Background: The fungi-specific alternative pathway of glutathione degradation requires DUG1, a Cys-Gly peptidase, and DUG2 and DUG3 of unknown function.
Results: (Dug2p-Dug3p)2 is a GATaseII enzyme that cleaves the γ-glutamyl linkage of glutathione; DUG2 and DUG3 are induced under sulfur limitation
Conclusion: The alternative pathway initiates cleavage of glutathione by a novel GATase.
Significance: This work delineates the mechanism of glutathione degradation by the alternative pathway.
Keywords: Enzyme Mechanisms, Glutathione, Peptidases, Sulfur, Yeast Metabolism, Alternative Pathway, DUG Pathway, Glutamine Amidotransferase Domain II, NTN Hydrolase, Sulfur Homeostasis
Abstract
The recently identified, fungi-specific alternative pathway of glutathione degradation requires the participation of three genes, DUG1, DUG2, and DUG3. Dug1p has earlier been shown to function as a Cys-Gly-specific dipeptidase. In the present study, we describe the characterization of Dug2p and Dug3p. Dug3p has a functional glutamine amidotransferase (GATase) II domain that is catalytically important for glutathione degradation as demonstrated through mutational analysis. Dug2p, which has an N-terminal WD40 and a C-terminal M20A peptidase domain, has no peptidase activity. The previously demonstrated Dug2p-Dug3p interaction was found to be mediated through the WD40 domain of Dug2p. Dug2p was also shown to be able to homodimerize, and this was mediated by its M20A peptidase domain. In vitro reconstitution assays revealed that Dug2p and Dug3p were required together for the cleavage of glutathione into glutamate and Cys-Gly. Purification through gel filtration chromatography confirmed the formation of a Dug2p-Dug3p complex. The functional complex had a molecular weight that corresponded to (Dug2p-Dug3p)2 in addition to higher molecular weight oligomers and displayed Michaelis-Menten kinetics. (Dug2p-Dug3p)2 had a Km for glutathione of 1.2 mm, suggesting a novel GATase enzyme that acted on glutathione. Dug1p activity in glutathione degradation was found to be restricted to its Cys-Gly peptidase activity, which functioned downstream of the (Dug2p-Dug3p)2 GATase. The DUG2 and DUG3 genes, but not DUG1, were derepressed by sulfur limitation. Based on these studies and the functioning of GATases, a mechanism is proposed for the functioning of the Dug proteins in the degradation of glutathione.
Introduction
Glutathione (GSH), l-γ-glutamyl-l-cysteinyl-l-glycine, is an essential thiol compound present in all eukaryotic systems with the exception of a few amitochondrial protozoans (1). The low redox potential of GSH, its ability to exist in reduced (GSH) and oxidized (GSSG) forms, the high intracellular concentrations achieved (up to 10 mm), and the high stability due to unusual γ linkage make it an important redox buffer of the cell. In addition, GSH plays important roles in iron metabolism, scavenging of free radicals, detoxification of metal ions and xenobiotics, and the regulation of the activity of many proteins by glutathionylation. It also functions as a source of sulfur and nitrogen (2, 3). Not surprisingly, the maintenance of glutathione homeostasis is crucial to the proper functioning of the cell.
Glutathione degradation in living cells has normally been considered to be initiated by γ-glutamyl transpeptidase, an enzyme of the γ-glutamyl cycle that is involved in the cyclic formation and degradation of glutathione (3). γ-Glutamyl transpeptidase is localized on the plasma membranes of mammalian cells (4, 5) and on the vacuolar membranes of yeasts and plants (6–8). Very recently, we have demonstrated the presence of an alternative pathway of GSH degradation, working independently of γ-glutamyl transpeptidase in yeasts and fungi (9, 10). GSH degradation by this novel pathway, which is cytosolic, was found to require the participation of three novel genes: DUG1, which encodes an M20A peptidase; DUG2, which encodes a protein with a WD40 repeat (11, 12) at its N terminus and an M20A peptidase domain at its C terminus; and DUG3, which encodes a protein with a glutamine amidotransferase II (GATaseII)4 domain. This alternative pathway, which was named the DUG pathway, is fungi-specific as the homologues of DUG2 and DUG3 are present only in fungal species. Genetic studies revealed that all the three proteins were required for GSH degradation. In contrast, the normal dipeptides and tripeptides (Cys-Gly and Glu-Cys-Gly) required only the participation of the DUG1 gene (10). However, the mechanism by which these proteins function in glutathione degradation has not yet been delineated.
The first step toward the understanding of the DUG pathway was made through investigations carried out on Dug1p. Biochemical characterization of the purified, recombinant Dug1p revealed that it functions as a Cys-Gly peptidase (with no activity seen toward glutathione or other tripeptides) and exists as a homodimer in the cell (13). However, whether Dug1p also functions as a part of the DUG complex (for glutathione degradation) in addition to its independent function as a Cys-Gly peptidase has not been investigated. The purification and roles of the other two proteins of the DUG pathway, Dug2p and Dug3p, have also not been described so far.
In this study, we continue our characterization of the DUG pathway and address the roles of the Dug2p and Dug3p proteins. Dug2p was found to interact with Dug3p via the WD40 domain of Dug2p. Dug2p also homodimerizes with itself, and this is mediated by its M20A peptidase domain. Dug2p, however, is devoid of catalytic activity, and thus, functions mostly as a scaffolding protein. Furthermore, no Dug1p-Dug2p heterodimerization was observed. Dug3p was found to function as a glutamine amidotransferase based on mutational analysis. In vitro reconstitution of the purified recombinant proteins, Dug2p and Dug3p, revealed that Dug2p and Dug3p come together to form a functional (Dug2p-Dug3p)2 GATase complex of ∼300 kDa that acts on the γ-Glu-Cys bond of glutathione. The kinetics of glutathione degradation revealed a Km of 1.2 ± 0.2 mm. The regulation of the DUG genes revealed that DUG3 and DUG2 expression was under strong sulfur regulation. Finally, a model is presented for the functioning of the DUG proteins in glutathione degradation.
EXPERIMENTAL PROCEDURES
Materials
All chemicals and reagents were of analytical reagent grade and were procured from different commercial sources. Protein molecular weight markers were purchased from MBI Fermentas. Oligonucleotide primers were synthesized from Biobasic Inc. and Sigma-Genosys. Medium components were purchased from BD Diagnostics (Difco). Restriction enzymes, DNA polymerases, and other DNA-modifying enzymes were obtained from New England Biolabs. A DNA sequencing kit (ABI PRISM 310 XL with dye termination cycle sequencing ready reaction kit) was obtained from PerkinElmer Life Sciences. Gel extraction kits, plasmid miniprep columns, and Ni-NTA resins were obtained from Qiagen or Sigma. Molecular weight standards for gel filtration were obtained from GE Healthcare. Metal salts were obtained from Sigma.
Strains, Media, and Growth Conditions
The Escherichia coli strain DH5α was used as a cloning host, and BL21 (DE3) was used as an expression host. Yeast strains used in the study are described in supplemental Table S1. Saccharomyces cerevisiae strains were regularly maintained on yeast extract, peptone, and dextrose (YPD) medium. Synthetic defined minimal medium contained yeast nitrogen base, ammonium sulfate, and dextrose supplemented with methionine, histidine, leucine, lysine, and uracil at 80 mg/liter (as per requirement). Complete Medium contained all the amino acids except those indicated. Yeast transformations were carried out by the lithium acetate method as described for S. cerevisiae (14). Growth, handling of bacteria, and yeast were according to the standard protocols (15).
Cloning and Gene Manipulations
All the molecular biology techniques used in this study were according to the standard protocols (16). Sequences of the primers used for cloning are given in supplemental Table S2.
Cloning of DUG3 Mutants
DUG3HA wild type cloned earlier in p416TEF was used as template to create the GATaseII mutants of DUG3. Site-directed mutagenesis was carried out by splice overlap extension method following standard PCR conditions using a specific set of primers. PCR products were spliced by DUG3EcoRIF and DUG3HAXhoIR primers and cloned at EcoRI-XhoI site of p416TEF. Mutation was confirmed by DNA sequencing.
Cloning of DUG1 and DUG2 Active Site Mutants
Wild type DUG1 and DUG2Myc cloned earlier in p416TEF were used as templates to create E171A and E586A mutants of DUG1 and DUG2, respectively. E171A mutant was created by the overlap extension method using DUG1E171AF and DUG1E171AR primers following standard PCR conditions. PCR products were spliced by DUG1BamHIF and DUG1HisSalIR primers and cloned at the BamHI-SalI site of p416TEF. E586A mutation was created by using the Stratagene site-directed mutagenesis kit using DUG2E586AF and DUG2E586AR primers. Mutations were confirmed by DNA sequencing.
