Abstract
The heparan sulfate 6-O-endosulfatase (SULF2) promotes growth and metastasis of solid tumors. We recently identified that cytosine methylation of the SULF2 promoter is associated with better survival of resected lung adenocarcinoma patients and now also demonstrate a marginal improvement in survival of advanced non-small cell lung cancer (NSCLC) patients receiving standard chemotherapy (HR = 0.63, p = 0.07). Subsequent studies focused on investigating the effect of methylation on SULF2 expression and its genome-wide impact. The genes and pathways modulated by epigenetic inactivation of SULF2 and the effects on sensitivity to chemotherapy were characterized in vitro and in vivo. Silencing SULF2 through siRNA or methylation primarily increased expression of interferon-inducible genes including ISG15, a marker for increased sensitivity to topoisomerase-1 inhibitors such as camptothecin. NSCLC cell lines with methylated SULF2 (SULF2M) express 60-fold higher ISG15 compared to SULF2 unmethylated (SULF2U) NSCLC cell lines and normal human bronchial epithelial cells. In vitro, SULF2M and high ISG15 (ISG15H) expressing NSCLC cell lines were 134-fold more sensitive to camptothecin than SULF2U and low ISG15 (ISG15L) expressing cell lines. Topotecan, a soluble analogue of camptothecin and FDA approved anti-cancer drug, dramatically arrested the growth of SULF2M-ISG15H, but not SULF2U-ISG15L lung tumors in nude mice (p < 0.002). Similarly, high ISG15 expression that is comparable to the topotecan sensitive NSCLC cell lines was found in tumors from 25% of NSCLC patients compared to normal lung indicating a potential to identify and target the most sensitive NSCLC subpopulation for personalized topotecan therapy.
Keywords: NSCLC, Camptothecin, SULF-2, Oncogene, Topotecan
Introduction
Heparan sulfate proteoglycans (HSPGs) are ubiquitously distributed on cell membrane and extracellular matrix (ECM) and play a critical role in the interaction between cells and their environment (Bishop et al., 2007). Cell surface HSPGs directly interact with the heparan sulfate (HS) domain of cell adhesion molecules in the ECM (e.g. fibronectin) and direct cell attachment and migration. Membrane-spanning HSPGs (e.g. syndicans) are composed of a transmembrane core protein covalently linked with an extracellular HS chain and serve as receptors or co-receptors for various signaling molecules. The plethora of ligands interacting with HSPGs and implicated in carcinogenesis include growth factors (GF) and cytokines such as fibroblast GF (FGF), vascular endothelial GF (VEGF), and Wnt proteins (Lai et al., 2010; Uchimura et al., 2006). The specificity of HSPG-ligand interaction is determined by the composition and sulfation pattern of the HS polysaccharide chain. HS sulfation is dynamically regulated, tissue specific, and precise positioning of the 6-O sulfates in particular is important for ligand interaction (Merry et al., 1999; Sugaya et al., 2008).
Two extracellular enzymes, SULF1 and SULF2, regulate the distribution of 6‐O sulfates in humans by selectively removing sulfate groups from the 6-O position (Morimoto-Tomita et al., 2002). Abnormalities in the expression of these 6-O-endosulfatases have been associated with various malignancies. SULF2 promotes the release of growth and angiogenic factors such as FGF-I, FGF-2, VEGF, and DSF-I from HS and is over expressed in some lung, breast, brain, and liver cancers (Johansson et al., 2005; Lai et al., 2008; Lemjabbar-Alaoui et al., 2010; Morimoto-Tomita et al., 2005; Uchimura et al., 2006). In hepatocellular carcinoma SULF2 expression increases with disease progression and patients with higher expression have lower survival and a more rapid rate of tumor recurrence after surgery (Lai et al., 2008). Inactivation of SULF2 using short hairpin RNA reduces cell proliferation and migration (Lemjabbar-Alaoui et al., 2010).
We recently identified that promoter CpG island methylation of SULF2, the gene encoding SULF2 endosulfatase, is highly prevalent in resected lung adenocarcinomas and is significantly associated with better survival (Tessema et al., 2009). The median overall survival (OS) of patients with methylated SULF2 (SULF2M) was 62.8 months compared to 35.1 months in SULF2 unmethylated (SULF2U) patients with a hazard ratio (HR) of 0.41. A more dramatic (> 4-fold) increase in median OS from 8.5 months in SULF2U to 36.2 months in SULF2M patients was found in resected advanced stage (Stage II-IV) patients (HR = 0.23; p < 0.001) (Tessema et al., 2009). However, the prognostic importance of SULF2 methylation for the majority of non-small cell lung cancer (NSCLC) patients (> 80%) who have advanced disease and treatment relies on chemo- and radiation-therapy is unknown.
The objective of this study was to determine the prevalence and prognostic value of SULF2 methylation in unresectable NSCLC patients receiving chemo- and radiation-therapy. The effect of SULF2 silencing on cancer phenotypes were characterized in vitro. Genome-wide changes in gene expression and the pathways regulated by epigenetic inactivation of SULF2 were also identified through a whole-genome screen. Finally the importance of SULF2 methylation and the genes it regulates as biomarkers for selecting the most effective chemotherapeutic agent was determined through in vitro and in vivo studies, and the potential use of identified markers for patient selection and personalized therapy was discussed.
Results
SULF2 methylation is common in lung cancer and silences gene expression
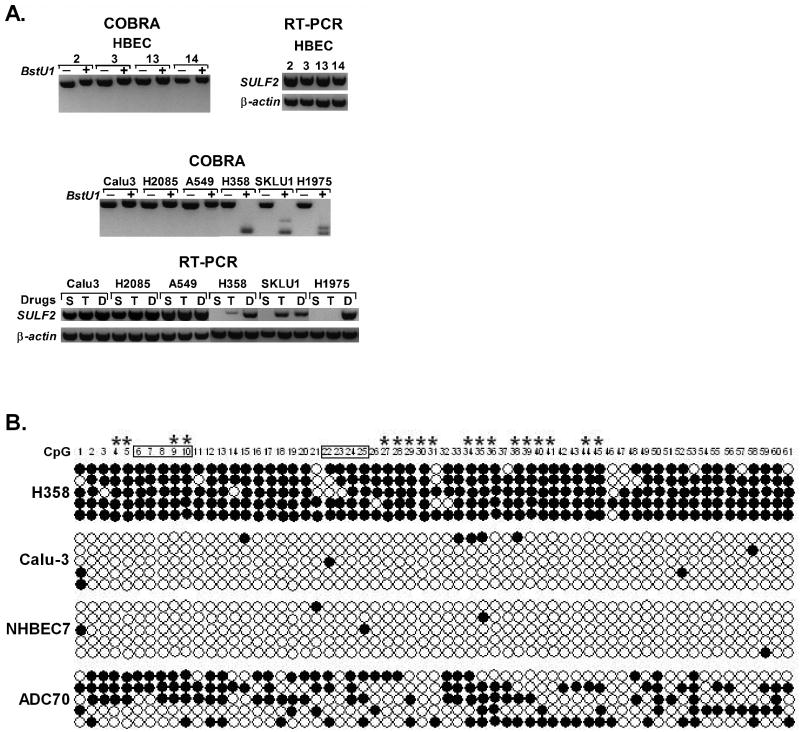
We recently reported that SULF2 methylation is highly prevalent and strongly associated with improved survival of resected lung adenocarcinoma patients independent of tumor stage (Tessema et al., 2009). To understand the impact of this epigenetic modification in lung cancer, we first evaluated its role on gene expression using normal human bronchial epithelial cells (HBEC) and NSCLC cell lines. SULF2 is unmethylated (SULF2U) and readily expressed in all HBEC and some NSCLC cell lines such as Calu-3, H2085, and A549 (Figure 1A). In contrast, SULF2 expression was completely silenced in NSCLC cell lines with densely methylated SULF2 promoter (SULF2M) such as H358, SKLU1 and H1975. Transcription could be restored in SULF2M cell lines primarily through treatment with the demethylating agent DAC and partial re-expression was induced in some TSA (a histone deacetylase inhibitor) treated cell lines. Among the NSCLC cell lines used in this study, expression of SULF2 in Calu-3, A549 and Calu-6, and its absence in H1975, H358, and H1299 has been recently demonstrated at transcript and protein levels (Lemjabbar-Alaoui et al., 2010). Our results confirmed these findings and revealed for the first time that promoter CpG island methylation is responsible for SULF2 silencing in these cell lines. Overall, SULF2 was unmethylated in NHBEC obtained from bronchoscopy of cancer free smokers (0/20) and HBEC (0/5) but methylated in 44% (8/18) of NSCLC cell lines. Similarly, primary lung tumors obtained from two independent groups of NSCLC patients, ECOG3598 and PCC, showed 60% (47/78) and 43% (25/58) methylation, respectively. The density and distribution of methylation across SULF2 promoter CpG island was assessed for selected samples representing the different sample types and degrees of methylation using bisulfite sequencing. The results were consistent with MSP and COBRA findings, and revealed that the methylation pattern across the promoter CpG island of a given sample was mostly similar (Figure 1B).
