Abstract
Head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer worldwide with about 600,000 new cases diagnosed each year. Understanding the molecular pathways that lead to HNSCC is crucial to identify new targets for anti-cancer drug development. Protein kinase Cε (PKCε) is elevated in HNSCC and regulates the activation of Akt, Stat3, and Rho GTPases. To date, the molecular mechanism of PKCε dysregulation in HNSCC remains to be elucidated. In silico analysis identified three putative microRNA-107 (miR-107) binding sites in the 3'-untranslated region (UTR) of PKCε. An inverse relationship was revealed between miR-107 and PKCε in HNSCC cell lines. Delivery of miR-107 reduced PKCε levels in SCC15, SCC25, and CAL27, three HNSCC cell lines with high PKCε and low miR-107. The activity of a luciferase reporter construct containing the 3'-UTR of PKCε was down-regulated by miR-107 and mutations in the three cognate miR-107 binding sites completely ablated the regulation by miR-107. Treatment with miR-107 significantly blocked cell proliferation, DNA replication, colony formation, and invasion in SCC25 and CAL27 cells. Ectopic expression of miR-resistant PKCε was sufficient to partially rescue the loss-of-function phenotype in miR-107-overexpressing SCC25 cells. Tumor growth in nude mice was retarded by 93 ± 7% in CAL27/miR-107 cells compared to CAL27/miR-control cells. Lastly, human primary HNSCC tumors with elevated PKCε had reduced miR-107 expression. Our results demonstrate that PKCε is directly regulated by miR-107 and moreover, suggest that miR-107 may be a potential anti-cancer therapeutic for HNSCC.
Keywords: Head and neck cancer, Protein kinase C, microRNA, miR-107
Introduction
Approximately 600,000 new cases of head and neck squamous cell carcinoma (HNSCC) are diagnosed worldwide each year (Kamangar et al, 2006; Leemans et al, 2011; Parkin, 2001). Curative treatment is often achieved in patients presented with early-stage disease through surgery or radiation. Palliative chemotherapy is the standard approach for patients with recurrent/metastatic disease. The majority, greater than two-thirds, of HNSCC patients present with locally advanced disease that requires a modern multi-disciplinary approach involving surgery, radiation, and pharmacotherapy. However, patients with locally advanced HNSCC frequently develop loco-regional recurrence and/or distant metastasis after definitive treatment. The risk of failure is particularly high in patients with inadequate resection margins, extra-nodal spread, or multiple lymph node involvement (Bonner et al, 2010). In these high-risk patients, primary surgery is often followed by adjuvant radiation therapy. In spite of this intense treatment regimen, loco-regional recurrence occurs in about 30% of patients, distant metastasis occurs in about 25% of patients, and the five-year survival rate is a dismal 40% (Laramore et al, 1992). Thus, a critical need remains to identify and characterize genes that are responsible for promoting an aggressive HNSCC phenotype prone to relapse and metastasis to allow for better clinical management.
Protein kinase C (PKC) is a family of serine/threonine kinases known to play critical roles in the signal transduction pathways involved in proliferation, differentiation, apoptosis, and migration (Aziz et al, 2010; Dempsey et al, 2000; Gutcher et al, 2003; Jaken and Parker, 2000). Published work showed that PKCα, ε, and ζ are dysregulated in HNSCC and contribute to HNSCC development and/or progression. PKCζ was reported to control EGFR-dependent proliferation of HNSCC cells (Cohen et al, 2006). A recent study showed that PKCα down-regulates miR-15a to enhance DNA synthesis and cell cycle progression in HNSCC (Cohen et al, 2009). Moreover, elevated PKCα expression was found to be associated with poor prognosis in HNSCC (Cohen et al, 2009). Our laboratory demonstrated that PKCε is elevated in HNSCC and promotes an invasive and motile phenotype through modulation of RhoA and RhoC GTPases (Pan et al, 2006). In primary HNSCC tumors, PKCα, β, ε, γ, and ζ levels were shown to be elevated but only PKCε was found to be a prognostic biomarker, even better than the traditional gold standard of TNM staging (Martinez-Gimeno et al, 1995). This prospective study indicated that elevated PKCε is significantly associated with an increase in disease recurrence and a decrease in overall survival (Martinez-Gimeno et al, 1995).
It is clear that PKCε is elevated in HNSCC; however, the molecular mechanisms leading to increased PKCε levels remain to be elucidated. In this study, we demonstrate that miR-107 directly regulates PKCε in HNSCC. An inverse association was found for miR-107 and PKCε in HNSCC cell lines and tumor specimens. Over-expression of miR-107 decreased PKCε levels and reduced the tumorigenic properties of HNSCC cells in vitro and in vivo. Moreover, enforced expression of miR-resistant PKCε partially reversed the loss-of-function phenotype in miR-107-over-expressing HNSCC cells. Our work revealed a novel mechanism for PKCε dysregulation in HNSCC and demonstrated miR-107 as a candidate tumor suppressor in this patient population.
