Abstract
DICER1 is essential for the generation of mature microRNAs (miRNAs) and other short noncoding RNAs. Several lines of investigation implicate DICER1 as a tumor suppressor. Reduced DICER1 levels and changes in miRNA abundance have been associated with aggressive tumor phenotypes. The global effects of reduced DICER1 on mRNA transcript abundance in tumor cells remain largely unknown. We used shRNA to stably knock down DICER1 in endometrial cancer cell lines to begin to determine how reduced DICER1 activity contributes to tumor phenotypes. DICER1 knockdown did not affect cell proliferation but caused enhanced cell migration and growth in soft agar. miRNA and mRNA profiling in KLE cells revealed overall decreases in miRNA levels and changes in the relative abundance of many mRNAs. One of the most striking changes in mRNA levels was the upregulation of interferon stimulated genes (ISGs), the majority of which lack known miRNA target sequences. IFNβ, a key upstream regulator of the interferon response, was significantly increased in DICER1 knockdowns in the AN3CA, Ishikawa, and KLE endometrial cancer cell lines and in the normal endometrial cell line EM-E6/E7/TERT. IFNβ secreted in media from KLE and EM-E6/E7/TERT shDcr cells was sufficient to activate an interferon response in HT29 cells. The reduced miRNA processing in DICER1 knockdowns was associated with increases in pre-miRNAs in the cytoplasm. Our findings suggest elevated pre-miRNA levels trigger the interferon response to double-stranded RNA. We thus report a novel effect of reduced DICER1 function in cancer cells.
Keywords: Endometrial cancer, DICER1, Interferon response, microRNA, RNA-Sequencing
Introduction
Endometrial cancer is the most common gynecological malignancy in the United States and approximately 15% of patients suffer from recurrent disease (1, 2). Discovery of the molecular lesions contributing to endometrial tumorigenesis will provide opportunities for targeted therapies.
DICER1 is an RNASE III helicase necessary to process double-stranded RNA (dsRNA) in mammalian cells, the predominant form of which is microRNA (miRNA). Primary miRNAs (pri-miRNAs) are cleaved by the enzyme DROSHA into pre-miRNAs. Pre-miRNAs are transported out of the nucleus by EXPORTIN-5 and processed in the cytoplasm by DICER1 and accessory proteins. Mature miRNAs go with AGO proteins to pair imperfectly with the 3′ UTRs of target mRNAs and either impede translation or degrade the mRNAs (3). About 60% of human genes may be regulated post-transcriptionally by miRNAs (4, 5). Given the key role of miRNAs in gene regulation it is not surprising DICER1, DROSHA, and other RNAi components have been implicated as “tumor suppressors” in solid tumors (6-10). Germline loss-of-function mutations in DICER1 are associated with the pleuropulmonary blastoma tumor susceptibility syndrome (8). The penetrance of inherited DICER1 mutations is, however, modest and it has been proposed that DICER1 is a haploinsufficient tumor suppressor (11). A recent report on somatic DICER1 mutations in nonepithelial ovarian tumors further supports the notion DICER1 is a haploinsufficient tumor suppressor (12). DICER1 is an essential gene. The Dicer1 homozygous knockout mouse is embryonic lethal (13, 14). Conditional deletion of Dicer1 in a mouse Kras lung cancer model caused homozygous knockout cells to die, but heterozygous tumors to be more aggressive than wild type tumors (7), consistent with Dicer’s role as a haploinsufficient tumor suppressor. Our group previously showed lower DICER1 mRNA levels in endometrial cancer are associated with recurrence and accelerated disease progression (15).
The interferon response is a component of the innate immune response to pathogens such as RNA viruses. Viral dsRNA binding to Toll-like receptor 3 (TLR3) on the cell membrane or IFIH1 (MDA5), PKR, or RIG-1 in the cytoplasm triggers IRF3 and NFκB translocation to the nucleus and transcription of early genes, specifically IFNβ. Secreted IFNβ activates cell surface receptors by autocrine and paracrine means to induce activation of STAT1 and expression of interferon stimulated genes (ISGs). Next, IFNα genes are transcribed leading to downstream effects including global inhibition of translation and apoptosis (16-19). Innate immunity and interferon responses in malignancies are context dependent and often paradoxical. An immune response may mediate tumor cell killing; interferons have been used to treat a variety of human cancers (20, 21). However, inflammatory cytokines downstream of the interferon response have been linked to cellular transformation (22). Cellular senescence can trigger an interferon response (23), but increases in ISGs such as ISG15 and IFI44 are prognostic for breast and lung cancer recurrence, respectively (24, 25). The interferon response and how it impacts tumor behavior is likely determined by a complex and context dependent interaction of tumor cell specific effects and humoral responses.
Using short hairpin RNAs (shRNAs) we reduced DICER1 levels in endometrial cancer and normal cell lines by greater than 50%. mRNA and miRNA profiling studies revealed global perturbations in RNA levels. The most striking change observed was an increase in transcription of IFNβ and ISGs characteristic of an interferon response. We demonstrate that the interferon response in endometrial cells with reduced DICER1 results from accumulation of pre-miRNAs in the cell cytoplasm.
Materials and Methods
Cell culture
Four endometrioid endometrial cancer cell lines were investigated. AN3CA and KLE were purchased from the American Type Culture Collection. The Ishikawa cell line was a gift from Dr. Stuart Adler (Washington University School of Medicine, Department of Internal Medicine). The MFE296 cell line was kindly provided by Dr. Pamela Pollock (Queensland University of Technology, Brisbane) and the HT29 cell line was kindly provided by Dr. Loren Michel (Washington University). The EM-E6/E7/TERT cell line was originally reported by Mizumoto and colleagues (26) and kindly provided by Dr. Pamela Pollock. Cell lines were grown as previously described (27) and authenticated as reported in (28).