Figure 1. Methylation of SULF2 promoter CpG island silences gene expression.
(A) COBRA results revealed that SULF2 is unmethylated (not digested by the BstU1 enzyme) in human bronchial epithelial cells (HBEC) (top left) and some lung cancer cell lines (middle). In contrast, complete methylation of SULF2 (shown by the completely digested bands) was found in some NSCLC cell lines such as H358, SKLU1, and H1975. In HBEC and lung cancer cell lines where SULF2 is unmethylated, the gene is readily expressed in sham treated (S) cells. In contrast, SULF2 expression was completely silenced in cell lines with methylated promoter and expression could be primarily restored by DAC (D) treatment. Partial restoration of expression was seen in some cell lines (e.g. H358 and SKLU1) after TSA treatment indicating epigenetic regulation through both methylation and histone modification. (B) For selected samples the degree and distribution of methylation at 61 CpGs (numbered 1–61) across SULF2 promoter CpG island was evaluated using bisulfite sequencing. Five clones were sequenced per sample and methylation is marked by filled (black) circle. CpGs within the primer binding sites for MSP (boxed numbers) and BstU1 enzyme recognition sites for COBRA (asterisk) are also indicated.
SULF2 methylation is associated with improved overall survival of NSCLC
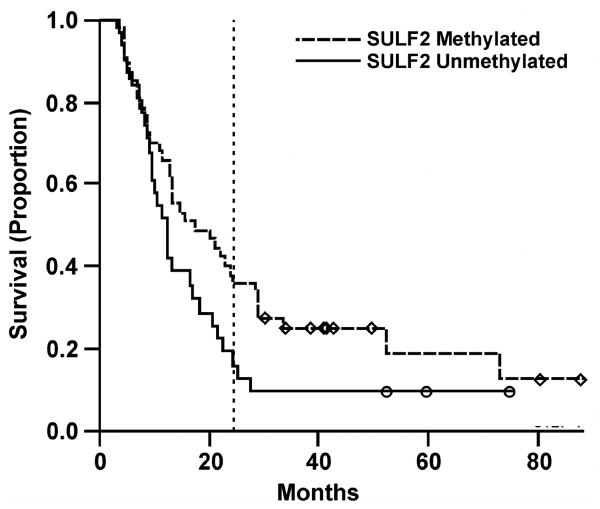
Survival data was available for ECOG3598 patients. Consistent with our previous observation that was obtained from an independent group of resected lung adenocarcinoma patients (Tessema et al., 2009), the overall survival (OS) of unresectable NSCLC patients with SULF2M tumor was marginally better than patients with SULF2U tumor independent of stage (p = 0.07). The median OS of SULF2M cases (n = 47) was 17.2 months (95% confidence interval [CI], 12.7 – 29.0) compared to 12.2 months (95% CI, 9.6 – 20.8) for SULF2U patients (n = 31) with a HR = 0.63 (95% CI, 0.39 – 1.04) (Figure 2). Adjustment for stage (IIIa vs. IIIb), histology (squamous vs. nonsquamous), performance status (0 vs. 1), gender, and weight loss (< 5% vs. other) have very little effect on the prognostic value of SULF2M (HR = 0.69, 95% CI, 0.41 – 1.15). While OS was marginally significant, more SULF2M patients (38.3%, 95% CI, 26.6 – 55.1%) survived beyond two years compared to SULF2U patients (19.4%, 95% CI, 9.4 – 39.7%; Figure 2). Although the median progression free survival (PFS) of 7.8 months for SULF2M (95% CI, 6.6 – 10.7) was slightly better than 7.2 months seen for SULF2U patients (95% CI, 5.1 – 11.4), it did not reach statistical significance (logrank test, p = 0.27; and HR = 0.77, 0.48 –1.23). The response rate (complete or partial remission, CR/PR) and disease control rate (stable disease/CR/PR) among SULF2M and SULF2U patients were also similar suggesting that SULF2 methylation does not directly affect sensitivity of ECOG3598 participants to carboplatin/paclitaxel combination therapy. Consistent with this premise, the IC50 of cisplatin, carboplatin, or paclitaxel for NSCLC cell lines in vitro was not associated with SULF2 methylation status (Table S1).
Figure 2. SULF2 methylation is marginally associated with better overall survival of NSCLC patients receiving chemotherapy for advanced disease.
Two-year overall survival is marked by dotted lines.
Epigenetic silencing of SULF2 activates interferon (INF)-inducible genes
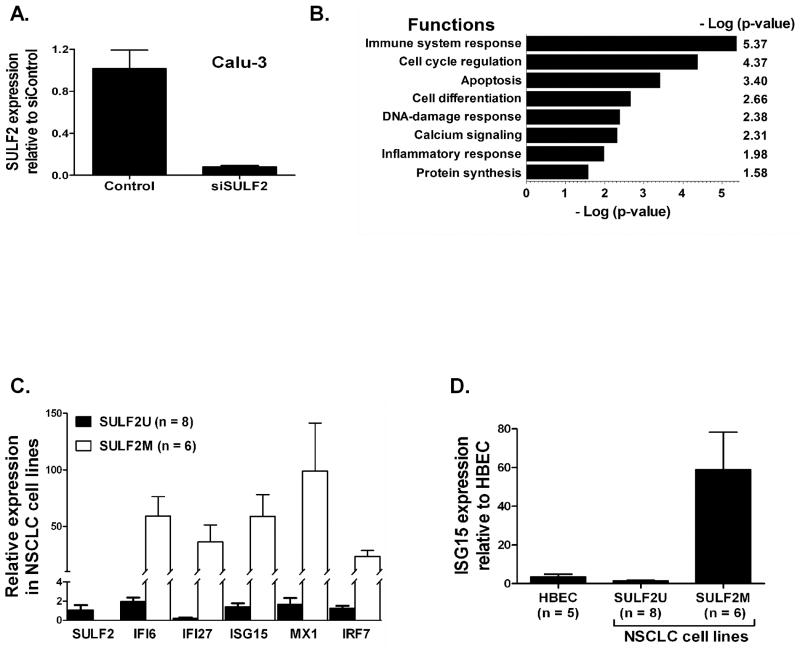
Calu-3 and A549 cell lines (where SULF2 is unmethylated and expressed) were transfected with control (siControl) or SULF2 specific siRNA (siSULF2) to evaluate the effect of epigenetic inactivation of SULF2 on cancer cell phenotypes (cell proliferation, survival, and migration). Compared to siControl, siSULF2 reduced SULF2 expression in Calu-3 cells by 80% (Figure 3A, p < 0.02) and significantly reduced wound closure (Figure S1A-B, p < 0.005). Similarly, slower wound closure was seen in A549 cells transfected with siSULF2 than siControl (Figure S1C, p < 0.005). However, SULF2 knockdown in both cell lines did not significantly change cell proliferation and survival (not shown).
