Abstract
Background
Cells avoid major mitochondrial damage and energy failure during systemic inflammatory states, such as severe acute infections, by specific targeting of the inflammatory response and by inducing anti-inflammatory and anti-oxidant defenses. Recent evidence indicates that these cell defenses also include mitochondrial biogenesis and the clearance of damaged mitochondria through autophagy.
Scope of Review
This review addresses a group of transcriptional signaling mechanisms that engage mitochondrial biogenesis, including energy-sensing and redox-regulated transcription factors and co-activators, after major inflammatory events.
Major Conclusions
Stimulation of the innate immune system by activation of toll-like receptors (TLR) generates pro-inflammatory mediators, such as tumor necrosis factor-α (TNF-α) and interleukin-1β, (IL-1β), necessary for optimal host defense, but which also contribute to mitochondrial damage through oxidative stress and other mechanisms. To protect its energy supply, host cells sense mitochondrial damage and initiate mitochondrial biogenesis under the control of an inducible transcriptional program that also activates anti-oxidant and anti-inflammatory gene expression. This multifunctional network not only increases cellular resistance to metabolic failure, oxidative stress, and cell death, but promotes immune tolerance as shown in the graphical abstract.
General Significance
The post-inflammatory induction of mitochondrial biogenesis supports metabolic function and cell viability while helping to control inflammation. In clinical settings, patients recovering from severe systemic infections may develop transient immune suppression, placing them at risk for recurrent infection, but there may be therapeutic opportunities to enhance mitochondrial quality control that would improve the resolution of life-threatening host responses to such infections.
Keywords: Infection, inflammasome, innate immunity, cell metabolism, oxidative stress
Introduction
The protection of mitochondrial function is essential not only to normal mammalian physiology, but also to the biological response to major inflammatory states, particularly when inflammation triggers significant cell stress. The innate and adaptive responses to intense, prolonged, or poorly-contained immunological stimuli produce mitochondrial damage in host cells that can impair their capacity to generate sufficient adenosine triphosphate (ATP) for homeostasis [1]. This may limit energy availability for cellular maintenance, and ultimately, compromise cell survival and organ function.
The damaging effects of inflammation on mitochondria are opposed by cellular programs that protect the quality of the cell’s mitochondria by disposing of badly damaged organelles and replacing them with fresh ones [2]. This process of mitochondrial quality control involves an intricate bi-genomic program that regulates mitochondrial DNA (mtDNA) transcription and replication and mitochondrial protein synthesis. The program is tailored to specific signals from the cell to rapidly adjust mitochondrial mass, number, size, distribution, and/or phenotype [3, 4]. Such changes in mitochondrial mass and function encompass structurally distinct events within the cell, including autophagy (mitophagy), changes in cytoskeletal morphology, the mitochondrial fusion and fission cycle, and mitochondrial biogenesis [5, 6].
Since mitochondria are not generated de novo, the cell must rely on processes that identify and target dysfunctional mitochondria for degradation while concomitantly stimulating healthy mitochondria to proliferate and repopulate it [5]. These processes effectively segregate mitochondria into functional and non-functional sub-populations and assure uninterrupted ATP provision for essential work and for survival. The molecular regulation of this sophisticated capability is still in the discovery phase, and this concise review focuses on regulatory aspects of the transcriptional network for mitochondrial biogenesis in mammalian systems that maintain and restore mitochondrial function after inflammation-induced cell and organ damage. It is focused on broadly translatable principles, but we have identified the known exceptions and information that is restricted to lineage-specific events.
The Transcriptional Program of Mitochondrial Biogenesis
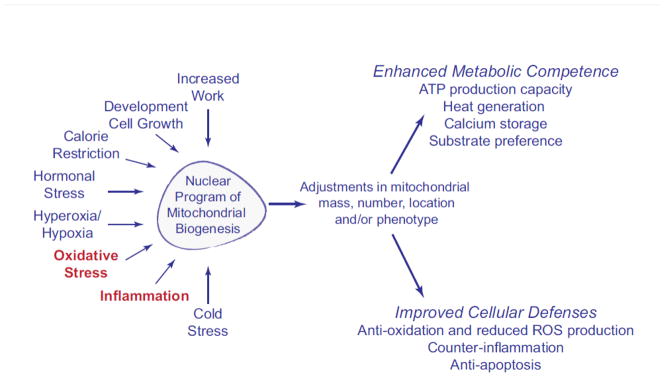
Mitochondrial biogenesis is activated by physiological and pathological stimuli including but not limited to cell division, development, exercise, thermogenesis, postnatal breathing, thyroid hormone and erythropoietin secretion, calorie restriction, oxidative stress, and inflammation [3, 4, 7–19]. Effective regulation of mitochondrial biogenesis during changing requirements for oxidative metabolism brought about by these factors provides ancillary or more efficient support for the indispensable energy-dependent functions of the cell (Figure 1). Such functions include muscle contraction, heat production, tissue growth and remodeling, and repair of cell and mitochondrial damage after exposure to stressors.
Figure 1.
The major physiological factors that are known to activate mitochondrial biogenesis and potential downstream effects on various mitochondrial functions.
Because mitochondria are adaptive organelles, about half of the 1,000 plus mitochondrial genes are expressed in a tissue- or lineage-specific manner, suggesting much of the mitochondrial proteome is dedicated to specialized functions [20]. The regulation of mitochondrial biogenesis presents a transcriptional challenge for the cell because the control mechanisms must coordinate a large set of inducible mitochondrial genes, while simultaneously enabling tissue- and signal-specific induction of gene subsets. Most of the genes required for mitochondrial biogenesis and respiration are under the control of a nuclear network of DNA–binding transcription factors and co-regulators that allows for vigorous activation in response to diverse physiological cues, as well as tissue- or signal-specific modifications of mitochondrial function and mass. Most of the nuclear-encoded genes for mitochondrial proteins, for instance, electron transport and oxidative phosphorylation proteins, contain conserved promoter binding motifs for nuclear respiratory factors-1 (NRF-1) and NRF-2 (also GA-binding protein or GABP). These transcription factors also activate nuclear genes that encode for proteins for mtDNA transcription and replication and mitochondrial protein importation, often operating in tandem as described in previous expert papers and reviews [4, 7, 21–23].
The identification of the peroxisome proliferator-activated receptor gamma (PPARγ) co-activator 1-α protein (PGC-1α) as a transcriptional co-activator of NRF-1, GABP, and the PPARs led to the recognition that PGC-1α integrates physiological signals with mitochondrial biogenesis and oxidative metabolism [7]. This recognition of PGC-1α also led to the identification of related co-activators (PGC-1β and the PGC-1-related co-activator, PRC) encoded by the Ppargc1a, Ppargc1b, and Pprc1 genes [23]. Other transcription factors, such as nuclear estrogen-related receptors (ERRs) operate along the same or related pathways [24]. Many aerobic tissues also express the orphan nuclear receptor ERRα, which regulates the machinery for fatty acid β-oxidation and is a PGC-1-α partner [25]. Other transcription factors are also important in the control of respiratory function and other genes of energy metabolism including the cyclic AMP response element binding protein (CREB) and the YY1 initiator binding factor, but details of their regulation are found elsewhere [26–31].
Among the nuclear-encoded regulatory genes for mitochondrial biogenesis are the proto-oncogene protein c-Myc and the myocyte-specific enhancer factor 2A (MEF2A) [32, 33]; the former is an activator of Ppargc1b (PGC-1β) and the latter, controlled in part by NRF-1, is a critical regulator of the oxidative capacity of skeletal and cardiac muscle [34–36]. MEF2A also activates growth factor and stress-induced genes and positively regulates cell growth and cell survival [36].
It has been increasingly appreciated that these elaborate and interactive bi-genomic transcriptional controls serve to match the mitochondrial mass and oxidative phenotype to the needs of various tissues, to changes in physiological environment, and to the need for organelle turnover in those tissues. Since mitochondrial biogenesis is controlled mainly at the level of transcription, the expression of the large number of requisite mitochondrial genes encoded in the nucleus must be synchronized with the few genes encoded in mitochondria. This bi-genomic coordination is achieved by nuclear-encoded mitochondrial proteins, including mitochondrial transcription factor A (TFAM), TFB1M, and TFB2M that control mtDNA transcription and replication and are synthesized in response to signals for mitochondrial biogenesis [3, 4].
Calcium and Mitochondrial Biogenesis
A great deal of work on the physiological induction of mitochondrial biogenesis, for instance in exercising muscle, has focused on the role of intracellular calcium (Ca2+). Calcium stimulates mitochondrial biogenesis in mammalian skeletal and cardiac muscle [37–43], but the role of Ca2+ in mitochondrial biogenesis in non-contractile cells, including those of the immune system is not clear [44]. In the post-inflammatory induction of mitochondrial biogenesis, calcium/calmodulin is required for the activation of two nitric oxide synthase (NOS) isoforms involved in mitochondrial biogenesis, NOS3 [16] and NOS1 [45], but NOS2, which is Ca2+-independent, is also a contributor [46].
In skeletal muscle, a Ca2+-sensitive PKC pathway [37] along with Ca2+-dependent regulation of nuclear genes for mitochondrial biogenesis have been found [40]. The calcium/calmodulin-dependent protein kinase (CaMK) family of serine/threonine kinases is involved in PGC-1 α regulation in skeletal muscle [43]. CaMK II [47, 48] and the Ca2+-activated phosphatase calcineurin (CaN) [39] are involved in combination with other processes that regulate muscle fiber plasticity [42].
Apart from CaMK II and CaN, mitogen-activated protein kinases (MAPK), for instance p38 [49] are activated by exercise, and certain p38 isoforms, such as p38γ (MAPK12), stimulate PGC-1α activity [50]. In the heart, p38 activates the nuclear peroxisome proliferator-activated receptor alpha (PPARα) through its PGC-1α co-activator [51]. In muscle (and fat) cells, the phosphorylation of ATF-2, a cyclic AMP response element binding (CREB) protein, activates PGC-1α gene expression [49, 52]. PGC-1α gene expression is also increased by MEF-2 binding to recognition sites on the promoter [35], and MEF2A and MEF2D in skeletal muscle contribute to regulation of the GLUT4 glucose transporter [41]. MEF2 also regulates Ca2+-dependent gene expression in developing cardiac muscle, which is repressed by class II histone deacetylases (HDACs). HDAC5, for instance, dissociates from MEF2 after serine phosphorylation in response to Ca2+, enabling transcriptional activation of PGC-1α in the heart [35]. Moreover, the MEF2/HDAC pathway is also linked by oxidative stress to pathological cardiac hypertrophy [53].