Cloning of M20A Domain of DUG2
DUG2-M20 domain (1275–2934 bp) was PCR-amplified from p416TEFDUG2 following standard PCR conditions using DUG2M20EcoRIF and DUG2XhoIR primers and cloned at the EcoRI-XhoI sites of the p416TEF vector. For purification of the DUG2-M20A domain from E. coli, M20 domain fragment was PCR-amplified using DUG2M20NdeIF and DUG2pETXhoIR primers and cloned at the NdeI-XhoI sites of the pET28c (+) vector.
Cloning of CNDP2 and LAP3
Human carnosinase 2 (CNDP2) cDNA was earlier cloned into the p416TEF vector (13). Rat LAP3 cDNA clones (IMAGE clone ID: 7111053) were obtained from the mammalian gene collection, Invitrogen. LAP3 cDNA was PCR-amplified from the LAP3 IMAGE clone using the LAP3XbaIF forward and LAP3EcoRIR reverse primers and cloned into pTEF416 at the XbaI and EcoRI restriction sites to yield p416TEF-LAP3.
Dilution Spotting
For dilution spotting assays, the cells were grown for 12–16 h at 30 °C and reinoculated into the desired medium at an initial A600 = 0.1. Cells were further grown at 30 °C and harvested at A600 = 0.6 by centrifugation at 6000 × g for 5 min. Cells were washed with water and resuspended in water at A600 = 2.0. Serial dilutions of A600 = 0.2, 0.02, 0.002, and 0.0002 were made in water, and 5 μl of the each dilution was spotted on the desired plates as mentioned in the results. Plates were incubated for 3–5 days at 30 °C.
Cloning, Expression, and Purification of DUG Proteins
Purification of Dug2p and Dug2pM20A
DUG2 was PCR-amplified from p416TEF DUG2 using the primers DUG2NdeIF and DUG2pETXhoIR and cloned at the NdeI-XhoI sites of the pET28c(+) vector. pET28c(+)DUG2 was transformed in BL21(DE3) E. coli cells. Expression of His-Dug2p was induced at A600 = 0.8 by 0.5 mm isopropyl-β-thiogalactopyranoside, and the induction was carried out at 16 °C for 16 h with shaking at 220 rpm. E. coli cultures induced with isopropyl-β-thiogalactopyranoside were harvested at 6000 × g for 5 min, 4 °C, and the pellet was resuspended in lysis buffer (300 mm NaCl and 50 mm Tris-HCl, pH 8.0). Cells were lysed by sonication for 10 min. The bulk of the induced protein was found in the soluble fraction. The soluble fraction was recovered by centrifugation at 10,000 × g for 20 min, 4 °C. The protein that had a His6 tag at the N terminus was purified using Ni-NTA affinity chromatography. Ni-NTA columns were washed with water and equilibrated with lysis buffer. Protein samples were loaded onto the column, and the column was washed with 5 column volumes of the lysis buffer. His-Dug2p was eluted with 2 column volumes of elution buffer (equilibration buffer containing 150 mm imidazole). The purified protein was dialyzed against the buffer (300 mm NaCl, 50 mm Tris-HCl, pH 8.0). The purity of the His-Dug2p was monitored by running SDS-PAGE followed by Coomassie Brilliant Blue R-250 staining. Expression and purification of Dug2p-M20A were essentially carried out similar to His-Dug2p.
Purification of Dug3p
DUG3 was PCR-amplified from p416TEF DUG3 by DUG3KpnIF and DUG3linkerpETXhoIR. PCR product was digested with KpnI, Klenow-filled, and digested with XhoI. pET23a(+) vector was digested with NdeI, Klenow-filled, digested with XhoI, and ligated to PCR product. For purification of Dug3p, DUG3 cloned in pET23(a) vector was expressed as C-terminally His-tagged protein having a glycine linker sequence (GGGGGGIP) between Dug3p and the His tag. In the absence of glycine linker, Dug3p was not able to bind to Ni-NTA resin. Purification of Dug3-linker-His was carried out similar to Dug2p, but 20 mm imidazole was included in the lysis and washing buffer.
Co-expression and Purification of Dug1p and Dug2p
For co-purification of Dug2p and Dug1p, pET28c(+)DUG2 and pET23a(+)DUG1 were co-transformed into BL21(DE3) cells, and protein was purified essentially as Dug2p. Co-purification of both was checked by SDS-PAGE and Coomassie Brilliant Blue R-250 staining. Gel filtration was carried out in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0.
Yeast Two-hybrid Analysis
pEG202-DUG2, pJG4-5-DUG2, and pEG202-DUG3 constructs were made earlier (10). For making pJG4-5-DUG2WD40 construct, WD40 domain of DUG2 (1–1274 bp) was PCR-amplified using the primers DUG2EcoRIF and DUG2WD40XhoIR and cloned at the EcoRI-XhoI site of the pJG4-5 vector. The duplex-A yeast two-hybrid system based on the system developed by Brent and co-workers was used in the study as described earlier (17). Briefly, pEG202 was used as bait plasmid, and pJG4-5 was used as prey plasmid. Interactions were studied in EGY48 strain using dual reports as described earlier (10) where interactions among two proteins results in the development of blue color on Complete Medium + X-gal plates due to expression of lacZ from the reporter plasmid pSH18-34 and growth on Complete Medium leucine plates. pSH17-4 and pRF HM1 were used as positive and negative controls, respectively.
γ-Glutamyl-p-Nitroanilide (γ-glu-pNA) Assay
Purified Dug2p and Dug3p were mixed in 1:1 stoichiometry and dialyzed in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0 (18). The given amount of Dug2p-Dug3p was incubated with 4 mm γ-glu-pNA in a 100-μl reaction in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, buffer in a 96-well plate. The reaction was carried out at 37 °C for different time intervals, and the amount of pNA released was monitored by absorbance at 410 nm in an ELISA reader. Nanomoles of pNA formed were calculated by considering the molar extinction coefficient of pNA at pH = 8.0 equal to 14,520 cm−1m−1.
Glutathione Degradation Assay
Purified Dug2p and Dug3p mixed in 1:1 stoichiometry, Dug2p alone, or Dug3p alone were dialyzed in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0. Given amounts of proteins were incubated with 10 mm glutathione in a 100-μl reaction in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, buffer. Reaction was carried out at 37 °C for 1 h and terminated by heating the reaction mixture at 95 °C for 5 min. Samples were centrifuged at 13,000 rpm for 15 min, and supernatants were used for HPLC analysis of reaction products. 20 μl of the supernatant was loaded on the C18 column, and 2% perchloric acid was used as solvent (19). HPLC was run at a flow rate of 1 ml/min. The area of the peaks was plotted as a function of protein concentration. Corresponding peaks were confirmed by spiking the reaction mixtures with purified glutathione, glutamate, or Cys-Gly. γ-Glu-Cys cleavage activity of Dug2p-Dug3p was monitored essentially as done for glutathione.
For kinetic analysis, the Cys-Gly produced by Dug2p-Dug3p activity was estimated by coupled assay using Dug1p. To the 100-μl reaction mixture, 5 μg of purified recombinant Dug1p and 20 μm MnCl2 were added, and volume was made up to 150 μl. Reaction was carried out at 37 °C for 1 h, and the amount of cysteine formed was estimated by acidic ninhydrin assay as described earlier (20).
For fractionation of the Dug2p-Dug3p complex, the purified proteins were mixed in 1:1 stoichiometry, and the mixture was dialyzed for 1 h in the 300 mm NaCl, 1 mm GSH, 50 mm Tris-HCl, pH 8.0, buffer. Dialyzed samples were analyzed by gel filtration using Superdex S200 column.
Promoter Cloning
The 600-bp upstream region of DUG3 and 1-kb upstream region of DUG1 and DUG2 promoter regions were PCR-amplified from genomic DNA of ABC733 (BY4741) using DUG1ProSalIF and DUG1ProBamHIR, DUG2ProXhoIF and DUG2ProBamHIR, and DUG3ProXhoIF and DUG3ProBamHIR primer sets, respectively. DUG2 and DUG3 promoters were cloned at the XhoI and BamHI sites, and DUG1 promoter was cloned at the SalI and BamHI sites upstream of β-galactosidase gene in the vector pLG699Z. Sequence of the insert was confirmed by DNA sequencing.