Lentiviral transduction to create stable knockdowns
DICER1 and GSK3β knockdowns were created in AN3CA, EM-E6/E7/TERT, Ishikawa, KLE, and MFE296 cell lines as previously described (29). Virus production and infections were carried out according to established methods (30). DROSHA knockdown was created with virus kindly provided by Michael Kuchenreuther in Dr. Jason Weber’s laboratory (Washington University).
The short hairpin sequences used were:
shDcrA 5′-GCTCGAAATCTTACGCAAATA-3′
shDcrC 5′-GCCAAGGAAATCAGCTAAATT-3′
shDro2 5′-CGAAGCTCTTTGGTGAATAAT-3′
shDro4 5′-CCAGCGTCCATTTGTACTATT-3′
shGSK3β 5′-AGCAAATCAGAGAAATGAAC-3′
shLuc 5′CCCTCTGAACATGAGCATCAA-3′ shRFP 5′-TGCTAAGGAGTTTGGAGACAA-3′ (31)
The shDcr3 hairpin construct was designed by Sigma-Aldrich (St. Louis, MO).
Reverse transcription polymerase chain reaction (RT-PCR)
Total cellular RNA was extracted utilizing the Trizol® method (Invitrogen, Carlsbad, CA). Nuclear and cytoplasmic fractions were prepared using the Norgen Biotek Cytoplasmic and Nuclear Purification Kit, according to the manufacturer’s instructions (Norgen Biotek, Thorold, Ontario, Canada). RNA concentration was determined with the NanoDrop machine and software (Thermo Fisher Scientific, Wilmington, DE). Complementary DNA (cDNA) was generated using 1 μg total RNA and the QuantiTect Reverse Transcription Kit (Qiagen, Valencia, CA). Quantitative RT-PCR of pre-miRNAs and the DUSP6 control was performed using SYBR Green (BioRad) methods. The primers used are listed below.
| Pre-microRNA Primers | Forward | Reverse |
| PRELET7D | 5′ TTTAGGGCAGGGATTTTGC 3′ | 5′ TAAGAAAGGCAGCAGGTCGT 3′ |
| PREMIR183 | 5′ CGCAGAGTGTGACTCCTGTT 3′ | 5′ TCGTGGATCTGTCTCTGCTC 3′ |
| PREMIR450A | 5′ AAACTATTTTTGCGATGTGTTCC 3′ | 5′ TGCAAAATGTCCCCAATACA 3′ |
| DUSP6 | 5′ CCCCTTCCAACCAGAATGTA 3′ | 5′ TGCCAAGAGAAACTGCTGAA 3′ |
Expression of DICER1, DROSHA, IFI44, IFI44L, IFI6, IFIH1, IFNβ1, MX1, and OAS3 mRNAs, and LET7B, LET7D, MIR107, MIR183, MIR450A, MIR542 pri-miRNAs was assessed by quantitative RT-PCR TaqMan® assays (Applied Biosystems, Foster City, CA) and the Applied Biosystems 7500 Fast real-time PCR system and software. Human β-actin was used as the endogenous control as previously described (32). Expression of let-7c, miR-10a, miR-16, miR-29b, and miR-126b mature miRNAs was assessed by quantitative TaqMan® microRNA assays (Applied Biosystems, Foster City, CA) and the Applied Biosystems 7500 Fast real-time PCR system and software. U6 was used as the endogenous control (9). Relative expression levels were calculated using the delta-delta Ct method (33).
All qPCR assays were performed in triplicate and then repeated with new cDNA synthesis. Minus RT controls (reverse transcriptase negative cDNA synthesis reactions) were carried out for at least one sample per plate.
MicroRNA profiling
KLE and AN3CA cell lines were subjected to global microRNA profiling with Nanostring™ technology (Seattle, WA). 749 miRNAs were evaluated using the nCounter Human miRNA Panel CodeSet®.
RNA-Sequencing
PolyA+ RNA was purified from total RNA using the Dynabeads mRNA Purification Kit (Invitrogen, Carlsbad, CA). Each sample was resuspended in 2 μl of 100 mM zinc acetate and heated at 60°C for 3 minutes to fragment the RNA by hydrolysis. The reaction was quenched by the addition of 2 μl volumes of 200 mM EDTA and purified with an Illustra Microspin G25 column (GE Healthcare). First strand cDNA was made using hexameric random primers and SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA), and the product was treated with E. coli DNA ligase, DNA polymerase I, and RNase H to prepare double stranded cDNA using standard methods. cDNA libraries were end-repaired with a Quick Blunting kit (New England BioLabs, Ipswich, MA) and A-tailed using Klenow exo- and dATP. Illumina adapters with four base barcodes were ligated to cDNA and fragments ranging from 150-250 bp were selected using gel electrophoresis. Libraries were enriched in a 10-cycle PCR with Phusion Hot Start II High-Fidelity DNA Polymerase (Thermo Fisher Scientific, Waltham, MA) and pooled in equimolar ratios for multiplex sequencing. Single read, 36-cycle runs were completed on the Illumina Genome Analyzer IIx.
Sequenced reads were aligned to the human reference sequence (hg19 / NCBI Build 37.1) using Tophat (34). Reads that aligned uniquely to the reference sequence were considered for gene expression quantification with Cufflinks (35). Gene expression was normalized using the Cufflinks provided option for quartile normalization.
Western blots
Western blot analysis of DICER1 was performed as previously described (27, 33). GAPDH was used as a loading control. Antibodies used were as follows: rabbit anti-DICER1 H212 (sc-30226, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, 1:200), goat anti-rabbit IgG-HRP (sc-2030, Santa Cruz Biotechnology, 1:2500), rabbit anti-DROSHA (ab12286, Abcam, 1:750), mouse anti-GAPDH (NB615, Novus Biologicals, Littleton, CO, 1:4000), goat anti-mouse IgG-HRP (sc-2005, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, 1:5000), rabbit polyclonal anti-STAT3 H-190 (sc-7179, Santa Cruz Biotechnology, 1:200), rabbit anti-phospho-STAT3 Ser727 (9134, Cell Signaling Technology, 1:500), rabbit anti-phospho-STAT3 Tyr705 EP2147Y (04-1059, Millipore, 1:500). Band intensities were quantified using the program ImageJ (National Institutes of Health).