Results
Inverse association between miR-107 and PKCε in HNSCC cell lines
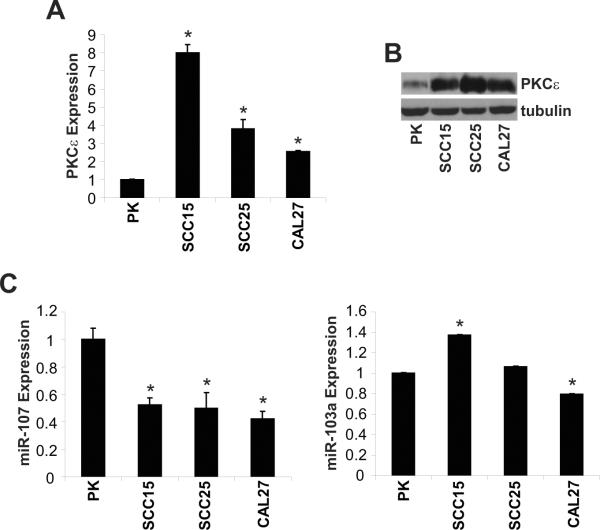
Our laboratory and others reported that PKCε is elevated in HNSCC (Cohen et al, 2006; Martinez-Gimeno et al, 1995; Pan et al, 2006). However, the molecular mechanism of this observation remains to be elucidated. Several miRs were predicted by TargetScan to bind to the 3'-UTR of PKCε suggesting that miRs may be a negative regulator of PKCε. We focused our initial experiments on miR-103a and miR-107 since both miRs have three putative binding sites in the 3'-UTR of the PKCε gene. As shown in Figure 1, three HNSCC cell lines, SCC15, SCC25, and CAL27, have significantly elevated PKCε protein levels and reduced miR-107 expression compared to primary keratinocytes. In contrast, the level of miR-103a, which has the same seed sequence as miR-107, was variable in the three HNSCC cell lines (Figure 1C). Compared to primary keratinocytes, miR-103a expression was higher in SCC15, unchanged in SCC25, and lower in CAL27. These results provide evidence to demonstrate an inverse association between miR-107 and PKCε in HNSCC.
Figure 1. miR-107 is down-regulated and inversely associated with PKCε in HNSCC cell lines.
A. PKCε mRNA expression is elevated in HNSCC cell lines. qPCR analysis of PKCε and GAPDH was performed with total RNA isolated from human primary keratinocytes and HNSCC cell lines using validated TaqMan assays. Data is presented and mean ± SEM. *p-value<0.005. B. PKCε protein levels are elevated in HNSCC cell lines. Western blot analysis of PKCε and tubulin was performed with whole cell lysates isolated from human primary keratinocytes and HNSCC cell lines. C. miR-107 expression levels are lower in HNSCC cell lines. qPCR analysis of miR-103 and miR-107 in primary keratinocytes and HNSCC cell lines was performed using validated TaqMan assays. Data is presented and mean ± SEM. *p-value<0.005.
PKCε is a target of miR-107
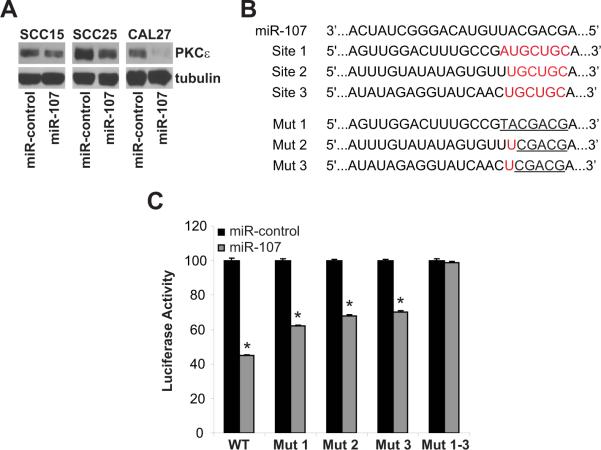
To further provide evidence that miR-107 regulates PKCε, HNSCC cells were transiently transfected with pre-miR-control or pre-miR-107 (50 nmol/L for 48 hours). Transfection with pre-miR-107 enhanced the expression of mature miR-107 by 329 ± 8-fold, 386 ± 4-fold, and 227 ± 11-fold in SCC15, SCC25 and CAL27 cells, respectively (p<0.001). In comparison to pre-miR-control, delivery of pre-miR-107 resulted in a dramatic decrease in PKCε levels in SCC15, SCC25, and CAL27 cells confirming that PKCε is a target of miR-107 (Figure 2A). Next, we determined if miR-107 directly binds to their predicted sites in the 3'-UTR of PKCε to control PKCε levels. In silico analysis with TargetScan, identified three putative miR-107 binding sites in the 3'-UTR of PKCε located at nucleotides 27–33 (Site 1), 1517–1523 (Site 2), and 1564–1570 (Site 3). CAL27 cells were co-transfected with pre-miR-control or pre-miR-107 and a reporter expression vector containing wildtype or mutant 3'-UTR of PKCε cloned downstream of a luciferase gene (Figure 2C). pre-miR-107 blocked luciferase activity by 55 ± 2% (p<0.01) in cells transfected with the full-length wildtype 3'-UTR of PKCε. Mutations in one of the three miR-107 sites dampened the inhibitory effect of pre-miR-107 on luciferase activity; 38 ± 2% inhibition for mutant 1, 32 ± 4% inhibition for mutant 2, and 30 ± 3% inhibition for mutant 3. Interestingly, pre-miR-107 had no effect on luciferase activity in cells transfected with the 3'-UTR of PKCε containing mutations in all three miR-107 sites (Mut 1–-3). Our work confirmed that miR-107 regulates PKCε through direct binding in the 3'-UTR of the PKCε mRNA and moreover, optimal suppression of PKCε requires occupation of all three miR-107 sites.