β-Galactosidase Assay
The reporter plasmids were transformed into the S. cerevisiae strains mentioned under “Results,” and the transformants were grown in minimal medium for 12–18 h. For the expression studies, the secondary inoculations were done at A600 = 0.1 into minimal medium having different sulfur source, and the cultures were grown for additional 6–8 h at 30 °C. 200 μm Cysteine or 200 μm methionine was used for repressing conditions, and 200 μm GSH, 20 μm methionine, or no methionine (no sulfur) was used for derepressing conditions. For the organic sulfur auxotroph strains for induction in no methionine conditions, reinoculation was done at A600 = 0.25. For DUG2 regulation studies in ABC733, we used A600 = 4.0 cells, and in all the other assays, A600 = 1.0 cells were used. β-Galactosidase activity was measured in the permeabilized cells by the protocol of Guarente et al. (21). Briefly, cells were harvested by centrifugation (6000 × g, 4 °C, 5 min) and washed with water and LacZ buffer (60 mm Na2HPO4·7H2O, 40 mm NaH2PO4·H2O, 10.06 mm KCl, 1 mm MgSO4·7H2O, 0.27% β-mercaptoethanol). The required number of cells was taken and permeabilized with 50 μl of chloroform and 20 μl of 1.0% SDS by vortexing for 10 s. The cell suspension was preincubated at 30 °C for 5 min, and the β-galactosidase activity was measured using 0.7 ml of ortho-nitrophenyl-β-galactoside (2 mg/ml of LacZ buffer). The reaction was allowed to proceed for 30–40 min at 30 °C and terminated with 0.5 ml of 1 m sodium carbonate. Cell debris was removed by centrifugation, and the absorbance of ortho-nitrophenol was taken at 420 nm. β-gal units were calculated as A420 × 1000 min−1 ml−1/A600. The experiment was repeated twice with two independent transformants.
RESULTS
DUG1 Role in Glutathione Degradation Can Be Replaced by Mammalian Cys-Gly Peptidase LAP3; Dug1p Role in GSH Degradation Is Restricted to Its Cys-Gly Peptidase Activity
Dug1p is a Cys-Gly peptidase and is also required for GSH degradation. Two possibilities exist in relation to its function. Firstly, it is possible that the function of Dug1p in GSH degradation may be limited to its Cys-Gly dipeptidase activity (and is independent of Dug2p and Dug3p). Alternatively, Dug1p may play two roles, the first as a part of a larger DUG protein complex involved in GSH degradation and the second as a Cys-Gly peptidase as suggested earlier (10, 13). To determine which of these possibilities reflected the true roles of Dug1p in the cell, we examined whether these dual functions in both Cys-Gly degradation and GSH degradation could be replaced by other Cys-Gly peptidases. Human CNDP2, which shares a similarity (54% identity, 64%, similarity) with Dug1p, has been shown from earlier studies from our laboratory as a Cys-Gly peptidase (13), but its ability to complement the GSH utilization defect in dug1Δ strain was never examined. LAP3, a leucine aminopeptidase from rat, is a member of the M17 family of metallopeptidase. LAP3 has no sequence similarity to Dug1p, but has been previously demonstrated to display Cys-Gly dipeptidase activity (22, 23). Human CNDP2 and rat LAP3 cDNAs were separately cloned and expressed in the p416TEF expression vector and transformed into the dug1Δ strain. Both LAP3 and CNDP2 complemented the dug1Δ deletion for utilization of GSH as the sole source of sulfur (data not shown). As these proteins, and particularly LAP3, which has no sequence similarity to Dug1p, were not expected to form a complex with Dug2p and Dug3p, their role in GSH degradation must be independent of such a complex and should be limited to Cys-Gly cleavage. Their ability to complement dug1Δ for GSH utilization suggests that the Dug1p role in GSH degradation is as a Cys-Gly peptidase. Dug2p and Dug3p may be acting upstream of Dug1p to release Cys-Gly from GSH.
Dug2p Interacts with Dug3p (via WD40 domain) and with Itself (via M20A Peptidase Domain)
We had previously shown strong interactions among Dug2p and Dug3p by yeast two-hybrid and co-immunoprecipitation studies (10). Dug2p has a WD40 domain at the N terminus (1–424 amino acids) and an M20A peptidase domain at the C terminus (425–878 amino acids). To identify the regions of Dug2p important for Dug2p-Dug3p interactions, the M20A and the WD40 domains of Dug2p were separately cloned into the yeast two-hybrid plasmids and evaluated for interactions. Expression of the WD40 domain of Dug2p (pJG4-5-DUG2WD40) along with Dug3p (pEG202-DUG3) produced a blue color on X-gal plates and growth on −leucine plates, although the interaction was weaker than seen with the full-length Dug2p (supplemental Fig. S1). This indicates that Dug2p-Dug3p interaction is mediated via the WD40 domain of Dug2p. No interaction was seen between Dug3p and the M20A domain of Dug2p (data not shown).
Dug1p, an M20A peptidase, exists as a homodimer in solution, and dimerization is predicted to be mediated by the M20A dimerization domain. As Dug2p also has an M20A dimerization domain similar to Dug1p, it appears possible that Dug2p might also self-dimerize like Dug1p. As seen from the yeast two-hybrid studies (supplemental Fig. S1), co-transformation of pJG4-5-DUG2 and pEG202-DUG2 along with reporter plasmid pSH18-34 produces blue color on X-Gal plates and growth on −leucine plates, indicating that a strong Dug2p-Dug2p interaction takes place in vivo.
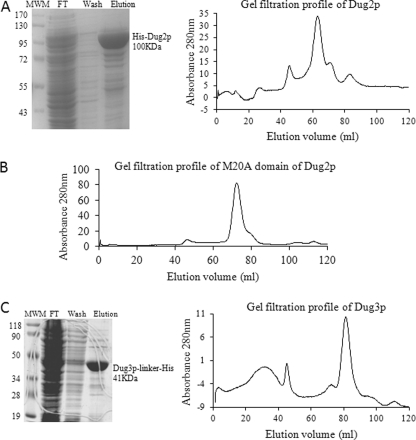
To further evaluate the Dug2p-Dug2p interaction through in vitro purification, the gene was cloned into an E. coli expression vector pET28a(+) and expressed as an N-terminally His6-tagged protein. His6-Dug2p was purified by Ni-NTA affinity chromatography. Purified Dug2p was about 90% pure and has a molecular mass of 100 kDa (including the mass of the tag) as seen on SDS-PAGE and Coomassie Brilliant Blue staining (Fig. 1A). The oligomeric state of the purified protein was analyzed by gel filtration chromatography using a Superdex S200 column. The elution profile of Dug2p shows three major peaks (Fig. 1A). The first peak corresponded to the aggregated form being eluted in the void volume. The majority of the protein eluted at the molecular mass of 243 kDa (elution volume, Ve = 62 ml). A very small proportion of the protein eluted at the molecular mass of 105 kDa (Ve = 72 ml). As the molecular mass of the Dug2 polypeptide is 100 kDa, it indicates that in solution, if present alone, Dug2p is primarily present as a dimer. To examine whether it was the M20A domain of Dug2p that was involved in the dimerization, the M20A domain of Dug2p was purified similar to Dug2p. The gel filtration profile of the M20A domain of Dug2p showed a single peak of 97 kDa (Ve = 73 ml) that corresponds to the molecular mass of the dimeric M20A domain (Fig. 1B).
FIGURE 1.
Purification and oligomeric status of Dug2p and Dug3p. A, expression of His-tagged Dug2p was induced in BL21 (DE3) cells by 0.5 mm isopropyl-β-thiogalactopyranoside for 16 h at 16 °C, and His-Dug2p was purified by Ni-NTA affinity chromatography. Samples were loaded on 10% SDS-PAGE, and gels were stained with Coomassie Brilliant Blue R-250. Lane 1, molecular weight marker (MWM); lane 2, flow-through; lane 3, wash; lane 4, eluted Dug2p. Ni-NTA-purified Dug2p was dialyzed against 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, and loaded onto a Superdex S200 column. The elution profile of the protein was monitored by absorbance at 280 nm. B, Ni-NTA-purified His-tagged Dug2p M20A domain was dialyzed against 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, and loaded onto a Superdex S200 column. The elution profile of the protein was monitored by absorbance at 280 nm. C, expression of His-tagged Dug3p was induced in BL21 (DE3) cells by 0.5 mm isopropyl-β-thiogalactopyranoside for 16 h at 16 °C, and Dug3p-Gly linker-His protein was purified by Ni-NTA affinity chromatography. Samples were loaded on 10% SDS-PAGE, and gels were stained with Coomassie Brilliant Blue R-250. Lane 1, molecular weight marker; lane 2, flow-through; lane 3, wash; lane 4, eluted Dug3p. Ni-NTA-purified Dug3p was dialyzed against 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, and loaded on the Superdex S200 column. Elution profile of the protein was monitored by absorbance at 280 nm.