ELISA
ELISA was performed with the Verikine-HS™ Human Interferon Beta Serum ELISA kit (PBL Interferon Source).
Cell proliferation, wound healing and colony formation assays
For cell proliferation assays 100,000 cells were plated in 6-well plates in triplicate. Cells were trypsinized and counted using trypan blue staining and a hemocytometer every 24 hours for 120 hours.
Wound healing assays were performed using AN3CA and KLE cells. Cells were grown to confluency then scratched down the middle of the plate. Cells were photographed every 4-6 hours for up to 96 hours (GE Healthcare IN Cell Analyzer 2000). The area of the “scratch” (area not filled in) was determined for each time point.
Growth of endometrial cancer cell lines in soft agar was determined as follows: First a base layer of 0.5% agar was plated in media, then a top layer of 0.3% agar in media with 30,000 cells per well was plated in 6-well dishes. After 4 weeks, cells were stained with crystal violet and imaged. Colonies were counted.
Interferon stimulation
PolyI:C (Invitrogen, Carlsbad, CA) was diluted into the media of cells or transfected using the Dharmafect reagent (Thermo Fisher Scientific, Waltham, MA).
Let-7 inhibition
Let-7 inhibition was performed as previously described (36). The CHECK-2 vector with the let-7b target site cloned into the 3′ UTR was a kind gift from Annaleen Vermeulen (Thermo Fisher Scientific).
Results and Discussion
Stable knockdown of DICER1
We used shRNA and lentiviral infection to stably knock down DICER1 in four endometrial cancer cell lines and a transformed normal endometrial epithelium cell line; AN3CA, Ishikawa, KLE, MFE296, and EM-E6/E7/TERT. Of five hairpins tested, two (shDcrA and shDcrC) resulted in substantial reductions in DICER1 protein levels (Figure 1A). Knockdowns were generated with shDcrA and shDcrC hairpins and shLuc and shRFP controls. Stable knockdown of DICER1 (<50% of controls) persisted for up to 30 passages for all cell lines, with the exception of MFE296, for which knockdown was unstable (Figure 1B and data not shown). In KLE, DICER1 was reduced to ~10% of controls, suggesting that sufficient shRNA processing can occur with substantially reduced DICER1 activity (Figure 1B). An additional shRNA targeting the DICER1 3′ UTR (shDcr3) was used in KLE cells leading to greater than 50% reduction in DICER1 protein levels (Supplemental Figure 1).
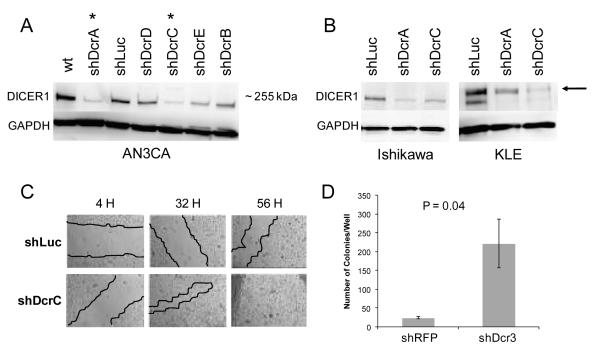
Figure 1.
Knockdown of DICER1 in endometrial cell lines. Western blot analysis of DICER1 expression in A) AN3CA (passage 5) with shRNA A-E against DICER1 or Luciferase control. * denotes hairpins showing greatest degree of knockdown. B) Representative knockdowns in additional cell lines, Ishikawa (passage 16) and KLE (passage 26). Arrow denotes nonspecific band of higher molecular weight above the ~255 kDa DICER1 band in KLE. C) Representative wound healing assay in AN3CA cells. shLuc control and shDcrC knockdown at 4, 32, and 56 hours. Red lines denote wound outlines. D) Increased soft agar colony formation in KLE cells with DICER1 knockdown. Results are one representative experiment of two performed in triplicate. KLE cells were plated and growth in soft agar was assessed by staining with crystal violet after 30 days.
Cell doubling times were similar in DICER1 knockdowns and control cells (Supplemental Figure 2). Cell migration was increased in AN3CA shDcr cells (Figure 1C) but no difference was seen in KLE shDcr cells. The Ishikawa and EM-E6/E7/TERT cells could not be evaluated in the cell migration assay because they did not grow as monolayers on glass slides (Supplemental Figure 2). In both KLE and EM-E6/E7/TERT, shDcr cells formed more colonies in soft agar than control cells (Figure 1D and Supplemental Figure 2). These in vitro assays for cancer-associated phenotypes suggest that reduced DICER1 in endometrial cancer cells can result in increased cell motility and anchorage independence. This increased cell motility was previously shown in breast cancer cell lines and attributed to a reduction in miR-200 and upregulation of genes involved in epithelial mesenchymal transition (37).