Physiologically, CaMK and CaN are activated by certain Ca2 + transients, specifically by prolonged transients befitting constant muscle stimulation, which promote nuclear translocation of transcription factors [54, 55] and activation of HDACs involved in muscle plasticity [55, 56]. Finally, these calcium signals interact in indistinct ways with ROS and NO in the control of muscle fiber phenotype [50].
Inflammation and Mitochondrial Biogenesis
During inflammation, the cell faces a unique set of challenges in the need to accelerate mitochondrial turnover. The first evidence of mitochondrial damage in inflamed tissues was reported some 40 years ago after the administration of bacterial lipopolysaccharide (LPS; endotoxin) and the experimental introduction of bacterial sepsis in animals [57–59]. Over the next decade or so, many post-inflammation abnormalities were reported in mitochondria in tissues such as liver, heart, and skeletal muscle including decreases in State 3 respiration, low respiratory control ratios, low rates of ATP synthesis, and cytochrome depletion. Similar effects were noted in hemorrhagic and cardiogenic shock, but a high degree of variably made it impossible to know whether these defects were caused primarily by LPS released from circulating bacteria, by secondary microbial or endogenous ligands, or by microcirculatory disturbances leading to tissue hypoxia or ischemia [60].
It was later recognized that mitochondria are susceptible to molecular damage from the actions of pro-inflammatory cytokines, like tumor necrosis factor alpha (TNF-α) and certain interleukins generated by innate immune cells including macrophages and Kupffer cells [61]. Damaged mitochondria generate high levels of reactive oxygen species (ROS) and release calcium and intrinsic apoptosis proteins during inflammation [62–64]. Moreover, rates of inflammatory ROS and NO production can be sufficient to compromise mitochondrial structure and function at the level of the respiratory chain, either by direct chemical oxidation of proteins and lipids or by the NO-superoxide reaction, which generates the powerful oxidant peroxynitrite (ONOO-) [65].
The prospect that the host response produces important cytotoxic effects on mitochondria, themselves likely ancient endosymbiotes, was bolstered in the 1990’s by the discovery of an evolutionarily-conserved system of pattern-recognition receptors (PRRs) that recognize pathogen-associated molecular patterns or PAMPs shared by classes of pathogens and identified by higher eukaryotes as “non-self” [66–68]. PAMPs can be entire molecules such as LPS or small conserved motifs within a molecule recognized by the host innate immune system as components of a class of pathogens. The first human PRRs were identified by cloning a human homolog of the Toll protein in Drosophila and the demonstration that it activates innate immunity through the classical NF-kB transcription factor pathway [66].
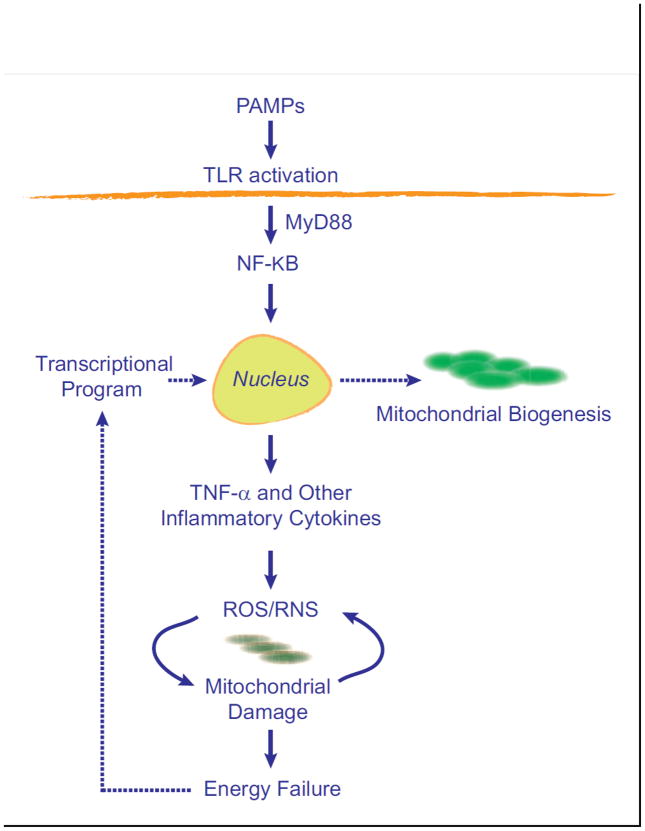
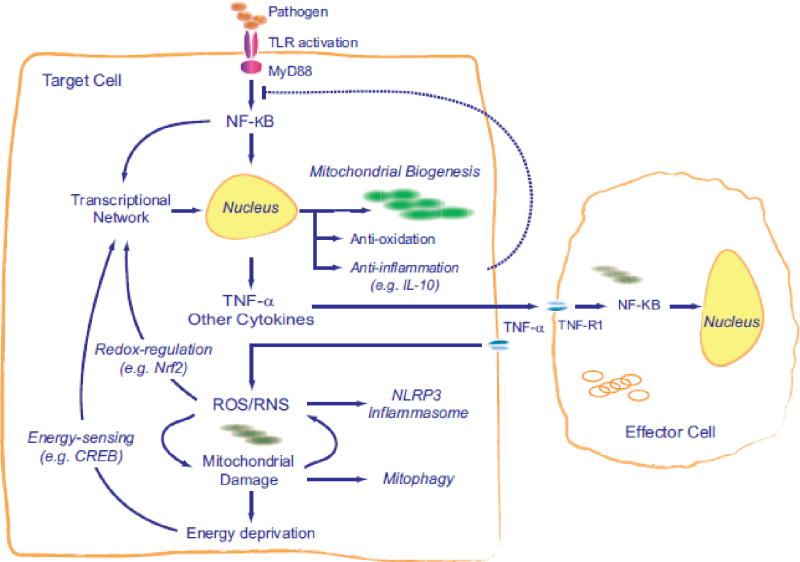
Toll-like receptors (TLR), located for instance on macrophages and many dendritic cells, recruit specific adaptor proteins, such as MyD88 and TRIF that initiate downstream signals for the synthesis of inflammatory cytokines, chemokines, antimicrobial peptides, and type I interferon [68]. TLR4 signaling through MyD88 to NF-kB is the most potent known LPS-response pathway and leads to rapid synthesis of TNF-α and interleukin-1 beta (IL-1β) and induction of nitric oxide synthase-2 (NOS2) [67]. Over-production of these and other effectors leads to mitochondrial damage that must be mitigated and corrected in the interest of cell survival. A minimal diagram of this idea is shown in Figure 2.
Figure 2.
A simplified concept of the induction of mitochondrial biogenesis by activation of the innate immune system. The production of pro-inflammatory cytokines creates a cycle of oxidative stress that causes mitochondrial damage and initiates retrograde signals for the transcriptional activation of mitochondrial biogenesis.
A dozen TLR family members (10 in the human) are known to operate on the plasma membrane or on the endosome to initiate the innate immune response to PAMPs from bacteria, fungi, parasites, and viruses. Early-phase cytokine and chemokine production promotes pathogen elimination by activation and recruitment of myeloid immune cells, such as neutrophils, macrophages, and lymphocytes to sites of infection, resulting in phagocytosis and stimulation of adaptive immune responses like antigen presentation and antibody production [69]. Roles for TLR signaling in dendritic cells, in somatic/structural cells, and in adaptive immunity [69], as well as the discovery of non-TLR PRRs, such as C-type lectin receptors, NOD-like and RIG-I-like receptors, and endogenous TLR activators generated by tissue damage all signify that this recognition system is more elaborate and ubiquitous than originally envisioned [70].
Despite these major advances, it remained unclear why hyper-activation of the acute inflammatory response did not consistently produce measurable mitochondrial damage, energy failure, or cell death despite evidence of organ dysfunction. Here, Figure 2 is deficient because it implies that post-inflammatory stress, mitochondrial damage and ATP limitation are isolated triggers for adaptive mitochondrial biogenesis, which builds in a lag in mitochondrial recovery that would predispose the cell to energy failure and apoptosis or necrosis. Moreover, no information existed on whether variable mitochondrial function simply involved stochastic differences in the degree of damage or reflected specific molecular mechanisms linking the immune system to mitochondrial damage, metabolic need, or mitochondrial quality control [71].
Severe inflammation induces ROS and NO stress in virtually all cell and animal models, but in pathological states, ROS are pro-inflammatory and activate NF-kB and other transcription factors that regulate inflammation [72, 73]. NF-kB activation by ROS is well known, often leading to enhanced TNF-α production, which further increases ROS production, for example by inflammatory and vascular cells [74, 75]. TNF-α is also a powerful catabolic agent, especially in skeletal muscle, where it inhibits respiration, mitochondrial biogenesis, and switching to the oxidative fiber phenotype [76]. In cardiomyocytes during inflammation, the NF-kB p65 subunit also appears to bind to the PGC-1α co-activator and block its activation of gene transcription [77].
Intense inflammation up-regulates many anti-oxidant defenses, but the transcriptional activity of both PGC-1α and NRF-1 also increases [78, 79]. PGC-1α promotes gene induction for certain ROS-detoxifying enzymes like mitochondrial superoxide dismutase (SOD2) and glutathione peroxidase-1 (GPx1), and data in neuronal cells suggest that CREB plays an important role in activating the Pargc1a (PGC-1α) gene promoter after oxidant exposure [78]. PGC-1α can also regulate myocardial SOD2 and thioredoxin (Trx2) expression and protect the heart against myocardial oxidative stress, hypertrophy, and contractile dysfunction (in mice) after transverse aortic banding [80]. Interestingly in macrophages, PGC-1β over-expression inhibits canonical NF-kB-dependent cytokine production, which could allay an acute inflammatory stress [81].
TNF-α is of further interest because TNF-α receptor 1 (TNF-R1) binding amplifies NF-kB activity and cytokine production, which increases oxidative stress and may lead to cell necrosis [82]. On the other hand, NF-kB is pro-survival and activates anti-oxidant enzyme genes, including SOD2 [83], and NF-kB inhibition increases TNF-mediated apoptosis in part by allowing recruitment of the FAS-associating death domain-containing protein (FADD) to a death-inducing signaling complex (DISC) that triggers caspase-8 activation [82]. The cell can avert both forms of premature death by the proteolytic shedding of TNF-R1 [84].
The TNF-R1-TNF-α ligand also causes cytosolic Ca2+ release and increases mitochondrial Ca2+ uptake [85]. The latter stimulates both respiration and mitochondrial ROS generation [86] . In lung vascular cells, when SOD2 activity is high, Complex III peroxide leak rate increases, activating the TNF-α converting enzyme (TACE or Adam17), a metalloproteinase that cleaves TNF-R1, allowing the receptor to be shed and curtailing the TNF-α mediated inflammatory response [87].