Figure 3. Epigenetic silencing of SULF2 increases interferon-inducible genes and modulate pathways associated with carcinogenesis.
(A) Calu-3 cells in which SULF2 is unmethylated and expressed at similar level to HBEC were transfected with control (siControl) or SULF2 specific siRNA (siSULF2). Compared to siControl transfected cells, SULF2 expression was reduced by 80% in siSULF2 transfected cells (p < 0.02). (B) Comparison of genome-wide changes in gene expression between siControl and siSULF2 cells revealed that SULF2 knockdown primarily activated the immune response, cell proliferation, differentiation, and DNA-damage responses pathways, log values are at base 10. (C) The mean expression of SULF2 in lung cancer cell lines with methylated SULF2 (SULF2M) was reduced 845-fold compared to cell lines with unmethylated SULF2 (SULF2U). In contrast, the mean expression of some INF-inducible genes was increased 25 – 125-fold in SULF2M cell lines (p < 0.01 each). (D) SULF2M NSCLC cell lines on average express 60-fold higher ISG15 compared to SULF2U NSCLC cell lines and HBEC. NSCLC cell lines used for Figures 3C and 3D are: SULF2U (H23, H1435, H1568, H2085, H2228, Calu-3, Calu-6, and A549) and SULF2M (H1993, H358, SKLU1, H1299, H1975, and HCC827).
The genome-wide impact of epigenetic silencing of SULF2 was compared between siControl and siSULF2 transfected cells using a whole-genome transcriptome array and the pathways modulated were identified using the MetaCore pathway analysis software from GeneGo Inc (St. Joseph, MI). Overall, SULF2 knockdown altered the expression of 286 genes by ≥ 1.5-fold and primarily affected (p < 0.005) the immune response, cell cycle regulation, apoptosis, cell differentiation, and DNA-damage response pathways (Figure 3B). The array data for SULF2 expression in siSULF2 transfected cells (0.23, Table S2) also confirmed the ∼80% knockdown of this gene seen from TaqMan assays (Figure 3A) and served as an internal control. Interestingly, one third (11/33) of the genes whose expression was increased by ≥ 2-fold and have known or predicted function were INF-inducible genes (Table 1). However, none of the INF-family genes were transcriptionally activated, suggesting an indirect regulation. Among the INF-inducible genes, expression of the top four (IFI6, IFI27, ISG15, and MX1) and the upstream regulator of INF-pathway (IRF7) was measured using quantitative TaqMan assays, normalized to β‐actin, quantified relative to expression in NHBEC as described (Livak and Schmittgen, 2001) and compared between SULF2U (n = 8) and SULF2M (n = 6) lung cancer cell lines (Figure 3C). The mean expression of these genes in SULF2M cancer cell lines was increased by 25- to 125-fold (p < 0.01 each). In contrast, consistent with the dense methylation throughout the promoter CpG island and complete silencing of SULF2 observed in these cell lines (Figure 1A and 1B), the mean SULF2 expression was reduced by 845-fold (Figure 3C).
Table 1. Genes with 2-fold or higher increased expression as a result of SULF2 knockdown.
| No. | Genes | Fold Δ | Function |
|---|---|---|---|
| 1 | KIAA1530 | 26.9 | Hypothetical protein coding gene, unknown function |
| 2 | C14orf118 | 21.1 | Hypothetical protein coding gene, unknown function |
| 3 | X58747 | 20.7 | T cell receptor V alpha gene segment, may function as splicing factor |
| 4 | ZRSR2 | 16.1 | Splicing factor, required for recognition of the 3′ splice site of pre-mRNA |
| 5 | A_24_P731000 | 10.2 | IGVK2-23, Ig kappa 2-23 pseudo-gene |
| 6 | UBE2B | 8.3 | An E2 ubiquitin-conjugating enzyme required for DNA damage repair |
| 7 | IFI6 (G1P3) | 7.7 | INF-α inducible anti-apoptotic protein |
| 8 | IFI27 (ISG12) | 7.3 | INF-α inducible pro-apoptotic protein, increases sensitivity to etoposide |
| 9 | ISG15 | 7.1 | INF-α and β inducible ubiquitin-like protein, has antiviral properties and inhibits ubiqutination and proteasomal degradation of proteins. |
| 10 | ZNF208 | 5.7 | Zink finger, may serve as a transcription factor |
| 11 | HIRA | 4.4 | Repress histone gene transcription during the cell cycle, formation of senescence-associated heterochromatin foci and efficient senescence-associated cell cycle exit. |
| 12 | CLEC3B | 3.9 | Involved in the packaging of molecules destined for exocytosis |
| 13 | MX1 | 3.4 | INF-inducible member of dynamin and large GTPases family, epigenetically inactivated in head and neck cancer. |
| 14 | MALT1 | 3.2 | May play a role in NF-kappa B activation and has ubiquitin ligase activity |
| 15 | CXCL1 | 3.0 | A chemokine involved in the trafficking of various types of leukocytes |
| 16 | IFI44 | 2.9 | INF-α inducible protein, suppresses cell proliferation |