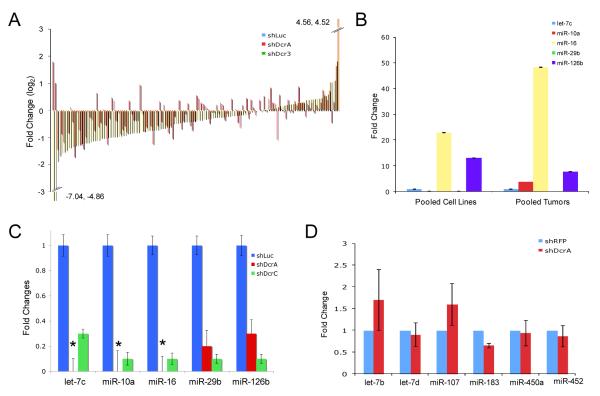
We profiled miRNAs globally in shDcr cells to identify reductions in particular miRNAs that might contribute to cancer-associated phenotypes. Nanostring™ miRNA profiling studies in AN3CA cells as well as KLE knockdowns and controls revealed 133 of 749 miRNAs interrogated were expressed at appreciable levels. When the average levels of miRNA expression in the two KLE knockdowns were compared with the KLE shLuc control, 64% of the 133 miRNAs showed reduced levels in the knockdowns (Supplemental Table 1 and Figure 2A). miR-200 was not expressed in endometrial cancer cell lines (Supplemental Table 1) so could not be responsible for the cancer-associated phenotypes mentioned above. We observed clear increases in a subset of miRNAs (Figure 2A) as previously described in colon cancer cells with reduced DICER1 protein (9). Similar effects on miRNA abundance were seen with both knockdowns in the KLE cell line; however, the magnitude of changes in miRNA levels seemed greater in the shDcr3 knockdown than in the shDcrA knockdown. For the shDcrA knockdown, 76/133 miRNAs were less than in shLuc control (average log2 fold change −.502). With the shDcr3 knockdown, 95/133 miRNAs were less abundant than in the shLuc control with an average −.828 fold change (log2). KLE shDcrA cells were evaluated at passage 15 and shDcr3 cells at passage 5. The more pronounced effect on miRNA levels seen with the shDcr3 knockdown could be attributable to more efficient targeting of DICER1 with the shDcr3 construct, greater reduction in DICER1 protein levels at earlier passages, or compensation for DICER1 as shDcrA cells were passaged (e.g. stabilization of miRNAs).
Figure 2.
miRNA expression in DICER1 knockdown cells. A) Nanostring™ miRNA profiling of 133 expressed miRNAs in the KLE cell line. B) Relative abundance of 5 miRNAs in endometrial cancer cell lines and tumors by Taqman qRT-PCR assays, normalized to U6 reference gene. RNA from 4 cell lines (AN3CA, KLE, Ishikawa, MFE296) and 4 tumors was pooled and converted to cDNA. C) Levels of 5 miRNAs in control and shDcr KLE cells by Taqman qRT-PCR assays, normalized to U6 reference gene. * denotes miRNAs that were undetectable in shDcrA cells. D) Pri-miRNAs in KLE cells measured by Taqman qRT-PCR and normalized to β-Actin reference gene. Fold change shDcrA/shRFP is plotted on the y-axis. For all qRT-PCR experiments, error bars are SD (data shown is average of two experiments performed in triplicate).
qRT-PCR of five miRNAs previously shown to be expressed in normal and cancerous endometrium (38) confirmed the relative abundance reported by Nanostring™ profiling in AN3CA and KLE cells. qRT-PCR in pooled endometrial cancers confirmed the rank order of five miRNAs reported by Nanostring™ (Supplemental Table 1 and Figure 2B). miR-16 was the highest expressed of the five miRNAs by profiling and qRT-PCR. miR-29b was the lowest expressed by both profiling and qRT-PCR. qRT-PCR confirmed the Nanostring™ profiling and the functional reduction of DICER1 processing, as five mature miRNAs were significantly decreased in KLE shDcr cells (Figure 2C). pri-miRNAs, the initial miRNA transcripts that are processed by DROSHA, were not significantly altered, showing that effects on mature miRNAs are due to a defect in miRNA processing, not transcription (Figure 2D).
DICER1 knockdown effects on mRNA expression: upregulation of interferon response genes
To further assess the functional consequences of DICER1 knockdown, we profiled mRNA expression using RNA-Sequencing (RNA-Seq) in KLE cells (35). Out of 9935 genes expressed in KLE by RNA-Seq, 584 were upregulated more than twofold in shDcr cells (Supplemental Table 2). Gene Ontology analysis showed enrichment for functions associated with response to virus or other pathogens when the upregulated gene set was analyzed (Supplemental Table 3). A striking number of interferon stimulated genes (ISGs) were upregulated (17 of the 28 present in the RNA-Seq data set) (Figure 3A). The probability of 17 genes at random being upregulated in this set is quite low (p<1.2 * 10E-14). qRT-PCR confirmed upregulation of six out of seven ISGs tested (Figure 3B). Similar increases in six ISG transcripts were seen in independent knockdowns, providing biologic validation of the effect of reduced DICER1 in KLE cells (shDcrA and shDcr3, Figure 3B; shDcrC, data not shown). To explore a possible mechanism for interferon response activation in shDcr cells, we evaluated mRNA levels of transcription factors that might target ISGs. No transcription factors predicted to bind upstream of the activated ISGs were overexpressed in shDcr cells by RNA-Seq (data not shown). Direct miRNA effects on ISG transcript levels were ruled out as the ISGs have no known targets in their 3′ UTRs for miRNAs expressed in KLE (Supplemental Table 4).
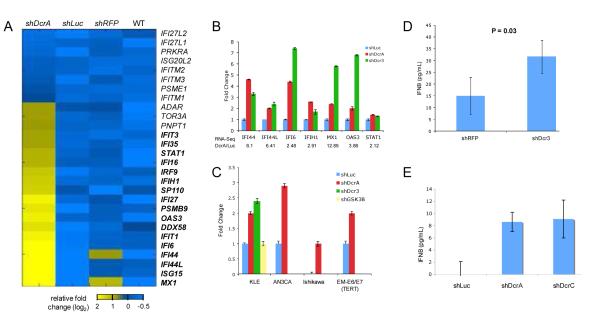
Figure 3.
Upregulation of interferon stimulated genes (ISGs) in shDcr cells. A) RNA-Seq heat map for ISGs. Genes indicated in bold revealed a fold change greater than two relative to the average expression of the three controls, shLuc, shRFP, and WT (untransfected KLE cells). B) qRT-PCR validation of ISGs upregulated in RNA-Seq. Fold change shDcr/shLuc is plotted on y-axis. C) IFNβ1 transcript levels are increased in shDcr cells in four cell lines. shDcr3 and shGSK3β were only performed in the KLE cell line. For all qRT-PCR experiments, mRNA levels were assessed using TaqMan qRT-PCR assays, normalized to β-Actin reference gene. Error bars are SD (data shown is average of two experiments performed in triplicate). IFNβ protein levels in cell culture media are increased in KLE (D) and EM-E6/E6/TERT (E) shDcr cells. IFNβ was assessed by ELISA. One representative experiment of three performed in triplicate is shown. Error bars represent SEM. Significance was determined by t-test.