The emphasis on protecting mitochondria against damage by immune mediators has not made a clear distinction between myeloid and somatic/structural cells, but the principles of protection are probably generalizable. For instance, the macrophage, a central immune cell, relies on respiration and tends to be resistant to apoptosis [88]. Macrophage apoptosis can be induced by certain microbes, especially bacteria, a disadvantage to the host by letting the organism survive. In other cases, macrophage apoptosis may amplify the immune response to infection [89]. Lymphocyte function, for example, the cytotoxicity of NK cells, also depends on respiration [90]. T cell function, including chemotaxis and cytotoxic T cell targeting [91, 92], is supported by respiration [93–95], and evidence indicates that mitochondrial dynamics support the formation of immunological synapses (IS) involving antigen-presenting cells and T helper cells [96, 97]. Mitochondrial re-localization to the IS maintains Ca2+ flux across the plasma membrane involved in T helper cell activation. Lymphocyte apoptosis is rapid and frequent in severe sepsis in lymphoid tissues and intestinal mucosa [98], as well as in lung endothelial and renal tubular cells [88]. Both the intrinsic and extrinsic apoptosis pathways contribute to lymphocyte depletion in sepsis, but the importance of mitochondrial quality control in the regulation, survival, and migration of various leukocytes and the clonal expansion of lymphocytes, is only now being explored.
In skeletal muscle, exaggerated local inflammatory reactions, higher circulating levels of TNF-α and IL-6, fiber damage, and muscle wasting occur during exercise in muscle-specific PGC-1α knockout mice [99–102]. In contrast, muscle-specific PGC-1α transgenic mice show preservation of mitochondrial function and muscle integrity with aging [103]. Increased skeletal muscle PGC-1α levels prevent muscle wasting by reducing apoptosis, autophagy, and proteasome activity. This preservation of muscle structure and function also slows the decline in bone density and reduces the chronic inflammation of aging. Some evidence implicates PGC-1α as a regulator of ROS generation, and Ppargc1a gene over-expression can suppress intracellular and mitochondrial ROS production induced by NF-kB activity and TNF-α in smooth muscle and endothelial cells [75].
In the liver, a role for TLR signaling in Ppargc1a and Ppargc1b gene expression in early Staphylococcal aureus peritonitis has been shown by the rapid up-regulation of hepatocyte Ppargc1a and Ppargc1b in wild type (WT) mice, whereas both genes are concordantly deregulated in TLR2−/− mice (not increased) and in TLR4−/− mice (highly increased) while PRC is up-regulated in all three strains [104]. Ppargc1a and Ppargc1b share microRNA binding sites for mmu-mir-202-3p, and mir-202-3p-mediated mRNA degradation has been implicated in Ppargc1a and Ppargc1b co-regulation during inflammation.
In non-sterile inflammation in animals, especially lethal poly-microbial infections (e.g. cecal ligation/puncture), long periods of intense inflammation lead to overlapping cycles of tissue damage and repair, obscuring the timing and consistency of metabolic gene expression. In models where sub-lethal LPS or heat-inactivated E. coli is administered, the innate immune response is accompanied by increased gene expression for mitochondrial biogenesis followed by a wave of mitochondrial proliferation throughout the tissue, e.g. in the liver and the heart [12, 105]. In both surrogate and authentic infections, observations have provided clear morphological evidence of accelerated mitochondrial turnover involving autophagy and mitochondrial biogenesis, which within days restores mitochondrial mass in the survivors while mitigating cell death [12, 106–108].
In mice, sub-lethal, but damaging LPS exposure transiently depletes hepatic mtDNA content and impairs mitochondrial transcription [12]. In response, redox-responsive mechanisms such as NRF- 1 phosphorylation and nuclear translocation directed by Akt/PKB increase the gene expression for Tfam and other proteins of the mitochondrial transcriptome. After translation and mitochondrial importation of these proteins, mtDNA copy number is restored [109]. The initial loss of mtDNA content depends on TLR4 and NF-kB activation and TNF-α and NO production leading to mtDNA depletion. Genetic ablation of TLR4 reduces, but does not eliminate this effect, and the recovery of mtDNA copy number is delayed in TLR4 null mice [110]. The cell responds to mtDNA depletion with the induction of the base excision repair glycosylase OGG1 by NRF-1 and NRF-2 binding to OGG1 promoter elements and mitochondrial importation of active enzyme [111]. Moreover, the loss of mtDNA copy number can be abrogated by increasing SOD2 in mitochondria, by inhibiting NOS activity, or by scavenging ONOO- [112].
Innate immune system activation by TLR2 and TLR4 ligands is involved in the early up-regulation of mitochondrial biogenesis [104, 110] through several transcription factors including NF-kB [113, 114], CREB [114], nuclear factor erythroid 2-related factor 2 (Nrf2) [115], and interferon response factors (IRF-3,IRF-7) [116]. TLR4-dependent activation of NF-kB and CREB co-regulate the NRF1 promoter with NF-kB intronic enhancement leading to NRF-1 synthesis and nuclear translocation, followed by target gene expression [114]. This also requires mitochondrial H2O2 production and identifies NRF-1 as an early-phase component of the host defense regulated by TLR signaling and by redox state.
Another key regulator of energy metabolism, the serine/threonine kinase AMP-activated protein kinase (AMPK) [117–123], is activated by ATP depletion and stimulates glucose and lipid catabolism and blocks energy utilizing pathways such as protein and fatty acid biosynthesis [119]. AMPK promotes mitochondrial biogenesis [117, 118, 121], NO production [124, 125], regulates autophagy [126] and opposes inflammation by interfering with NF-κB–dependent cytokine expression [127–134]. Conversely, a decrease in AMPK activity is associated with increased inflammation. The mechanism of NF-kB inhibition is not clear, but this may occur indirectly through PGC-1α, Forkhead box O (FoxO) transcription factors, and sirtuin-1 (SIRT1) [23, 122, 135, 136].
SIRT1 is an NAD-dependent deacetylase that regulates energy homeostasis in response to changing nutrient availability [122, 137–140]. As nutrients are depleted, NAD+ increases and enhances SIRT1 activity; this deacetylates PGC-1α and increases transcription of its target genes [141]. Hepatic SIRT1 regulates lipid homeostasis by positively regulating nuclear PPARα, which cooperates in PGC-1α activation and mediates adaptation to fasting and starvation [138, 142]. Hepatocyte-specific SIRT1 deletion blocks PPARα and decreases β-oxidation, whereas SIRT1 over-expression induces PPARα target genes. Liver-specific SIRT1−/− mice develop steatosis, ER stress, and hepatic inflammation. SIRT1 thus not only regulates lipid homeostasis, but is anti-inflammatory in liver and fat cells, as well as in cells of the immune system [143]. Gene expression studies in peritoneal macrophages indicate that SIRT1 silencing increases pro-inflammatory genes while SIRT1 activation inhibits LPS-stimulated inflammation, including TNF-α production [144].
The transcriptional activity of nuclear hormone receptors, particularly estrogen-related receptor ERRα, is repressed by the nuclear receptor interacting protein-1 (RIP140) [145], which also suppresses metabolic gene expression and mitochondrial biogenesis [146]. In macrophages, RIP140 co-activates certain cytokine genes, and RIP140 deficiency leads to the inhibition of the inflammatory response [147]. RIP140 interacts with the RelA subunit of NF-kB and the histone acetylase CBP (CREB-binding protein) and cooperates with CBP co-activator complex on RelA-regulated promoters. RIP140 modulation of inflammatory gene expression is thus a nice example of the cell-specific integration of control pathways for metabolism and inflammation [147].
Mitochondria and the Inflammasome
Many host danger-signals, infection and tissue damage included, lead to the assembly of inflammasomes needed for fully-developed tissue inflammation. The NLRP3 inflammasome and caspase-1 (and caspase-11 in mice) are required to generate IL-1β by the cleavage of its pro-form, and assembly of the NLRP3 inflammasome also influences metabolic pathways such as glycolysis and lipogenesis. Mitochondrial ROS production can activate the NLRP3 inflammasome, whereas removal of damaged or dysfunctional mitochondria by autophagy negatively regulates the NLRP3 inflammasome. Two lines of evidence suggest mitochondria as the main source of ROS for NLRP3 inflammasome activation and a signal-integrating organelle for inflammasome activation [148–150]. In macrophages, inflammasome activation is impaired by inactivation of the outer membrane voltage dependent anion channel, VDAC, or by depletion of mitochondrial DNA [150]. Complex I inhibitor, rotenone, and Complex III inhibitor, antimycin A, also activate the NLRP3 inflammasome [149].
To avoid destructive inflammation, the cell removes ROS-generating mitochondria by a specialized form of autophagy called mitophagy [151]. Inhibition of mitophagy leads to the retention of ROS-generating mitochondria and activation of the NLRP3 inflammasome [149, 150]. ROS-producing mitochondria removed by mitophagy must be replaced through mitochondrial biogenesis in order to avoid persistent inflammasome activation, chronic inflammation, and energy failure. In chronic conditions that display mitochondrial dysfunction, including certain neurodegenerative diseases, cancers, and cardiovascular diseases, understanding the links between inflammation, mitochondrial biogenesis, and metabolism, is important because malfunctions of any of the regulatory branches may be a root cause of disease.
Mitochondrial Biogenesis, Anti-oxidation, and Counter-inflammation
In addition to roles for innate immune activation and subsequent redox stress in up-regulating mitochondrial biogenesis, the transcriptional network coordinately up-regulates mitochondrial anti-oxidant and anti-apoptotic genes along with specific counter-inflammatory genes. Eukaryote cells balance ROS production with anti-oxidant enzyme activity to maintain redox homeostasis. Many anti-oxidant enzyme genes are regulated by the Nrf2 transcription factor through Keap1, a redox-sensitive cytosolic adaptor protein for a cullin-based ubiquitin ligase that targets Nrf2 for proteasomal degradation and prevention of Nrf2-dependent gene expression [152]. A mitochondrial Keap1-binding protein, PGAM5, a phosphoglycerate mutase (PGM) family member has been identified that is targeted to the outer mitochondrial membrane by an N-terminal localization sequence. PGAM5 has no PGM activity, but forms a ternary complex with Keap1 and Nrf2 in which a Keap1 dimer simultaneously binds PGAM5 and Nrf2 through conserved E (S/T) GE motifs [153]. Knockdown of either Keap1 or PGAM5 activates Nrf2-dependent gene expression.
Nrf2 binds to antioxidant response elements (ARE) in the promoter regions of xenobiotic- and oxidative stress-responsive genes, including heme oxygenase-1 (HO-1; Hmox1) [154]. HO-1 scavenges toxic heme, generates physiological carbon monoxide (CO), and increases the cellular iron-handling defenses [155]. HO-1 induction mainly through Nrf2 protects against inflammation, oxidative stress, heavy metals, and many other stresses [152, 156]. Most electrophiles and oxidants, including the pro-oxidant effects of CO, activate Nrf2, leading to the amplification of ARE-containing genes, including Nrf2 itself [157]. In the heart, HO-1 stimulates mitochondrial biogenesis and protects against the progressive toxic cardiomyopathy of the anthracycline chemotherapeutic drug, doxorubicin [158]. This agent disrupts mitochondrial biogenesis and causes intrinsic apoptosis, necrosis, inflammation, and myocardial fibrosis that can be prevented by low level CO administration or by over-expression of active HO-1 [158].