Thus, both yeast two-hybrid and gel filtration studies revealed that Dug2p interacts with itself and that the dimeric form is the dominant form of the protein. Furthermore, the dimerization was mediated by the M20A domain of Dug2p.
As Dug1p also contains an M20A peptidase domain, and in a previous study, it was observed that Dug1p interacts with Dug2p (although weakly); in vitro purification and yeast two-hybrid studies were undertaken to evaluate this more carefully. Co-expression and purification of both Dug1p and Dug2p, each of which have a single dimerization domain, reveals that the dominant population consists of Dug1p-Dug1p and Dug2p-Dug2p homodimer, with no detectable heterodimer. (supplemental Fig. S2). In yeast two-hybrid studies, independent WD40 and M20A peptidase domains of Dug2p were also checked for their interaction with Dug1p. None of the domains showed any significant interaction with Dug1p (data not shown), further confirming that Dug2p interacts only with itself and with Dug3p.
Mutational Analysis of DUG3 Reveals That a Functional Glutamine Amidotransferase Type II Domain Is Required for Glutathione Degradation
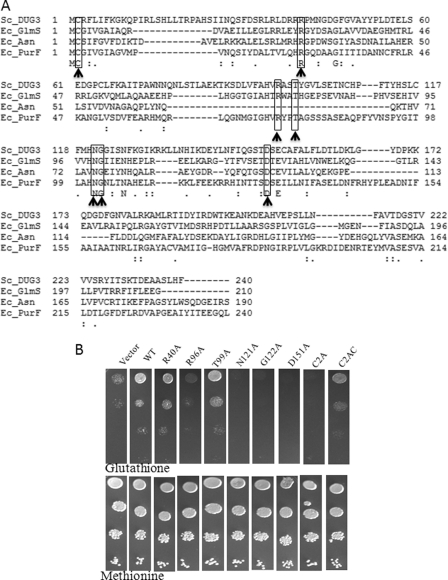
Preliminary sequence analysis had revealed that the Dug3p had a weak similarity to the GATaseII domain (24). This class of proteins has two domains in the single polypeptide chain with an N-terminal GATaseII domain and a C-terminal synthase domain (25). The GATaseII domain of different amidotransferases shares weak homology; for example, the GATaseII domain of glutamine phosphoribosyl amidotransferase of Bacillus subtilis and asparagine synthetase of E. coli share an identity of only 25% and a similarity of 41%. However, the residues important for catalysis and substrate binding as identified from the crystal structures of these enzymes are fully conserved among all the class II family members (26–31).
Multiple sequence alignments revealed that these residues were also conserved in Dug3p (Fig. 2A) despite an otherwise weak similarity with known GATaseII domains. To elucidate the role of these conserved residues in GSH degradation, these residues were mutated to alanine, and the activity of the mutant proteins was checked by complementation in a dug3Δ deletion strain. N120A, G121A, and D150A mutants showed complete loss in activity (Fig. 2B). The corresponding Asn and Gly residues in other GATases were found to be important in stabilization of the oxyanion intermediate in catalysis, whereas the Asp is important for stabilization of the α-NH2 of glutamine. The R96A mutation shows a partial defect, whereas the R40A and T99A mutants did not display any defect and behaved like wild type. The Arg-96 was predicted to be important for stabilization of the carboxylate (COO-) group of glutamine (29). Interestingly, the mutants that did not show any defect in GSH degradation (R40A and T99A) were also those that were not found to be important in catalysis in the majority of the GATaseII domain enzymes with the exception of phosphoribosyl amidotransferase (Arg-40) and glucosamine-6-phosphate synthase (Arg-40 and Thr-99). None of the mutant DUG3 constructs (tagged with HA) had any significant defect on protein expression as seen in Western blots (supplemental Fig. S3). All these results clearly indicate the functional importance of the GATaseII domain of Dug3p in GSH degradation.
FIGURE 2.
Dug3p has functional GATaseII domain. A, multiple sequence alignment of GATaseII domain of Dug3p with the GATaseII domain of glutamine phosphoribosyl amidotransferase (PurF), glucosamine-6-phosphate synthase (Glms), and asparagine synthetase (Asn). Multiple sequence alignment was carried out by the ClustalW program. Residues predicted to be important for activity are highlighted and indicated by a bold arrow. Ec, E. coli; Sc, S. cerevisiae. B, DUG3 wild type and mutant constructs in p416TEF vector were transformed into met15Δ dug3Δ strains of S. cerevisiae, and transformants were grown in minimal supplemented medium. Cells were harvested, washed, and resuspended to an A600 = 2. Serial dilutions of A600 = 0.2, 0.02, 0.002, and 0.0002 were made, and 5 μl of each dilution was spotted on minimal medium having methionine or glutathione as sulfur source. Plates were incubated at 30 °C for 3 days.
A feature of the GATaseII proteins is that they belong to the N-terminal hydrolase family of enzymes, in which the N-terminal exposed residue functions as both a nucleophile and a proton donor in catalysis and hence is absolutely essential for the activity (32). In the GATaseII domain, the N-terminal conserved cysteine residue is shown to be essential for GATase activity (25). In Dug3p, the N-terminal residue after methionine is also cysteine. To validate the importance of this cysteine as a catalytic residue, Cys-2 of Dug3p was mutated to alanine. The C2A mutation in Dug3p led to complete loss in the activity as the mutant protein was not able to complement the dug3Δ deletion phenotype for GSH utilization as the sole source of sulfur. To further confirm the importance of the cysteine at the extreme N terminus of the protein, an alanine residue was inserted before cysteine, thereby making the cysteine the second amino acid in the Dug3p polypeptide (C2AC mutation). The C2AC mutant showed a defect in the utilization of GSH, although the defect was less severe than C2A mutation (Fig. 2B). Taken together these results demonstrate that Dug3p has a functional GATaseII domain.
As most of the known members of the GATaseII family are oligomeric in nature, we evaluated the oligomeric state of Dug3p alone in the absence of Dug2p. Initial attempts at Dug3p (His6-tgged) purification were unsuccessful, and we subsequently observed that Dug3p could only be purified when a GGGGGGIP glycine linker sequence was inserted between the C terminus of Dug3p and the His tag. The linker did not affect Dug3p function (data not shown). Dug3p was expressed in BL21 (DE3) cells and purified by Ni-NTA affinity chromatography. The molecular mass of the protein was 41 kDa, and it was nearly 90% pure as seen on SDS-PAGE with Coomassie Brilliant Blue R250 staining (Fig. 1C). On gel filtration, the protein eluted as a major peak at 48 kDa, indicating that purified Dug3p, when expressed alone, primarily exists as a monomer as opposed to other GATaseII family members. However, it is possible that the association with Dug2p allows it to achieve the desired oligomeric status.
Dug2p Lacks Functional Peptidase Activity
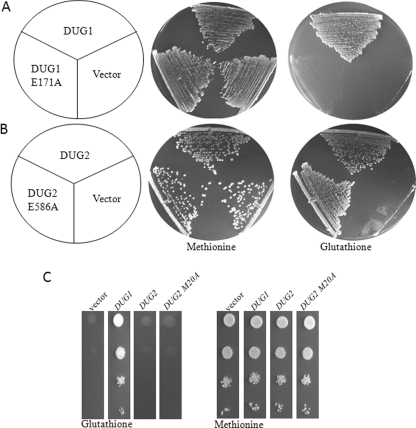
The M20A peptidase domain of Dug2p has 32% identity and 50% similarity with Dug1p. In the M20A family, a set of amino acids required for metal ion coordination and catalysis is conserved in different members of the family despite low sequence similarity (33). A glutamic acid residue is the catalytic residue in all the M20A family peptidases. On multiple sequence alignment, a glutamate residue corresponding to the catalytic glutamate is found to be conserved in both Dug1p (Glu-171) and Dug2p (Glu-586). To validate the importance of these residues and hence the Dug1p and Dug2p catalytic activity in GSH degradation, the respective glutamates were mutated to alanine, and the functionality of the mutant proteins was analyzed by the complementation in the respective deletion backgrounds. E171A mutation in Dug1p leads to complete loss in activity as assayed by growth on GSH plates (Fig. 3A). It indicates that Dug1p is catalytically important in GSH utilization by the alternative pathway. However, a similar mutation in Dug2p (E586A) complemented dug2Δ for GSH degradation like wild type Dug2p (Fig. 3B). This indicates that Dug2p may not have any direct catalytic role in GSH degradation and is possibly functioning only as a scaffold protein. To further confirm the in vivo results, we examined the peptidase activity of the in vitro purified Dug2p dimer. The activity was checked against Cys-Gly peptide. However, purified Dug2p showed negligible cleavage of Cys-Gly peptide (only 4%) when compared with the Cys-Gly peptidase activity of purified Dug1p under similar conditions (data not shown).