This increase in ISGs appeared to be a canonical interferon response (17, 18). To determine if the upstream IFNβ gene was upregulated and activating ISGs, we assessed IFNβ mRNA and protein levels in DICER1 knockdowns. RNA-Seq did not detect expression of IFNβ1 in any of the cell lines investigated, as would be expected for a low abundance transcript. IFNβ1 transcript was, however, detectable using qRT-PCR. Two shDcr hairpins caused upregulated IFNβ1 transcript compared to shLuc (Figure 3C). The control shRFP hairpin did not significantly upregulate IFNβ1 while the shDcrC hairpin did (data not shown). shRNA alone does not trigger the interferon response (39, 40). We tested the possibility that knockdown of a cell-essential gene might activate the interferon response by measuring IFNβ1 transcript levels in KLE shGSK3β cells. Because neither the shGSK3β nor the control shLuc and shRFP hairpins activate the interferon response, we conclude that the interferon response seen is a DICER1-specific effect. The IFNβ1 transcript was upregulated at least twofold in DICER1 knockdowns in two additional endometrial cancer cell lines, AN3CA and Ishikawa, and an immortalized normal endometrial cell line, EM-E6/E7/TERT (Figure 3C). The increase in IFNβ1 transcript due to reduced DICER1 led to increased IFNβ protein levels in the media of KLE shDcr cells (Figure 3D). A similar increase in IFNβ protein was observed in EM-E6/E7/TERT shDcr cell media (Figure 3E), showing that reduced DICER1 leads to increased IFNβ expression in both normal and cancer endometrial cell lines.
DICER1 knockdown causes a canonical interferon response
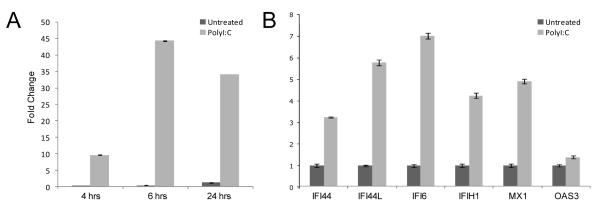
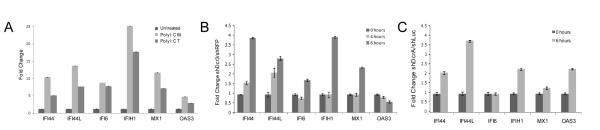
As some cancer cell lines have abrogated interferon responses (19), we postulated that activation of the interferon response in KLE might be an artifact of a mutated interferon response pathway. However, the interferon response is intact in the KLE endometrial cancer cell line. Transfection with polyI:C, a dsRNA analog, activated the interferon response (Figure 4). IFNβ1 transcript levels rose rapidly and peaked at six hours, with concomitant increases in ISGs (Figure 4B). In addition, a cytoplasmic receptor sensing dsRNA (IFIH1) was overexpressed in KLE cells with low DICER1 (Figure 3B). The interferon response in KLE shDcr cells upregulates the same genes as that in KLE cells transfected with polyI:C, albeit with a smaller magnitude (Figure 3B, 4B).
Figure 4.
Interferon response in the KLE cell line. (A) Upregulation of IFNβ1 in polyI:C-treated KLE 4, 6 and 24 hours post transfection. (B) ISGs upregulation 24 hours post polyI:C transfection. Transcript abundance was measured by Taqman qRT-PCR assays, normalized to β-Actin reference gene. Error bars are SD (data shown is average of two experiments performed in triplicate).
We used media transfer to determine the biological activity of secreted IFNβ protein in the media of shDcr cells. HT29 colon carcinoma cells exhibit a strong interferon response (40, 41), activating IFNβ and ISGs when polyI:C is either added to the cell culture media or transfected (Figure 5A, Supplemental Figure 3). Transfer of media from KLE shDcr3 cells to HT29 cells stimulated an interferon response, while shRFP cell media did not (Figure 5B). Media from EM-E6/E7/TERT shDcr cells similarly stimulated 4/6 ISGs (Figure 5C), indicating DICER1 knockdown causes an interferon response in both normal and cancerous endometrial cells. When media from KLE shDcr3 cells was transferred to KLE shRFP cells, no appreciable interferon response was seen (Supplemental Figure 4). This difference could be due to the relative strength of interferon responses in KLE and HT29 cells (Figure 4B, 5A). KLE shDcr3 cells in culture reflect long-term, continual IFNβ stimulation and KLE controls may not respond to a short stimulus with conditioned medium as HT29s do.
Figure 5.
KLE and EM-E6/E7/TERT shDcr media stimulate an interferon response in HT29 cells. A) PolyI:C stimulates a canonical interferon response in HT29 cells. PolyI:C was diluted into media (PolyI:C M) or transfected (PolyI:C T) into HT29 cells and RNA was isolated. B) shDcr3 but not shRFP media stimulates a canonical interferon response in HT29 cells. Media was transferred from KLE shRFP and shDcr3 cells to HT29s and RNA was isolated. Y-axis represents fold change of shDcr3/shRFP media. C) shDcrA but not shLuc media stimulates a canonical interferon response in HT29 cells. Media was transferred from EM-E6/E7/TERT shLuc and shDcrA cells to HT29s and RNA was isolated. Y-axis represents fold change of shDcrA/shLuc media. For all qRT-PCR experiments, transcript abundance was measure by Taqman qRT-PCR assays, normalized to β-Actin reference gene. Error bars are SD (data shown is average of two experiments performed in triplicate).