Figure 2. PKCε is a downstream target of miR-107.
A. Ectopic expression of miR-107 reduced PKCε levels in HNSCC. SCC15, SCC25, and CAL27 cells were transfected with premiR-control or pre-miR-107 (50 nmol/L) using Lipofectamine 2000 for 48 hours. Whole cell lysates were extracted and western blot analysis was performed using anti-PKCε and anti-tubulin antibodies. B. Wildtype and mutant miR-107 binding sites in the 3'-UTR of PKCε. C. Luciferase activity controlled by 3'-UTR of PKCε is inhibited by ectopic expression of miR-107. HEK293T cells were co-transfected with wildtype or mutant PKCε-3'UTR and pre-miR-control or pre-miR-107. Renilla and Firefly luciferase activities were measured using the Dual-Luciferase Reporter assay system. Renilla luciferase is normalized to Firefly luciferase and data is presented as mean ± SEM of four independent experiments. *p-value < 0.01.
miR-107 reduces the tumorigenic potential of HNSCC in vitro and in vivo
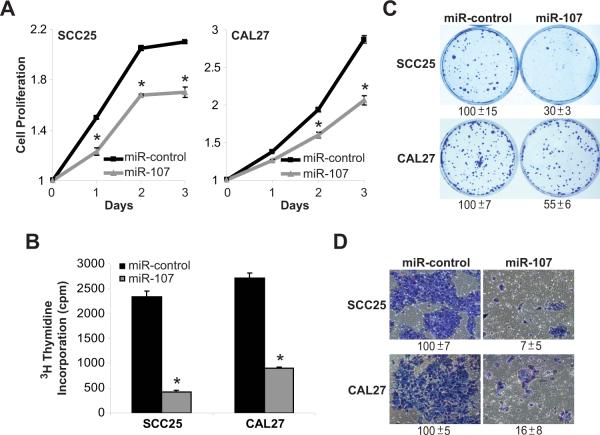
Our previous work demonstrated that PKCε plays a critical role in promoting an aggressive phenotype in HNSCC (Pan et al, 2006). Since miR-107 was sufficient to dramatically reduced PKCε levels, the anti-cancer effects of miR-107 on HNSCC cells were assessed. As shown in Figure 3, cell proliferation, DNA replication, colony formation, and cell invasion were determined in SCC25 and CAL27 cells following treatment with pre-miR-control or pre-miR-107 (50 nmol/L). Cell proliferation was reduced in SCC25 cells by 20 ± 2% (p<0.01) and in CAL27 cells by 28 ± 2% (p<0.01) following pre-miR-107 treatment. pre-miR-107 blocked DNA replication and colony formation by 72 ± 5% and 70 ± 3% in SCC25 cells and 67 ± 3% and 45 ± 6% in CAL27 cells, respectively (p<0.01). Treatment with pre-miR-107 inhibited cell invasion by 93 ± 5% in SCC25 cells and 84 ± 8% in CAL27 cells (p<0.01).
Figure 3. Ectopic expression of miR-107 reduces the tumorigenic properties of SCC25 and CAL27 cells in vitro.
SCC25 and CAL27 cells were transfected with pre-miR-control or pre-miR-107 (50 nmol/L) using Lipofectamine 2000. After 48 hours, cells were used for the phenotypic assays. A. Cell proliferation. Cell proliferation was measured using the MTT assay. Data is presented as mean ± SEM. *p-value<0.01, n=3. B. DNA synthesis. Incorporation of 3H-thymidine into DNA was monitored after 4 hours in a scintillation counter. Data is presented as mean ± SEM. *p-value<0.01, n=3. C. Colony formation. Colonies were stained with crystal violet and counted. Data is presented as mean ± SEM. *p-value<0.01, n=3. D. Cell invasion. Cell invasion was assessed using the Modified Boyden chamber invasion assay with Matrigel basement membrane. Invasive cells were counted. Data is presented as mean ± SEM. *p-value<0.01, n=3.
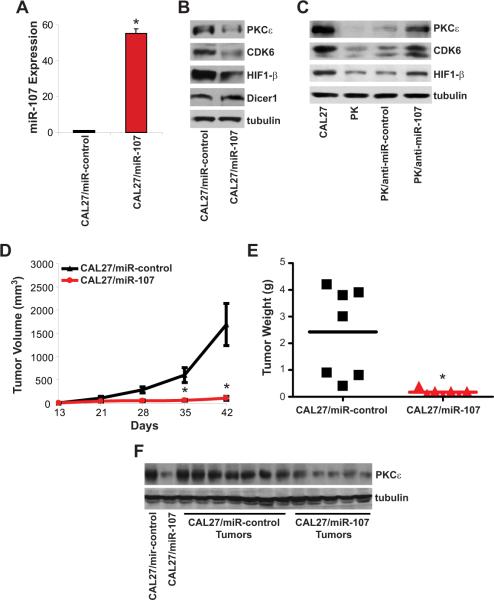
Stable polyclonal miR-control or miR-107 over-expressing CAL27 cells were implanted in athymic nude mice to determine the effect of miR-107 on in vivo tumorigenicity (Figure 4). In Figure 4A, CAL27/miR-107 cells had a dramatic increase in miR-107 expression compared to CAL27/miR-control cells (p<0.005). The levels of PKCε and two other confirmed miR-107 targets, hypoxia inducible factor 1-β (HIF1-β) and cyclin-dependent kinase 6 (CDK6), were decreased in miR-107 over-expressing CAL27 cells. Interestingly, Dicer1, a confirmed miR-107 target in breast cancer, was unchanged in CAL27/miR-107 cells compared to CAL27-miR-control cells. Compared to anti-miR-control transfection, primary keratinocytes transfected with anti-miR-107 (PK/anti-miR-107) had lower mature miR-107 expression (70 ± 1% inhibition, p<0.001). Importantly, PK/anti-miR-107 cells demonstrated an increase in PKCε, HIF1-β, and CDK6 protein levels providing further evidence that miR-107 is a key regulator of these genes (Figure 4C). Tumor growth was significantly reduced in the mice implanted with CAL27/miR-107 cells compared to CAL27/miR-control cells. Mice bearing CAL27/miR-control tumors had a mean tumor volume of 1691 ± 455 mm3 at 6 weeks post-implantation. In contrast, CAL27/miR-107 cells were less tumorigenic and had a mean tumor volume of 108 ± 39 mm3 (p<0.005, n=7). Moreover, mean tumor weight was significantly lowered by 93 ± 2% in CAL/miR-107 tumors compared to CAL27/miR-control tumors (p<0.005, n=7). Importantly, PKCε protein levels were dramatically lower in tumors from CAL27/miR-107 cells than in tumors from CAL27/miR-control cells. Taken together, these results indicate that down-regulation of PKCε by miR-107 is an effective approach to suppress the tumorigenicity of HNSCC in vitro and in vivo.