FIGURE 3.
Dug1p, but not Dug2p, has functional peptidase domain. A and B, conserved glutamic acid residues that had a potential catalytic role were mutated to alanine; DUG1 wild type and DUG1 E171A mutant constructs in p416TEF vector were transformed into met15Δ dug1Δ strains of S. cerevisiae (A), and DUG2 wild type and DUG2 E586A mutant construct in p416TEF vector were transformed into met15Δ dug2Δ strains of S. cerevisiae and streaked on methionine or glutathione plates (B). C, DUG1, DUG2, and DUG2 M20A domain constructs in p416TEF vector were transformed into met15Δ dug1Δ strains of S. cerevisiae. Transformants were grown in minimal supplemented medium. Cells were harvested, washed, and resuspended to an A600 = 2. Serial dilutions of A600 = 0.2, 0.02, 0.002, and 0.0002 were made, and 5 μl of each dilution was spotted on minimal medium containing either methionine or glutathione as sulfur source. Plates were incubated at 30 °C for 3 days.
Full-length Dug2p was not able to complement dug1Δ deletion phenotype despite having an M20A domain with high similarity with Dug1p (data not shown). As it was possible that the presence of the WD40 domain at the N terminus may be hindering the catalytic activity of the M20A domain of Dug2p, the M20A domain of the Dug2p was expressed separately and checked for functionality in the dug1Δ strains by growth on GSH as the sole source of sulfur. However, overexpression of the DUG2 M20A domain separately also did not complement the dug1Δ defect (Fig. 3C).
Dug2p and Dug3p Are Required Together to Cleave γ-Glutamyl Linkage
As Dug1p was only playing a role as a Cys-Gly peptidase in glutathione degradation, we needed to examine the possible γ-glutamyl cleavage activity of the Dug2p and Dug3p proteins in vitro. Dug2p and Dug3p were independently purified from E. coli as described above and were examined independently as well as together (the latter by mixing in a 1:1 stoichiometry and further dialyzing into 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, buffer at 4 °C; Dug2p and Dug3p were expected to form a complex of correct stoichiometry under these conditions, and this complex is designated as Dug2p-Dug3p). γ-Glutamyl cleavage activity of the Dug2p, Dug3p, and Dug2p-Dug3p proteins was monitored using γ-glu-pNA as artificial substrate. γ-glu-pNA cleavage activity was carried out at 37 °C in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, buffer, and pNA released was monitored at 410 nm. When Dug2p or Dug3p was present alone, they did not cleave γ-glu-pNA even at the highest protein concentrations used in the reaction. However, when both the proteins were present together, the γ-glutamyl bond of γ-glu-pNA was efficiently cleaved (data not shown). The amount of the pNA released increased proportionally upon increasing the amount of the Dug2p-Dug3p protein complex from 0.25 to 2 μg. The pNA release is also seen to be time-dependent from 15 min to 1 h when using a fixed amount of protein (1 μg).
We also needed to determine whether these proteins functioned together or in a sequential manner where activity of one protein is followed by the activity of the second protein. To test these two possibilities, the substrate was incubated initially with either 5 μg of purified Dug2p or Dug3p alone for 1 h, and protein was denatured at 95 °C for 5 min. To the supernatant of the above reaction, 5 μg of the purified second protein was added, and the reaction was again carried out for an additional 1 h at 37 °C. Interestingly, Dug2p and Dug3p were able to cleave the substrate only when both the proteins were present in the active from at the same time. Denaturing one protein before the addition of the other protein did not allow substrate cleavage (data not shown). This result strongly suggested that Dug2p and Dug3p function together most likely as a complex in the cleavage of the γ-glutamyl linkage and that this activity is independent of Dug1p.
Dug2p and Dug3p Function as Complex to Degrade GSH into Glutamate and Cys-Gly
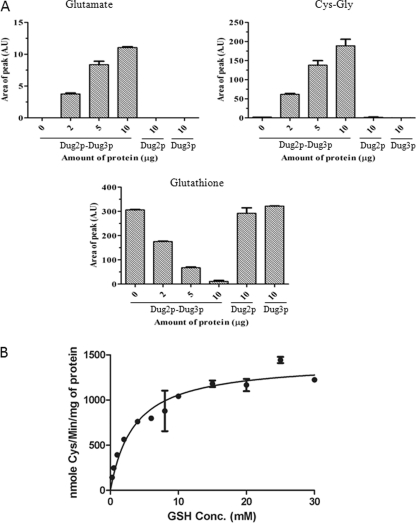
The above studies clearly demonstrated that Dug2p and Dug3p together were essential, as well as sufficient to cleave the γ-glutamyl linkage in γ-glu-pNA. We also wished to examine whether the putative complex was also responsible for the cleavage of γ-glutamyl linkage of GSH. To study this activity, the Dug2p, Dug3p, and Dug2p-Dug3p proteins (the latter mixed in 1:1 stoichiometry as described above) were incubated with 10 mm GSH for 1 h in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, buffer at 37 °C, and products were analyzed by HPLC. No degradation was observed when the Dug2p or Dug3p proteins were used separately even at high protein concentrations (10 μg). However, by increasing the amount of the Dug2p-Dug3p protein complex from 2 to 10 μg, the peak corresponding to GSH decreased with the corresponding increase in the peaks of glutamate and Cys-Gly dipeptide. At 10 μg of protein, there was complete disappearance of the peak corresponding to GSH (Fig. 4A). A similar observation was observed using γ-Glu-Cys as the substrate (supplemental Fig. S4). The peaks were further confirmed by spiking the reaction mixtures with purified GSH, glutamate, and Cys-Gly. As the appearance of glutamate (rather than oxoproline that is seen in the case of γ-glutamyl transpeptidase) was unusual, this was further confirmed using the sensitive Amplex Red fluorescent assay for glutamate (data not shown).
FIGURE 4.
Dug2p-Dug3p together cleaves glutathione into glutamate and Cys-Gly. A, purified recombinant Dug2p and Dug3p were mixed in 1:1 stoichiometry and dialyzed in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, to form a Dug2p-Dug3p complex. Dug2p and Dug3p alone were also dialyzed in the same buffer. Increasing amounts of Dug2p-Dug3p from 2 to 10 μg, 10 μg of Dug2p, or 10 μg of Dug3p were incubated with 10 mm glutathione in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, at 30 °C for 1 h, and the reaction was terminated by heating at 95 °C for 5 min. Products of the reaction were analyzed by HPLC using a C18 column in 2% perchloric acid as solvent. The area of the peak is plotted as function of the protein amount. Values are plotted as mean ± S.E. (A.U, arbitrary units × 105). B, 1 μg of the Dug2p-Dug3p complex was incubated with increasing amounts of glutathione (0.25 mm to 30 mm) in 50 mm Tris-HCl, 300 mm NaCl, pH 8.0, for 1 h at 37 °C, and the reaction was terminated at 95 °C for 5 min. The amount of the Cys-Gly produced was estimated as described under ”Experimental Procedures.“ Nanomoles of cysteine formed were plotted as a function of glutathione concentration (GSH Conc.), and Km was estimated by Michaelis-Menten graph using the GraphPad Prism software. Values are plotted as mean ± S.E.
GSH degradation by the Dug2p-Dug3p follows a classical Michaelis-Menten kinetics as analyzed by a coupled assay using Dug1p (Cys-Gly peptidase) and monitoring the cysteine formed using acidic ninhydrin (20). The unpurified complex has a Km of 3.83 ± 0.46 mm measured for GSH (Fig. 4B).