The role of HO-1 in the induction of mitochondrial biogenesis, e.g. in cardiac myocytes, requires CO production [159]. HO-1 activity increases endogenous CO levels and stimulates SOD2 up-regulation and mitochondrial H2O2 production, which activates Akt/PKB and deactivates glycogen synthase kinase-3β, permitting Nrf2 nuclear translocation and occupancy of AREs in the NRF-1 promoter [160]. This is the first clear transcriptional mechanism for the expansion of cardiac mitochondrial mass linked to inducible xenobiotic and antioxidant defenses.
HO-1 induction by inflammation, for instance by LPS, is associated not only with the induction of mitochondrial biogenesis, but with subsequent counter-inflammatory and anti-apoptotic responses mediated in part by anti-inflammatory cytokines such as IL-10 [161], and anti-apoptotic proteins such as BclXL [162]. HO-1 promotes the expression of IL-1 receptor antagonist (IL-1Ra), and suppressor of cytokine synthesis-3 (SOCS3) [115]. Mechanistically, HO-1 acting through CO enables the Nrf2, Gabpa, and MEF2 transcription factors to bind to the IL10 promoter and NRF-1 and MEF2 to bind to the IL1Ra promoter. In liver cells and in macrophages, RNA silencing of either Nrf2 or Hmox1 blocks IL-10 and IL-1Ra up-regulation, and post-inflammatory hepatic HO-1 induction fails in Nrf2−/− mice.
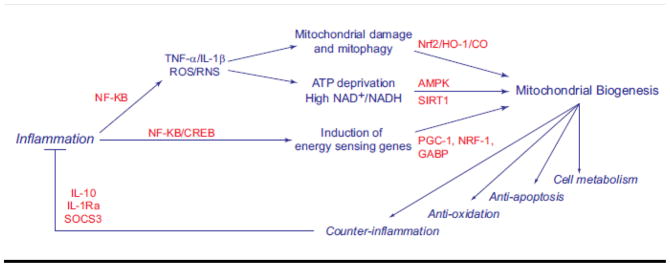
In short, mitochondrial ROS generate signals that lead to Nrf2 activation and HO-1 induction and result in co-induction of genes for mitochondrial biogenesis and counter-inflammation. This information together with the work above showing that PPARs, PCG-1 proteins, AMPK, and SIRT1 have anti-inflammatory effects implicates mitochondrial biogenesis in connecting the cell’s anti-oxidant, anti-apoptotic, and counter-inflammatory defenses to cell metabolism into an integrated regulatory transcriptional network (Figure 3).
Figure 3.
Interrelationships among the innate immune response, oxidative mitochondrial damage and the induction of mitochondrial biogenesis, counter-inflammation, and the anti-oxidant defenses.
Mitochondrial Biogenesis and Cell Survival and Repair during Inflammation
Cell substrate metabolism is implicitly linked to survival under both homeostatic and conditions involving high energy demands, implying the existence of common regulatory processes. Nutrient and oxygen availability also determine the competency of the cell to proliferate and differentiate as well as to avoid programmed cell death in the face of imminent cytotoxic threats. The cell normally maintains a high ratio of ATP/ADP/AMP, and a rising AMP/ATP is a sensitive indicator of impending energy limitation that triggers AMPK activation, which blocks ATP-consuming pathways and switches on ATP-generating pathways [119].
AMPK blocks the mTOR pathway [163], a central growth regulator responsive to mitogens and carbon substrate [164] by phosphorylating the TSC2 (hamartin) tumor suppressor, which co-operates with TSC1 (tuberin) [163]. Signaling by mTOR is activated by the Akt/PKB survival kinase, which phosphorylates and inactivates TSC2 [165]. Akt/PKB promotes mitochondrial biogenesis through NRF-1 phosphorylation and nuclear translocation [79] and supports cell survival in part by increasing mitochondrial hexokinase (HK) activity, committing glucose to glucose-6-phosphate using ATP generated by mitochondria and thereby coupling glycolysis to oxidative phosphorylation [166]. This may be the tip of an iceberg for convergence of nutrient sensing with survival signaling.
These survival pathways also impinge on proteins involved in programmed cell death. For instance, the pro-apoptotic function of BAD, a BH3-only domain pro-apoptotic member of the Bcl-2 family is normally inactivated by phosphorylation by survival kinases resulting in cytoplasmic localization [167]. At the mitochondrion, BAD is involved in the assembly of a large protein complex that also catalyzes the first step of glycolysis through HK-4 activity (glucokinase). The absence of BAD restricts respiration in response to glucose, while glucose deprivation leads to BAD dephosphorylation and apoptosis [168]. These findings too highlight the interplay between energy metabolism and regulation of apoptosis.
It would seem self-evident that apposite regulation of mitochondrial biogenesis is pro-survival, but pathological acceleration of mitochondrial biogenesis may imply mitochondrial incompetence [169]. Although interference with mitochondrial biogenesis usually exacerbates inflammation and promotes oxidation and apoptosis, the need to support rapid mitochondrial turnover due to continual mitochondrial damage increases the likelihood of immune suppression by the production of counter-inflammatory mediators, as mentioned for IL-10 [170, 171]. Although greater than normal immune tolerance during periods of active mitochondrial biogenesis should improve the fidelity of the bi-genomic program, actual immune suppression is often encountered in patients recovering from severe sepsis [172–174]. This so-called immune paralysis puts patients at risk for recurrent and secondary infections after their initial episode of sepsis. Still, there is unexplored therapeutic potential for agents to activate mitochondrial biogenesis and allied metabolic pathways in the resolution of dangerous immunological reactions to serious infections [175].
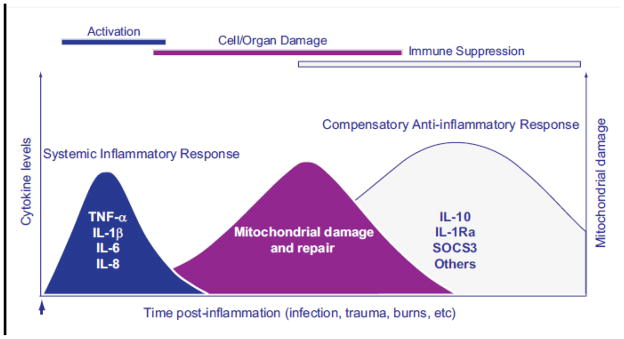
The key events are diagrammed in Figure 4, where a scale-free time profile of mitochondrial damage and recovery is interposed between an acute inflammatory response and a long period of immune suppression. The figure sets out the usual nomenclature for the initial state of innate immune hyper-activation and the period of immune suppression following serious infections, but in practice, the situation is rarely so clear cut.
Figure 4.
A mitochondrial damage and recovery profile interposed on a hypothetical timeline between an acute inflammatory insult and a subsequent period of immune suppression. The diagram indicates the nomenclature for the systemic inflammatory response and the compensatory anti-inflammatory response used most conspicuously in bacterial sepsis to describe hyper-activation of innate immunity followed by an extended period of immune suppression. The risk of secondary or recurrent infection is increased during the period of immune suppression.
Summary and Conclusions
Activation of the innate immune response leads to rapid synthesis and release of soluble mediators that establish competency of the innate and adaptive immune systems, but a hyperactive response compromises mitochondrial function and cell metabolism both in long-lived leukocytes and in somatic cells. The over-production of effectors like TNF-α and IL-1β promotes oxidative stress, NO over-production, and calcium deregulation that injure mitochondria and initiate cycles of mitochondrial damage, cell death, and deregulation of inflammation. In response, the recovering cell induces mitochondrial biogenesis, removes damaged mitochondria by autophagy, and up-regulates anti-oxidant and counter-inflammatory defense genes. This multifaceted protective system involves a highly-integrated nuclear transcriptional network controlled by energy-sensing and inflammation- and redox-sensitive transcription factors and co-activators involved in mitochondrial biogenesis, anti-oxidant enzyme induction, cell survival, and immune tolerance as illustrated in the graphical abstract for this paper. These mechanisms support mitochondrial function and minimize cell death and organ dysfunction until the agent of inflammation can be brought under control.
Graphical abstract.
Highlights.
