Abstract
Polyadenylation of mitochondrial RNAs in higher eukaryotic organisms have diverse effects on their function and metabolism. Polyadenylation completes the UAA stop codon of a majority of mitochondrial mRNAs in mammals, regulates the translation of the mRNAs, and has diverse effects on their stability. In contrast, polyadenylation of most mitochondrial mRNAs in plants leads to their degradation, consistent with the bacterial origin of this organelle. PAPD1 (mtPAP, TUTase1), a noncanonical poly(A) polymerase (ncPAP), is responsible for producing the poly(A) tails in mammalian mitochondria. The crystal structure of human PAPD1 was reported recently, offering molecular insights into its catalysis.
Keywords: mRNA processing, mRNA stability, RNA editing, noncanonical poly(A) polymerase, polynucleotide phosphorylase, cytoplasmic polyadenylation, trypanosome
Introduction
Polyadenylation has important roles in regulating the function and metabolism of RNAs. In bacteria, the addition of a poly(A) tail to an mRNA generally leads to its degradation, and therefore bacterial polyadenylation is a destabilizing modification. In contrast, polyadenylation in eukaryotes produces much more diverse effects, which depend on the cellular organelle (nucleus, cytoplasm, mitochondrion, chloroplast) as well as the organism in which the modification takes place. Nuclear polyadenylation of mRNAs is important for their export, stability, and translation, and is part of the 3′-end modifications for most mRNA precursors (pre-mRNAs), and this area has been reviewed extensively over the years [1-5]. Cytoplasmic polyadenylation is involved in regulation of mRNA translation [6, 7]. Polyadenylation in chloroplasts generally leads to RNA degradation, consistent with the bacterial origin of this organelle [8, 9].
The addition of a poly(A) tail is catalyzed by poly(A) polymerases (PAPs). Nuclear PAPs that are involved in the 3′-end processing of pre-mRNAs have been studied extensively over the years. More recently, a collection of non-canonical PAPs (ncPAPs) has been identified, which are involved in polyadenylation in other organelles (as well as in the nucleus). Some of these ncPAPs have relaxed base selectivity and can also synthesize oligo- or poly-uridylate tails, and hence are also known as poly(U) polymerases (PUPs) or terminal uridylate transferases (TUTs or TUTases).
Degradation of the poly(A) tail in the mitochondria is carried out by polynucleotide phosphorylase (PNPase) in plants [8]. In humans, a recent report identifies 2′-phosphodiesterase (PDE12) as a deadenylating enzyme in the mitochondria [10]. Human PNPase, on the other hand, is localized in the intermembrane space [11, 12], while mitochondrial mRNA degradation occurs in the matrix. Other studies showed however that PNPase forms a stable complex with the helicase hSuv3p, which is a matrix protein [13, 14]. Depletion of PNPase increased the poly(A) tail length of some mitochondrial mRNAs, but did not affect the steady-state levels [15]. The exact function of human PNPase in regulating poly(A) tails remains to be demonstrated. PNPase has an important role in maintaining mitochondrial homeostasis, and is also required for importing nuclear-encoded RNA into the mitochondria [16]. PNPase also has PAP activity in some organisms, although it produces 3′-end tails with mixed base composition since it does not have a stringent substrate selectivity.
This review will focus on the current knowledge on mitochondrial polyadenylation and the enzyme that adds the poly(A) tail to mitochondrial RNAs. Mitochondrial polyadenylation was first reported nearly 40 years ago [17-19]. It is required for the completion of the UAA stop codon of many of the mitochondrial mRNAs in mammals. It is also involved in regulating the stability of mRNAs, although the actual effects may be rather complicated in mammals. Among the seven ncPAPs in humans, only one, known as PAPD1, mtPAP or TUTase1, has been found to function in the mitochondria [15, 20]. The crystal structure of this ncPAP was reported recently [21], offering molecular insights into its catalysis.
Mitochondrial polyadenylation in animals
Human mitochondrial DNA (mtDNA) exists in a double-stranded circular form with 16,569 base pairs. Both strands of the mtDNA are transcribed to generate large, polycistronic RNA molecules, which are processed by endoribonucleases to produce RNAs involved in replication and transcription initiation, along with 2 rRNAs, 22 tRNAs and mRNAs (with no introns) encoding 13 proteins required for oxidative phosphorylation [22, 23].
Human mitochondrial mRNAs carry a poly(A) tail of ∼50 adenylates. One function of polyadenylation is to complete the UAA stop codon, as endoribonucleolytic processing of seven of the mRNAs leave only the U or UA of the stop codon at the 3′-end [22, 24-27]. A pathogenic microdeletion in the ATP6 mRNA removes two bases of the stop codon, and the non-stop mRNA is deadenylated and then degraded in a translation-dependent manner [28]. On the other hand, the initial round of translation appears to be able to produce functional ATPase 6 enzyme [29], before this aberrant mRNA is degraded.
