Abstract
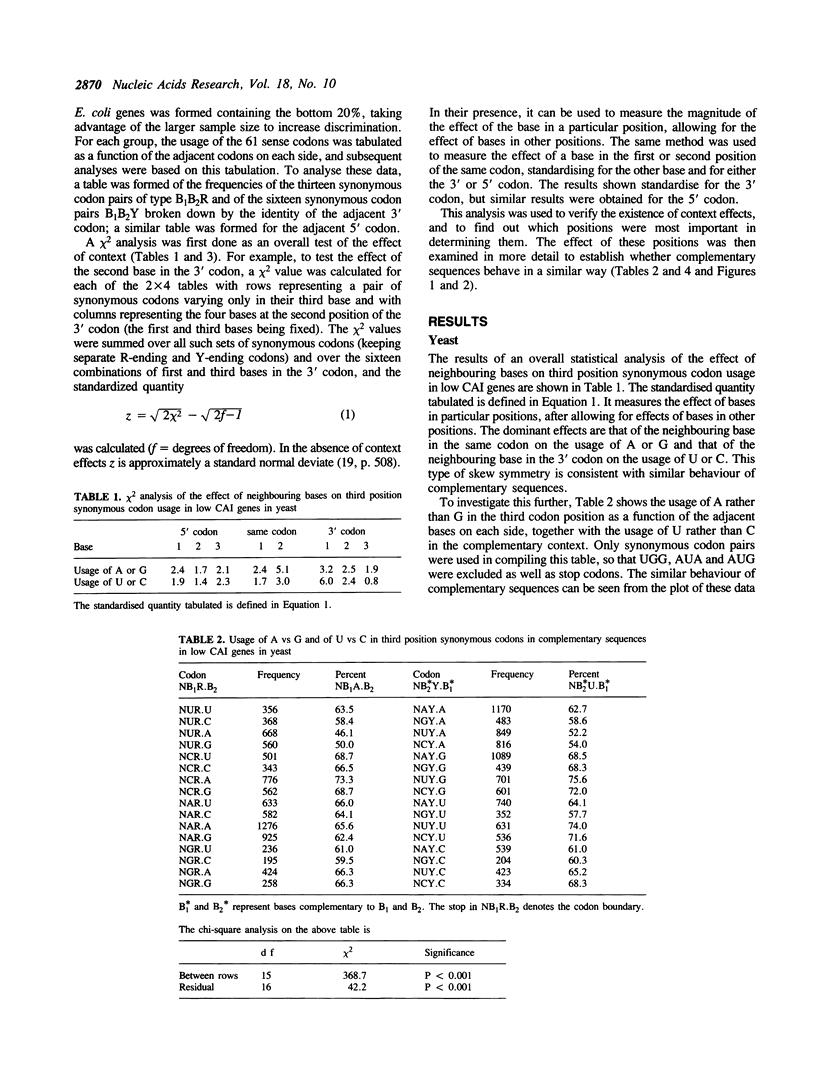
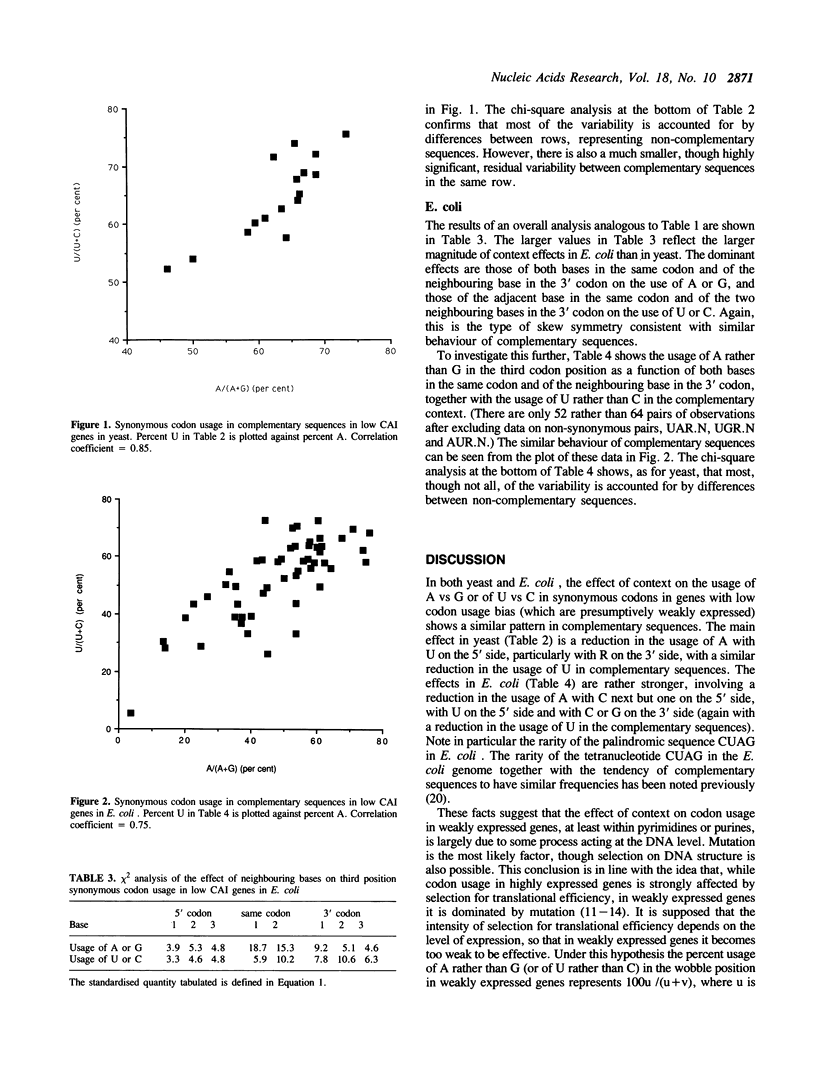
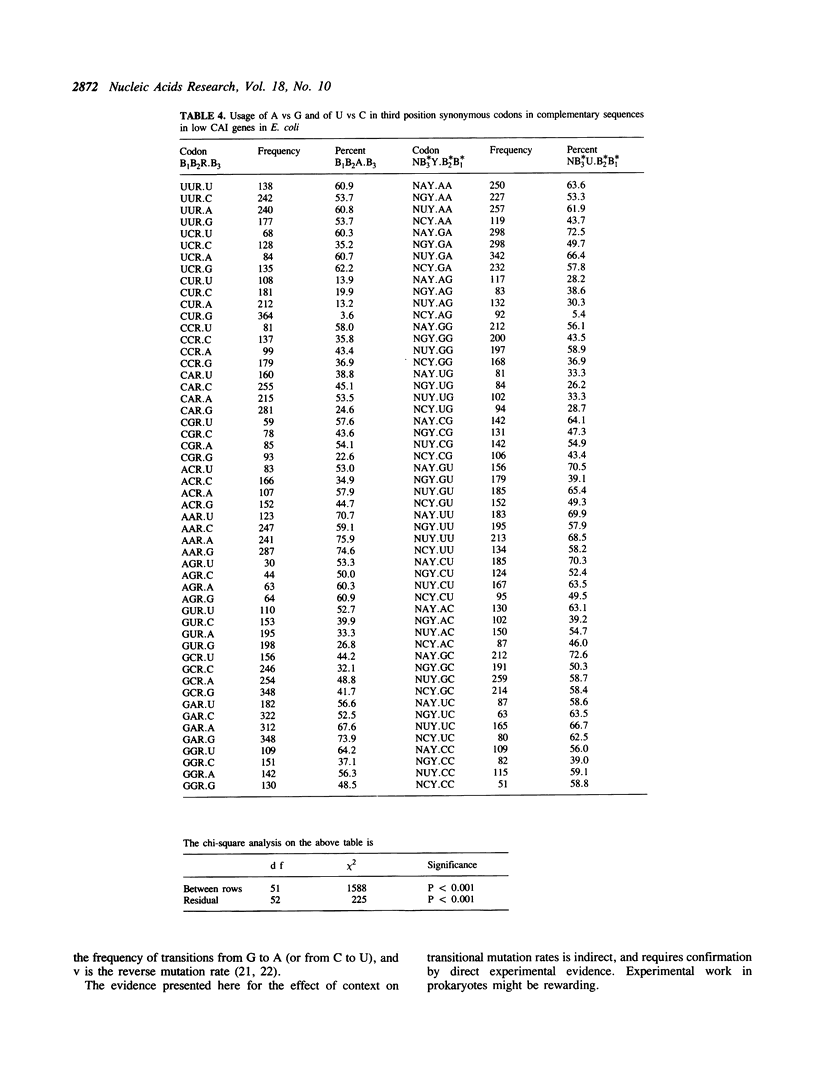
The effect of neighbouring bases on the usage of synonymous codons in genes with low codon usage bias in yeast and E. coli is examined. The codon adaptation index is employed to identify a group of genes in each organism with low codon usage bias, which are likely to be weakly expressed. A similar pattern is found in complementary sequences with respect to synonymous usage of A vs G or of U vs C. It is suggested that this may reflect an effect of context on mutation rates in weakly expressed genes.
Full text
PDF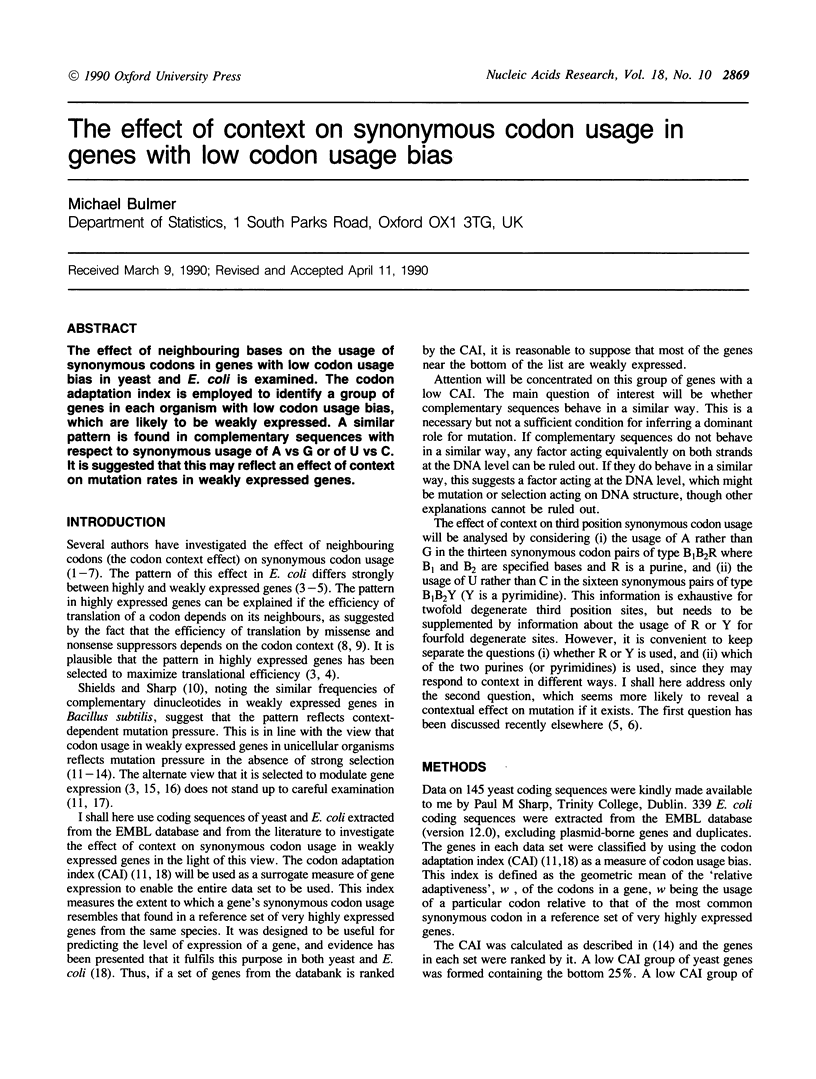

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blaisdell B. E. Choice of base at silent codon site 3 is not selectively neutral in eucaryotic structural genes: it maintains excess short runs of weak and strong hydrogen bonding bases. J Mol Evol. 1983;19(3-4):226–236. doi: 10.1007/BF02099970. [DOI] [PubMed] [Google Scholar]
- Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J Mol Biol. 1983 Feb 15;164(1):73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987 Feb 19;325(6106):728–730. doi: 10.1038/325728a0. [DOI] [PubMed] [Google Scholar]
- Gouy M. Codon contexts in enterobacterial and coliphage genes. Mol Biol Evol. 1987 Jul;4(4):426–444. doi: 10.1093/oxfordjournals.molbev.a040450. [DOI] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Gutman G. A., Hatfield G. W. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989 May;86(10):3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanai R., Wada A. Novel third-letter bias in Escherichia coli codons revealed by rigorous treatment of coding constraints. J Mol Biol. 1989 Jun 20;207(4):655–660. doi: 10.1016/0022-2836(89)90235-0. [DOI] [PubMed] [Google Scholar]
- Konigsberg W., Godson G. N. Evidence for use of rare codons in the dnaG gene and other regulatory genes of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Feb;80(3):687–691. doi: 10.1073/pnas.80.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipman D. J., Wilbur W. J. Contextual constraints on synonymous codon choice. J Mol Biol. 1983 Jan 25;163(3):363–376. doi: 10.1016/0022-2836(83)90063-3. [DOI] [PubMed] [Google Scholar]
- Murgola E. J., Pagel F. T., Hijazi K. A. Codon context effects in missense suppression. J Mol Biol. 1984 May 5;175(1):19–27. doi: 10.1016/0022-2836(84)90442-x. [DOI] [PubMed] [Google Scholar]
- Phillips G. J., Arnold J., Ivarie R. Mono- through hexanucleotide composition of the Escherichia coli genome: a Markov chain analysis. Nucleic Acids Res. 1987 Mar 25;15(6):2611–2626. doi: 10.1093/nar/15.6.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUEOKA N. On the genetic basis of variation and heterogeneity of DNA base composition. Proc Natl Acad Sci U S A. 1962 Apr 15;48:582–592. doi: 10.1073/pnas.48.4.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. An evolutionary perspective on synonymous codon usage in unicellular organisms. J Mol Evol. 1986;24(1-2):28–38. doi: 10.1007/BF02099948. [DOI] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986 Oct 10;14(19):7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp P. M., Li W. H. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987 Feb 11;15(3):1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields D. C., Sharp P. M. Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases. Nucleic Acids Res. 1987 Oct 12;15(19):8023–8040. doi: 10.1093/nar/15.19.8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpaer E. G. Constraints on codon context in Escherichia coli genes. Their possible role in modulating the efficiency of translation. J Mol Biol. 1986 Apr 20;188(4):555–564. doi: 10.1016/s0022-2836(86)80005-5. [DOI] [PubMed] [Google Scholar]
- Sueoka N. Directional mutation pressure and neutral molecular evolution. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2653–2657. doi: 10.1073/pnas.85.8.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus M., Folley L. S. Sense codons are found in specific contexts. J Mol Biol. 1985 Apr 20;182(4):529–540. doi: 10.1016/0022-2836(85)90239-6. [DOI] [PubMed] [Google Scholar]