| 17 | LMNB1 | 2.9 | Nuclear lamina involved in nuclear stability, chromatin structure and gene expression |
| 18 | LRWD1 | 2.9 | Modulates chromatin association of origin recognition complex |
| 19 | IFIT1 (ISG56) | 2.6 | INF-α inducible, inhibits DNA replication of virus infected cells |
| 20 | DLSTP | 2.5 | Dihydrolipoamide S-succinyltransferase pseudogene 1 |
| 21 | TFPI | 2.5 | Inhibits coagulation factors |
| 22 | SRPR | 2.5 | Molecular trafficking of vesicles to endoplasmic reticulum |
| 23 | SOD2 | 2.5 | An enzyme that converts superoxide radicals to H2O2 and then to H2O |
| 24 | RELB | 2.4 | Component of NF-Kappa B |
| 25 | IRF7 | 2.4 | INF regulatory factor, increases INF expression |
| 26 | BE672039 | 2.2 | Unknown function |
| 27 | DUSP6 | 2.1 | Negatively regulate MAPK, specific for ERK2 |
| 28 | NFKB2 | 2.1 | NF-kappaB activaton, has ubiquitin ligase activity |
| 29 | A_32_P140501 | 2.1 | Unknown function |
| 30 | FZD5 | 2.0 | WNT5A receptor, regulates inflammatory responses including INF-γ production |
| 31 | SAA1 | 2.0 | Acute phase protein induced during inflammation and tissue injury |
| 32 | DDAH1 | 2.0 | Inhibitor of nitric oxides |
| 33 | IFI35 | 2.0 | INF-α inducible protein involved in apoptosis. |
| 34 | C15orf48 | 2.0 | A candidate tumor suppressor gene in esophageal and cervical cancer |
| 35 | AKT3 | 2.0 | Serine/threonine protein kinase functions in cell survival |
| 36 | PLSCR1 | 2.0 | INF-stimulated gene, interacts with EGFR in plasma membrane lipid rafts |
| 37 | IFI30 | 2.0 | INF-α and β inducible, help recognition of tumor cells by immune system |
| 38 | IFITM1 | 2.0 | INF-α inducible, down-regulated in cervical cancer |
SULF2 methylation sensitizes NSCLC cells to camptothecin via ISG15
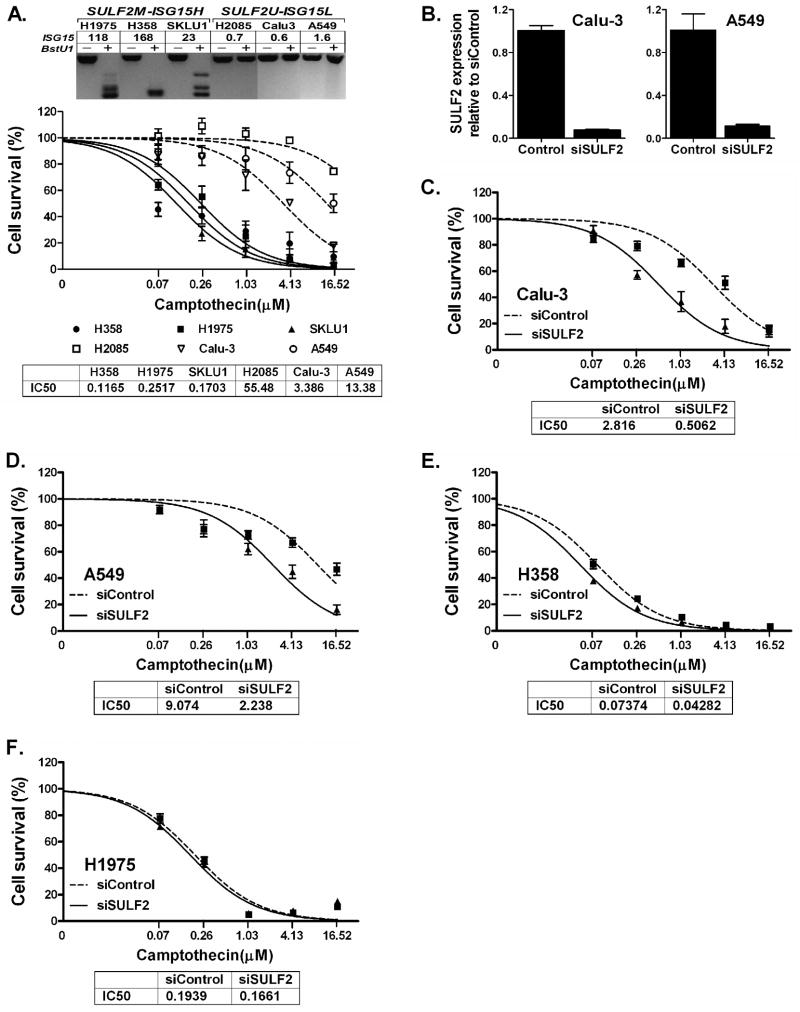
Previous studies have demonstrated that INF-inducible regulator of ubiquitination (ISG15) interferes with the ubiquitin/26S proteasome pathway and increases the sensitivity of breast cancer cells to camptothecin (CPT), a topoisomerase-I (TOPO1) inhibitor (Desai et al., 2006; Desai et al., 2008; Lin et al., 2008). Our data revealed that SULF2U NSCLC cell lines (n = 8) express similar level of ISG15 to HBEC (n = 5). In contrast, SULF2M NSCLC cell lines (n = 6) was on average express 60-fold higher ISG15 than SULF2U cells (Figure 3D). The strong association between SULF2M and high ISG15 (ISG15H) phenotypes prompted us to test if SULF2M and ISG15H NSCLC cells could also show increased sensitivity to CPT and its analogues. The average dose of CPT required to kill 50% (IC50) of SULF2M-ISG15H cell lines H358, SKLU1 and H1975 was 0.12, 0.17, and 0.25 μM, respectively. In contrast, the IC50 for SULF2U-ISG15low (ISG15L) cell lines Calu-3, A549, and H2085 was 3.39, 13.38, and 55.48 μM, respectively (Figure 4A). This indicates that SULF2M-ISG15H cell lines are on average 134-fold (IC50 = 0.18 vs. 24.08 μM) more sensitive to CPT than SULF2U-ISG15L NSCLC cell lines. Similarly, knockdown of 90% and 85% of SULF2 in Calu-3 and A549 increased the sensitivity of these cells to CPT by 5.6- and 4.1-fold, respectively (Figure 4B-D). In contrast, SULF2 expression remained undetectable in both siControl and siSULF2 transfected H358 and H1975 (not shown) and the sensitivity of both cell lines to CPT did not change substantially following transfection (Figure 4E and F). Whereas knockdown of SULF2 in Calu-3 and A549 increased ISG15 expression by 6.7- and 3.1-fold, respectively, no significant difference was found between siSULF2 or siControl transfected H358 and H1975 cells (Figure S2).
Figure 4. SULF2 methylated and high ISG15 expressing lung cancer cells are more sensitive to camptothecin in vitro.
(A) Top panel: SULF2 methylation and level of ISG15 expression (fold changes relative to expression in NHBEC) are shown for six NSCLC cell lines used for drug sensitivity assays. Lower Panel: the average dose of camptothecin (CPT) required to kill 50% (IC50) of SULF2M-ISG15H cell lines, 0.18 μM (H358 = 0.12 μM, SKLU1 = 0.17 μM, and H1975 = 0.25 μM) was more than 134-fold lower than the 24.08 μM required for SULF2U-ISG15L cell lines (Calu-3 = 3.39 μM, A549 = 13.38 μM, and H2085 = 55.48 μM). All assays were done in triplicate and results are described as mean ± SEM. Compared to siControl, transfection of Calu-3 and A549 cells with siSULF2 reduced SULF2 expression by 90% and 85% (B) and lead to 5.6-fold (C) and 4.1-fold (D) increased sensitivity of these cells to CPT, respectively. In contrast, similar transfection of H358 (E) and H1975 (F) cells with these siRNAs did not significantly change the sensitivity of these cells to CPT.