Figure 4. miR-107 reduces tumorigenicity in nude mice.
A. miR-107 expression. Total RNA was isolated from CAL27/miR-control and CAL27/miR-107 cells. miR-107 expression was measured using qPCR. Data is presented as mean ± SEM. *p-value<0.005, n=3. B. Protein levels of miR-107 target genes in miR-107 overexpressing CAL27 cells. Whole cell lysates were extracted and western blot analysis was performed. C. Protein levels of miR-107 target genes in primary keratinocytes transfected with anti-miR-107. Whole cell lysates were extracted and western blot analysis was performed. D. Tumor growth. CAL27/miR-control or CAL27/miR-107 cells were injected subcutaneously to the flanks of the nude mice. Tumors were measured weekly using a caliper and tumor volumes were calculated. Data is presented as mean ± SEM. *p-value<0.005, n=7. E. Tumor weight. Tumors were weighed and data is presented as mean ± SEM for each group. *p-value<0.005, n=7. F. Tumor PKCε levels. Whole cell lysates were isolated from the resected tumors and PKCε levels were measured by western blot analysis.
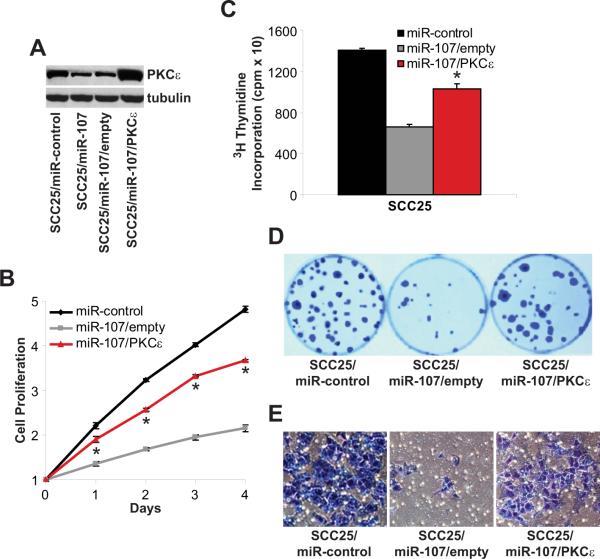
miR-resistant PKCε reverses the anti-tumorigenic effects of miR-107
In addition to targeting PKCε, other groups have reported that miR-107 directly regulates HIF1-β and CDK6 (Feng et al, 2011; Lee et al, 2009; Yamakuchi et al, 2010). To provide direct evidence that down-regulation of PKCε is required for the anti-tumorigenic effects of miR-107, miR-resistant PKCε was over-expressed in miR-107 expressing SCC25 (SCC25/miR-107) cells to determine if restoring PKCε would be sufficient to rescue the miR-107 loss-of-function phenotype. SCC25/miR-107 cells were transfected with a wild type PKCε expression or empty vector without the 3'-UTR of PKCε. In Figure 5A, the SCC25/miR-107/PKCε cells had the proper genetic alterations and had elevated PKCε levels in a high miR-107 background. Empty vector-transfected SCC25/miR-107 cells had minimal effect on cell phenotype as these cells maintained their miR-107 loss-of-function defect compared to SCC25/miR-controls cells. Importantly, ectopic over-expression of PKCε partially rescues the defect of SCC25/miR-107 cells and resulted in a significant increase in cell proliferation (p<0.005), DNA replication (56 ± 8%, p<0.005), colony formation (130 ± 18%, p<0.005), and cell invasion (392 ± 39%, p<0.005) compared with SCC25/miR-107/empty cells. These results reveal that the tumor-suppressive actions of miR-107 are, at least, partially through modulation of PKCε levels.
Figure 5. miR-resistant PKCε reverses the tumor suppressive actions of miR-107.