The innate immune response triggers a cycle of oxidative mitochondrial damage
The host cell activates the transcriptional program for mitochondrial biogenesis
The program concurrently activates anti-oxidant and anti-inflammatory defense genes
This network limits metabolic failure and apoptosis and promotes immune tolerance
Acknowledgments
NIH HL090679 and GM084116
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Singer M. Mitochondrial function in sepsis: acute phase versus multiple organ failure. Crit Care Med. 2007;35:S441–448. doi: 10.1097/01.CCM.0000278049.48333.78. [DOI] [PubMed] [Google Scholar]
- 2.Okamoto K, Kondo-Okamoto N. Mitochondria and autophagy: Critical interplay between the two homeostats. Biochim Biophys Acta. 2011 doi: 10.1016/j.bbagen.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 4.Scarpulla RC. Transcriptional paradigms in Mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–638. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- 5.Kowald A, Kirkwood TB. Evolution of the mitochondrial fusion-fission cycle and its role in aging. Proc Natl Acad Sci U S A. 2011;108:10237–10242. doi: 10.1073/pnas.1101604108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palmer CS, Osellame LD, Stojanovski D, Ryan MT. The regulation of mitochondrial morphology: Intricate mechanisms and dynamic machinery. Cell Signal. 2011;23:1534–1545. doi: 10.1016/j.cellsig.2011.05.021. [DOI] [PubMed] [Google Scholar]
- 7.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 8.Xia Y, Buja LM, Scarpulla RC, McMillin JB. Electrical stimulation of neonatal cardiomyocytes results in the sequential activation of nuclear genes governing mitochondrial proliferation and differentiation. Proc Natl Acad Sci U S A. 1997;94:11399–11404. doi: 10.1073/pnas.94.21.11399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 10.Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;286:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 11.Guidot DM. Endotoxin pretreatment in vivo increases the mitochondrial respiratory capacity in rat hepatocytes. Arch Biochem Biophys. 1998;354:9–17. doi: 10.1006/abbi.1998.0699. [DOI] [PubMed] [Google Scholar]
- 12.Suliman HB, Carraway MS, Piantadosi CA. Postlipopolysaccharide oxidative damage of mitochondrial DNA. Am J Respir Crit Care Med. 2003;167:570–579. doi: 10.1164/rccm.200206-518OC. [DOI] [PubMed] [Google Scholar]
- 13.Carraway MS, Suliman HB, Jones WS, Chen CW, Babiker A, Piantadosi CA. Erythropoietin activates mitochondrial biogenesis and couples red cell mass to mitochondrial mass in the heart. Circ Res. 2010;106:1722–1730. doi: 10.1161/CIRCRESAHA.109.214353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Himms-Hagen J. Cellular thermogenesis. Annu Rev Physiol. 1976;38:315–351. doi: 10.1146/annurev.ph.38.030176.001531. [DOI] [PubMed] [Google Scholar]
- 15.Mutvei A, Husman B, Andersson G, Nelson BD. Thyroid hormone and not growth hormone is the principle regulator of mammalian mitochondrial biogenesis. Acta Endocrinol (Copenh) 1989;121:223–228. doi: 10.1530/acta.0.1210223. [DOI] [PubMed] [Google Scholar]
- 16.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial biogenesis in mammals: the role of endogenous nitric oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 19.Lee HC, Yin PH, Chi CW, Wei YH. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci. 2002;9:517–526. doi: 10.1007/BF02254978. [DOI] [PubMed] [Google Scholar]
- 20.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 21.Vercauteren K, Pasko RA, Gleyzer N, Marino VM, Scarpulla RC. PGC-1-related coactivator: immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu Z, Huang X, Feng Y, Handschin C, Feng Y, Gullicksen PS, Bare O, Labow M, Spiegelman B, Stevenson SC. Transducer of regulated CREB-binding proteins (TORCs) induce PGC-1alpha transcription and mitochondrial biogenesis in muscle cells. Proc Natl Acad Sci U S A. 2006;103:14379–14384. doi: 10.1073/pnas.0606714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scarpulla RC. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim Biophys Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 25.Schreiber SN, Emter R, Hock MB, Knutti D, Cardenas J, Podvinec M, Oakeley EJ, Kralli A. The estrogen-related receptor alpha (ERRalpha) functions in PPARgamma coactivator 1alpha (PGC-1alpha)-induced mitochondrial biogenesis. Proc Natl Acad Sci U S A. 2004;101:6472–6477. doi: 10.1073/pnas.0308686101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spiegelman BM, Puigserver P, Wu Z. Regulation of adipogenesis and energy balance by PPARgamma and PGC-1. Int J Obes Relat Metab Disord. 2000;24(Suppl 4):S8–10. doi: 10.1038/sj.ijo.0801492. [DOI] [PubMed] [Google Scholar]
- 27.Wang YX, Zhang CL, Yu RT, Cho HK, Nelson MC, Bayuga-Ocampo CR, Ham J, Kang H, Evans RM. Regulation of muscle fiber type and running endurance by PPARdelta. PLoS Biol. 2004;2:e294. doi: 10.1371/journal.pbio.0020294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baar K. Involvement of PPAR gamma co-activator-1, nuclear respiratory factors 1 and 2, and PPAR alpha in the adaptive response to endurance exercise. Proc Nutr Soc. 2004;63:269–273. doi: 10.1079/PNS2004334. [DOI] [PubMed] [Google Scholar]
- 29.Andersen G, Wegner L, Yanagisawa K, Rose CS, Lin J, Glumer C, Drivsholm T, Borch-Johnsen K, Jorgensen T, Hansen T, Spiegelman BM, Pedersen O. Evidence of an association between genetic variation of the coactivator PGC-1beta and obesity. J Med Genet. 2005;42:402–407. doi: 10.1136/jmg.2004.026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barish GD, Narkar VA, Evans RM. PPAR delta: a dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madrazo JA, Kelly DP. The PPAR trio: regulators of myocardial energy metabolism in health and disease. J Mol Cell Cardiol. 2008;44:968–975. doi: 10.1016/j.yjmcc.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 32.Morrish F, Giedt C, Hockenbery D. c-MYC apoptotic function is mediated by NRF-1 target genes. Genes Dev. 2003;17:240–255. doi: 10.1101/gad.1032503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dang CV, Li F, Lee LA. Could MYC induction of mitochondrial biogenesis be linked to ROS production and genomic instability? Cell Cycle. 2005;4:1465–1466. doi: 10.4161/cc.4.11.2121. [DOI] [PubMed] [Google Scholar]
- 34.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 35.Czubryt MP, McAnally J, Fishman GI, Olson EN. Regulation of peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1 alpha ) and mitochondrial function by MEF2 and HDAC5. Proc Natl Acad Sci U S A. 2003;100:1711–1716. doi: 10.1073/pnas.0337639100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramachandran B, Yu G, Gulick T. Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J Biol Chem. 2008;283:11935–11946. doi: 10.1074/jbc.M707389200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freyssenet D, Di Carlo M, Hood DA. Calcium-dependent regulation of cytochrome c gene expression in skeletal muscle cells. Identification of a protein kinase c-dependent pathway. J Biol Chem. 1999;274:9305–9311. doi: 10.1074/jbc.274.14.9305. [DOI] [PubMed] [Google Scholar]
- 38.Freyssenet D, Irrcher I, Connor MK, Di Carlo M, Hood DA. Calcium-regulated changes in mitochondrial phenotype in skeletal muscle cells. Am J Physiol Cell Physiol. 2004;286:C1053–1061. doi: 10.1152/ajpcell.00418.2003. [DOI] [PubMed] [Google Scholar]
- 39.Guerfali I, Manissolle C, Durieux AC, Bonnefoy R, Bartegi A, Freyssenet D. Calcineurin A and CaMKIV transactivate PGC-1alpha promoter, but differentially regulate cytochrome c promoter in rat skeletal muscle. Pflugers Arch. 2007;454:297–305. doi: 10.1007/s00424-007-0206-6. [DOI] [PubMed] [Google Scholar]
- 40.Ojuka EO, Jones TE, Han DH, Chen M, Wamhoff BR, Sturek M, Holloszy JO. Intermittent increases in cytosolic Ca2+ stimulate mitochondrial biogenesis in muscle cells. Am J Physiol Endocrinol Metab. 2002;283:E1040–1045. doi: 10.1152/ajpendo.00242.2002. [DOI] [PubMed] [Google Scholar]
- 41.Ojuka EO, Jones TE, Nolte LA, Chen M, Wamhoff BR, Sturek M, Holloszy JO. Regulation of GLUT4 biogenesis in muscle: evidence for involvement of AMPK and Ca(2+) Am J Physiol Endocrinol Metab. 2002;282:E1008–1013. doi: 10.1152/ajpendo.00512.2001. [DOI] [PubMed] [Google Scholar]
- 42.Tavi P, Westerblad H. The role of in vivo Ca(2) signals acting on Ca(2)-calmodulin-dependent proteins for skeletal muscle plasticity. J Physiol. 2011;589:5021–5031. doi: 10.1113/jphysiol.2011.212860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu H, Kanatous SB, Thurmond FA, Gallardo T, Isotani E, Bassel-Duby R, Williams RS. Regulation of mitochondrial biogenesis in skeletal muscle by CaMK. Science. 2002;296:349–352. doi: 10.1126/science.1071163. [DOI] [PubMed] [Google Scholar]
- 44.Rasbach KA, Schnellmann RG. Signaling of mitochondrial biogenesis following oxidant injury. J Biol Chem. 2007;282:2355–2362. doi: 10.1074/jbc.M608009200. [DOI] [PubMed] [Google Scholar]
- 45.Gutsaeva DR, Carraway MS, Suliman HB, Demchenko IT, Shitara H, Yonekawa H, Piantadosi CA. Transient hypoxia stimulates mitochondrial biogenesis in brain subcortex by a neuronal nitric oxide synthase-dependent mechanism. J Neurosci. 2008;28:2015–2024. doi: 10.1523/JNEUROSCI.5654-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds CM, Suliman HB, Hollingsworth JW, Welty-Wolf KE, Carraway MS, Piantadosi CA. Nitric oxide synthase-2 induction optimizes cardiac mitochondrial biogenesis after endotoxemia. Free Radic Biol Med. 2009;46:564–572. doi: 10.1016/j.freeradbiomed.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chin ER. The role of calcium and calcium/calmodulin-dependent kinases in skeletal muscle plasticity and mitochondrial biogenesis. Proc Nutr Soc. 2004;63:279–286. doi: 10.1079/PNS2004335. [DOI] [PubMed] [Google Scholar]
- 48.Chin ER. Role of Ca2+/calmodulin-dependent kinases in skeletal muscle plasticity. J Appl Physiol. 2005;99:414–423. doi: 10.1152/japplphysiol.00015.2005. [DOI] [PubMed] [Google Scholar]
- 49.Akimoto T, Pohnert SC, Li P, Zhang M, Gumbs C, Rosenberg PB, Williams RS, Yan Z. Exercise stimulates Pgc-1alpha transcription in skeletal muscle through activation of the p38 MAPK pathway. J Biol Chem. 2005;280:19587–19593. doi: 10.1074/jbc.M408862200. [DOI] [PubMed] [Google Scholar]