Polyadenylation of mitochondrial mRNAs may also regulate their stability, although the actual effects can be rather complicated and may depend on the RNA or other factors [24-27, 30]. Adenylated truncated RNA transcripts, some with internal polyadenylation, are observed in human mitochondria [30]. Degradation intermediates and aberrant RNAs with extended poly(A) tails also accumulate in the absence of hSuv3p [14]. Reduction of the length of the poly(A) tail by knocking down PAPD1 in HeLa cells decreased the stability of the CO1, CO2, CO3 and ATP6 mRNAs but had no effect on the stability of the ND3 mRNA [15]. In another study, however, PAPD1 knock-down had no effect on the stability of ATP6/8, CO3 and ND3 mRNAs [20]. Removal of the poly(A) tails by targeting the cytosolic poly(A)-specific 3′-5′ exoribonuclease (PARN) into the mitochondria made one group of mRNAs more stable, a second group of mRNAs less stable, and had no effect on a third group [31]. Increased abundance of the ND1 mRNA was observed when the poly(A) tail was shortened or removed [10].
Interestingly, oligoadenylate tails are observed in the absence of PAPD1, both in knock-down experiments and in patients with mutated PAPD1 (see below) [32]. Therefore, there may be two classes of adenylated RNAs in mammalian mitochondria, with poly(A) or oligo(A) tails. It has been suggested that there may be a separate mitochondrial enzyme that can produce the oligo(A) tails [33, 34].
The poly(A) tail may also be involved in regulating translation in the mitochondria [10]. This is further supported by the observation that targeting the cytosolic poly(A)-binding protein (PABPC1) into the mitochondria caused strong inhibition of translation [31].
Polyadenylation may also play a role in the editing of the 3′-end acceptor region of some mitochondrial tRNAs [35, 36].
Mitochondrial polyadenylation in plants
Similar to what happens in chloroplasts, polyadenylation of mitochondrial RNAs in plants generally lead to their degradation [8, 9]. In comparison, most of the mature stable mRNAs are not polyadenylated, while levels of polyadenylated RNAs are low due to their rapid degradation. Under heat stress conditions, polyadenylated (and presumably stabilized) mRNAs accumulate in Arabidopsis [37], through an unknown mechanism. Mitochondrial polyadenylation in plants may be carried out by bacterial-type PAPs [38], whereas polyadenylation-assisted RNA degradation is carried out by the mitochondrial PNPase [8, 9].
Mitochondrial polyadenylation in trypanosomes
Polycistronic mitochondrial precursor RNA transcripts in trypanosomes are processed by endoribonucleases. Some of the released pre-mRNAs are then edited extensively by the insertion or deletion of uridines, defined by small, guide RNAs, while other pre-mRNAs do not require editing (named never-edited pre-mRNAs) [39, 40]. Polyadenylation is related to the editing status and has diverse effects on these mRNAs at various stages of processing. Pre-edited, partially edited and fully edited transcripts are stabilized by the addition of a short (20-25 nt) poly(A) tail, catalyzed by KPAP1 (kinetoplast PAP1) [41-43]. Fully edited transcripts that lack an adenylate tail are degraded much faster than its precursor in an in vitro turnover assay [42].
Never-edited and fully edited mRNAs acquire a long (200-300 nt) poly(A/U) tail by extending the short poly(A) tail, making them translationally competent [44]. The poly(A/U) tails are added by the enzymes KPAP1 and RET1 (RNA editing TUTase1) [45], and this process is stimulated by a heterodimer of two pentatricopeptide repeat (PPR) proteins KPAF1 and KPAF2 (kinetoplast polyadenylation/uridylation factor) [44].
Mitochondrial mRNAs in yeast
Budding yeast (S. cerevisiae) mitochondrial mRNAs are not polyadenylated, and no PAP activity has been identified in this organelle in yeast. Instead, they carry a conserved dodecamer sequence, AAUAA(U/C)AUUCUU, at their 3′-ends [46]. Similarly, mitochondrial mRNAs in fission yeast (S. pombe) are not polyadenylated either, and they have a C-rich motif near the 3′-end [47]. Therefore, polyadenylation of mitochondrial mRNAs may be restricted to the higher eukaryotes.
It is believed that the dodecamer sequence may recruit protein factor(s) that can protect the mRNAs from the mitochondrial degradosome (mtExo) in budding yeast [48]. The mtExo degradosome is a protein complex with two subunits, the SUV3 helicase and the DSS1 exoribonuclease [49-51]. Deficient degradosome activity can be rescued by a reduction in the transcription rate of the mitochondrial RNA polymerase, indicating the importance of maintaining proper mRNA levels in the mitochondria [49, 50]. Proteins that are homologous to the components of the budding yeast mtExo have been identified in the fission yeast, although they may be more involved in mitochondrial mRNA processing than degradation [52].