SCC25/miR-107 cells were transfected with an empty vector or a PKCε expression vector to generate SCC25/miR-107/empty and SCC25/miR-107/PKCε cells. A. PKCε levels. Whole cell lysates were extracted and western blot analysis was performed using anti-PKCε and anti-tubulin antibodies. B. Cell proliferation. Cell proliferation was measured using the MTT assay. Data is presented as mean ± SEM. *p-value<0.005, n=3. C. DNA synthesis. Incorporation of 3H-thymidine into DNA was monitored after 4 hours in a scintillation counter. Data is presented as mean ± SEM. *p-value<0.005, n=3. D. Colony formation. Colonies were stained with crystal violet and counted. E. Cell invasion. Cell invasion was assessed using the Modified Boyden chamber invasion assay with Matrigel basement membrane. Invasive cells were counted.
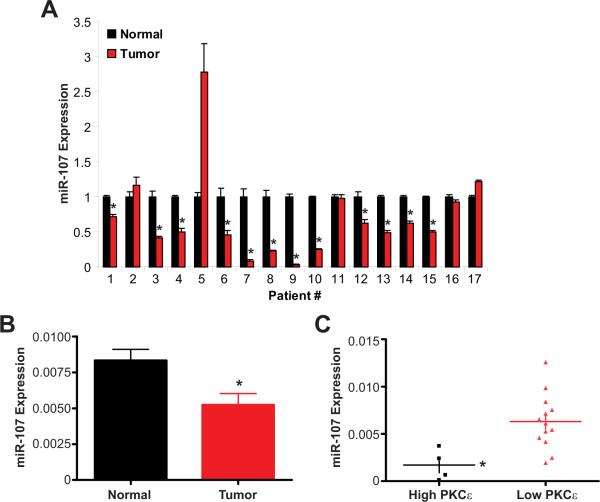
miR-107 is reduced and inversely associated with PKCε in primary HNSCC tumors
Our studies with HNSCC cell lines demonstrated an inverse association between miR-107 expression and PKCε protein levels. Next, we examined miR-107 and PKCε expression in primary tumors and matched normal adjacent epithelium from previously untreated HNSCC patients by qPCR. As shown in Figure 6A, a majority (12/17) of HNSCC patients had significantly lower miR-107 expression in the tumor compared to normal tissue. A 37.5% reduction in miR-107 expression in the tumor compared to the normal tissue was demonstrated for this patient cohort (p<0.008, n=17); mean miR-107 expression was 0.005 ± 0.0008 for the primary HNSCC tumors and 0.008 ± 0.0007 for the adjacent normal epithelium. These observations are consistent and further support our results demonstrating that miR-107 has tumor suppressive actions in HNSCC. In this cohort, four patients were found to have significantly higher PKCε expression in their primary tumor compared to normal adjacent epithelium. All four patients with high PKCε were part of the cohort of twelve patients with reduced miR-107 expression in the tumor. We were unable to determine PKCε protein levels in this population due to insufficient and/or unavailable patient specimen. Since miRs are known to regulate mRNA stability and translation, assessment of PKCε protein may reveal additional patients in the low miR-107 population with elevated PKCε levels in the tumor. Nonetheless, even with this small sample size, miR-107 expression was found to be significantly lower (73 ± 13% reduction, p<0.01) in patients with high PKCε expression than in patients with low PKCε (Figure 6C). Our results demonstrate that miR-107 is reduced in HNSCC tumors and associated with elevated PKCε expression in HNSCC.
Figure 6. miR-107 is significantly down-regulated and associated with elevated PKCε in HNSCC tumors.
A. miR-107 expression in individual HNSCC patients. Primary tumors and adjacent normal tissue from HNSCC patients were measured for miR-107 expression using qPCR. *p<0.05, n=3. B. miR-107 expression is reduced in HNSCC tumors. Data is presented as mean ± SEM for each group. *p<0.008, n=17. C. HNSCC patients with elevated PKCε expression have reduced miR-107 expression. Data is presented as mean ± SEM for each group. *p<0.01.
Discussion
miRs are short (20–25 nucleotides) single stranded noncoding RNAs that play a critical role in the regulation of global gene expression. miRs negatively regulate target protein levels by suppressing gene mRNA translation and/or enhancing gene mRNA degradation (Filipowicz et al 2008). Published work has clearly shown that miRs are involved in the regulation of different signaling pathways including cellular differentiation, proliferation, migration and apoptosis (Avissar et al, 2009; Calin and Croce, 2006; Croce, 2009; Datta et al, 2008; Hwang and Mendell, 2006; Liu et al, 2009; Nasser et al, 2008). Thus, it is not surprising that aberrant expression of certain miRs is associated with cancer development and progression, including HNSCC (Avissar et al, 2009; Calin and Croce, 2006; Croce, 2009; Datta et al, 2008; Hwang and Mendell, 2006; Liu et al, 2009; Nasser et al, 2008). In the present study, our results revealed that miR-107 is under-expressed and functions as a tumor suppressor in HNSCC. Ectopic expression of miR-107 is sufficient to dampen the tumorigenic potential of HNSCC cells in vitro and in vivo. PKCε was confirmed as a direct target of miR-107 and had an inverse relationship with miR-107. Our study indicates that suppression of miR-107 leads to an increase in PKCε resulting in the development of HNSCC.