SULF2M-ISG15H tumors are sensitive to topotecan in vivo
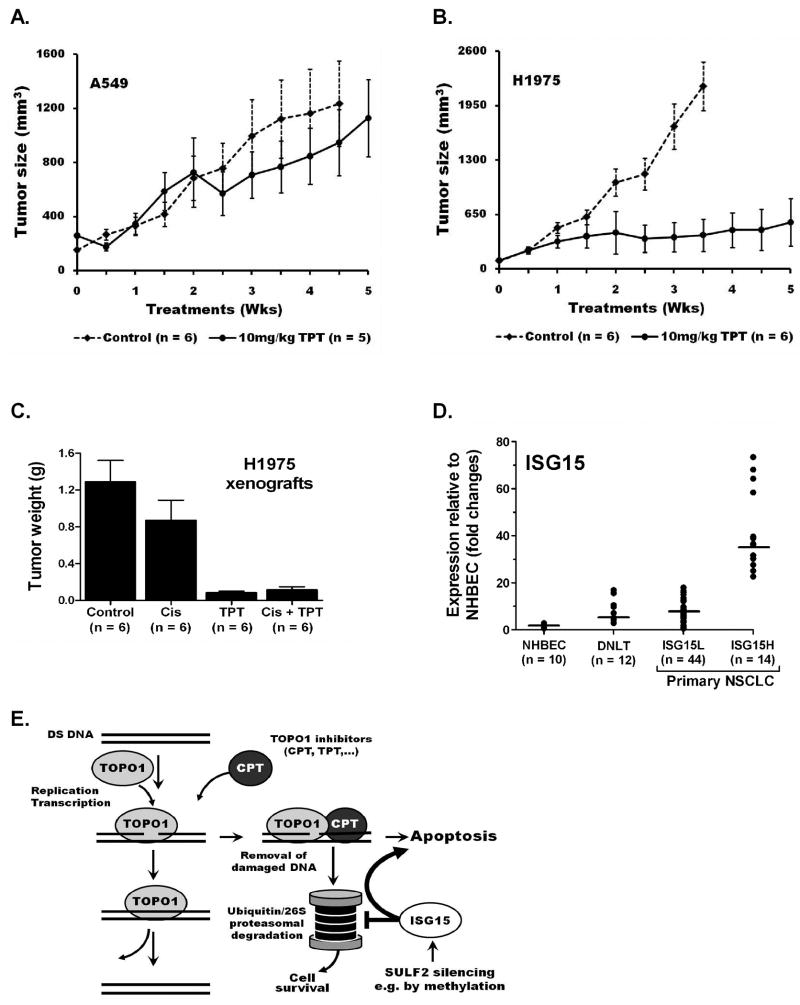
The in vivo effectiveness of TOPO1 inhibitors to suppress the growth of human NSCLC xenografts was evaluated using the soluble CPT analogue, topotecan (TPT). The SULF2M-ISG15H cell line H1975 that is resistant to both platinum therapy and tyrosine kinase inhibitors (due to an activating L858R and drug resistance T790M EGFR mutations, respectively) and the K‐ras mutant SULF2U-ISG15L A549 were selected for this study due to the commonality of these mutations in primary NSCLC (Helfrich et al., 2006; Kalikaki et al., 2008). Once-a-week 10 mg/kg TPT therapy of nude mice carrying subcutaneous xenografts of these cell lines was well tolerated and dramatically arrested the growth of H1975 tumors (p < 0.002), but had little effect (p = 0.58) on A549 tumors (Figure 5A and B). A separate experiment comparing the efficacy of TPT, cisplatin, and TPT-cisplatin combination therapy revealed that cisplatin has marginal impact (p = 0.054) on reducing the growth of H1975 xenografts. In contrast, TPT alone, or in combination with cisplatin significantly arrested the growth of H1975 tumors (p = 0.001). TPT therapy alone was superior to cisplatin and adding cisplatin to TPT treatments had no additional benefit in suppressing H1975 xenografts (Figure 5C).
Figure 5. High ISG15 expressing tumors are sensitive to topotecan in vivo.
Once a week 10 mg/kg topotecan (TPT) therapy had (A) no effect (p = 0.58) on the growth of tumors derived from A549 (SULF2U-ISG15L cell line) but (B) dramatically arrested the growth of tumors derived from H1975 (SULF2M-ISG15H cell line) in nude mice (p < 0.002). (C) TPT is more effective than cisplatin in suppressing SULF2M-ISG15H NSCLC xenografts. Once a week 3mg/kg cisplatin (Cis) treatment of nude mice had moderate (p = 0.054) effect on the growth of tumors derived from H1975 (SULF2M-ISG15H). In contrast, once a week 10 mg/kg TPT strongly suppressed the growth of H1975 derived tumors (p < 0.001) and adding 3 mg/kg Cis two days after each TPT therapy had no additional benefit (p = 0.85). The average tumor weight in TPT treated mice (0.085g) was 15× smaller than untreated tumors (1.289g) and 10× smaller than Cis treated tumors (0.872g). (D) ISG15 expression is elevated in a subset of primary NSCLC. Distant normal lung tissue (DNLT) obtained from NSCLC patients on average express 7.1-fold (2.9 – 15.6) higher ISG15 than NHBEC (0.37 – 1.64) and showed more variation between samples. Lines indicate median expression. The variation was much larger (0.5 – 73-fold) among primary tumors. NSCLC patients can be classified into two distinct groups, ISG15L and ISG15H, based on the level of ISG15 expression. Tumors from 75% of NSCLC patients express lower levels of ISG15 (ISG15L) that was similar to DNLT (mean 7.7- vs. 7.1-fold and median 7.5- vs. 5.2-fold). In contrast, tumors from 25% of NSCLC patients express much higher ISG15 (mean 42-fold and median 36-fold) that was comparable to the TPT sensitive NSCLC cell lines, representing an ISG15H subpopulation that could benefit from personalized topotecan therapy. (E) A schematic representation of the mechanism by which SULF2 methylation and the associated high ISG15 expression could sensitize cancer cells to topoismerase-1 inhibitors (detailed description is incorporated in the discussion).
ISG15 expression is elevated in a subset of primary lung tumors
RNA for gene expression analysis was available for 10 NHBEC, 58 tumors and 12 distant normal lung tissues (DNLT) obtained from NSCLC patients. To address the relationship between EGFR mutation and high ISG15 expression observed in some lung adenocarcinoma cell lines (e.g. H1975 and HCC827), 29 EGFR wild-type (EGFRwt) and 29 mutant (EGFRmut) primary tumors were analyzed. The prevalence for SULF2 methylation was similar between EGFRwt (13/29) and EGFRmut (12/29) tumors. Gene expression assays revealed that the ISG15 level in DNLT was on average 7.1-fold (2.9 – 15.6) higher and showed more variation between samples than in NHBEC (0.37 – 1.64), likely due to different levels of expression within the multiple cell types comprising the lung parenchyma (Figure 5D). The mean expression of ISG15 in the primary tumors was 15.6 and 2.2-fold higher than NHBEC and DNLT, respectively. A much larger variation in ISG15 expression ranging from 0.5 – 73-fold was also found between tumors. Expression did not differ by EGFR mutation status and surprisingly, was not significantly associated with SULF2 methylation indicating other regulators of ISG15 expression [e.g. cytokines from infiltrating inflammatory cells (Andersen et al., 2006)] potentially contribute to the discrepancy between the pure cell population (NHBEC, HBEC, and NSCLC cell lines) and primary tissue (DNLC and tumors from patients). However, the level of ISG15 expression within the tumors clearly separates NSCLC patients into two distinct subpopulations, ISG15L and ISG15H (Figure 5A). Tumors from 75% of NSCLC patients (44/58) express lower levels of ISG15 (ISG15L) (mean 7.7-fold and median 7.5-fold relative to NHBEC) that was similar to DNLT (mean 7.1-fold and median 5.2-fold). In contrast, tumors from 25% of NSCLC patients (14/58) express much higher ISG15 (mean 42-fold and median 36-fold) that was comparable to the TPT sensitive NSCLC cell lines, representing an ISG15H subpopulation that could benefit from personalized therapy with TOPO1 inhibitors approved for cancer therapy such as topotecan (Hycamtin®, GlaxoSmithKline) and Irinotecan (Camptosar®, Pfizer).