Poly(A) polymerases (PAPs)
Canonical PAPs contain three domains (Fig. 2). The N-terminal (palm) domain shows high sequence homology with the Polβ superfamily, which contains a signature helix-turn motif that is conserved from yeast to humans [53]. In contrast to DNA polymerases, however, PAPs incorporate rNTPs in a template-independent manner. A conserved catalytic triad of Asp residues in the palm domain is critical for activity. This triad coordinates two divalent metal ions, which in turn interact with the incoming nucleotide substrates [54-56]. The middle (fingers) domain is tightly associated with the palm domain, and the active site is located at the interface between the two domains (Fig. 3A). A C-terminal, RNA-binding domain (RBD) follows immediately after the fingers domain, which contributes to the binding of the RNA substrate [56] (Fig. 3A).
Figure 2.
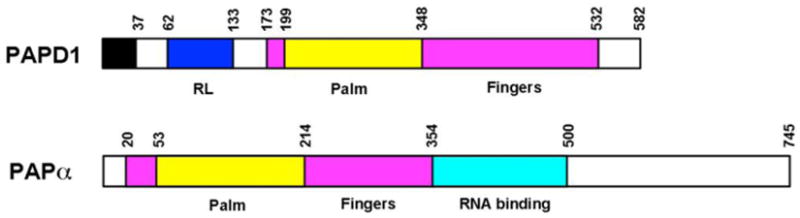
Schematic drawing of the domain organization of yeast canonical PAP and human PAPD1, a noncanonical PAP (ncPAP). The palm and fingers domains are shown in yellow and magenta, respectively. The RBD of yeast PAP is shown in cyan. The mitochondrial targeting sequence and RL domain of PAPD1 are shown in black and blue, respectively.
Figure 3.
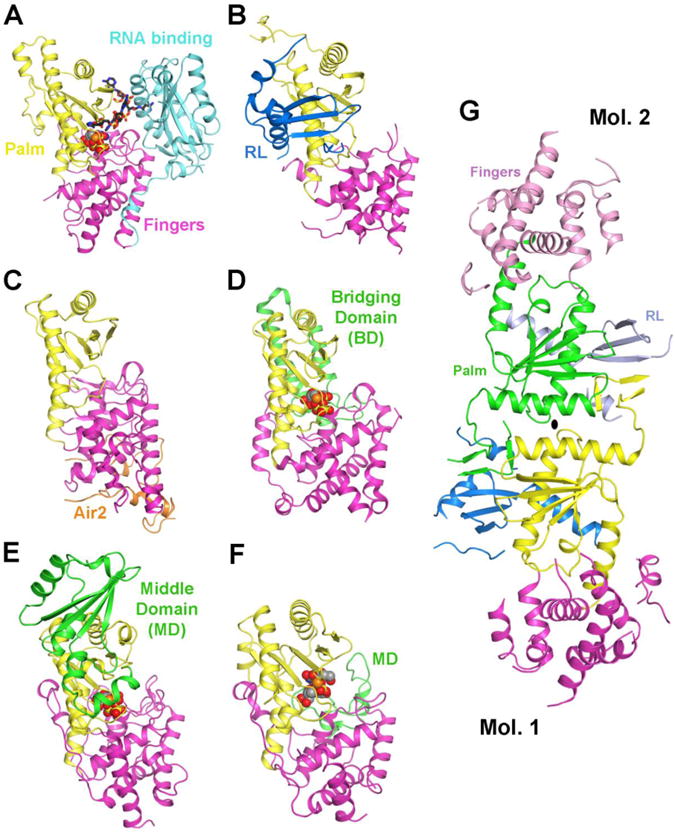
Structural comparisons of PAPD1 with other PAPs and TUTases. (A). Schematic drawing of the structure of yeast PAP (D154A mutant) in complex with ATP and oligoadenylate [56], colored as in Fig. 2. ATP is shown in a cpk model, and the oligo(A) in a stick model. The divalent metal ion is shown as a sphere in orange. (B). Structure of human PAPD1 monomer [21]. (C). Yeast Trf4 in complex with a fragment of Air2 (in orange) [64]. (D). T. brucei MEAT1 in complex with UTP [65]. The bridging domain (BD) is colored in green. (E). T. brucei RNA-editing TUTase2 in complex with UTP [66]. The middle domain (MD) is colored in green. (F). T. brucei TUTase4 in complex with dinucleotide, UpU [68]. The palm and fingers domains of each structure are shown in the same orientation. (G). Structure of human PAPD1 dimer [21]. All structure figures were produced with PyMOL (http://www.pymol.org).
A family of non-canonical PAPs (ncPAPs) has been identified over the recent years, from yeast to humans [57-62]. One common feature of the ncPAPs is that they lack the RBD that directly follows the middle (finger) domain (Fig. 2). As a result, some of the ncPAPs require RNA-binding protein cofactors for activity. In addition, many of the ncPAPs have uridylate transferase activity, and hence are also known as TUTs or TUTases. Moreover, the sequence conservation between canonical and ncPAPs is quite low (10-15% identity) for their palm and fingers domains.