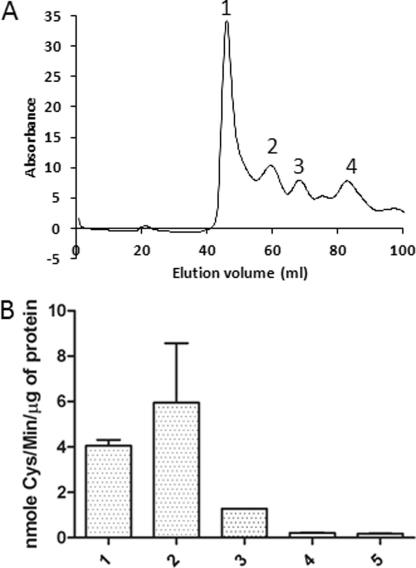
To purify and fractionate the Dug2p-Dug3p complex, Dug2p and Dug3p were mixed in 1:1 stoichiometry and fractionated on gel filtration. 10 mm GSH was added into the dialysis and gel filtration buffers to stabilize the complex. Interestingly, on mixing Dug2p and Dug3p in 1:1 stoichiometry, we could detect the higher molecular complexes of Dug2p and Dug3p with the complete absence of individual Dug2p or Dug3p protein peaks. Each of these fractions was run on SDS-PAGE to confirm the presence of both Dug2p and Dug3p proteins (Fig. 5A and supplemental Fig. S5). We could detect complexes corresponding to 156–159 (peak 3) and 298–346 kDa (peak 2), along with a higher molecular mass peak. The estimated molecular mass of the higher molecular mass fraction (∼899 kDa, peak 1) is not in the resolution limit of the column as it is above 600 kDa. Peak 4, corresponding to 53 kDa, does not have either Dug2p or Dug3p and is a contaminant protein being co-purified with Dug2p.
FIGURE 5.
Dug2p-Dug3p functions as a complex to cleave GSH. A, gel filtration profile of the Dug2p-Dug3p complex. Recombinant purified Dug2p and Dug3p were mixed in 1:1 stoichiometry, dialyzed in 300 mm NaCl, 50 mm Tris-HCl, pH 8.0, 1 mm GSH buffer for 1 h, and loaded on a Superdex S200 gel filtration column. Elution profile was monitored by absorbance at 280 nm, and four peaks were seen. Peak 1, high molecular mass fraction; peak 2, 346 kDa; peak 3, 156 kDa; peak 4, 53 kDa. B, relative activity of different Dug2p-Dug3p fractions obtained from gel filtration. GSH degradation activity of different fractions is measured by using a coupled assay using Dug1p and monitoring the cysteine formed. 1, unfractionated Dug2p-Dug3p complex; 2, high molecular mass fraction; 3, 346-kDa fraction; 4, 156-kDa fraction; 5, 53-kDa fraction. Values are plotted as mean ± S.E.
As a single heterodimer of the Dug2p-Dug3p complex including the molecular mass of purification tags corresponds to about 142 kDa, it appears that we can detect the single heterodimeric complex, as well as the dimeric (Dug2p-Dug3p)2 complex of expected molecular mass of 282 kDa and also higher order associations. It is possible that the high molecular mass fraction represents the higher molecular association of the (Dug2p-Dug3p)2 complex. The minimal functional complex was the 298–346-kDa fraction, although the higher molecular mass fraction also has GSH cleavage activity. Interestingly, the 156-kDa Dug2p-Dug3p complex lacks any significant GSH cleavage activity above the background levels, indicating that formation of the (Dug2p-Dug3p)2 complex is required for GSH degradation (Fig. 5B). GSH degradation by (Dug2p-Dug3p)2 follows a classical Michaelis-Menten kinetics as analyzed by a coupled assay using Dug1p and monitoring cysteine formed. The Km of the 346-kDa fraction was found to be 1.2 mm, whereas the higher molecular mass fractions had a Km of about 1.9 mm for GSH, which is in the physiological range of glutathione concentration. In some of the fractionation experiments, we could also detect an additional peak of 615 kDa, which also displayed GSH cleavage activity (data not shown).
Regulation of DUG Genes; DUG3 and DUG2 Are under Sulfur Regulation, whereas DUG1 Is Expressed Constitutively at Basal Levels
As GSH is an essential metabolite in the cell, the pathways of GSH degradation would need to be either compartmentalized or tightly regulated. The γ-glutamyl transpeptidase pathway had been shown to be localized in the vacuole of yeast, and it is also regulated by the nitrogen source in the medium (9, 34). We had earlier shown that DUG proteins, Dug1p, Dug2p, and Dug3p, were localized to cytosol (10). As their presence in the cytosol would deplete GSH levels in the cytosol, this suggests that their activity might be regulated. To study the regulation of the DUG1, DUG2, and DUG3 genes, their promoter regions were fused in-frame upstream to lacZ gene in plasmid pLG699z. These fusion constructs were transformed into BY4741 (met15Δ) and BY47412 (MET15) strains, and the expression was checked under different sulfur and nitrogen conditions by lacZ assay as described earlier (21).
The DUG1, DUG2, and DUG3 genes did not appear to be regulated by the nitrogen content in the medium because no significant difference in lacZ reporter activity was seen in different nitrogen conditions (repressing nitrogen sources such as glutamine/ammonium or nonrepressing nitrogen sources such as glutamate) for any of the DUG1, DUG2, and DUG3 reporter constructs. The DUG1 gene was found to be very minimally expressed in the cell, and the expression levels were also not affected by the sulfur source used in the study (data not shown).
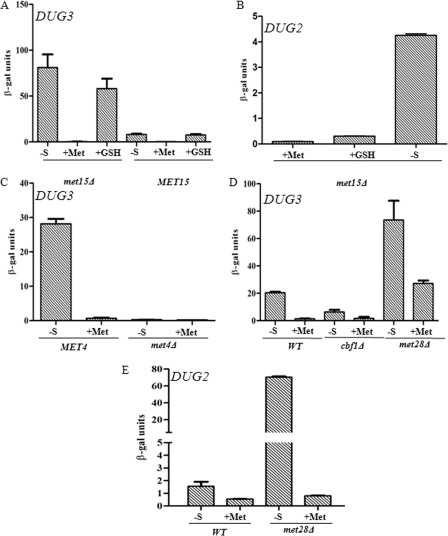
The expression of the DUG3 and DUG2 genes was strongly derepressed under sulfur limitation. We also examined the expression in a met15Δ strain. This strain is an organic sulfur auxotroph, and sulfur starvation conditions tend to be more accentuated in these strains. The effect of sulfur repression and derepression was more evident in the met15Δ strain than in a WT (MET15). (Fig. 6, A and B).
FIGURE 6.
DUG2 and DUG3 are under sulfur regulation. A, C, and D, β-galactosidase reporter assay plasmid (pLG699Z) having a 600-bp upstream region of DUG3, fused upstream of the lacZ gene, was transformed in MET15 (BY4742) and met15Δ (BY4741) strains (A), met4Δ strains (C), and cbf1Δ and met28Δ strains (D). B and E, β-galactosidase reporter assay plasmid (pLG699Z) having a 1-kb upstream region of DUG2, fused upstream of lacZ gene, was transformed in met15Δ strains (B) and met28Δ strains (E). Transformants were grown under different sulfur sources (-S, 0 μm methionine; +Met, 200 μm methionine; +GSH, 200 μm glutathione). β-Galactosidase activity is assayed for each condition as described under ”Experimental Procedures.“ The experiment was repeated twice in duplicate, and mean ± S.E. is plotted.
When we analyzed the promoter of DUG3, we detected a classical S-regulatory motif AAANTGTGG to be present at the −174 to −166 region of the DUG3 promoter (supplemental Fig. S6). This was suggestive of the pathway being regulated by the classical sulfur regulatory network (35). To check whether DUG3 was regulated by the classical sulfur regulatory networks, we carried out lacZ reporter assays in different mutant backgrounds defective in sulfur regulation. As shown in Fig. 6C, MET4 was absolutely required for the expression of DUG3 because the met4Δ strain failed to derepress DUG3 expression even under no-sulfur conditions (Fig. 6C) (36). Expression of DUG3 was reduced in the cbf1Δ background when compared with wild type even under no-sulfur conditions, whereas in met28Δ strains, the expression of DUG3 was induced in both no-sulfur (derepressing) and methionine (repressing) conditions over wild type (Fig. 6D) (37). These data indicate the role of Cbf1p as activator and Met28p as repressor of DUG3 expression. met31Δ and met32Δ had no effect on the expression of DUG3 (data not shown) (38).