Discussion
These studies confirm that SULF2 methylation is a common abnormality and a prognostic biomarker for better survival of lung cancer patients. In addition, epigenetic silencing of SULF2 via promoter methylation or siRNA in cancer cell lines significantly increased expression of multiple INF-inducible genes, one of which, ISG15 sensitizes NSCLC cells to the topoisomerase-1 inhibitors, CPT and TPT. In nude mice, TPT therapy is associated with a dramatic reduction in the growth of tumors derived from the SULF2M-ISG15H line H1975 that harbors EGFR mutations rendering it resistant to both platinum-based and tyrosine kinase inhibitory drugs (Kobayashi et al., 2005; Pao et al., 2005). Importantly, although SULF2 methylation was not strongly associated with increased expression of ISG15 in primary tumors, 25% of NSCLC patients had tumors with elevated expression of this gene at levels comparable to cell lines sensitive to topoisomerase inhibitors, suggesting that this subset of patients could benefit from personalized TPT therapy.
The improved survival of SULF2M patients in ECOG3598 did not appear to be mediated through increased sensitivity to carboplatin or paclitaxel. Rather, epigenetic suppression of the oncogenic pathways triggered through this enzyme is the likely cause. This assessment is based on multiple findings. First, both the response and disease control rates among NSCLC patients receiving a combination of carboplatin and paclitaxel therapy as well as the in vitro sensitivity of lung cancer cell lines to these drugs were not associated with SULF2 methylation status. Second, as shown in our previous study, SULF2 methylation was also strongly associated with survival of lung adenocarcinoma patients after surgery (Tessema et al., 2009). Finally, higher SULF2 expression in hepatocellular carcinoma is also associated with worse prognosis and higher recurrence after surgery (Lai et al., 2008). SULF2 expression in tumors exhibits the behavior of an oncogene by mobilizing heparin/heparan sulfate bound growth factors and promoting pathways such as FGF and Wnt signaling leading to increased tumor growth and angiogenesis (Lai et al., 2008; Lemjabbar-Alaoui et al., 2010; Uchimura et al., 2006). In vitro inactivation of SULF2 suppresses these pathways and decreases growth of lung, liver, breast, and pancreatic tumors (Lai et al., 2008; Lemjabbar-Alaoui et al., 2010; Morimoto-Tomita et al., 2005; Nawroth et al., 2007), effects that should reduce the aggressiveness of tumors a supposition supported by the more significant improvement in overall survival seen in patients with resected compared to unresected lung cancer. Currently, the mechanism or selective advantage of silencing SULF2 in cancer cells remains to be identified.
Epigenetic inactivation of SULF2, as demonstrated by the siRNA knockdown and genome-wide array, primarily activated INF-inducible genes. Consistent with the function of the INF-pathway to trigger immune response against tumors, six of the eight most significantly affected pathways regulate cell-differentiation, proliferation, apoptosis, and immune response. However, this increase in the expression of INF-inducible genes occurred with no change in the transcription of the INF family genes suggesting SULF2 enzyme may negatively regulate the activity and/or bioavailability of these cytokines. HSPGs and HS present on the cell surface and ECM bind INF-γ (and possibly INF-α/β) with high affinity (Kd = 5 nM) using two highly sulfated sequences each binding to one INF-γ monomer (Lortat-Jacob et al., 1991). Thus, SULF2-mediated removal of sulfate groups from these sequences could prevent INF‐γ binding and suppress the downstream effects without changing the transcription of the cytokine. Furthermore, unbound INF-γ is rapidly degraded through C-terminal processing to prevent undesired effects of the cytokine to the surrounding tissue (Lortat-Jacob et al., 1991). Epigenetic silencing of SULF2 could prevent the removal of the sulfate groups from INF-binding sites, maintain the high affinity binding and activity, prevent degradation, increase bioavailability of the cytokine, and as shown in this study increase transcription of multiple INF-inducible genes.
The increased sensitivity of SULF2M-ISG15H lung cancer cell lines and xenograft to topoisomerase-1 (TOPO1) inhibitors appears to be mediated through ISG15, one of the genes induced by epigenetic inactivation of SULF2. ISG15 has also been demonstrated to increase the sensitivity of breast cancer cell lines to CPT in vitro through affecting the removal of DNA damage induced by this chemotherapeutic agent (Desai et al., 2008). Specifically, during DNA replication and gene transcription TOPO1 opens the double helix by nicking one DNA strand, unwinds, and re-ligates it back (Figure 5E). This process is more frequent in highly replicating cells such as cancer cells. CPT and its analogues form a ternary complex (DNA-TOPO1-CPT) that prevents the re-ligation process, and induces DNA damage leading to cell cycle arrest until the damage is repaired or the cell undergoes apoptosis. A key step in the repair process is the degradation and removal of the DNA adducts through the ubiquitin/26S proteasomal pathway that in turn, can render cells resistant to CPT and its analogues (Desai et al., 2006; Lin et al., 2008). ISG15 blocks the ubiquitin/26S proteasomal pathway leading to accumulation of CPT induced DNA damage that results in increased apoptosis (Desai et al., 2006; Desai et al., 2008).
Topoisomerase-I inhibitors, TPT and irinotecan have been used for cancer therapy as a single agent or in combination with other drugs. TPT is FDA approved as a second-line drug for the treatment of advanced ovarian, cervical, and small cell lung cancers (McGuire et al., 2000; Monk et al., 2009; O'Brien et al., 2007). The limited number of clinical trials using TPT as a single agent for NSCLC have demonstrated response rates (RR) and median OS ranging from 0 – 25% and 40 – 74 weeks as first-line and 2.8 – 8% and 10 – 34 weeks in second-line settings, respectively (Gonzalez et al., 2010; Jones et al., 2008). Two recent phase II and phase III clinical trials, demonstrated that oral TPT in unselected NSCLC patients is as effective as docetaxel, the FDA approved second-line drug for NSCLC (Gonzalez et al., 2010; Jones et al., 2008; Ramlau et al., 2006; Weitz et al., 2000). Large intra-study differences in OS ranging from 4 – 59 and 2 – 46 weeks were also seen when TPT was used as a first- or second-line therapy for NSCLC patients, respectively (Gonzalez et al., 2010; White et al., 2000). Thus, it is provocative to hypothesize that the most responsive NSCLC patients to TPT are those whose tumors have elevated expression of ISG15. Furthermore, the fact that 25% of primary tumors had elevated expression of ISG15 comparable to levels that led to increased in vitro and in vivo sensitivity to TOPO1 inhibitors supports extending these findings to a Phase II clinical trial of TPT with patient selection driven by ISG15 expression.