Among the seven ncPAPs in humans, only PAPD1 (mtPAP, TUTase1) has a mitochondrial targeting sequence (residues 1-37) at the N-terminus (Fig. 2), and is known to be localized in mitochondria [15, 20, 26]. In contrast to other ncPAPs, PAPD1 appears to be active on its own, without the need for a cofactor [15], and mediates the polyadenylation of mitochondrial mRNAs. On the other hand, structural and biochemical studies suggest that PAPD1 may be active only as a dimer (see below) [21], and therefore its monomer may not be functional.
PAPD1 has also been identified as one of two ncPAPs that can oligouridylate histone mRNAs (which are not polyadenylated), thereby initiating their degradation (hence the name TUTase1) [60]. However, it is not clear how PAPD1 becomes localized in the cytoplasm for this modification, and how it can distinguish between the A and U substrates in different cellular compartments. A recent report identifies another ncPAP as being responsible for histone mRNA oligo-uridylation [63].
RNAs with poly(U) tails have also been observed in human mitochondria under certain conditions [14, 27, 30, 34]. It is not clear whether this process is related to the oligouridylation of histone mRNAs, or whether PAPD1 has a role in mitochondrial mRNA polyuridylation.
Structure of the mitochondrial poly(A) polymerase (PAPD1)
The structure of PAPD1 contains three domains: the canonical palm and fingers domains and an N-terminal RBD-like (RL) domain (Figs. 2, 3B) [21]. The overall structure of the palm and fingers domains of PAPD1 is similar to that in the canonical PAPs (Fig. 3A), the yeast ncPAP Trf4 (Fig. 3C) [64], as well as the mitochondrial editosome-like complex associated TUTase1 (MEAT1, Fig. 3D) [65], the RNA editing TUTase2 (Fig. 3E) [66] and the minimal TUTase4 (Fig. 3F) [67, 68] from T. brucei. These other enzymes do not contain an RNA binding domain following the fingers domain either, although they carry insertions in other parts of the structure.
Residues 62-133 in the N-terminal region of PAPD1 forms a separate domain (Fig. 2), with a backbone fold that is similar to RBDs [21]. This domain has been named the RBD-like (RL) domain (Fig. 3B), as it is probably unlikely to be involved in binding the RNA substrate in PAPD1. It has tight interactions with the palm domain, is on the opposite face from the active site, and its position is entirely different from that of the RBD in canonical PAPs (Fig. 3A). Instead, the domain helps to mediate the dimerization of PAPD1 (Fig. 3G), which appears to be required for the catalytic activity of this enzyme [21].
Despite lacking the RBD of canonical PAPs, a large pocket at the interface between the palm and fingers domains of PAPD1 is sufficient to accommodate the ATP and the last nucleotide of the RNA substrate [21]. On the other hand, the adenine base of ATP does not appear to be specifically recognized by PAPD1 in this model of the active site. Data from in vitro studies indicate that PAPD1 can utilize all four nucleotides as substrates, although it is much more active with ATP or UTP [21]. It remains to be seen how PAPD1 achieves specificity for a poly(A) tail in the mitochondria.
Different mechanisms for substrate preference have been established from studies on other PAPs. The canonical PAP from yeast is an induced-fit enzyme, and this behavior is important for its specificity for ATP [56, 69]. Studies with the T. brucei TUTases suggest that base stacking and specific hydrogen-bonding with the enzyme, direct and water-mediated, may be important for substrate selectivity [65, 67, 68].
Disease-associated mutations in PAPD1
Studies with autosomal-recessive spastic ataxia with optic atrophy identified an N478D missense mutation in PAPD1 that is associated with the disease phenotype [32]. The poly(A) tails of mitochondrial mRNAs are very short in affected individuals. This residue, located in the fingers domain, is highly conserved among PAPD1 homologs. It is disordered in the structure of PAPD1, but may be involved in binding the RNA substrate of the enzyme [21].
A single-nucleotide polymorphism, resulting in a D39G mutation, was found to be associated with intramuscular fat deposit and extreme obesity in cattle [70]. This residue is located just after the mitochondrial targeting sequence of PAPD1. How this mutation can affect PAPD1 function is currently unknown.
Concluding remarks
Polyadenylation in the mitochondria is an important regulatory mechanism for RNA function and stability in higher eukaryotes. Mitochondrial polyadenylation in plants leads to the degradation of mRNAs, consistent with the bacterial origin of this organelle. Polyadenylation in the mitochondria of other higher eukaryotes has evolved new functions, and can stabilize (as well as destabilize) mRNAs. In addition, polyadenylation may be involved in regulating mitochondrial translation. In mammals, polyadenylation is required to complete the UAA stop codon of a majority of the mitochondrial mRNAs. In trypanosomes, poly(adenylation/uridylation) is coupled to the extensive editing and stability of the mitochondrial mRNAs. On the other hand, mitochondrial polyadenylation appears to have been lost in the fungal species during evolution.