Although no known sulfur regulatory motif was found to be present upstream of DUG2, we still examined whether DUG2 expression was also regulated by the classical sulfur regulatory network. Interestingly, despite the absence of a discernible motif, like DUG3, expression of the DUG2 promoter was strongly induced in met28Δ background when compared with wild type control in sulfur limitation (no methionine) and methionine (200 μm methionine) conditions (Fig. 6E).
Together these results suggest that the activity of the alternative pathway of glutathione degradation (DUG pathway) is regulated at the transcriptional level via regulation of DUG2 and DUG3, where low levels of sulfur in the media induce the expression of these proteins and thereby the formation of the competent (Dug2p-Dug3p)2 complex. DUG1 is expressed in low levels under all the conditions tested, suggesting basal, but constitutive, expression of DUG1.
DISCUSSION
In the present study, we have carried out studies on the Dug2p and Dug3p proteins that along with Dug1p are required for degradation of glutathione in yeast cells by the recently discovered alternative (DUG) pathway of glutathione degradation. The studies have delineated domains responsible for the interactions between the Dug2p and Dug3p proteins, and together with mutational analysis of key residues in these proteins and our ability to purify the Dug2p-Dug3p complex and reconstitute glutathione degradation in vitro, these results have allowed us to arrive at a model of how the DUG proteins function in glutathione degradation in this pathway.
The Dug2p protein has two distinct domains, WD40 and peptidase M20A. However, no catalytic activity could be ascribed to it.
The Dug3p protein, which showed weak similarity to the glutamine amidotransferase type II domain, is a member of the N-terminal hydrolase family of enzymes. Mutational analysis confirmed the presence of a functional GATaseII domain. As N-terminal hydrolase family members, GATaseII enzymes use the N-terminal cysteine residue as the nucleophile, and the mechanism of catalysis is general acid-base. The importance of the N-terminal cysteine of Dug3p suggests that it acts as a nucleophile for the cleavage of the γ-glutamyl bond of GSH to release glutamate and Cys-Gly.
Most of the known GATaseII enzymes are oligomeric proteins. The gel filtration profiles of purified Dug3p (when expressed alone) revealed that the protein exists as a monomer, unlike other members of the GATaseII family. However, as Dug3p interacts with Dug2p, which can also self-dimerize, the interaction of Dug3p with Dug2p can provide the oligomeric status to the otherwise monomeric Dug3p. The minimal Dug2p-Dug3p complex is thus likely to exist as a (Dug2p-Dug3p)2 because Dug2p interacts with itself by the M20A domain and each molecule of Dug2p further interacts with a molecule of Dug3p via its WD40 domain.
Purification of Dug2p and Dug3p together confirmed the minimal functional complex that corresponded to the dimer of the heterodimer, (Dug2p-Dug3p)2. Higher molecular weight oligomers were also observed and were not only functional, but had a similar Km as the (Dug2p-Dug3p)2 complex. It is difficult to determine whether the higher molecular weight oligomers also form in vivo and whether they have any physiological significance. However, what is interesting is that the Km of the complex for glutathione is within the physiological range of glutathione that is present in the cytosol, and it may explain why the complex needs to be tightly regulated by the sulfur pathway, a finding that has also been demonstrated in this study. Furthermore, in addition to demonstrating that the Dug2p and Dug3p were sufficient to cleave the γ-glutamyl linkage of GSH, we could also demonstrate that the products of the reaction were glutamate and Cys-Gly.
Earlier studies had suggested that Dug1p interacts with Dug2p. However, more detailed in vitro purification studies reported in this study clearly revealed the absence of Dug1p-Dug2p heterodimer formation. This suggested that the role of Dug1p might be limited to its Cys-Gly peptidase activity.
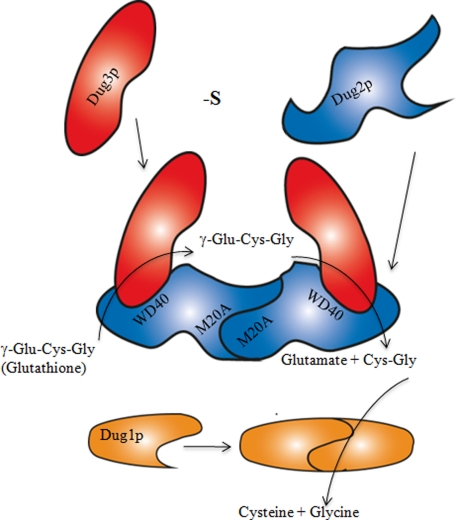
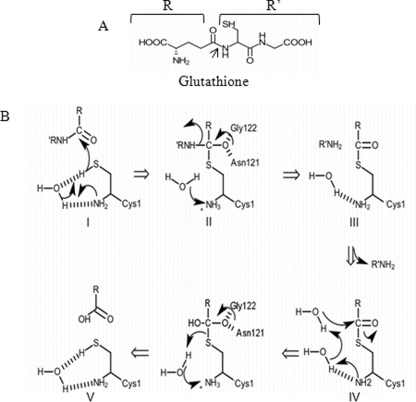
Based on these observations, we propose the following model of glutathione degradation by the DUG pathway. The Dug2p and Dug3p proteins are derepressed during sulfur limitation conditions and come together to form a (Dug2p-Dug3p)2 complex, a novel GATaseII, that may exist in vivo as such or as higher multimers. The complex is sufficient to cleave the γ-glutamyl linkage (Fig. 7). As the residues important for activity of the GATaseII proteins are shown to be essential for Dug3p activity, the catalytic mechanism for the Dug3p for cleavage of the γ-glutamyl linkage of glutathione is schematically depicted in Fig. 8. A nucleophile is activated by abstraction of proton from the thiol group of Cys-1 via the α-amino group of Cys-1, probably through a bridging water molecule. Hence, Cys-1 at the N terminus is absolutely important for Dug3p activity. The activated nucleophile attacks the amide carbon in the γ-glutamyl bond of glutathione, resulting in the formation of the tetrahedral intermediate, which is stabilized via oxyanion hole through residues Asn-121 and Gly-122. Further collapse of the tetrahedral intermediate results in the formation of γ-glutamyl thioester intermediate with simultaneous release of Cys-Gly. The γ-glutamyl thioester intermediate is deacylated through nucleophilic attack by a water molecule followed by the release of glutamate and regeneration of enzyme involving residues Asn-121 and Gly-122 (Fig. 8). The Cys-Gly then released would be acted upon by the homodimeric Dug1p to yield cysteine and glycine.
FIGURE 7.
Schematic diagram showing formation of (Dug2p-Dug3p)2 complex and role of Dug1p, Dug2p, and Dug3p proteins in glutathione degradation. Dug2p self-dimerizes to form a homodimer via its M20A peptidase domain at the N terminus. Both the subunits of the Dug2p homodimer interact with Dug3p by the WD40 domains at the C terminus of Dug2p. (Dug2p-Dug3p)2 complex can bind glutathione and cleave its γ-glutamyl linkage, thereby releasing Cys-Gly and glutamate. Cys-Gly released by the Dug2p-Dug3p complex is further acted upon by the Dug1p homodimer to form free cysteine and glycine. -S, 0 μm methionine.
FIGURE 8.
Proposed catalytic mechanism for cleavage of γ-glutamyl linkage in glutathione by GATaseII domain of Dug3p. A, structure of glutathione with representations of R and R′ groups. The arrow indicates the bond being cleaved. B, detailed catalytic mechanism proposed on the basis of the reaction mechanism of other GATaseII enzymes. Structures were drawn using the ChemDoodle software. I, activation of nucleophile by abstraction of proton from the thiol group via α-amino group of Cys-1 through bridging water molecule. Activated nucleophile attacks the amide carbon of glutathione. II, stabilization of the tetrahedral intermediate via oxyanion hole through Asn-121 and Gly-122. III, collapse of the tetrahedral intermediate to form γ-glutamyl thioester intermediate and release of Cys-Gly. IV, deacylation through nucleophilic attack by water molecule. V, release of glutamate and regeneration of enzyme.
In conclusion, the findings described in this study have enabled us to delineate the mechanism of glutathione degradation by the DUG proteins, and the results should be of great significance in understanding glutathione and sulfur homeostasis in yeasts and fungi.
Supplementary Material
This work was supported in part by a grant-in-aid project from the Department of Science and Technology and Department of Biotechnology, Government of India (to A. K. B.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S6.
