Background: The caseinolytic protease P (ClpP) degrades proteins within a chamber.
Results: An arginine sensor links ClpP oligomerization with activity.
Conclusion: The tetradecameric oligomerization state is necessary for proper formation of the active site.
Significance: The results reveal unprecedented insights into ClpP assembly, regulation, and substrate release.
Keywords: Bacteria, Enzyme Mechanisms, Mutagenesis, Protease, X-ray Crystallography, ClpP, Oligomerization, Virulence
Abstract
The barrel-shaped caseinolytic protease P (ClpP) is a main virulence regulator in the bacterial pathogen Staphylococcus aureus (SaClpP). It consists of two heptameric rings forming a homotetradecamer with an inner chamber that houses the 14 active sites. We recently showed that SaClpP is able to adopt a compressed, inactive conformation. We present here the 2.3 Å resolution structure of SaClpP in its closed, active conformation as well as the structure of the S98A mutant. Comprehensive mutational analysis aiming at destabilizing one or the other or both conformations was able to pinpoint key residues involved in this catalytic switch and in the heptamer-heptamer interaction. By probing the active site serine with a covalently modifying β-lactone probe, we could show that the tetradecameric organization is essential for a proper formation of the active site. Structural data suggest that a highly conserved hydrogen-bonding network links oligomerization to activity. A comparison of ClpP structures from different organisms provides suggestive evidence for the presence of a universal mechanism regulating ClpP activity in which binding of one subunit to the corresponding subunit on the other ring interface is necessary for the functional assembly of the catalytic triad and thus for protease function. This mechanism ensures controlled access to the active sites of a highly unspecific protease.
Introduction
The caseinolytic protease P (ClpP)5 protein is a highly conserved serine protease with homologs in most bacterial and eukaryotic organisms (1). It has been assigned roles in stress response, protein quality control, and transcriptional regulation, and it is one of the major machineries involved in cellular protein degradation (2–8). In pathogenic bacteria such as Staphylococcus aureus it has furthermore been attributed functions associated with virulence regulation which makes it an interesting target for antivirulence treatment of bacterial infections (9–12).
ClpP consists of two heptameric rings forming a cylindrically shaped homotetradecamer with an inner chamber in which 14 active sites align in two rings (13). ClpP alone shows only moderate and unspecific peptidase activity (8). The proteolytically active complex is intracellularly formed by interaction of ClpP with ATP-driven chaperones from the AAA+ family of proteins such as ClpX or ClpC yielding the ClpXP or ClpCP complexes, respectively (14). The chaperone recognizes, binds, unfolds, and then threads proteins prone to degradation into the inner chamber of the protease where they are subsequently degraded (15, 16).
The crystal structures of several ClpP proteins have been determined, including those of Escherichia coli (13), Bacillus subtilis (17), and Plasmodium falciparum (18). The structures show a common fold with three distinct features: (i) flexible N-terminal loops protrude on the axial side ends of the cylinders and facilitate the interaction with assisting chaperones (14); (ii) a large head domain comprises the active site residues in the inner side of the cylinder and highly hydrophobic surfaces responsible for the intra-ring subunit-subunit interface (17); (iii) moreover, a handle domain (helix E) interacts with its counterpart on the opposite ring. Surprisingly, deletion of the handle domain does not lead to dissociation into heptamers, but yields proteolytically inactive tetradecamers (19). This led to the assumption that the interaction between the two rings is mainly stabilized by charge-charge interactions between residues of the head domain (20).
Although much work has been carried out to characterize the chaperone and the chaperone-protease interaction (14), the core protease function on the molecular level is rather poorly understood. It is generally assumed that equatorial side pores formed by the handle region are responsible for peptide release (21, 22). An NMR-based study demonstrated that this helical part of the handle domain is highly dynamic in solution and is able to adopt two distinct conformations that rapidly exchange at elevated temperatures (23). Moreover, a normal mode analysis based on an artificially cross-linked E. coli ClpP mutant structure suggests that ClpP samples different conformations (24).
We recently showed that ClpP from S. aureus (SaClpP) is able to adopt a compressed, inactive conformation (21). Although a similar conformation was observed before in the structures of Mycobacterium tuberculosis, P. falciparum, Streptococcus pneumonia (A153P), the full handle domain in this compressed state could be observed for the first time in the SaClpP structure. This region shows no defined electron density in the other structures including a recent structure of the B. subtilis ClpP in the compressed state (25). Looking through the ClpP entries in the Protein Data Bank, one notes that all ClpP structures fall into two categories: either they show an extended E helix and a catalytic triad in its active rearrangement, or they show a compressed cylinder ∼1 nm smaller in height with unaligned active site residues (for a complete list, see supplemental Table 2).