Mitochondrial polyadenylation is catalyzed by the noncanonical poly(A) polymerase (ncPAP) PAPD1 (mtPAP, TUTase1) in mammals. This enzyme is able to function on its own, without the need for an RNA-binding domain (as in canonical PAPs) or an RNA-binding protein cofactor (as for some other ncPAPs). The crystal structure of PAPD1 reveals a well-defined active site. However, the molecular basis for the substrate preference of this enzyme is still not known, and it appears to have strong activity with both ATP and UTP in vitro. It will be of great interest to elucidate how PAPD1 and other mammalian ncPAPs can produce homogeneous poly-/oligo-adenylate or uridylate tails in vivo.
Figure 1.
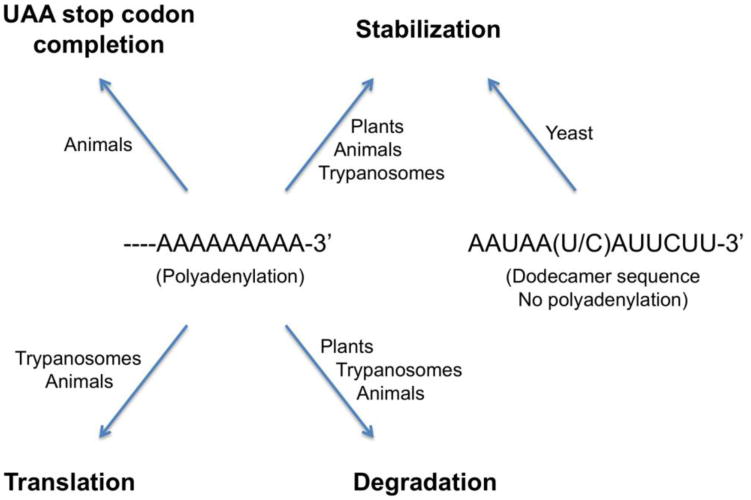
Diverse effects of mitochondrial polyadenylation in higher eukaryotes. Yeast mitochondrial mRNAs are not polyadenylated. Instead, their stability is controlled by a dodecamer sequence at the 3′-end.
Highlights.
Mitochondrial polyadenylation regulates RNA function and metabolism.
Mitochondrial polyadenylation occurs in higher eukaryotes.
Human mitochondrial poly(A) polymerase (PAPD1) is a noncanonical PAP.
The structure of PAPD1 provides molecular insights into its catalysis.
Acknowledgments
This research is supported in part by a grant from the NIH (GM077175) to LT.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zhao J, Hyman L, Moore CL. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis, Microbiol. Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nature Struct Biol. 2000;7:838–842. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 3.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog Nucl Acid Res Mol Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 4.Mandel CR, Bai Y, Tong L. Protein factors in pre-mRNA 3′-end processing. Cell Mol Life Sci. 2008;65:1099–1122. doi: 10.1007/s00018-007-7474-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X, Virtanen A, Kleiman FE. To polyadenylate or to deadenylate: that is the question. Cell Cycle. 2010;9:4437–4449. doi: 10.4161/cc.9.22.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnard DC, Ryan K, Manley JL, Richter JD. Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell. 2004;119:641–651. doi: 10.1016/j.cell.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 7.Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genetics Develop. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 8.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 9.Lang H, Sement FM, Canaday J, Gagliardi D. Polyadenylation-assisted RNA degradation processes in plants. Trends Plant Sci. 2009;14:497–504. doi: 10.1016/j.tplants.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 10.Rorbach J, Nicholls TJJ, Minczuk M. PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucl Acid Res. 2011;39:7750–7763. doi: 10.1093/nar/gkr470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen HW, Rainey RN, Balatoni CE, Dawson DW, Troke JJ, Wasiak S, Hong JS, McBride HM, Koehler CM, Teitell MA, French SW. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol Cell Biol. 2006;26:8475–8487. doi: 10.1128/MCB.01002-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen HW, Koehler CM, Teitell MA. Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 2007;17:600–608. doi: 10.1016/j.tcb.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Wang DD, Zhu Z, Lieser SA, Chen PL, Lee WH. Human mitochondrial SUV3 and polynucleotide phosphorylase form a 330-kDa heteropentamer to cooperatively degrade double-stranded RNA with a 3′-to-5′ directionality. J Biol Chem. 2009;284:20812–20821. doi: 10.1074/jbc.M109.009605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szczesny RJ, Borowski LS, Brzezniak LK, Dmochowska A, Gewartowski K, Bartnik E, Stepien PP. Human mitochondrial RNA turnover caught in flagranti: involvement of hSuv3p helicase in RNA surveillance. Nucl Acid Res. 2009;38:279–298. doi: 10.1093/nar/gkp903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagaike T, Suzuki T, Katoh T, Ueda T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J Biol Chem. 2005;280:19721–19727. doi: 10.1074/jbc.M500804200. [DOI] [PubMed] [Google Scholar]