- 50.Lira VA, Benton CR, Yan Z, Bonen A. PGC-1alpha regulation by exercise training and its influences on muscle function and insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;299:E145–161. doi: 10.1152/ajpendo.00755.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barger PM, Browning AC, Garner AN, Kelly DP. p38 mitogen-activated protein kinase activates peroxisome proliferator-activated receptor alpha: a potential role in the cardiac metabolic stress response. J Biol Chem. 2001;276:44495–44501. doi: 10.1074/jbc.M105945200. [DOI] [PubMed] [Google Scholar]
- 52.Cao W, Daniel KW, Robidoux J, Puigserver P, Medvedev AV, Bai X, Floering LM, Spiegelman BM, Collins S. p38 mitogen-activated protein kinase is the central regulator of cyclic AMP-dependent transcription of the brown fat uncoupling protein 1 gene. Mol Cell Biol. 2004;24:3057–3067. doi: 10.1128/MCB.24.7.3057-3067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oka S, Ago T, Kitazono T, Zablocki D, Sadoshima J. The role of redox modulation of class II histone deacetylases in mediating pathological cardiac hypertrophy. J Mol Med. 2009;87:785–791. doi: 10.1007/s00109-009-0471-2. [DOI] [PubMed] [Google Scholar]
- 54.Chen SL, Wang SC, Hosking B, Muscat GE. Subcellular localization of the steroid receptor coactivators (SRCs) and MEF2 in muscle and rhabdomyosarcoma cells. Mol Endocrinol. 2001;15:783–796. doi: 10.1210/mend.15.5.0637. [DOI] [PubMed] [Google Scholar]
- 55.Liu Y, Randall WR, Schneider MF. Activity-dependent and -independent nuclear fluxes of HDAC4 mediated by different kinases in adult skeletal muscle. J Cell Biol. 2005;168:887–897. doi: 10.1083/jcb.200408128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen T, Liu Y, Randall WR, Schneider MF. Parallel mechanisms for resting nucleo-cytoplasmic shuttling and activity dependent translocation provide dual control of transcriptional regulators HDAC and NFAT in skeletal muscle fiber type plasticity. J Muscle Res Cell Motil. 2006;27:405–411. doi: 10.1007/s10974-006-9080-7. [DOI] [PubMed] [Google Scholar]
- 57.Mela L, Bacalzo LV, Jr, Miller LD. Defective oxidative metabolism of rat liver mitochondria in hemorrhagic and endotoxin shock. Am J Physiol. 1971;220:571–577. doi: 10.1152/ajplegacy.1971.220.2.571. [DOI] [PubMed] [Google Scholar]
- 58.Mela L. Direct and indirect effects of endotoxin on mitochondrial function. Prog Clin Biol Res. 1981;62:15–21. [PubMed] [Google Scholar]
- 59.Mela L, Miller LD. Efficacy of glucocorticoids in preventing mitochondrial metabolic failure in endotoxemia. Circ Shock. 1983;10:371–381. [PubMed] [Google Scholar]
- 60.Tavakoli H, Mela L. Alterations of mitochondrial metabolism and protein concentrations in subacute septicemia. Infect Immun. 1982;38:536–541. doi: 10.1128/iai.38.2.536-541.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schulze-Osthoff K, Bakker AC, Vanhaesebroeck B, Beyaert R, Jacob WA, Fiers W. Cytotoxic activity of tumor necrosis factor is mediated by early damage of mitochondrial functions. Evidence for the involvement of mitochondrial radical generation. J Biol Chem. 1992;267:5317–5323. [PubMed] [Google Scholar]
- 62.Llesuy S, Evelson P, Gonzalez-Flecha B, Peralta J, Carreras MC, Poderoso JJ, Boveris A. Oxidative stress in muscle and liver of rats with septic syndrome. Free Radic Biol & Med. 1994;16:445–451. doi: 10.1016/0891-5849(94)90121-x. [DOI] [PubMed] [Google Scholar]
- 63.Taylor DE, Ghio AJ, Piantadosi CA. Reactive oxygen species produced by liver mitochondria of rats in sepsis. Arch Biochem Biophys. 1995;316:70–76. doi: 10.1006/abbi.1995.1011. [DOI] [PubMed] [Google Scholar]
- 64.Kantrow SP, Taylor DE, Carraway MS, Piantadosi CA. Oxidative metabolism in rat hepatocytes and mitochondria during sepsis. Arch Biochem Biophys. 1997;345:278–288. doi: 10.1006/abbi.1997.0264. [DOI] [PubMed] [Google Scholar]
- 65.Kurose I, Miura S, Fukumura D, Yonei Y, Saito H, Tada S, Suematsu M, Tsuchiya M. Nitric oxide mediates Kupffer cell-induced reduction of mitochondrial energization in hepatoma cells: a comparison with oxidative burst. Cancer Res. 1993;53:2676–2682. [PubMed] [Google Scholar]
- 66.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 67.Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- 68.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 69.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 70.Chtarbanova S, Imler JL. Microbial sensing by toll receptors: a historical perspective. Arterioscler Thromb Vasc Biol. 2011;31:1734–1738. doi: 10.1161/ATVBAHA.108.179523. [DOI] [PubMed] [Google Scholar]
- 71.Kuznetsov AV, Janakiraman M, Margreiter R, Troppmair J. Regulating cell survival by controlling cellular energy production: novel functions for ancient signaling pathways? FEBS Lett. 2004;577:1–4. doi: 10.1016/j.febslet.2004.10.021. [DOI] [PubMed] [Google Scholar]
- 72.Schreck R, Rieber P, Baeuerle PA. Reactive oxygen intermediates as apparently widely used messengers in the activation of the NF-kappa B transcription factor and HIV-1. Embo J. 1991;10:2247–2258. doi: 10.1002/j.1460-2075.1991.tb07761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koay MA, Christman JW, Segal BH, Venkatakrishnan A, Blackwell TR, Holland SM, Blackwell TS. Impaired pulmonary NF-kappaB activation in response to lipopolysaccharide in NADPH oxidase-deficient mice. Infect Immun. 2001;69:5991–5996. doi: 10.1128/IAI.69.10.5991-5996.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Figari IS, Mori NA, Palladino MA., Jr Regulation of neutrophil migration and superoxide production by recombinant tumor necrosis factors-alpha and -beta: comparison to recombinant interferon-gamma and interleukin-1 alpha. Blood. 1987;70:979–984. [PubMed] [Google Scholar]
- 75.Kim HJ, Park KG, Yoo EK, Kim YH, Kim YN, Kim HS, Kim HT, Park JY, Lee KU, Jang WG, Kim JG, Kim BW, Lee IK. Effects of PGC-1alpha on TNF-alpha-induced MCP-1 and VCAM-1 expression and NF-kappaB activation in human aortic smooth muscle and endothelial cells. Antioxid Redox Signal. 2007;9:301–307. doi: 10.1089/ars.2006.1456. [DOI] [PubMed] [Google Scholar]
- 76.Remels AH, Gosker HR, Schrauwen P, Hommelberg PP, Sliwinski P, Polkey M, Galdiz J, Wouters EF, Langen RC, Schols AM. TNF-alpha impairs regulation of muscle oxidative phenotype: implications for cachexia? Faseb J. 2010;24:5052–5062. doi: 10.1096/fj.09-150714. [DOI] [PubMed] [Google Scholar]
- 77.Alvarez-Guardia D, Palomer X, Coll T, Davidson MM, Chan TO, Feldman AM, Laguna JC, Vazquez-Carrera M. The p65 subunit of NF-kappaB binds to PGC-1alpha, linking inflammation and metabolic disturbances in cardiac cells. Cardiovasc Res. 2010;87:449–458. doi: 10.1093/cvr/cvq080. [DOI] [PubMed] [Google Scholar]
- 78.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 79.Piantadosi CA, Suliman HB. Mitochondrial transcription factor A induction by redox activation of nuclear respiratory factor 1. J Biol Chem. 2006;281:324–333. doi: 10.1074/jbc.M508805200. [DOI] [PubMed] [Google Scholar]
- 80.Lu Z, Xu X, Hu X, Fassett J, Zhu G, Tao Y, Li J, Huang Y, Zhang P, Zhao B, Chen Y. PGC-1 alpha regulates expression of myocardial mitochondrial antioxidants and myocardial oxidative stress after chronic systolic overload. Antioxid Redox Signal. 2010;13:1011–1022. doi: 10.1089/ars.2009.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vats D, Mukundan L, Odegaard JI, Zhang L, Smith KL, Morel CR, Wagner RA, Greaves DR, Murray PJ, Chawla A. Oxidative metabolism and PGC-1beta attenuate macrophage-mediated inflammation. Cell Metab. 2006;4:13–24. doi: 10.1016/j.cmet.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen G, Goeddel DV. TNF-R1 signaling: a beautiful pathway. Science. 2002;296:1634–1635. doi: 10.1126/science.1071924. [DOI] [PubMed] [Google Scholar]
- 83.Kiningham KK, Xu Y, Daosukho C, Popova B, St Clair DK. Nuclear factor kappaB-dependent mechanisms coordinate the synergistic effect of PMA and cytokines on the induction of superoxide dismutase 2. Biochem J. 2001;353:147–156. [PMC free article] [PubMed] [Google Scholar]
- 84.Ogura H, Tsukumo Y, Sugimoto H, Igarashi M, Nagai K, Kataoka T. Ectodomain shedding of TNF receptor 1 induced by protein synthesis inhibitors regulates TNF-alpha-mediated activation of NF-kappaB and caspase-8. Exp Cell Res. 2008;314:1406–1414. doi: 10.1016/j.yexcr.2008.01.019. [DOI] [PubMed] [Google Scholar]
- 85.Kim BC, Kim HT, Mamura M, Ambudkar IS, Choi KS, Kim SJ. Tumor necrosis factor induces apoptosis in hepatoma cells by increasing Ca(2+) release from the endoplasmic reticulum and suppressing Bcl-2 expression. J Biol Chem. 2002;277:31381–31389. doi: 10.1074/jbc.M203465200. [DOI] [PubMed] [Google Scholar]
- 86.Balaban RS. The role of Ca(2+) signaling in the coordination of mitochondrial ATP production with cardiac work. Biochim Biophys Acta. 2009;1787:1334–1341. doi: 10.1016/j.bbabio.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rowlands DJ, Islam MN, Das SR, Huertas A, Quadri SK, Horiuchi K, Inamdar N, Emin MT, Lindert J, Ten VS, Bhattacharya S, Bhattacharya J. Activation of TNFR1 ectodomain shedding by mitochondrial Ca2+ determines the severity of inflammation in mouse lung microvessels. J Clin Invest. 2011;121:1986–1999. doi: 10.1172/JCI43839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Stearns-Kurosawa DJ, Osuchowski MF, Valentine C, Kurosawa S, Remick DG. The pathogenesis of sepsis. Annu Rev Pathol. 2011;6:19–48. doi: 10.1146/annurev-pathol-011110-130327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.DeLeo FR. Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis. 2004;9:399–413. doi: 10.1023/B:APPT.0000031448.64969.fa. [DOI] [PubMed] [Google Scholar]
- 90.Abarca-Rojano E, Muniz-Hernandez S, Moreno-Altamirano MM, Mondragon-Flores R, Enriquez-Rincon F, Sanchez-Garcia FJ. Re-organization of mitochondria at the NK cell immune synapse. Immunol Lett. 2009;122:18–25. doi: 10.1016/j.imlet.2008.10.008. [DOI] [PubMed] [Google Scholar]
- 91.Bielawska-Pohl A, Crola C, Caignard A, Gaudin C, Dus D, Kieda C, Chouaib S. Human NK cells lyse organ-specific endothelial cells: analysis of adhesion and cytotoxic mechanisms. J Immunol. 2005;174:5573–5582. doi: 10.4049/jimmunol.174.9.5573. [DOI] [PubMed] [Google Scholar]