Pre-miRNAs build up in the cytoplasm and may cause an interferon response
To determine a mechanism for activation of the interferon response, we focused on a candidate miRNA. Members of the let-7 miRNA family, known for their tumor-suppressive roles (42, 43), were significantly reduced in shDcr cells (Figure 2C and Supplemental Table 1). The let-7 family downregulates the cytokine IL6, which when activated leads to phosphorylation of STAT3 by NFκB, resulting in an inflammatory response linked to cellular transformation (22). To determine whether let-7 was responsible for the interferon response, we inhibited let-7 in KLE cells (Supplemental Figure 5A). No increase in IFNβ1 was observed when let-7 was inhibited (Supplemental Figure 5B). Thus, let-7 alone is not responsible for the activation of the interferon response.
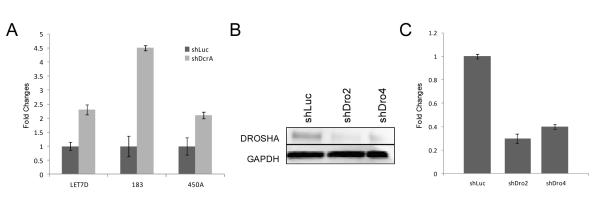
dsRNA (usually viral) activates the interferon response in mammalian cells. Our studies suggest a possible mechanism for interferon response upregulation by reduced DICER1. Mature miRNAs are too short (averaging 22 nt) to elicit the interferon response through viral dsRNA sensors (17, 44). Their precursor molecules, pre-miRNAs, are larger (~70 nt) and as such could be recognized by cytoplasmic dsRNA sensors IFIH1, PKR, or RIG-1 (45). We determined the subcellular location of pre-miRNAs in control and shDcr cells (Figure 6A). Pre-let7d, pre-miR183, and pre-miR450a were increased in the cytoplasmic fraction of shDcr cells. The corresponding mature miRNAs were decreased in shDcr cells (Supplemental Table 5), reflecting reduced DICER1 processing that results in buildup of pre-miRNAs and reduction of processed, mature miRNAs. Buildup of pre-miRNAs in the cytoplasm may elicit the canonical interferon response. To determine the specificity of this effect, we knocked down DROSHA in the KLE cell line (Figure 6B). Knockdown of DICER1 or DROSHA causes a reduction in mature miRNAs because of reduced processing. However, only DICER1 knockdown results in a buildup of pre-miRNAs. Lower levels of DROSHA did not trigger an interferon response as indicated by IFNβ1 levels (Figure 6C). Interestingly, DROSHA knockdown appeared to decrease IFNβ1 transcript levels. This could point to a role for pre-miRNAs in modulating the interferon response. Because DROSHA knockdown results in fewer pre-miRNAs (due to reduced pri-miRNA processing), this finding demonstrates that pre-miRNA buildup, rather than a decrease in mature miRNAs, causes the interferon response.
Figure 6.
Pre-miRNAs build up in the cytoplasm of shDcr cells. A) Pre-miRNAs are increased in shDcr cytoplasm. qRT-PCR was performed on LET7D, MIR183, and MIR450A pre-miRNAs in shLuc and shDcrA cytoplasmic RNA, using DUSP6 mRNA as an endogenous control. B) KLE whole cell lysates were probed for DROSHA using GAPDH as a reference gene. One representative experiment (of two). C) IFNβ1 transcript is not increased in shDrosha cells. IFNβ1 was measured by Taqman qRT-PCR and normalized to β-Actin reference gene. For all qRT-PCR experiments, error bars are SD (data shown is average of two experiments performed in triplicate).
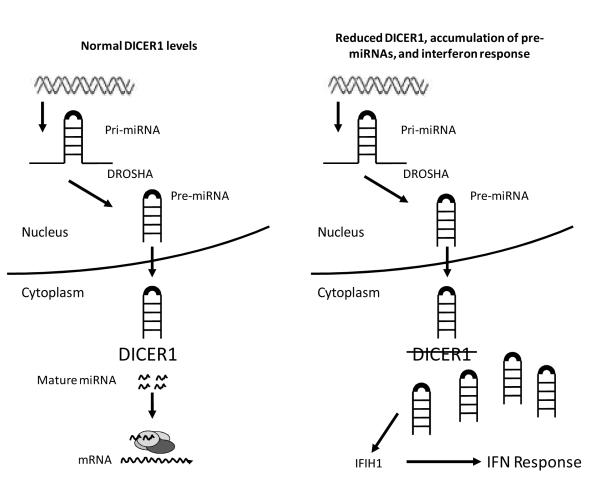
Our data point to the accumulation of pre-miRNAs in the cytoplasm as the trigger for the interferon response we observed in cells with reduced DICER1 activity (Figure 7). This is unlikely to be an effect of the system used; while siRNAs may activate the interferon response in mammalian cells (46), shRNAs do not (40). The immune response to dsRNA is highly conserved, with organisms such as plants and fungi enacting an RNAi-based response to viral RNA (47). Prior studies demonstrating that overexpression of pre-miRNAs can activate the interferon response in zebrafish (48) and that so-called “long hairpin RNAs” similarly activate the innate immune response (49) are consistent with our findings that build-up of pre-miRNAs elicits an interferon response. It remains unclear whether or how the interferon response is related to cancer phenotypes such as increased cell migration and growth in soft agar we observed in our DICER1 knockdown cells.
Figure 7.
Model for pre-miRNA buildup and interferon stimulation. In cells with normal levels of DICER1 (left panel), pre-miRNAs are processed to mature miRNAs and mRNAs are targeted for silencing. In cells with reduced DICER1 (right panel), pre-miRNA processing is inhibited and pre-miRNAs build up in the cytoplasm. This dsRNA can be sensed by cytoplasmic sensors such as IFIH1 and activate the interferon response.
While DICER1 homologs are required for the immune response in many eukaryotes including D. melanogaster (50), several lines of investigation indicate DICER1 may not be necessary for the interferon response in mammals (17). Li and Tainsky evaluated the effects of increased DICER1 in Li-Fraumeni fibroblasts with and without an intact interferon response and showed that overexpression of DICER1 can activate the interferon response (19). The difference in responses seen in fibroblasts in which DICER1 levels were increased and epithelial cells with reduced DICER1 could reflect cell-type specific differences or potentially opposing functional consequences of excess and deficient DICER1 activity in mammalian cells. DICER1 knockdown cell lines have increased susceptibility to influenza virus infection, implying DICER1 is necessary for recognizing viral dsRNA (51). However, the cancer cell lines we studied were not challenged by virus. In the absence of viral infection, pre-miRNAs have a stimulatory effect on the interferon response. The relationship between alterations in the miRNA processing machinery and the mammalian interferon response may point to a previously unrecognized role for DICER1 in tumorigenesis.