Consistent with our work, other groups have reported that miR-107 functions as a tumor suppressor in bladder, colon, and pancreatic cancer. miR-107 was dramatically reduced in bladder carcinoma in situ compared to normal bladder (Ayala de la Pena et al, 2011). An inverse association between miR-107 and HIF1-β was demonstrated in human colon cancer specimens (Yamakuchi et al, 2010). Over-expression of miR-107 inhibits HIF1-β levels and retards tumor growth in HCT116 colon cancer cells (Yamakuchi et al, 2010). Enforced expression of miR-107 in pancreatic cancer cells, MiaPACA-2 and PANC-1, repressed CDK6 levels and inhibited cell proliferation (Lee et al, 2009). In contrast, several reports have shown that miR-107 functions as an oncogene in breast and gastric cancer. Elevated miR-107 levels were associated with distant metastasis and poorer clinical outcome in breast cancer (Martello et al, 2010). Over-expression of miR-107 in MCF10A mammary epithelial cells promotes epithelial-to-mesenchymal transition resulting in a highly metastatic phenotype thorough modulation of Dicer1 (Martello et al, 2010). In line with this report, knockdown of miR-107 leads to an increase in Dicer1 and inhibition of cell invasion and migration in gastric cancer cells (Li et al, 2010). Taken together, these results indicate that the biology of miR-107 is complex and highly cell-type dependent.
Our data showed that CDK6 and HIF1-β levels are decreased whereas Dicer1 levels were unchanged in CAL27/miR-107 cells compared to CAL27/miR-control cells. This indicates that miR-107 target genes are not regulated in concert and raises the possibility that other factors, including additional genetic alterations, may dictate which set of miR-107 targets is regulated in a particular cell type. Moreover, it is possible that down-regulation of Dicer1 may be a prerequisite for miR-107 to function as an oncogene instead of a tumor suppressor. Expression of miR-resistant PKCε was sufficient, at least partially, to rescue the loss-of-function phenotype observed for SCC25/miR-107 cells. This key observation indicates that a critical event for the tumor suppressive actions of miR-107 in HNSCC is to negatively control the levels of PKCε.
Global miR expression analysis showed that miR-107 was significantly down-regulated in HNSCC tumors compared to matched normal tissue (Wong et al, 2008). In addition, miR expression profiling between 18 HNSCC cell lines and oral keratinocytes indicate that miR-107 is decreased at high frequency in HNSCC (Kozaki et al, 2008). Our work showed that a majority (12/17) of HNSCC patients have reduced miR-107 expression in the primary tumor and thus, provide additional support that loss of miR-107 is a frequent event in the development of HNSCC. In this cohort, HNSCC patients with high PKCε mRNA expression had a dramatic 73% reduction in miR-107 expression compared to patients with low PKCε mRNA expression. This observation is in line with our in vitro results and provide further evidence that miR-107 is inversely associated with PKCε in HNSCC.
In summary, our results showed that miR-107 functions as a tumor suppressor in HNSCC through regulation of PKCε. Reconstitution of miR-107 is sufficient to retard the tumorigenicity of HNSCC cells. Our study indicates that reduction in miR-107 expression is a pathogenetic event in HNSCC and suggests that miR-107 may be a potential anti-cancer therapeutic for this patient population.
Materials and Methods
Cell Culture
Human head and neck squamous cell carcinoma cell lines (SCC15, SCC25, and CAL27) were obtained from ATCC (Rockville, MD). CAL27 cells were cultured in DMEM medium containing 10% fetal bovine serum. SCC25 and SCC15 cells were grown in DMEM/F12 (1:1) medium containing 10% fetal bovine serum. Primary keratinocytes were obtained from ATCC and cultured in EpiLife medium (Invitrogen, Carlsbad, CA) supplemented with Ca2+ and other growth factors in a 5% CO2 incubator at 37°C. SCC15, SCC25, and CAL27 cell lines were authenticated using short tandem repeat profiling (ATCC).
Generation of transient and stable cell lines
Transient miR-107 and miR-control overexpressing SCC15, SCC25, and CAL27 cells were generated by transfection with pre-miR-107 or pre-miR-control (50 nmol/L) using Lipofectamine 2000. Cells were harvest after 48 hours for gene expression, protein levels, and functional assays. Stable CAL27/miR-107, CAL27/miR-control, SCC25/miR-107 and SCC25/miR-control were generated by transduction using shMIMIC miR-107 or miR-control lentiviral particles (Thermo Fisher Scientific, Waltham, MA). Cells were incubated with viral particles (MOI=1:20) mixed with polybrene (at a final concentration of 10μg/ml) for 12 hours. Subsequently, cells were cultured in complete growth media for 72 hours and polyclonal populations were selected with 1μg/ml puromycin. SCC25/miR-107 cells were transiently transfected with an empty vector or a PKCε expression vector (Ex-A1280-MO2; GeneCopoeia, Rockville, MD) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were harvested after 48 hours for protein levels and functional assays. Primary keratinocytes were transiently transfected with anti-miR-control or anti-miR-107 (100 nmol/L) using Lipofectamine 2000. Cells were harvested after 48 hours for gene expression and protein levels.
Real-time PCR
Total RNA was isolated from HNSCC cells and primary keratinocytes with TRIzol (Invitrogen, Carlsbad, CA). Expression of PKCε, GAPDH, mature miR-103a, mature miR-107, and RNU44 were determined using the Applied Biosystems 7900HT Fast Real-Time PCR System with validated TaqMan gene expression assays (Applied Biosystems, Foster City, CA). PKCε expression was normalized to GAPDH and mature miR-103a/miR-107 expression was normalized to RNU44 using the ΔΔCt method.