Materials and Methods
Samples and drugs
Seventy-eight pre-therapy lung tumor biopsies were obtained from patients participating in a Phase III clinical trial of carboplatin, paclitaxel, and radiation through the Eastern Cooperative Oncology Group (ECOG3598). Demographic variables and disease characteristics of patents are shown in Table S3. NHBEC isolated from bronchoscopy of 20 cancer-free smokers, five HBEC immortalized as described (Ramirez et al., 2004), and 18 NSCLC cell lines (Table S4) obtained from and authenticated by the American Type Culture Collection (Manassas, VA) were also studied. Experiments were conducted in cell lines passed for a maximum of 6 months post-resuscitation. Primary lung tumor-distant normal pairs (n = 12) obtained from NSCLC patients at the University of New Mexico Cancer Center and NSCLC biopsies (n = 58) obtained from the University of Pittsburgh Cancer Center (PCC) were also evaluated. Camptothecin (CPT), topotecan (TPT), cisplatin, carboplatin, and paclitaxel were purchased from Sigma-Aldrich (Saint Louis, MO).
DNA methylation and gene expression
DNA extraction, modification, and methylation analysis using combined bisulfite modification and restriction analysis (COBRA), methylation specific PCR (MSP), and bisulfite sequencing were done as described (Tessema et al., 2008). SULF2 methylation in HBEC and cancer cell lines was determined using COBRA; positive and negative controls were selected and used to optimize the MSP assays. SULF2 methylation in primary tumors was determined using MSP and results were validated for selected samples through COBRA and bisulfite sequencing. For gene expression assays lung cancer cell lines were treated with sham (vehicle), TSA (300nM trichostatin A for 18 h), or DAC (500nM 5-aza-2′-deoxycytidine for 4 days), harvested and RNA extracted as described (Tessema et al., 2008). RNA from primary tumor biopsies was isolated using RecoverAll™ Total Nucleic Acid Isolation Kit for Formalin Fixed Paraffin Embedded Samples from Applied Biosystems (Foster City, CA) following the manufacturers' instruction. RNA was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit from Applied Biosystems. Target genes were amplified in multiplex with an endogenous control (β‐actin) using primer probe mixes for quantitative TaqMan assays from Applied Biosystems. Gene expression was normalized to the housekeeping gene β‐actin and quantified relative to expression in NHBEC as described (Livak and Schmittgen, 2001). The effect of sham, TSA, and DAC treatments on SULF2 expression was also assessed using a gel-based assay as described (Tessema et al., 2008). Primers and PCR conditions for all methylation and gel-based gene expression assays are shown in Table S5.
SULF2 knockdown and genome-wide expression
Cells were transfected with SULF2 specific s31806 (siSULF2) or negative control #1 (siControl) siRNA (Applied Biosystems) using Lipofectamine 2000 (Invitrogen, Santa Clara, CA). For expression assays, cells were harvested 48h post-transfection, SULF2 knockdown was confirmed by TaqMan, and effects on genome-wide gene expression was compared between siControl and siSULF2 Calu-3 cells using the Agilent whole genome transcriptome array as described (Tessema et al., 2010).
Cell survival, migration, and drug sensitivity
Cell survival and migration were compared between parental, siControl, and siSULF2 transfected cells using MTT and wound closure assays, respectively as described (Duan et al., 2004). Briefly, equal size wounds were created 24h post-transfection and pictures of wound size taken at 0, 24, 48, 72, and 96h post-wounding. The distance between the edges of the wounds were measured at ten regions across the wound for each time point, the changes in wound size were compared to the measurements taken at time 0h, and the differences are shown as wound closure (%). For changes in survival and/or proliferation, equal number of cells were plated, transfected the next day, and the number of live parental, siControl, and siSULF2 cells were compared at 24, 48, 72, and 96h post-transfection using MTT. For drug sensitivity, NSCLC cell lines were plated 20,000 cells/well in 24-well plates, treated the next day or 24 h post-siRNA tansfection with a range of drug concentration; CPT or TPT (0 – 16.52 μM), cisplatin (0 – 100 μM), carboplatin (0 – 500 μM), or paclitaxel (0 – 100 nM) for 4 days, and cell survival was determined by MTT. Each experiment was conducted in triplicate and the results are shown as mean ± SEM.
Tumor growth in nude mice
All animal experiments were approved by the Lovelace Respiratory Research Institute animal study review board and were conducted according to the institute's Animal Care guidelines. One-to-one mixture of BD Matrigel™ Basement Membrane Matrix (BD Biosciences, San Jose, CA) with H1975 (SULF2M) or A549 (SULF2U) cells was subcutaneously injected (2.5 × 106 cells/site) into both sides of the dorso-lateral abdomen of 12 female athymic (NCr-nu/nu) nude mice (Frederick, MD). When tumor growth was visible, mice were grouped into control and treatment groups, and received once-a-week intra-peritoneal injection of 0.1 ml PBS (control) or 10 mg/kg TPT for 4 weeks. To compare the efficacy of TPT to cisplatin, another 12 mice with H1975 tumors were similarly treated weekly with PBS (control), 10 mg/kg TPT, 3 mg/kg cisplatin, or 10 mg/kg TPT followed by 3 mg/kg cisplatin two days later. Tumor size and animal weight were measured twice a week until the mice were sacrificed after 4 weeks of treatment. Mice in the control groups were sacrificed earlier due to extensive tumor growth. Tumor volume was calculated as (a × b2)/2, a (longer) and b (shorter) tumor dimensions.
Data analysis
SULF2 methylation and patient characteristics including age, gender, smoking status, tumor histology, and performance were summarized with mean and standard deviation for continuous variables and proportions for categorical variables. Survival time was calculated from time of diagnosis until death from any cause or last follow-up. The association between methylation and patient characteristics was assessed by Fisher's exact test. Kaplan-Meier plots, the log-rank test, and proportional hazards models were also employed. The effect of SULF2 methylation (SULF2U vs. SULF2M) and siRNA knockdown (siControl vs. siSULF2) on gene expression, and the effect of treatment on tumor weight (control, Cis, TPT, or Cis+TPT) were compared using one way analysis of variance (ANOVA). Tukey's and Dunnett's method were used for pair wise and treatment control comparison adjustments, respectively. The impact of potential outliers on the one way ANOVA values was controlled using nonparametric Wilcoxon Rank-sum test. The effects of SULF2 knockdown on wound-closure, and drugs on tumor-size over time were compared using a two-way mixed effect repeated measurement model.
Supplementary Material
Acknowledgments
Grant Support: This study is supported by NIH grants R01 ES008801 and CA089551 to SAB and P50 CA090440 to JMS.
Footnotes
Disclosure of Potential Conflict of Interest: Authors have no relevant conflict of interest related to this manuscript.
