C. After equilibration, the increase in the fluorescence anisotropy of the fluorescently labeled ligand was recorded and fitted against a quadratic binding equation in Kaleidagraph (Synergy Software) to yield the dissociation constant (KD).
Supplementary Material
Acknowledgments
3T3 (src−/−) cells transfected with a plasmid encoding c-Src Y529F or null vector were a gift from Professor Jonathan Cooper (Fred Hutchinson Cancer Research Center, Seattle, Washington). We thank the beamline staff at X29A at the National Synchrotron Light Source at Brookhaven National Laboratory, the use of which was supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences contract DE-AC02-98CH10886. G.G. and M.A.S. thank Lisa Malone for her assistance. This research was supported by the Howard Hughes Medical Institute (D.R.L.) and NIH/NIGMS grants GM065865 (D.R.L.) and GM080097 (M.A.S.). R.E.K. acknowledges NIH training grant support to the Harvard University Training Program in Molecular, Cellular, and Chemical Biology (MCCB). G.G. acknowledges NIH training grant support to the Graduate Program in Molecular and Cellular Pharmacology at Stony Brook University.
Footnotes
Authorship contribution
D.R.L and R.E.K. developed and synthesized the compounds, performed Z-LYTE assays and tested activity in 3T3 cells. G.G., M.P.-G., and M.A.S. prepared proteins and performed the structural studies and biochemical assays. All authors analyzed the data and wrote the manuscript.
Competing Financial Interest
D.R.L. is a consultant for Ensemble Therapeutics, a company that uses DNA-templated synthesis in drug discovery and development.
References
- 1.Cohen P. Protein kinases--the major drug targets of the twenty-first century? Nat Rev Drug Discov. 2002;1:309–15. doi: 10.1038/nrd773. [DOI] [PubMed] [Google Scholar]
- 2.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–34. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 3.Cools J, et al. PKC412 overcomes resistance to imatinib in a murine model of FIP1L1-PDGFR[alpha]-induced myeloproliferative disease. Cancer Cell. 2003;3:459–469. doi: 10.1016/s1535-6108(03)00108-9. [DOI] [PubMed] [Google Scholar]
- 4.Bikker JA, Brooijmans N, Wissner A, Mansour TS. Kinase domain mutations in cancer: implications for small molecule drug design strategies. J Med Chem. 2009;52:1493–509. doi: 10.1021/jm8010542. [DOI] [PubMed] [Google Scholar]
- 5.Capdeville R, Buchdunger E, Zimmermann J, Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat Rev Drug Disc. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Yang PL, Gray NS. Targeting cancer with small molecule kinase inhibitors. Nat Rev Cancer. 2009;9:28–39. doi: 10.1038/nrc2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adrian FJ, et al. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 8.Ohren JF, et al. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat Struct Mol Biol. 2004;11:1192–7. doi: 10.1038/nsmb859. [DOI] [PubMed] [Google Scholar]
- 9.Shan Y, et al. How Does a Drug Molecule Find Its Target Binding Site? Journal of the American Chemical Society. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levitzki A. Protein tyrosine kinase inhibitors as novel therapeutic agents. Pharmacol Ther. 1999;82:231–9. doi: 10.1016/s0163-7258(98)00066-7. [DOI] [PubMed] [Google Scholar]
- 11.Gazit A, Yaish P, Gilon C, Levitzki A. Tyrphostins I: synthesis and biological activity of protein tyrosine kinase inhibitors. J Med Chem. 1989;32:2344–52. doi: 10.1021/jm00130a020. [DOI] [PubMed] [Google Scholar]
- 12.Krishnamurty R, Maly DJ. Biochemical mechanisms of resistance to small-molecule protein kinase inhibitors. ACS Chem Biol. 2010;5:121–38. doi: 10.1021/cb9002656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soverini S, et al. Contribution of ABL Kinase Domain Mutations to Imatinib Resistance in Different Subsets of Philadelphia-Positive Patients: By the GIMEMA Working Party on Chronic Myeloid Leukemia. Clinical Cancer Research. 2006;12:7374–7379. doi: 10.1158/1078-0432.CCR-06-1516. [DOI] [PubMed] [Google Scholar]
- 14.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–18. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc. 2001;123:6961–3. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gartner ZJ, et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–5. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X, Liu DR. DNA-templated organic synthesis: nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem Int Ed Engl. 2004;43:4848–70. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 18.Doyon JB, Snyder TM, Liu DR. Highly sensitive in vitro selections for DNA-linked synthetic small molecules with protein binding affinity and specificity. J Am Chem Soc. 2003;125:12372–3. doi: 10.1021/ja036065u. [DOI] [PubMed] [Google Scholar]