- GATaseII
- glutamine amidotransferase domain II
- Cys-Gly
- cysteinyl-glycine
- γ-Glu-pNA
- γ-glutamyl-p-nitroanilide
- pNA
- p-nitroanilide
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Fahey R. C., Newton G. L. (1987) Determination of low molecular weight thiols using monobromobimane fluorescent labeling and high-performance liquid chromatography. Methods Enzymol. 143, 85–96 [DOI] [PubMed] [Google Scholar]
- 2. Meister A., Anderson M. E. (1983) Glutathione. Annu. Rev. Biochem. 52, 711–760 [DOI] [PubMed] [Google Scholar]
- 3. Penninckx M. J. (2002) An overview on glutathione in Saccharomyces versus non-conventional yeasts. FEMS Yeast Res. 2, 295–305 [DOI] [PubMed] [Google Scholar]
- 4. Barouki R., Finidori J., Chobert M. N., Aggerbeck M., Laperche Y., Hanoune J. (1984) Biosynthesis and processing of γ-glutamyl transpeptidase in hepatoma tissue culture cells. J. Biol. Chem. 259, 7970–7974 [PubMed] [Google Scholar]
- 5. Griffith O. W., Meister A. (1979) Translocation of intracellular glutathione to membrane-bound γ-glutamyl transpeptidase as a discrete step in the γ-glutamyl cycle: glutathionuria after inhibition of transpeptidase. Proc. Natl. Acad. Sci. U.S.A. 76, 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Nakano Y., Okawa S., Prieto R., Sekiya J. (2006) Subcellular localization and possible functions of γ-glutamyltransferase in the radish (Raphanus sativus L.) plant. Biosci. Biotechnol. Biochem. 70, 1790–1793 [DOI] [PubMed] [Google Scholar]
- 7. Mehdi K., Thierie J., Penninckx M. J. (2001) γ-Glutamyl transpeptidase in the yeast Saccharomyces cerevisiae and its role in the vacuolar transport and metabolism of glutathione. Biochem. J. 359, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jaspers C. J., Penninckx M. J. (1984) Glutathione metabolism in yeast Saccharomyces cerevisiae: evidence that γ-glutamyl transpeptidase is a vacuolar enzyme. Biochimie 66, 71–74 [DOI] [PubMed] [Google Scholar]
- 9. Kumar C., Sharma R., Bachhawat A. K. (2003) Utilization of glutathione as an exogenous sulfur source is independent of γ-glutamyl transpeptidase in the yeast Saccharomyces cerevisiae: evidence for an alternative glutathione degradation pathway. FEMS Microbiol. Lett. 219, 187–194 [DOI] [PubMed] [Google Scholar]
- 10. Ganguli D., Kumar C., Bachhawat A. K. (2007) The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics 175, 1137–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stirnimann C. U., Petsalaki E., Russell R. B., Müller C. W. (2010) WD40 proteins propel cellular networks. Trends Biochem. Sci. 35, 565–574 [DOI] [PubMed] [Google Scholar]
- 12. Smith T. F., Gaitatzes C., Saxena K., Neer E. J. (1999) The WD repeat: a common architecture for diverse functions. Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 13. Kaur H., Kumar C., Junot C., Toledano M. B., Bachhawat A. K. (2009) Dug1p is a Cys-Gly peptidase of the γ-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J. Biol. Chem. 284, 14493–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ito S., Miyahara R., Takahashi R., Nagai S., Takenaka K., Wada H., Tanaka F. (2009) Stromal aminopeptidase N expression: correlation with angiogenesis in non-small cell lung cancer. Gen. Thorac. Cardiovasc. Surg. 57, 591–598 [DOI] [PubMed] [Google Scholar]
- 15. Guthrie C., Fink G. R. (1991) Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–863 [PubMed] [Google Scholar]
- 16. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: a Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17. Gyuris J., Golemis E., Chertkov H., Brent R. (1993) Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell 75, 791–803 [DOI] [PubMed] [Google Scholar]
- 18. Penninckx M., Jaspers C., Wiame J. M. (1980) Glutathione metabolism in relation to the amino acid permeation systems of the yeast Saccharomyces cerevisiae. Occurrence of γ-glutamyl transpeptidase: its regulation and the effects of permeation mutations on the enzyme cellular level. Eur. J. Biochem. 104, 119–123 [DOI] [PubMed] [Google Scholar]
- 19. Ohkama-Ohtsu N., Oikawa A., Zhao P., Xiang C., Saito K., Oliver D. J. (2008) A γ-glutamyl transpeptidase-independent pathway of glutathione catabolism to glutamate via 5-oxoproline in Arabidopsis. Plant Physiol. 148, 1603–1613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gaitonde M. K. (1967) A spectrophotometric method for the direct determination of cysteine in the presence of other naturally occurring amino acids. Biochem. J. 104, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guarente L. (1983) Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 101, 181–191 [DOI] [PubMed] [Google Scholar]
- 22. Jösch C., Klotz L. O., Sies H. (2003) Identification of cytosolic leucyl aminopeptidase (EC 3.4.11.1) as the major cysteinyl-glycine-hydrolyzing activity in rat liver. Biol. Chem. 384, 213–218 [DOI] [PubMed] [Google Scholar]
- 23. Cappiello M., Lazzarotti A., Buono F., Scaloni A., D'Ambrosio C., Amodeo P., Méndez B. L., Pelosi P., Del Corso A., Mura U. (2004) New role for leucyl aminopeptidase in glutathione turnover. Biochem. J. 378, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zalkin H. (1993) The amidotransferases. Adv. Enzymol. Relat. Areas Mol. Biol. 66, 203–309 [DOI] [PubMed] [Google Scholar]
- 25. Massière F., Badet-Denisot M. A. (1998) The mechanism of glutamine-dependent amidotransferases. Cell. Mol. Life Sci. 54, 205–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Muchmore C. R., Krahn J. M., Kim J. H., Zalkin H., Smith J. L. (1998) Crystal structure of glutamine phosphoribosylpyrophosphate amidotransferase from Escherichia coli. Protein Sci. 7, 39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mouilleron S., Badet-Denisot M. A., Golinelli-Pimpaneau B. (2006) Glutamine binding opens the ammonia channel and activates glucosamine-6P synthase. J. Biol. Chem. 281, 4404–4412 [DOI] [PubMed] [Google Scholar]
- 28. Larsen T. M., Boehlein S. K., Schuster S. M., Richards N. G., Thoden J. B., Holden H. M., Rayment I. (1999) Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product. Biochemistry 38, 16146–16157 [DOI] [PubMed] [Google Scholar]
- 29. Milewski S. (2002) Glucosamine-6-phosphate synthase: the multi-facets enzyme. Biochim. Biophys. Acta 1597, 173–192 [DOI] [PubMed] [Google Scholar]
- 30. van den Heuvel R. H., Curti B., Vanoni M. A., Mattevi A. (2004) Glutamate synthase: a fascinating pathway from l-glutamine to l-glutamate. Cell. Mol. Life Sci. 61, 669–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smith J. L. (1998) Glutamine PRPP amidotransferase: snapshots of an enzyme in action. Curr. Opin. Struct. Biol. 8, 686–694 [DOI] [PubMed] [Google Scholar]
- 32. Oinonen C., Rouvinen J. (2000) Structural comparison of Ntn hydrolases. Protein Sci. 9, 2329–2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rawlings N. D., Morton F. R., Barrett A. J. (2006) MEROPS: the peptidase database. Nucleic Acids Res. 34, D270–D272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Springael J. Y., Penninckx M. J. (2003) Nitrogen source regulation of yeast glutamyl transpeptidase-glutamyl transpeptidase synthesis involves the regulatory network including the GATA zinc-finger factors Gln3, Nil1/Gat1, and Gzf3. Biochem. J. 371, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thomas D., Surdin-Kerjan Y. (1997) Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 61, 503–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomas D., Jacquemin I., Surdin-Kerjan Y. (1992) MET4, a leucine zipper protein, and centromere-binding factor 1 are both required for transcriptional activation of sulfur metabolism in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 1719–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kuras L., Cherest H., Surdin-Kerjan Y., Thomas D. (1996) A heteromeric complex containing the centromere-binding factor 1 and two basic leucine zipper factors, Met4 and Met28, mediates the transcription activation of yeast sulfur metabolism. EMBO J. 15, 2519–2529 [PMC free article] [PubMed] [Google Scholar]
- 38. Blaiseau P. L., Isnard A. D., Surdin-Kerjan Y., Thomas D. (1997) Met31p and Met32p, two related zinc finger proteins, are involved in transcriptional regulation of yeast sulfur amino acid metabolism. Mol. Cell. Biol. 17, 3640–3648 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.