- 16.Wang G, Chen Hw, Oktay Y, Zhang Y, Allen EL, Smith GM, Fan KC, Hong JS, French SW, McCaffery JM, Lightowlers RN, Morse HC, 3rd, Koehler CM, Teitell MA. PNPase regulates RNA import into mitochondria. Cell. 2010;142:456–467. doi: 10.1016/j.cell.2010.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacob ST, Schindler DG, Morris HP. Mitochondrial polyriboadenylate polymerase: relative lack of activity in hepatomas. Science. 1972;178:639–640. doi: 10.1126/science.178.4061.639. [DOI] [PubMed] [Google Scholar]
- 18.Perlman S, Abelson HT, Penman S. Mitochondrial protein synthesis: RNA with the properties of eukaryotic messenger RNA. Proc Natl Acad Sci USA. 1973;70:350–353. doi: 10.1073/pnas.70.2.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ojala D, Attardi G. Identification of discrete polyadenylate-containing RNA components transcribed from HeLa cell mitochondrial DNA. Proc Natl Acad Sci USA. 1974;71:563–567. doi: 10.1073/pnas.71.2.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomecki R, Dmochowska A, Gewartowski K, Dziembowski A, Stepien PP. Identification of a novel human nuclear-encoded mitochondrial poly(A) polymerase. Nucl Acid Res. 2004;32:6001–6014. doi: 10.1093/nar/gkh923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai Y, Srivastava SK, Chang JH, Manley JL, Tong L. Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Mol Cell. 2011;41:311–320. doi: 10.1016/j.molcel.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Human mitochondrial mRNAs-like members of all families, similar but different. Biochim Biophys Acta. 2010;1797:1081–1085. doi: 10.1016/j.bbabio.2010.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mercer TR, Neph S, Dinger ME, Crawford J, Smith MA, Shearwood AMJ, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, Filipovska A, Mattick JS. The human mitochondrial transcriptome. Cell. 2011;146:645–658. doi: 10.1016/j.cell.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gagliardi D, Stepien PP, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Messenger RNA stability in mitochondria: different means to an end. Trends Genet. 2004;20:260–267. doi: 10.1016/j.tig.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 25.Bobrowicz AJ, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Polyadenylation and degradation of mRNA in mammalian mitochondria: a missing link? Biochem Soc Trans. 2008;36:517–519. doi: 10.1042/BST0360517. [DOI] [PubMed] [Google Scholar]
- 26.Nagaike T, Suzuki T, Ueda T. Polyadenylation in mammalian mitochondria: insights from recent studies. Biochim Biophys Acta. 2008;1779:266–269. doi: 10.1016/j.bbagrm.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Borowski L, Szczesny RJ, Brzezniak LK, Stepien PP. RNA turnover in human mitochondria: more questions than answers? Biochim Biophys Acta. 2010;1797:1066–1070. doi: 10.1016/j.bbabio.2010.01.028. [DOI] [PubMed] [Google Scholar]
- 28.Temperley RJ, Seneca SH, Tonska K, Bartnik E, Bindoff LA, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Investigation of a pathogenic mtDNA microdeletion reveals a translation-dependent deadenylation decay pathway in human mitochondria. Human Mol Gen. 2003;12:2341–2348. doi: 10.1093/hmg/ddg238. [DOI] [PubMed] [Google Scholar]
- 29.Chrzanowska-Lightowlers ZMA, Temperley RJ, Smith PM, Seneca SH, Lightowlers RN. Functional polypeptides can be synthesized from human mitochondrial transcripts lacking termination codons. Biochem J. 2004;377:725–731. doi: 10.1042/BJ20031556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slomovic S, Laufer D, Geiger D, Schuster G. Polyadenylation and degradation of human mitochondrial RNA: the prokaryotic past leaves its mark. Mol Cell Biol. 2005;25:6427–6435. doi: 10.1128/MCB.25.15.6427-6435.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wydro M, Bobrowicz AJ, Temperley RJ, Lightowlers RN, Chrzanowska-Lightowlers ZMA. Targeting of the cytosolic poly(A) binding protein PABPC1 to mitochondria causes mitochondrial translation inhibition. Nucl Acid Res. 2010;38:3732–3742. doi: 10.1093/nar/gkq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crosby AH, Patel H, Chioza BA, Proukakis C, Gurtz K, Patton MA, Sharifi R, Harlalka G, Simpson MA, Dick K, Reed JA, Al-Memar A, Chrzanowska-Lightowlers ZMA, Cross HE, Lightowlers RN. Defective mitochondrial mRNA maturation is associated with spastic ataxia. Amer J Hum Genet. 2010;87:655–660. doi: 10.1016/j.ajhg.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piechota J, Tomecki R, Gewartowski K, Szczesny R, Dmochowska A, Kudla M, Dybczynska L, Stepien PP, Bartnik E. Differential stability of mitochondrial mRNA in HeLa cells. Acta Biochim Polonica. 2006;53:157–167. [PubMed] [Google Scholar]