- 19.Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chem Soc Rev. 2011;40:5707–17. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tse BN, Snyder TM, Shen Y, Liu DR. Translation of DNA into a library of 13,000 synthetic small-molecule macrocycles suitable for in vitro selection. J Am Chem Soc. 2008;130:15611–26. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR. In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J Am Chem Soc. 2010;132:11779–91. doi: 10.1021/ja104903x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knight ZA, Shokat KM. Features of selective kinase inhibitors. Chem Biol. 2005;12:621–37. doi: 10.1016/j.chembiol.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Biron E, Chatterjee J, Kessler H. Optimized selective N-methylation of peptides on solid support. J Pept Sci. 2006;12:213–9. doi: 10.1002/psc.711. [DOI] [PubMed] [Google Scholar]
- 24.Abbott L, et al. Discovery of thienopyridines as Src-family selective Lck inhibitors. Bioorg Med Chem Lett. 2007;17:1167–71. doi: 10.1016/j.bmcl.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Burchat AF, et al. Pyrazolo[3,4-d]pyrimidines containing an extended 3-substituent as potent inhibitors of Lck -- a selectivity insight. Bioorg Med Chem Lett. 2002;12:1687–90. doi: 10.1016/s0960-894x(02)00196-8. [DOI] [PubMed] [Google Scholar]
- 26.Maly DJ, Choong IC, Ellman JA. Combinatorial target-guided ligand assembly: identification of potent subtype-selective c-Src inhibitors. Proc Natl Acad Sci U S A. 2000;97:2419–24. doi: 10.1073/pnas.97.6.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamborough P, Drewry D, Harper G, Smith GK, Schneider K. Assessment of chemical coverage of kinome space and its implications for kinase drug discovery. J Med Chem. 2008;51:7898–914. doi: 10.1021/jm8011036. [DOI] [PubMed] [Google Scholar]
- 28.Karaman MW, et al. A quantitative analysis of kinase inhibitor selectivity. Nat Biotechnol. 2008;26:127–32. doi: 10.1038/nbt1358. [DOI] [PubMed] [Google Scholar]
- 29.Songyang Z, Cantley LC. Recognition and specificity in protein tyrosine kinase-mediated signalling. Trends Biochem Sci. 1995;20:470–5. doi: 10.1016/s0968-0004(00)89103-3. [DOI] [PubMed] [Google Scholar]
- 30.Songyang Z, et al. Catalytic specificity of protein-tyrosine kinases is critical for selective signalling. Nature. 1995;373:536–9. doi: 10.1038/373536a0. [DOI] [PubMed] [Google Scholar]
- 31.Levinson NM, et al. A Src-like inactive conformation in the abl tyrosine kinase domain. PLoS Biol. 2006;4:e144. doi: 10.1371/journal.pbio.0040144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–81. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barouch-Bentov R, et al. A Conserved Salt Bridge in the G Loop of Multiple Protein Kinases Is Important for Catalysis and for In Vivo Lyn Function. Molecular Cell. 2009;33:43–52. doi: 10.1016/j.molcel.2008.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Laszlo GS, Cooper JA. Restriction of Src activity by Cullin-5. Curr Biol. 2009;19:157–62. doi: 10.1016/j.cub.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu W, Harrison SC, Eck MJ. Three-dimensional structure of the tyrosine kinase c-Src. Nature. 1997;385:595–602. doi: 10.1038/385595a0. [DOI] [PubMed] [Google Scholar]
- 36.Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the Src family tyrosine kinase Hck. Nature. 1997;385:602–9. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- 37.Seeliger MA, et al. High yield bacterial expression of active c-Abl and c-Src tyrosine kinases. Protein Sci. 2005;14:3135–9. doi: 10.1110/ps.051750905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seeliger MA, et al. c-Src binds to the cancer drug imatinib with an inactive Abl/c-Kit conformation and a distributed thermodynamic penalty. Structure. 2007;15:299–311. doi: 10.1016/j.str.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 40.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 41.Cowan-Jacob SW, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Src activation. Structure. 2005;13:861–71. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–54. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 44.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography. 1992 [Google Scholar]
- 45.Xu W, Doshi A, Lei M, Eck MJ, Harrison SC. Crystal structures of c-Src reveal features of its autoinhibitory mechanism. Mol Cell. 1999;3:629–38. doi: 10.1016/s1097-2765(00)80356-1. [DOI] [PubMed] [Google Scholar]
- 46.Barker SC, et al. Characterization of pp60c-src tyrosine kinase activities using a continuous assay: autoactivation of the enzyme is an intermolecular autophosphorylation process. Biochemistry. 1995;34:14843–51. doi: 10.1021/bi00045a027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.