Luciferase reporter assay
Wildtype or mutant PKCε-3′UTR psiCHECK-2 plasmid was transiently transfected into H293T cells. Cells were transfected with 50 ng of psiCHECK-2 along with pre-miR-107 or pre-miR-control (100 nmol/L) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). After 48 hours, cells were washed with PBS, resuspended in the lysis buffer (100 mM potassium phosphate pH 7.8, 0.2% Triton X-100, 0.5 mM dithiothreitol), and measured for Firefly/Renilla luciferase activities in a luminometer using the Dual-Light System (Applied Biosystems, Foster City, CA). Renilla luciferase activities were normalized to Firefly luciferase activities to control for transfection efficiency.
Western blot analysis
Whole cell lysates were mixed with Laemmli loading buffer, boiled, separated by SDS-PAGE, and transferred to a nitrocellulose membrane. Subsequently, immunoblot analyses were performed using antibodies specific to PKCε (Santa Cruz Biotechnology, Santa Cruz, CA), CDK6 (Thermo Fisher Scientific), HIF1-β (Cell Signaling Technology, Danvers, MA), Dicer1 (Abcam, Cambridge, MA) or tubulin (Cell Signaling Technology). The signal was developed with ECL (Thermo Fisher Scientific) after incubation with appropriate secondary antibodies.
Cell invasion and proliferation
Cell invasion was determined as described from the cell invasion assay kit (BD Biosciences, Bedford, MA). Cells were harvested and resuspended in serum-free medium. An aliquot (1 × 105 cells) of the prepared cell suspension was added into the chamber and incubated for 24 hours at 37°C in a 10% CO2 tissue culture incubator. Non-invading cells were gently removed from the interior of the inserts with a cotton-tipped swab. Invasive cells were stained and counted. Cell proliferation was assessed using the MTT reagent to detect metabolic active cells (Roche Molecular Biochemicals, Nutley, NJ). Absorbance was measured at 570nm in the Spectra Max 190 ELISA reader (Molecular Devices, Sunnyvale, CA) after overnight incubation.
Colony formation
Cells (500) were plated in complete growth media and allowed to grow until visible colonies formed (10–12 days). Cell colonies were fixed with cold methanol, stained with 0.25% crystal violet in 25% methanol, washed and air dried.
DNA replication
Cells (10,000 cells per well) were plated in 24-well plates and allowed to grow for 24 hours. Subsequently, cells were incubated with serum-free media for 16 hours and then incubated with complete growth media containing 1 μCi of 3H-thymidine for 6 hours. Cells were washed with PBS and 3H-thymidine incorporation into DNA was measured using a Hitachi scintillation counter.
Tumor growth in nude mice
CAL27/miR-control or CAL27/miR-107 cells (1×106 cells) were implanted into the flanks of nude mice (6–8 weeks). After two weeks, tumors were measured once a week using a digital caliper and tumor volumes were calculated using the formula d1×d2×d3×0.5236, where d represents the three orthogonal diameters. In addition, tumors were resected and weighed at the end of the 6-week study protocol. Whole cell lysates were extracted from tumor tissues and determined for PKCε protein levels.
Analysis of primary HNSCC tissues
Seventeen primary tumors were collected at The Ohio State University James Cancer Hospital from HNSCC patients at the time of surgical resection between 1997 and 2000. All tissues were diagnosed histologically as HNSCC by a board certified pathologist. Written informed consent, as required by the institutional review board, was obtained from all patients. Collected samples were stored immediately in liquid nitrogen at −80°C until analysis. Total RNA was isolated from the frozen tumors with TRIzol (Invitrogen, Carlsbad, CA). Expression of PKCε, GAPDH, mature miR-107, and RNU44 were determined using the Applied Biosystems 7900HT Fast Real-Time PCR System with validated TaqMan gene expression assays (Applied Biosystems, Foster City, CA). PKCε and mature miR-107 expression were normalized to GAPDH and RNU44, respectively, using the ΔΔCt method.
Acknowledgements
This work was supported in part by the National Cancer Institute at the National Institutes of Health (R01CA135096); American Cancer Society (RSG0821901); The Michelle Theado Memorial Grant and John Young Memorial Grant from the Joan Bisesi Fund for Head and Neck Oncology Research; and Arthur G. James Cancer Hospital and Richard J. Solove Research Institute, The Ohio State University Comprehensive Cancer Center.
Footnotes
Conflict of Interest The authors declare no conflict of interest.