- 92.Kaspar AA, Okada S, Kumar J, Poulain FR, Drouvalakis KA, Kelekar A, Hanson DA, Kluck RM, Hitoshi Y, Johnson DE, Froelich CJ, Thompson CB, Newmeyer DD, Anel A, Clayberger C, Krensky AM. A distinct pathway of cell-mediated apoptosis initiated by granulysin. J Immunol. 2001;167:350–356. doi: 10.4049/jimmunol.167.1.350. [DOI] [PubMed] [Google Scholar]
- 93.Krauss S, Brand MD, Buttgereit F. Signaling takes a breath--new quantitative perspectives on bioenergetics and signal transduction. Immunity. 2001;15:497–502. doi: 10.1016/s1074-7613(01)00205-9. [DOI] [PubMed] [Google Scholar]
- 94.Beltran B, Mathur A, Duchen MR, Erusalimsky JD, Moncada S. The effect of nitric oxide on cell respiration: A key to understanding its role in cell survival or death. Proc Natl Acad Sci U S A. 2000;97:14602–14607. doi: 10.1073/pnas.97.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rathmell JC, Fox CJ, Plas DR, Hammerman PS, Cinalli RM, Thompson CB. Akt-directed glucose metabolism can prevent Bax conformation change and promote growth factor-independent survival. Mol Cell Biol. 2003;23:7315–7328. doi: 10.1128/MCB.23.20.7315-7328.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campello S, Lacalle RA, Bettella M, Manes S, Scorrano L, Viola A. Orchestration of lymphocyte chemotaxis by mitochondrial dynamics. J Exp Med. 2006;203:2879–2886. doi: 10.1084/jem.20061877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Quintana A, Schwindling C, Wenning AS, Becherer U, Rettig J, Schwarz EC, Hoth M. T cell activation requires mitochondrial translocation to the immunological synapse. Proc Natl Acad Sci U S A. 2007;104:14418–14423. doi: 10.1073/pnas.0703126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hotchkiss RS, Osmon SB, Chang KC, Wagner TH, Coopersmith CM, Karl IE. Accelerated lymphocyte death in sepsis occurs by both the death receptor and mitochondrial pathways. J Immunol. 2005;174:5110–5118. doi: 10.4049/jimmunol.174.8.5110. [DOI] [PubMed] [Google Scholar]
- 99.Handschin C, Chin S, Li P, Liu F, Maratos-Flier E, Lebrasseur NK, Yan Z, Spiegelman BM. Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals. J Biol Chem. 2007;282:30014–30021. doi: 10.1074/jbc.M704817200. [DOI] [PubMed] [Google Scholar]
- 100.Handschin C, Choi CS, Chin S, Kim S, Kawamori D, Kurpad AJ, Neubauer N, Hu J, Mootha VK, Kim YB, Kulkarni RN, Shulman GI, Spiegelman BM. Abnormal glucose homeostasis in skeletal muscle-specific PGC-1alpha knockout mice reveals skeletal muscle-pancreatic beta cell crosstalk. J Clin Invest. 2007;117:3463–3474. doi: 10.1172/JCI31785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Handschin C, Spiegelman BM. The role of exercise and PGC1alpha in inflammation and chronic disease. Nature. 2008;454:463–469. doi: 10.1038/nature07206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Handschin C. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha in muscle links metabolism to inflammation. Clin Exp Pharmacol Physiol. 2009;36:1139–1143. doi: 10.1111/j.1440-1681.2009.05275.x. [DOI] [PubMed] [Google Scholar]
- 103.Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106:20405–20410. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 104.Sweeney TE, Suliman HB, Hollingsworth JW, Piantadosi CA. Differential regulation of the PGC family of genes in a mouse model of Staphylococcus aureus sepsis. PLoS ONE. 2010;5:e11606. doi: 10.1371/journal.pone.0011606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Suliman HB, Welty-Wolf KE, Carraway M, Tatro L, Piantadosi CA. Lipopolysaccharide induces oxidative cardiac mitochondrial damage and biogenesis. Cardiovasc Res. 2004;64:279–288. doi: 10.1016/j.cardiores.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 106.Crouser ED, Julian MW, Huff JE, Struck J, Cook CH. Carbamoyl phosphate synthase-1: a marker of mitochondrial damage and depletion in the liver during sepsis. Crit Care Med. 2006;34:2439–2446. doi: 10.1097/01.CCM.0000230240.02216.21. [DOI] [PubMed] [Google Scholar]
- 107.Watanabe E, Muenzer JT, Hawkins WG, Davis CG, Dixon DJ, McDunn JE, Brackett DJ, Lerner MR, Swanson PE, Hotchkiss RS. Sepsis induces extensive autophagic vacuolization in hepatocytes: a clinical and laboratory-based study. Lab Invest. 2009;89:549–561. doi: 10.1038/labinvest.2009.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Haden DW, Suliman HB, Carraway MS, Welty-Wolf KE, Ali AS, Shitara H, Yonekawa H, Piantadosi CA. Mitochondrial biogenesis restores oxidative metabolism during Staphylococcus aureus sepsis. Am J Respir Crit Care Med. 2007;176:768–777. doi: 10.1164/rccm.200701-161OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Suliman HB, Carraway MS, Welty-Wolf KE, Whorton AR, Piantadosi CA. Lipopolysaccharide stimulates mitochondrial biogenesis via activation of nuclear respiratory factor-1. J Biol Chem. 2003;278:41510–41518. doi: 10.1074/jbc.M304719200. [DOI] [PubMed] [Google Scholar]
- 110.Suliman HB, Welty-Wolf KE, Carraway MS, Schwartz DA, Hollingsworth JW, Piantadosi CA. Toll-like receptor 4 mediates mitochondrial DNA damage and biogenic responses after heat-inactivated E. coli. Faseb J. 2005;19:1531–1533. doi: 10.1096/fj.04-3500fje. [DOI] [PubMed] [Google Scholar]
- 111.Bartz RR, Suliman HB, Fu P, Welty-Wolf K, Carraway MS, Macgarvey NC, Withers CM, Sweeney TE, Piantadosi CA. Staphylococcus Aureus Sepsis and Mitochondrial Accrual of OGG1 DNA Repair Enzyme in Mice. Am J Respir Crit Care Med. 2010 doi: 10.1164/rccm.200911-1709OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Choumar A, Tarhuni A, Letteron P, Reyl-Desmars F, Dauhoo N, Damasse J, Vadrot N, Nahon P, Moreau R, Pessayre D, Mansouri A. Lipopolysaccharide-Induced Mitochondrial DNA Depletion. Antioxid Redox Signal. 2011 doi: 10.1089/ars.2010.3713. [DOI] [PubMed] [Google Scholar]
- 113.Bakkar N, Wang J, Ladner KJ, Wang H, Dahlman JM, Carathers M, Acharyya S, Rudnicki MA, Hollenbach AD, Guttridge DC. IKK/NF-kappaB regulates skeletal myogenesis via a signaling switch to inhibit differentiation and promote mitochondrial biogenesis. J Cell Biol. 2008;180:787–802. doi: 10.1083/jcb.200707179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Suliman HB, Sweeney TE, Withers CM, Piantadosi CA. Co-regulation of nuclear respiratory factor-1 by NF{kappa}B and CREB links LPS-induced inflammation to mitochondrial biogenesis. J Cell Sci. 2010;123:2565–2575. doi: 10.1242/jcs.064089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Piantadosi CA, Withers CM, Bartz RR, Macgarvey NC, Fu P, Sweeney TE, Welty-Wolf KE, Suliman HB. Heme oxygenase-1 couples activation of mitochondrial biogenesis to anti-inflammatory cytokine expression. J Biol Chem. 2011 doi: 10.1074/jbc.M110.207738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sweeney TE, Suliman HB, Hollingsworth JW, Welty-Wolf KE, Piantadosi CA. A toll-like receptor 2 pathway regulates the Ppargc1a/b metabolic co-activators in mice with Staphylococcal aureus sepsis. PLoS ONE. 2011;6:e25249. doi: 10.1371/journal.pone.0025249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Bergeron R, Ren JM, Cadman KS, Moore IK, Perret P, Pypaert M, Young LH, Semenkovich CF, Shulman GI. Chronic activation of AMP kinase results in NRF-1 activation and mitochondrial biogenesis. Am J Physiol Endocrinol Metab. 2001;281:E1340–1346. doi: 10.1152/ajpendo.2001.281.6.E1340. [DOI] [PubMed] [Google Scholar]
- 118.Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A. 2002;99:15983–15987. doi: 10.1073/pnas.252625599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hardie DG. The AMP-activated protein kinase pathway--new players upstream and downstream. J Cell Sci. 2004;117:5479–5487. doi: 10.1242/jcs.01540. [DOI] [PubMed] [Google Scholar]
- 120.Atherton PJ, Babraj J, Smith K, Singh J, Rennie MJ, Wackerhage H. Selective activation of AMPK-PGC-1alpha or PKB-TSC2-mTOR signaling can explain specific adaptive responses to endurance or resistance training-like electrical muscle stimulation. Faseb J. 2005;19:786–788. doi: 10.1096/fj.04-2179fje. [DOI] [PubMed] [Google Scholar]
- 121.Reznick RM, Shulman GI. The role of AMP-activated protein kinase in mitochondrial biogenesis. J Physiol. 2006;574:33–39. doi: 10.1113/jphysiol.2006.109512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Canto C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature. 2009;458:1056–1060. doi: 10.1038/nature07813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Steinberg GR, Kemp BE. AMPK in Health and Disease. Physiol Rev. 2009;89:1025–1078. doi: 10.1152/physrev.00011.2008. [DOI] [PubMed] [Google Scholar]
- 124.Chen ZP, Mitchelhill KI, Michell BJ, Stapleton D, Rodriguez-Crespo I, Witters LA, Power DA, Ortiz de Montellano PR, Kemp BE. AMP-activated protein kinase phosphorylation of endothelial NO synthase. FEBS Lett. 1999;443:285–289. doi: 10.1016/s0014-5793(98)01705-0. [DOI] [PubMed] [Google Scholar]
- 125.Morrow VA, Foufelle F, Connell JM, Petrie JR, Gould GW, Salt IP. Direct activation of AMP-activated protein kinase stimulates nitric-oxide synthesis in human aortic endothelial cells. J Biol Chem. 2003;278:31629–31639. doi: 10.1074/jbc.M212831200. [DOI] [PubMed] [Google Scholar]
- 126.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13:132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Giri S, Nath N, Smith B, Viollet B, Singh AK, Singh I. 5-aminoimidazole-4-carboxamide-1-beta-4-ribofuranoside inhibits proinflammatory response in glial cells: a possible role of AMP-activated protein kinase. J Neurosci. 2004;24:479–487. doi: 10.1523/JNEUROSCI.4288-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cacicedo JM, Yagihashi N, Keaney JF, Jr, Ruderman NB, Ido Y. AMPK inhibits fatty acid-induced increases in NF-kappaB transactivation in cultured human umbilical vein endothelial cells. Biochem Biophys Res Commun. 2004;324:1204–1209. doi: 10.1016/j.bbrc.2004.09.177. [DOI] [PubMed] [Google Scholar]
- 129.Hallows KR, Fitch AC, Richardson CA, Reynolds PR, Clancy JP, Dagher PC, Witters LA, Kolls JK, Pilewski JM. Up-regulation of AMP-activated kinase by dysfunctional cystic fibrosis transmembrane conductance regulator in cystic fibrosis airway epithelial cells mitigates excessive inflammation. J Biol Chem. 2006;281:4231–4241. doi: 10.1074/jbc.M511029200. [DOI] [PubMed] [Google Scholar]