However, it has not been demonstrated to date that both conformations are relevant to the catalytic cycle of the ClpP protease. Moreover, it is presently unclear whether the different conformations in the handle domain also impact on the oligomeric state of the protease. Contradicting statements regarding the link between oligomeric organization and activity are found in the literature (22, 26). Moreover, the contributions of the residues forming the inter-ring interface have not yet been fully experimentally validated. This is important because some of the residues that are assumed to be involved in mediating this connection show drastically changed interaction partners in the two states of the protease.
We therefore set out to characterize comprehensively this general mechanism underlying ClpP protease function on a molecular basis. We report here structural and mutational studies that provide unique insights into ClpP protease function establishing a link between activity and structural organization.
EXPERIMENTAL PROCEDURES
Strain Construction
Primers used are listed in supplemental Table 1. The expression strain of C-terminally STREP-II-tagged SaClpP from S. aureus NCTC 8325 (protein ID: YP_499347) was constructed using primers 1 and 2 according to the Invitrogen Gateway cloning protocol with pDonr207Gen as donor vector and pET301Amp as destination vector. Point-mutated versions of SaClpP were constructed using the QuikChange II site-directed mutagenesis protocol (Stratagene) with pDonr207-SaClpP-wt serving as template. Amplification reactions were carried out using Phusion High-Fidelity DNA Polymerase (NEB) with individually optimized reaction conditions and primers designed by the Stratagene QuikChange PrimerDesign program. Residual wild-type plasmid was digested with DpnI, and PCR products were transformed into chemically competent XL1-Blue cells for ring closing ligation. Expression plasmids were transformed into chemically competent BL21(DE3) cells (Invitrogen). All plasmids were verified by single-strand sequencing (GATC Biotec). Antibiotics were used at 100 μg/ml (ampicillin) and 15 μg/ml (gentamicin).
Protein Expression
For purification of SaClpP variants, overnight cultures of the appropriate BL21(DE3) strain were diluted 1:100 in 1 liter of LB medium and grown shaking at 37 °C. When an A600 of 0.6 was reached, isopropyl-1-thio-β-d-galactopyranoside was added to 500 μm final concentration, and cultures were grown for 16 h. Cultures were pelleted, washed with PBS, resuspended in 20 ml of lysis buffer (150 mm NaCl, 100 mm Tris-HCl, pH 8.0, 1 mm EDTA) and lysed with a Constant Cell Disruption System. Following sonification for 60 s, the lysates were cleared by centrifugation at 36,000 × g for 30 min. The supernatant was loaded on a 5-ml preequilibrated StrepTrap column (GE Healthcare), which was washed with 40 ml of binding buffer. Protein was then eluted into elution buffer (binding buffer + 2.5 mm desthiobiotin) using an ÄKTA Purifier 10 (GE Healthcare). Protein containing fractions were pooled and concentrated. Buffer exchange was performed using HiTrap Desalting columns (GE Healthcare). The protein obtained was found to be sufficiently pure for biochemical assays judged by SDS-PAGE. Concentrations were measured on a Tecan infinite M200Pro plate reader by absorption at 280 nm (E(SaClpP-wt) = 14,440 m−1 cm−1). Yields were between 10 and 80 mg of purified protein per liter of culture. Protein samples for crystallization trials were concentrated to 1 ml and further purified by size exclusion chromatography using a HiLoad 16/60 Superdex 200-pg column (GE Healthcare) with buffer A (100 mm NaCl, 20 mm Tris-HCl, pH 7.0).
Crystallization and Structure Determination
SaClpP was concentrated to 10 mg/ml in buffer A (100 mm NaCl, 20 mm Tris-HCl, pH 7.0). Crystals of ClpP were grown at 20 °C within 2 days to their final size of about 0.2 × 0.2 × 0.3 μm3 by using the hanging drop vapor diffusion method. Drops contained equal volumes of protein solution and reservoir solution (200 mm sodium malonate, containing 38% MPD for the wild-type protein and 180 mm magnesium acetate, 100 mm sodium cacodylate, pH 7.0, 12% MPD for the S98A mutant protein). Crystals were soaked for about 30 s in mother liquor and were subsequently cooled in a stream of nitrogen gas at 100 K (Oxford Cryo Systems). Data were processed using the program package XDS (27).