Supplementary Material
Acknowledgements
We thank Jayne Marasa in the High Throughput Core and the Molecular Imaging Center, Mallinckrodt Institute of Radiology, and BRIGHT Institute for assistance with imaging the wound healing assay (supported by P50 CA94056P30 CA091842 and an Anheuser-Busch/Emerson challenge gift).
We thank Sarah Spencer for assistance in preparing libraries and Francesco Vallania for help analyzing the RNA-Sequencing results. We thank Dr. Deborah Lenschow for assistance in interpreting the results of ISG experiments.
Grant Support Katherine Chiappinelli is supported by the Siteman Cancer Center Cancer Biology Pathway Fellowship and Molecular Oncology Training Grant T32 CA113275. The experimental work was supported by R01CA071754, P50CA134254 and a grant from the Foundation for Barnes-Jewish Hospital (PJG). Brian Haynes is supported by T32 HG000045 from the NHGRI.
Footnotes
The authors declare that there are no conflicts of interest.
References
- 1.Creutzberg CL, van Putten WL, Koper PC, Lybeert ML, Jobsen JJ, Warlam-Rodenhuis CC, et al. Surgery and postoperative radiotherapy versus surgery alone for patients with stage-1 endometrial carcinoma: multicentre randomised trial. PORTEC Study Group. Post Operative Radiation Therapy in Endometrial Carcinoma. Lancet. 2000;355(9213):1404–11. doi: 10.1016/s0140-6736(00)02139-5. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R, Ward E, Brawley O, Jemal A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J Clin. 2011;61(4):212–36. doi: 10.3322/caac.20121. [DOI] [PubMed] [Google Scholar]
- 3.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10(2):126–39. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 4.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 5.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19(1):92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359(25):2641–50. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kumar MS, Pester RE, Chen CY, Lane K, Chin C, Lu J, et al. Dicer1 functions as a haploinsufficient tumor suppressor. Genes Dev. 2009;23(23):2700–4. doi: 10.1101/gad.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hill DA, Ivanovich J, Priest JR, Gurnett CA, Dehner LP, Desruisseau D, et al. DICER1 mutations in familial pleuropulmonary blastoma. Science. 2009;325(5943):965. doi: 10.1126/science.1174334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Melo SA, Ropero S, Moutinho C, Aaltonen LA, Yamamoto H, Calin GA, et al. A TARBP2 mutation in human cancer impairs microRNA processing and DICER1 function. Nat Genet. 2009;41(3):365–70. doi: 10.1038/ng.317. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Melo SA, Moutinho C, Ropero S, Calin GA, Rossi S, Spizzo R, et al. A genetic defect in exportin-5 traps precursor microRNAs in the nucleus of cancer cells. Cancer Cell. 2010;18(4):303–15. doi: 10.1016/j.ccr.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Slade I, Bacchelli C, Davies H, Murray A, Abbaszadeh F, Hanks S, et al. DICER1 syndrome: clarifying the diagnosis, clinical features and management implications of a pleiotropic tumour predisposition syndrome. J Med Genet. 2011;48(4):273–8. doi: 10.1136/jmg.2010.083790. [DOI] [PubMed] [Google Scholar]
- 12.Heravi-Moussavi A, Anglesio MS, Cheng SW, Senz J, Yang W, Prentice L, et al. Recurrent Somatic DICER1 Mutations in Nonepithelial Ovarian Cancers. N Engl J Med. 2011 doi: 10.1056/NEJMoa1102903. [Epub ahead of print.] [DOI] [PubMed] [Google Scholar]
- 13.Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, et al. Dicer is essential for mouse development. Nat Genet. 2003;35(3):215–7. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 14.Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19(4):489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zighelboim I, Reinhart AJ, Gao F, Schmidt AP, Mutch DG, Thaker PH, et al. DICER1 expression and outcomes in endometrioid endometrial adenocarcinoma. Cancer. 2011;117(7):1446–53. doi: 10.1002/cncr.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–8. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q, Carmichael GG. Effects of length and location on the cellular response to double-stranded RNA. Microbiol Mol Biol Rev. 2004;68(3):432–52. doi: 10.1128/MMBR.68.3.432-452.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–86. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 19.Li Q, Tainsky MA. Higher miRNA tolerance in immortal Li-Fraumeni fibroblasts with abrogated interferon signaling pathway. Cancer Res. 2011;71(1):255–65. doi: 10.1158/0008-5472.CAN-10-1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caraglia M, Marra M, Tagliaferri P, Lamberts SW, Zappavigna S, Misso G, et al. Emerging strategies to strengthen the anti-tumour activity of type I interferons: overcoming survival pathways. Curr Cancer Drug Targets. 2009;9(5):690–704. doi: 10.2174/156800909789056980. [DOI] [PubMed] [Google Scholar]
- 21.Krejcova D, Prochazkova J, Kubala L, Pachernik J. Modulation of cell proliferation and differentiation of human lung carcinoma cells by the interferon-alpha. Gen Physiol Biophys. 2009;28(3):294–301. doi: 10.4149/gpb_2009_03_294. [DOI] [PubMed] [Google Scholar]
- 22.Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novakova Z, Hubackova S, Kosar M, Janderova-Rossmeislova L, Dobrovolna J, Vasicova P, et al. Cytokine expression and signaling in drug-induced cellular senescence. Oncogene. 2010;29(2):273–84. doi: 10.1038/onc.2009.318. [DOI] [PubMed] [Google Scholar]