References
- Avissar M, Christensen BC, Kelsey KT, Marsit CJ. MicroRNA expression ratio is predictive of head and neck squamous cell carcinoma. Clin Cancer Res. 2009;15:2850–2855. doi: 10.1158/1078-0432.CCR-08-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala de la Pena F, Kanasaki K, Kanasaki M, Tangirala N, Maeda G, Kalluri R. Loss of p53 and acquisition of angiogenic microRNA profile is insufficient to facilitate progression of bladder urothelial carcinoma in situ to invasive carcinoma. J Biol Chem. 2011 doi: 10.1074/jbc.M110.198069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz MH, Hafeez BB, Sand JM, Pierce DB, Aziz SW, Dreckschmidt NE, et al. Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2) Oncogene. 2010;29:3100–3109. doi: 10.1038/onc.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Cohen RB, Jones CU, Sur RK, et al. Radiotherapy plus cetuximab for locoregionally advanced head and neck cancer: 5-year survival data from a phase 3 randomised trial, and relation between cetuximab-induced rash and survival. Lancet Oncol. 2010;11:21–28. doi: 10.1016/S1470-2045(09)70311-0. [DOI] [PubMed] [Google Scholar]
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Cohen EE, Lingen MW, Zhu B, Zhu H, Straza MW, Pierce C, et al. Protein kinase C zeta mediates epidermal growth factor-induced growth of head and neck tumor cells by regulating mitogen-activated protein kinase. Cancer Res. 2006;66:6296–6303. doi: 10.1158/0008-5472.CAN-05-3139. [DOI] [PubMed] [Google Scholar]
- Cohen EE, Zhu H, Lingen MW, Martin LE, Kuo WL, Choi EA, et al. A feed-forward loop involving protein kinase Calpha and microRNAs regulates tumor cell cycle. Cancer Res. 2009;69:65–74. doi: 10.1158/0008-5472.CAN-08-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dempsey EC, Newton AC, Mochly-Rosen D, Fields AP, Reyland ME, Insel PA, et al. Protein kinase C isozymes and the regulation of diverse cell responses. Am J Physiol Lung Cell Mol Physiol. 2000;279:L429–438. doi: 10.1152/ajplung.2000.279.3.L429. [DOI] [PubMed] [Google Scholar]
- Feng L, Xie Y, Zhang H, Wu Y. miR-107 targets cyclin-dependent kinase 6 expression, induces cell cycle G1 arrest and inhibits invasion in gastric cancer cells. Med Oncol. 2011 doi: 10.1007/s12032-011-9823-1. [DOI] [PubMed] [Google Scholar]
- Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- Gutcher I, Webb PR, Anderson NG. The isoform-specific regulation of apoptosis by protein kinase C. Cell Mol Life Sci. 2003;60:1061–1070. doi: 10.1007/s00018-003-2281-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaken S, Parker PJ. Protein kinase C binding partners. Bioessays. 2000;22:245–254. doi: 10.1002/(SICI)1521-1878(200003)22:3<245::AID-BIES6>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- Kozaki K, Imoto I, Mogi S, Omura K, Inazawa J. Exploration of tumor-suppressive microRNAs silenced by DNA hypermethylation in oral cancer. Cancer Res. 2008;68:2094–2105. doi: 10.1158/0008-5472.CAN-07-5194. [DOI] [PubMed] [Google Scholar]
- Laramore GE, Scott CB, al-Sarraf M, Haselow RE, Ervin TJ, Wheeler R, et al. Adjuvant chemotherapy for resectable squamous cell carcinomas of the head and neck: report on Intergroup Study 0034. Int J Radiat Oncol Biol Phys. 1992;23:705–713. doi: 10.1016/0360-3016(92)90642-u. [DOI] [PubMed] [Google Scholar]
- Lee KH, Lotterman C, Karikari C, Omura N, Feldmann G, Habbe N, et al. Epigenetic silencing of MicroRNA miR-107 regulates cyclin-dependent kinase 6 expression in pancreatic cancer. Pancreatology. 2009;9:293–301. doi: 10.1159/000186051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leemans CR, Braakhuis BJ, Brakenhoff RH. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11:9–22. doi: 10.1038/nrc2982. [DOI] [PubMed] [Google Scholar]
- Li X, Zhang Y, Shi Y, Dong G, Liang J, Han Y, et al. MicroRNA-107, an Oncogene MicroRNA that Regulates Tumor Invasion and Metastasis By Targeting DICER1 in Gastric Cancer: MiR-107 promotes gastric cancer invasion and metastasis. J Cell Mol Med. 2010 doi: 10.1111/j.1582-4934.2010.01194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141:1195–1207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- Martinez-Gimeno C, Diaz-Meco MT, Dominguez I, Moscat J. Alterations in levels of different protein kinase C isotypes and their influence on behavior of squamous cell carcinoma of the oral cavity: epsilon PKC, a novel prognostic factor for relapse and survival. Head Neck. 1995;17:516–525. doi: 10.1002/hed.2880170609. [DOI] [PubMed] [Google Scholar]
- Nasser MW, Datta J, Nuovo G, Kutay H, Motiwala T, Majumder S, et al. Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression of tumorigenic property of lung cancer cells and their sensitization to doxorubicin-induced apoptosis by miR-1. J Biol Chem. 2008;283:33394–33405. doi: 10.1074/jbc.M804788200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Pan Q, Bao LW, Teknos TN, Merajver SD. Targeted disruption of protein kinase C epsilon reduces cell invasion and motility through inactivation of RhoA and RhoC GTPases in head and neck squamous cell carcinoma. Cancer Res. 2006;66:9379–9384. doi: 10.1158/0008-5472.CAN-06-2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkin DM. Global cancer statistics in the year 2000. Lancet Oncol. 2001;2:533–543. doi: 10.1016/S1470-2045(01)00486-7. [DOI] [PubMed] [Google Scholar]
- Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP, Wei WI. Mature miR-184 as Potential Oncogenic microRNA of Squamous Cell Carcinoma of Tongue. Clin Cancer Res. 2008;14:2588–2592. doi: 10.1158/1078-0432.CCR-07-0666. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lotterman CD, Bao C, Hruban RH, Karim B, Mendell JT, et al. P53-induced microRNA-107 inhibits HIF-1 and tumor angiogenesis. Proc Natl Acad Sci U S A. 2010;107:6334–6339. doi: 10.1073/pnas.0911082107. [DOI] [PMC free article] [PubMed] [Google Scholar]