Wild-type ClpP crystallized in the space group P21 with cell parameters of a = 117 Å, b = 95 Å, c = 139 Å, β = 98° (see Table 1). S98A mutant ClpP crystallized in the space group P1 with cell parameters of a = 98 Å, b = 110 Å, c = 171 Å, α = 73°, β = 79°, γ = 71°. Crystal structure analysis was performed by molecular replacement using the program PHASER (28) and coordinates of ClpP from S. aureus deposited at the Protein Data Bank (code 3QWD) (21). Model building was performed with the graphic program MAIN (29). The models were refined by REFMAC (30) using conventional crystallographic rigid body, positional, and isotropic temperature factor refinements yielding current crystallographic values of Rwork 20.1%, Rfree 23.1%, r.m.s.d. bond length 0.005 Å, and r.m.s.d. bond angle 0.86° for wild-type ClpP and Rwork 21.6%, Rfree 24.0%, r.m.s.d. bond length 0.012 Å, and r.m.s.d. bond angle 1.36° for S98A mutant ClpP.
TABLE 1.
Data collection and refinement statistics
| Statistics | SaClpP-wt | SaClpP-S98A |
|---|---|---|
| Crystal parameters | ||
| Space group | P21 | P1 |
| Cell constants | a = 117 Å, b = 95 Å, c = 139 Å, β = 98° | a = 98 Å, b = 110 Å, c = 171 Å, α = 73°, β = 79°, γ = 71° |
| Molecules per AUa | 1 | 2 |
| Data collection | ||
| Preirradiation | No | No |
| SLS, X06DA | SLS, X06DA | SLS, X06DA |
| Wavelength (Å) | 1.0 | 1.0 |
| Resolution range (Å)b | 48–2.3 (2.4–2.3) | 25–2.8 (2.9–2.8) |
| No. unique reflectionsc | 129,058 | 150,830 |
| Completeness (%)b | 96.1 (98.6) | 95.6 (96.5) |
| Rmerge (%)b,d | 6.5 (51.0) | 9.1 (39.3) |
| I/σ (I)b | 11.21 (2.23) | 5.55 (1.67) |
| Refinement (REFMAC5) | ||
| Resolution range (Å) | 15–2.3 | 15–2.8 |
| No. atoms | ||
| Protein | 19,908 | 39,789 |
| Water | 1,119 | 676 |
| Rwork/Rfree (%)e | 20.1/23.1 | 21.6/24.0 |
| R.m.s. deviationsf | ||
| Bond lengths (Å) | 0.005 | 0.012 |
| Bond angles (°) | 0.860 | 1.357 |
| Average B factor (Å2) | 48.15 | 86.52 |
| Ramachandran plot (%)g | 98.7/1.3/0.0 | 97.8/2.2/0.0 |
| PDB accession code | 3V5E | 3V5I |
a Asymmetric unit.
b The values in parentheses of resolution range, completeness, Rmerge, and I/σ (I) correspond to the last resolution shell.
c Friedel pairs were treated as different reflections.
d Rmerge(I) = ΣhklΣj |[I(hkl)j − II(hkl)]|/[Σhkl Ihkl, where I(hkl)j is the measurement of the intensity of reflection hkl and <I(hkl)> is the average intensity.
e r = Σhkl | |Fobs| − |Fcalc| |/ Σhkl |Fobs|, where Rfree is calculated without a sigma cutoff for a randomly chosen 5% of reflections, which were not used for structure refinement, and Rwork is calculated for the remaining reflections.
f Deviations from ideal bond lengths/angles.
g Number of residues in favored region/allowed region/outlier region.
Coordinates were confirmed to have good stereochemistry indicated by the Ramachandran plot with 98.7% (97.8% for the S98A mutant protein) of residues in the most favored region and 1.3% (2.2%) of residues in the additionally allowed regions. In the asymmetric unit of the wild-type protein, the refined structure contains one ClpP molecule (the N-terminal 17 amino acids being structurally disordered) and 1211 water molecules. The asymmetric unit of the S98A mutant protein structure contains two practically identical ClpP molecules that are superimposable with an r.m.s.d. of 0.2 Å.