- 24.Bektas N, Noetzel E, Veeck J, Press MF, Kristiansen G, Naami A, et al. The ubiquitin-like molecule interferon-stimulated gene 15 (ISG15) is a potential prognostic marker in human breast cancer. Breast Cancer Res. 2008;10(4):R58. doi: 10.1186/bcr2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee ES, Son DS, Kim SH, Lee J, Jo J, Han J, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14(22):7397–404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 26.Mizumoto Y, Kyo S, Ohno S, Hashimoto M, Nakamura M, Maida Y, et al. Creation of tumorigenic human endometrial epithelial cells with intact chromosomes by introducing defined genetic elements. Oncogene. 2006;25(41):5673–82. doi: 10.1038/sj.onc.1209575. [DOI] [PubMed] [Google Scholar]
- 27.Byron SA, Gartside MG, Wellens CL, Mallon MA, Keenan JB, Powell MA, et al. Inhibition of activated fibroblast growth factor receptor 2 in endometrial cancer cells induces cell death despite PTEN abrogation. Cancer Res. 2008;68(17):6902–7. doi: 10.1158/0008-5472.CAN-08-0770. [DOI] [PubMed] [Google Scholar]
- 28.Dewdney SB, Rimel BJ, Thaker PH, Thompson DM, Jr., Schmidt A, Huettner P, et al. Aberrant methylation of the X-linked ribosomal S6 kinase RPS6KA6 (RSK4) in endometrial cancers. Clin Cancer Res. 2011;17(8):2120–9. doi: 10.1158/1078-0432.CCR-10-2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramsingh G, Koboldt DC, Trissal M, Chiappinelli KB, Wylie T, Koul S, et al. Complete characterization of the microRNAome in a patient with acute myeloid leukemia. Blood. 2010;116(24):5316–26. doi: 10.1182/blood-2010-05-285395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9(4):493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124(6):1283–98. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- 32.Poliseno L, Salmena L, Zhang J, Carver B, Haveman WJ, Pandolfi PP. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiappinelli KB, Rimel BJ, Massad LS, Goodfellow PJ. Infrequent methylation of the DUSP6 phosphatase in endometrial cancer. Gynecol Oncol. 2010;119(1):146–50. doi: 10.1016/j.ygyno.2010.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25(9):1105–11. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robertson B, Dalby AB, Karpilow J, Khvorova A, Leake D, Vermeulen A. Specificity and functionality of microRNA inhibitors. Silence. 2010;1(1):10. doi: 10.1186/1758-907X-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martello G, Rosato A, Ferrari F, Manfrin A, Cordenonsi M, Dupont S, et al. A MicroRNA targeting dicer for metastasis control. Cell. 2010;141(7):1195–207. doi: 10.1016/j.cell.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 38.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 39.Gondai T, Yamaguchi K, Miyano-Kurosaki N, Habu Y, Takaku H. Short-hairpin RNAs synthesized by T7 phage polymerase do not induce interferon. Nucleic Acids Res. 2008;36(3):e18. doi: 10.1093/nar/gkm1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.The RNAi Consortium (TRC) The RNAi Consortium shRNA Library, Date and Figures: Interferon response genes are not induced following transduction with TRC Lentiviral shRNAs. MA Broad Institute; Cambridge: [July 14, 2006]. 2010. Available from: http://www.broadinstitute.org/science/projects/rnai-consortium/rnai-consortium-shrna-library. [Google Scholar]
- 41.Chelbi-Alix MK, Boissard C, Sripati CE, Rosselin G, Thang MN. VIP induces in HT-29 cells 2′5′oligoadenylate synthetase and antiviral state via interferon beta/alpha synthesis. Peptides. 1991;12(5):1085–93. doi: 10.1016/0196-9781(91)90064-v. [DOI] [PubMed] [Google Scholar]
- 42.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120(5):635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 43.Kumar MS, Erkeland SJ, Pester RE, Chen CY, Ebert MS, Sharp PA, et al. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc Natl Acad Sci U S A. 2008;105(10):3903–8. doi: 10.1073/pnas.0712321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar M, Carmichael GG. Antisense RNA: function and fate of duplex RNA in cells of higher eukaryotes. Microbiol Mol Biol Rev. 1998;62(4):1415–34. doi: 10.1128/mmbr.62.4.1415-1434.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang S, Tutton S, Pierce E, Yoon K. Specific double-stranded RNA interference in undifferentiated mouse embryonic stem cells. Mol Cell Biol. 2001;21(22):7807–16. doi: 10.1128/MCB.21.22.7807-7816.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sledz CA, Holko M, de Veer MJ, Silverman RH, Williams BR. Activation of the interferon system by short-interfering RNAs. Nat Cell Biol. 2003;5(9):834–9. doi: 10.1038/ncb1038. [DOI] [PubMed] [Google Scholar]
- 47.Choudhary S, Lee HC, Maiti M, He Q, Cheng P, Liu Q, et al. A double-stranded-RNA response program important for RNA interference efficiency. Mol Cell Biol. 2007;27(11):3995–4005. doi: 10.1128/MCB.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dang LT, Kondo H, Aoki T, Hirono I. Engineered virus-encoded pre-microRNA (pre-miRNA) induces sequence-specific antiviral response in addition to nonspecific immunity in a fish cell line: convergence of RNAi-related pathways and IFN-related pathways in antiviral response. Antiviral Res. 2008;80(3):316–23. doi: 10.1016/j.antiviral.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 49.Gantier MP, Baugh JA, Donnelly SC. Nuclear transcription of long hairpin RNA triggers innate immune responses. J Interferon Cytokine Res. 2007;27(9):789–97. doi: 10.1089/jir.2006.0152. [DOI] [PubMed] [Google Scholar]
- 50.Ding SW. RNA-based antiviral immunity. Nat Rev Immunol. 2010;10(9):632–44. doi: 10.1038/nri2824. [DOI] [PubMed] [Google Scholar]
- 51.Matskevich AA, Moelling K. Dicer is involved in protection against influenza A virus infection. J Gen Virol. 2007;88(Pt 10):2627–35. doi: 10.1099/vir.0.83103-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.