- 34.Slomovic S, Schuster G. Stable PNPase RNAi silencing: its effect on the processing and adenylation of human mitochondrial RNA. RNA. 2008;14:310–323. doi: 10.1261/rna.697308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomita K, Ueda T, Watanabe K. RNA editing in the acceptor stem of squid mitochondrial tRNA(Tyr) Nucl Acid Res. 1996;24:4987–4991. doi: 10.1093/nar/24.24.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yokobori S, Paabo S. Polyadenylation creates the discriminator nucleotide of chicken mitochondrial tRNA(Tyr) J Mol Biol. 1997;265:95–99. doi: 10.1006/jmbi.1996.0728. [DOI] [PubMed] [Google Scholar]
- 37.Adamo A, Pinney JW, Kunova A, Westhead DR, Meyer P. Heat stress enhances the accumulation of polyadenylated mitochondrial transcripts in Arabidopsis thaliana. PLoS One. 2008;3:e2889. doi: 10.1371/journal.pone.0002889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zimmer SL, Schein A, Zipor G, Stern DB, Schuster G. Polyadenylation in Arabidopsis and Chlamydomonas organelles: the input of nucleotidyltransferases, poy(A) polymerases and polynucleotide phosphorylase. Plant J. 2009;59:88–99. doi: 10.1111/j.1365-313X.2009.03853.x. [DOI] [PubMed] [Google Scholar]
- 39.Schnaufer A, Wu M, Park YJ, Nakai T, Deng J, Proff R, Hol WGJ, Stuart KD. A protein-protein interaction map of trypanosome ∼20S editosomes. J Biol Chem. 2010;285:5282–5295. doi: 10.1074/jbc.M109.059378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aphasizhev R, Aphasizhev I. Mitochondrial RNA processing in trypanosomes. Res Microbiol. 2011;162:655–663. doi: 10.1016/j.resmic.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Etheridge RD, Aphasizhev I, Gershon PD, Aphasizhev R. 3′ adenylation determines mRNA abundance and monitors completion of RNA editing in T. brucei mitochondria. EMBO J. 2008;27:1596–1608. doi: 10.1038/emboj.2008.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kao CY, Read LK. Opposing effects of polyadenylation on the stability of edited and unedited mitochondrial RNA in Trypanosoma brucei. Mol Cell Biol. 2005;25:1634–1644. doi: 10.1128/MCB.25.5.1634-1644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kao CY, Read LK. Targeted depletion of a mitochondrial nucleotidyltransferase suggests the presence of multiple enzymes that polymerize mRNA 3′ tails in Trypanosoma brucei mitochondria. Mol Biochem Parasitol. 2007;154:158–169. doi: 10.1016/j.molbiopara.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aphasizhev I, Maslov D, Wang X, Huang L, Aphasizhev R. Pentatricopeptide repeat proteins stimulate mRNA adenylation/uridylation to activate mitochondrial translation in trypanosomes. Mol Cell. 2011;42:106–117. doi: 10.1016/j.molcel.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aphasizhev R. RNA uridylyltransferases. Cell Mol Life Sci. 2005;62:2194–2203. doi: 10.1007/s00018-005-5198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Butow RA, Zhu H, Perlman P, Conrad-Webb H. The role of a conserved dodecamer sequence in yeast mitochondrial gene expression. Genome. 1989;31:757–760. doi: 10.1139/g89-134. [DOI] [PubMed] [Google Scholar]
- 47.Schafer B, Hansen M, Lang BF. Transcription and RNA-processing in fission yeast mitochondria. RNA. 2005;11:785–795. doi: 10.1261/rna.7252205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, Zassenhaus HP. Purification and characterization of an RNA dodecamer sequence binding protein from mitochondria of Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1999;261:740–745. doi: 10.1006/bbrc.1999.1085. [DOI] [PubMed] [Google Scholar]
- 49.Dziembowski A, Piwowarski J, Hoser R, Minczuk M, Dmochowska A, Siep M, van der Spek H, Grivell L, Stepien PP. The yeast mitochondrial degradosome. Its composition, interplay between RNA helicase and RNase activities and the role in mitochondrial RNA metabolism. J Biol Chem. 2003;278:1603–1611. doi: 10.1074/jbc.M208287200. [DOI] [PubMed] [Google Scholar]