References
- Andersen JB, Aaboe M, Borden EC, Goloubeva OG, Hassel BA, Orntoft TF. Stage-associated overexpression of the ubiquitin-like protein, ISG15, in bladder cancer. Br J Cancer. 2006;94:1465–71. doi: 10.1038/sj.bjc.6603099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature. 2007;446:1030–7. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- Desai SD, Haas AL, Wood LM, Tsai YC, Pestka S, Rubin EH, et al. Elevated expression of ISG15 in tumor cells interferes with the ubiquitin/26S proteasome pathway. Cancer Res. 2006;66:921–8. doi: 10.1158/0008-5472.CAN-05-1123. [DOI] [PubMed] [Google Scholar]
- Desai SD, Wood LM, Tsai YC, Hsieh TS, Marks JR, Scott GL, et al. ISG15 as a novel tumor biomarker for drug sensitivity. Mol Cancer Ther. 2008;7:1430–9. doi: 10.1158/1535-7163.MCT-07-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Brakora KA, Seiden MV. Inhibition of ABCB1 (MDR1) and ABCB4 (MDR3) expression by small interfering RNA and reversal of paclitaxel resistance in human ovarian cancer cells. Mol Cancer Ther. 2004;3:833–8. [PubMed] [Google Scholar]
- Gonzalez EE, Villanueva N, Fra J, Berros JP, Jimenez P, Luque M, et al. Activity of topotecan given intravenously for 5 days every three weeks in patients with advanced non-small cell lung cancer pretreated with platinum and taxanes: a phase II study. Invest New Drugs. 2010 May 13; doi: 10.1007/s10637-010-9442-2. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Helfrich BA, Raben D, Varella-Garcia M, Gustafson D, Chan DC, Bemis L, et al. Antitumor activity of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitor gefitinib (ZD1839, Iressa) in non-small cell lung cancer cell lines correlates with gene copy number and EGFR mutations but not EGFR protein levels. Clin Cancer Res. 2006;12:7117–25. doi: 10.1158/1078-0432.CCR-06-0760. [DOI] [PubMed] [Google Scholar]
- Johansson FK, Goransson H, Westermark B. Expression analysis of genes involved in brain tumor progression driven by retroviral insertional mutagenesis in mice. Oncogene. 2005;24:3896–905. doi: 10.1038/sj.onc.1208553. [DOI] [PubMed] [Google Scholar]
- Jones S, Thompson D, Barton J, Patton J, Shipley D, Greco FA, et al. A randomized phase II trial of oral topotecan versus docetaxel in the second-line treatment of non-small-cell lung cancer. Clin Lung Cancer. 2008;9:154–9. doi: 10.3816/CLC.2008.n.023. [DOI] [PubMed] [Google Scholar]
- Kalikaki A, Koutsopoulos A, Trypaki M, Souglakos J, Stathopoulos E, Georgoulias V, et al. Comparison of EGFR and K-RAS gene status between primary tumours and corresponding metastases in NSCLC. Br J Cancer. 2008;99:923–9. doi: 10.1038/sj.bjc.6604629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- Lai JP, Oseini AM, Moser CD, Yu C, Elsawa SF, Hu C, et al. The oncogenic effect of sulfatase 2 in human hepatocellular carcinoma is mediated in part by glypican 3-dependent Wnt activation. Hepatology. 2010;52:1680–9. doi: 10.1002/hep.23848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, et al. Sulfatase 2 up-regulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology. 2008;47:1211–22. doi: 10.1002/hep.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemjabbar-Alaoui H, van Zante A, Singer MS, Xue Q, Wang YQ, Tsay D, et al. Sulf-2, a heparan sulfate endosulfatase, promotes human lung carcinogenesis. Oncogene. 2010;29:635–46. doi: 10.1038/onc.2009.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Ban Y, Lyu YL, Desai SD, Liu LF. A ubiquitin-proteasome pathway for the repair of topoisomerase I-DNA covalent complexes. J Biol Chem. 2008;283:21074–83. doi: 10.1074/jbc.M803493200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lortat-Jacob H, Kleinman HK, Grimaud JA. High-affinity binding of interferon-gamma to a basement membrane complex (matrigel) J Clin Invest. 1991;87:878–83. doi: 10.1172/JCI115093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire WP, Blessing JA, Bookman MA, Lentz SS, Dunton CJ. Topotecan has substantial antitumor activity as first-line salvage therapy in platinum-sensitive epithelial ovarian carcinoma: A Gynecologic Oncology Group Study. J Clin Oncol. 2000;18:1062–7. doi: 10.1200/JCO.2000.18.5.1062. [DOI] [PubMed] [Google Scholar]
- Merry CL, Lyon M, Deakin JA, Hopwood JJ, Gallagher JT. Highly sensitive sequencing of the sulfated domains of heparan sulfate. J Biol Chem. 1999;274:18455–62. doi: 10.1074/jbc.274.26.18455. [DOI] [PubMed] [Google Scholar]
- Monk BJ, Sill MW, McMeekin DS, Cohn DE, Ramondetta LM, Boardman CH, et al. Phase III trial of four cisplatin-containing doublet combinations in stage IVB, recurrent, or persistent cervical carcinoma: a Gynecologic Oncology Group study. J Clin Oncol. 2009;27:4649–55. doi: 10.1200/JCO.2009.21.8909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Bistrup A, Lum DH, Egeblad M, Boudreau N, et al. Sulf-2, a proangiogenic heparan sulfate endosulfatase, is upregulated in breast cancer. Neoplasia. 2005;7:1001–10. doi: 10.1593/neo.05496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mice and humans. J Biol Chem. 2002;277:49175–85. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawroth R, van Zante A, Cervantes S, McManus M, Hebrok M, Rosen SD. Extracellular sulfatases, elements of the Wnt signaling pathway, positively regulate growth and tumorigenicity of human pancreatic cancer cells. PLoS One. 2007;2:e392. doi: 10.1371/journal.pone.0000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien M, Eckardt J, Ramlau R. Recent advances with topotecan in the treatment of lung cancer. Oncologist. 2007;12:1194–204. doi: 10.1634/theoncologist.12-10-1194. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez RD, Sheridan S, Girard L, Sato M, Kim Y, Pollack J, et al. Immortalization of human bronchial epithelial cells in the absence of viral oncoproteins. Cancer Res. 2004;64:9027–34. doi: 10.1158/0008-5472.CAN-04-3703. [DOI] [PubMed] [Google Scholar]
- Ramlau R, Gervais R, Krzakowski M, von Pawel J, Kaukel E, Abratt RP, et al. Phase III study comparing oral topotecan to intravenous docetaxel in patients with pretreated advanced non-small-cell lung cancer. J Clin Oncol. 2006;24:2800–7. doi: 10.1200/JCO.2005.03.6491. [DOI] [PubMed] [Google Scholar]
- Sugaya N, Habuchi H, Nagai N, Ashikari-Hada S, Kimata K. 6-O-sulfation of heparan sulfate differentially regulates various fibroblast growth factor-dependent signalings in culture. J Biol Chem. 2008;283:10366–76. doi: 10.1074/jbc.M705948200. [DOI] [PubMed] [Google Scholar]
- Tessema M, Klinge DM, Yingling CM, Do K, Van Neste L, Belinsky SA. Re-expression of CXCL14, a common target for epigenetic silencing in lung cancer, induces tumor necrosis. Oncogene. 2010;29:5159–70. doi: 10.1038/onc.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessema M, Willink R, Do K, Yu YY, Yu W, Machida EO, et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–14. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- Tessema M, Yu YY, Stidley CA, Machida EO, Schuebel KE, Baylin SB, et al. Concomitant promoter methylation of multiple genes in lung adenocarcinomas from current, former and never smokers. Carcinogenesis. 2009;30:1132–8. doi: 10.1093/carcin/bgp114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, et al. HSulf-2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparin-bound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7 doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitz JJ, Marschke RF, Jr, Sloan JA, Grill JP, Jett JR, Knost JA, et al. A randomized phase II trial of two schedules of topotecan for the treatment of advanced stage non-small cell lung cancer. Lung Cancer. 2000;28:157–62. doi: 10.1016/s0169-5002(99)00128-2. [DOI] [PubMed] [Google Scholar]
- White SC, Cheeseman S, Thatcher N, Anderson H, Carrington B, Hearn S, et al. Phase II study of oral topotecan in advanced non-small cell lung cancer. Clin Cancer Res. 2000;6:868–73. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.