- 130.Hattori Y, Suzuki K, Hattori S, Kasai K. Metformin inhibits cytokine-induced nuclear factor kappaB activation via AMP-activated protein kinase activation in vascular endothelial cells. Hypertension. 2006;47:1183–1188. doi: 10.1161/01.HYP.0000221429.94591.72. [DOI] [PubMed] [Google Scholar]
- 131.Zhao X, Zmijewski JW, Lorne E, Liu G, Park YJ, Tsuruta Y, Abraham E. Activation of AMPK attenuates neutrophil proinflammatory activity and decreases the severity of acute lung injury. Am J Physiol Lung Cell Mol Physiol. 2008;295:L497–504. doi: 10.1152/ajplung.90210.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Okayasu T, Tomizawa A, Suzuki K, Manaka K, Hattori Y. PPARalpha activators upregulate eNOS activity and inhibit cytokine-induced NF-kappaB activation through AMP-activated protein kinase activation. Life Sci. 2008;82:884–891. doi: 10.1016/j.lfs.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 133.Ko HJ, Zhang Z, Jung DY, Jun JY, Ma Z, Jones KE, Chan SY, Kim JK. Nutrient stress activates inflammation and reduces glucose metabolism by suppressing AMP-activated protein kinase in the heart. Diabetes. 2009;58:2536–2546. doi: 10.2337/db08-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bai A, Ma AG, Yong M, Weiss CR, Ma Y, Guan Q, Bernstein CN, Peng Z. AMPK agonist downregulates innate and adaptive immune responses in TNBS-induced murine acute and relapsing colitis. Biochem Pharmacol. 2010;80:1708–1717. doi: 10.1016/j.bcp.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 135.Salminen A, Hyttinen JM, Kaarniranta K. AMP-activated protein kinase inhibits NF-kappaB signaling and inflammation: impact on healthspan and lifespan. J Mol Med (Berl) 2011;89:667–676. doi: 10.1007/s00109-011-0748-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Barroso E, Eyre E, Palomer X, Vazquez-Carrera M. The peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) agonist GW501516 prevents TNF-alpha-induced NF-kappaB activation in human HaCaT cells by reducing p65 acetylation through AMPK and SIRT1. Biochem Pharmacol. 2011;81:534–543. doi: 10.1016/j.bcp.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 137.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J Biol Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 138.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 140.Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie Restriction Increases Muscle Mitochondrial Biogenesis in Healthy Humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Dominy JE, Jr, Lee Y, Gerhart-Hines Z, Puigserver P. Nutrient-dependent regulation of PGC-1alpha's acetylation state and metabolic function through the enzymatic activities of Sirt1/GCN5. Biochim Biophys Acta. 2010;1804:1676–1683. doi: 10.1016/j.bbapap.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gillum MP, Erion DM, Shulman GI. Sirtuin-1 regulation of mammalian metabolism. Trends Mol Med. 2010 doi: 10.1016/j.molmed.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoshizaki T, Schenk S, Imamura T, Babendure JL, Sonoda N, Bae EJ, Oh DY, Lu M, Milne JC, Westphal C, Bandyopadhyay G, Olefsky JM. SIRT1 inhibits inflammatory pathways in macrophages and modulates insulin sensitivity. Am J Physiol Endocrinol Metab. 2010;298:E419–428. doi: 10.1152/ajpendo.00417.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.L'Horset F, Dauvois S, Heery DM, Cavailles V, Parker MG. RIP-140 interacts with multiple nuclear receptors by means of two distinct sites. Mol Cell Biol. 1996;16:6029–6036. doi: 10.1128/mcb.16.11.6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Powelka AM, Seth A, Virbasius JV, Kiskinis E, Nicoloro SM, Guilherme A, Tang X, Straubhaar J, Cherniack AD, Parker MG, Czech MP. Suppression of oxidative metabolism and mitochondrial biogenesis by the transcriptional corepressor RIP140 in mouse adipocytes. J Clin Invest. 2006;116:125–136. doi: 10.1172/JCI26040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zschiedrich I, Hardeland U, Krones-Herzig A, Diaz M Berriel, Vegiopoulos A, Muggenburg J, Sombroek D, Hofmann TG, Zawatzky R, Yu X, Gretz N, Christian M, White R, Parker MG, Herzig S. Coactivator function of RIP140 for NFkappaB/RelA-dependent cytokine gene expression. Blood. 2008;112:264–276. doi: 10.1182/blood-2007-11-121699. [DOI] [PubMed] [Google Scholar]
- 148.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 149.Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
- 150.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, Ryter SW, Choi AM. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zhang J, Ney PA. Role of BNIP3 and NIX in cell death, autophagy, and mitophagy. Cell Death Differ. 2009;16:939–946. doi: 10.1038/cdd.2009.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008;314:1789–1803. doi: 10.1016/j.yexcr.2008.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap'n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 155.Ryter SW, Alam J, Choi AM. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol Rev. 2006;86:583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 156.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, Yamamoto M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13:76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lee BS, Heo J, Kim YM, Shim SM, Pae HO, Kim YM, Chung HT. Carbon monoxide mediates heme oxygenase 1 induction via Nrf2 activation in hepatoma cells. Biochem Biophys Res Commun. 2006;343:965–972. doi: 10.1016/j.bbrc.2006.03.058. [DOI] [PubMed] [Google Scholar]
- 158.Suliman HB, Carraway MS, Ali AS, Reynolds CM, Welty-Wolf KE, Piantadosi CA. The CO/HO system reverses inhibition of mitochondrial biogenesis and prevents murine doxorubicin cardiomyopathy. J Clin Invest. 2007;117:3730–3741. doi: 10.1172/JCI32967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Suliman HB, Carraway MS, Tatro LG, Piantadosi CA. A new activating role for CO in cardiac mitochondrial biogenesis. J Cell Sci. 2007;120:299–308. doi: 10.1242/jcs.03318. [DOI] [PubMed] [Google Scholar]
- 160.Piantadosi CA, Carraway MS, Babiker A, Suliman HB. Heme oxygenase-1 regulates cardiac mitochondrial biogenesis via Nrf2-mediated transcriptional control of nuclear respiratory factor-1. Circ Res. 2008;103:1232–1240. doi: 10.1161/01.RES.0000338597.71702.ad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Lee TS, Chau LY. Heme oxygenase-1 mediates the anti-inflammatory effect of interleukin-10 in mice. Nat Med. 2002;8:240–246. doi: 10.1038/nm0302-240. [DOI] [PubMed] [Google Scholar]
- 162.Kruger AL, Peterson SJ, Schwartzman ML, Fusco H, McClung JA, Weiss M, Shenouda S, Goodman AI, Goligorsky MS, Kappas A, Abraham NG. Up-regulation of heme oxygenase provides vascular protection in an animal model of diabetes through its antioxidant and antiapoptotic effects. J Pharmacol Exp Ther. 2006;319:1144–1152. doi: 10.1124/jpet.106.107482. [DOI] [PubMed] [Google Scholar]
- 163.Li Y, Corradetti MN, Inoki K, Guan KL. TSC2: filling the GAP in the mTOR signaling pathway. Trends Biochem Sci. 2004;29:32–38. doi: 10.1016/j.tibs.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 164.Fingar DC, Blenis J. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene. 2004;23:3151–3171. doi: 10.1038/sj.onc.1207542. [DOI] [PubMed] [Google Scholar]
- 165.Manning BD, Tee AR, Logsdon MN, Blenis J, Cantley LC. Identification of the tuberous sclerosis complex-2 tumor suppressor gene product tuberin as a target of the phosphoinositide 3-kinase/akt pathway. Mol Cell. 2002;10:151–162. doi: 10.1016/s1097-2765(02)00568-3. [DOI] [PubMed] [Google Scholar]
- 166.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes Dev. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cory S, Huang DC, Adams JM. The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene. 2003;22:8590–8607. doi: 10.1038/sj.onc.1207102. [DOI] [PubMed] [Google Scholar]
- 168.Danial NN, Gramm CF, Scorrano L, Zhang CY, Krauss S, Ranger AM, Datta SR, Greenberg ME, Licklider LJ, Lowell BB, Gygi SP, Korsmeyer SJ. BAD and glucokinase reside in a mitochondrial complex that integrates glycolysis and apoptosis. Nature. 2003;424:952–956. doi: 10.1038/nature01825. [DOI] [PubMed] [Google Scholar]
- 169.Russell LK, Mansfield CM, Lehman JJ, Kovacs A, Courtois M, Saffitz JE, Medeiros DM, Valencik ML, McDonald JA, Kelly DP. Cardiac-specific induction of the transcriptional coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha promotes mitochondrial biogenesis and reversible cardiomyopathy in a developmental stage-dependent manner. Circ Res. 2004;94:525–533. doi: 10.1161/01.RES.0000117088.36577.EB. [DOI] [PubMed] [Google Scholar]
- 170.Brandtzaeg P, Osnes L, Ovstebo R, Joo GB, Westvik AB, Kierulf P. Net inflammatory capacity of human septic shock plasma evaluated by a monocyte-based target cell assay: identification of interleukin-10 as a major functional deactivator of human monocytes. J Exp Med. 1996;184:51–60. doi: 10.1084/jem.184.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lin CY, Tsai IF, Ho YP, Huang CT, Lin YC, Lin CJ, Tseng SC, Lin WP, Chen WT, Sheen IS. Endotoxemia contributes to the immune paralysis in patients with cirrhosis. J Hepatol. 2007;46:816–826. doi: 10.1016/j.jhep.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 172.Wang TS, Deng JC. Molecular and cellular aspects of sepsis-induced immunosuppression. J Mol Med (Berl) 2008;86:495–506. doi: 10.1007/s00109-007-0300-4. [DOI] [PubMed] [Google Scholar]
- 173.Biswas SK, Lopez-Collazo E. Endotoxin tolerance: new mechanisms, molecules and clinical significance. Trends Immunol. 2009;30:475–487. doi: 10.1016/j.it.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 174.Reddy RC, Standiford TJ. Effects of sepsis on neutrophil chemotaxis. Curr Opin Hematol. 2010;17:18–24. doi: 10.1097/MOH.0b013e32833338f3. [DOI] [PubMed] [Google Scholar]
- 175.Fedson DS. Meeting the challenge of influenza pandemic preparedness in developing countries. Emerg Infect Dis. 2009;15:365–371. doi: 10.3201/eid1503.080857. [DOI] [PMC free article] [PubMed] [Google Scholar]