- 50.Rogowska AT, Puchta O, Czarnecka AM, Kaniak A, Stepien PP, Golik P. Balance between transcription and RNA degradation is vital for Saccharomyces cerevisiae mitochondria: reduced transcription rescues the phenotype of deficient RNA degradation. Mol Biol Cell. 2006;17:1184–1193. doi: 10.1091/mbc.E05-08-0796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malecki M, Jedrzejczak R, Stepien PP, Golik P. In vitro reconstitution and characterization of the yeast mitochondrial degradosome complex unravels tight functional interdependence. J Mol Biol. 2007;372:23–36. doi: 10.1016/j.jmb.2007.06.074. [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann B, Nickel J, Speer F, Schafer B. The 3′ ends of mature transcripts are generated by a processosome complex in fission yeast mitochondria. J Mol Biol. 2008;377:1024–1037. doi: 10.1016/j.jmb.2008.01.038. [DOI] [PubMed] [Google Scholar]
- 53.Aravind L, Koonin EV. DNA polymerase b-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucl Acid Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bard J, Zhelkovsky AM, Helmling S, Earnest TN, Moore CL, Bohm A. Structure of yeast poly(A) polymerase alone and in complex with 3′-dATP. Science. 2000;289:1346–1349. doi: 10.1126/science.289.5483.1346. [DOI] [PubMed] [Google Scholar]
- 55.Martin G, Keller W, Doublie S. Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. EMBO J. 2000;19:4193–4203. doi: 10.1093/emboj/19.16.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Balbo PB, Bohm A. Mechanism of poly(A) polymerase: structure of the enzyme-MgATP-RNA ternary complex and kinetic analysis. Structure. 2007;15:1117–1131. doi: 10.1016/j.str.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stevenson AL, Norbury CJ. The CidI family of non-canonical poly(A) polymerases. Yeast. 2006;23:991–1000. doi: 10.1002/yea.1408. [DOI] [PubMed] [Google Scholar]
- 58.Kwak JE, Wickens M. A family of poly(U) polymerases. RNA. 2007;13:830–837. doi: 10.1261/rna.514007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin G, Keller W. RNA-specific ribonuleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Develop. 2008;22:50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wilusz CJ, Wilusz J. New ways to meet your (3′) end-oligouridylation as a step on the path to destruction. Genes Develop. 2008;22:1–7. doi: 10.1101/gad.1634508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schmidt H, Norbury CJ. Polyadenylation and beyond: emerging roles for noncanonical poly(A) polymerases. WIREs RNA. 2010;1:142–151. doi: 10.1002/wrna.16. [DOI] [PubMed] [Google Scholar]
- 63.Schmidt MJ, West S, Norbury CJ. The human cytoplasmic RNA terminal U-transferase ZCCHC11 targets histone mRNAs for degradation. RNA. 2011;17:39–44. doi: 10.1261/rna.2252511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hamill S, Wolin SL, Reinisch KM. Structure and function of the polymerase core of TRAMP, a RNA surveillance complex. Proc Natl Acad Sci USA. 2010;107:15045–15050. doi: 10.1073/pnas.1003505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stagno J, Aphasizhev I, Bruystens J, Luecke H, Aphasizhev R. Structure of the mitochondrial editosome-like complex associated TUTase 1 reveals divergent mechanisms of UTP selection and domain organization. J Mol Biol. 2010;399:464–475. doi: 10.1016/j.jmb.2010.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Deng J, Ernst NL, Turley S, Stuart KD, Hol WGJ. Structural basis for UTP specificity of RNA editing TUTases from Trypanosoma brucei. EMBO J. 2005;24:4007–4017. doi: 10.1038/sj.emboj.7600861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stagno J, Aphasizheva I, Rosengarth A, Luecke H, Aphasizhev R. UTP-bound and apo structures of a minimal RNA uridylyltransferase. J Mol Biol. 2007;366:882–899. doi: 10.1016/j.jmb.2006.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stagno J, Aphasizheva I, Aphasizhev R, Luecke H. Dual role of the RNA substrate in selectivity and catalysis by terminal uridylyl transferases. Proc Natl Acad Sci USA. 2007;104:14634–14639. doi: 10.1073/pnas.0704259104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balbo PB, Meinke G, Bohm A. Kinetic studies of yeast polyA polymerase indicate an induced fit mechanism for nucleotide specificity. Biochem. 2005;44:7777–7786. doi: 10.1021/bi050089r. [DOI] [PubMed] [Google Scholar]
- 70.Xiao Q, Wu XL, Michal JJ, Reeves JJ, Busboom JR, Thorgaard GH, Jiang Z. A novel nuclear-encoded mitochondrial poly(A) polymerase PAPD1 is a potential candidate gene for the extreme obesity related phenotypes in mammals. Int J Biol Sci. 2006;2:171–178. doi: 10.7150/ijbs.2.171. [DOI] [PMC free article] [PubMed] [Google Scholar]