Peptidase Activity Assay
Peptidase activity of SaClpP WT and mutant proteins was measured using a fluorescent substrate assay. Processing of Suc-LeuTyr-AMC leads to the release of fluorescent AMC which can be quantified by spectroscopic readout. In a typical experiment, 10 μl of diluted protein solution (0.25 mg/ml) was added to 40 μl of Suc-LeuTyr-AMC (different concentrations ranging from 25 to 940 μm in buffer A) in black 96-well flat bottom plates (Cellstar). Fluorescence was measured with a Tecan infinite M200Pro plate reader at 37 °C (excitation, 380 nm; emission, 440 nm) for 30 cycles every 60 s. All data were recorded in triplicate. Kinetic fitting was performed with OriginPro 8 software. Mutants were defined as inactive if their activity was <1% of the wild-type activity. Basal activity is used to describe mutants that show between 1 and 10% of the respective wild-type activity.
Gel Filtration Experiments
Gel filtration experiments were performed on an ÄKTA Purifier 10 coupled to an SLS TDA 305 triple detector array (Malvern-Viscotek) with a calibrated Superdex 200 10/300 GL column (GE Healthcare) in buffer A. For calibration, a set of standard proteins (GE Healthcare) was used: ferritin (440 kDa), aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), ribonuclease A (13.7 kDa) and aprotinin (6.5 kDa). Protein masses were estimated from the retention times by linear regression according to the column manufacturer's manual.
Activity-based Labeling Assay
Protein samples were diluted to 0.25 mg/ml into buffer A in a total volume of 43 μl. 1 μl of ClpP-specific lacton probe D3 was added (5 mm in dimethyl sulfoxide, 10 μm final concentration in 50 μl). Following 60 min of incubation at room temperature (22 °C), rhodamine azide (1 μl, 5 mm, 100 μm final concentration) was added followed by the addition of TCEP (1 μl, 50 mm in ddH2O, 1 mm final concentration) and TBTA (3 μl, 1.67 mm in dimethyl sulfoxide, 100 μm final concentration). Samples were gently mixed, and the cycloaddition was initiated by the addition of CuSO4 (1 μl, 50 mm in ddH2O, 1 mm final concentration). The reactions were incubated at 22 °C for 1 h. For SDS-PAGE, 50 μl of 2× SDS loading buffer was added and 50 μl of the resulting solution applied to the gel. Fluorescence was recorded in a Fujifilm Las-4000 luminescent image analyzer with a Fujinon VRF43LMD3 lens and a 575DF20 filter.
Thermal Shift Assays
Thermal shift assays were performed on a Bio-Rad CFX 96 Real Time Cycler. In a typical experiment, 5 μl of 45× SYPRO Orange (Sigma-Aldrich, diluted from 5000× stock into buffer A), 15 μl of buffer A, and 5 μl of the respective protein sample (0.25 mg/ml, diluted into buffer A) were mixed on ice in a white 96-well PCR plate (Brand), and fluorescence was measured from 20 °C to 80 °C in 0.5 °C steps (excitation, 450–490 nm; detection, 560–580 nm). All measurements were carried out in triplicate. Data evaluation and melting point determination were performed using the Bio-Rad CFX Manager software (see supplemental Table 3).
Circular Dichroism Spectroscopy
Protein samples for CD analysis were diluted into 10 mm potassium phosphate, pH 7.0, 100 mm NaCl at 0.1 mg/ml protein concentration. Circular dichroism was recorded on a JASCO J-715 spectropolarimeter from 188 to 260 nm at 20 °C.
RESULTS
X-ray Structures of SaClpP in Its Active Conformation
To prove that the compressed state of SaClpP reflects a conformational state rather than a species-specific fold, we sought a crystal structure of SaClpP in its active, extended conformation. We cloned, expressed, and purified SaClpP with a C-terminal Strep Tag II to >99% purity as determined by SDS-PAGE. Because the crystals of SaClpP in the inactive compressed state were obtained at low pH of 4.5, we first determined a pH profile of SaClpP activity. We therefore made use of a fluorogenic substrate assay in which a peptide-AMC conjugate is cleaved by the protease to release 7-amino-4-methylcoumarin whose increase was quantified by spectroscopic readout. SaClpP shows highest activity at pH 7.0 with half-maximal activity at 6.5 and 8.0 (see supplemental Fig. 1). We therefore focused in our crystallization trials for the active, extended conformation on conditions with pH values within this region.
Crystals were obtained at pH 7.0 that diffracted to 2.3 Å resolution, and the structure was solved by molecular replacement using the coordinates of the compressed SaClpP state (21). The refined coordinates fulfill all geometric restraints (see Table 1). The structure shows two rings of heptamers stacked on top of each other consistent with the general topology of ClpP proteases. The handle domain is well defined in electron density and connects the two heptameric rings, resulting in a cylindrical shape with a height of 10 nm. The residues forming the catalytic triad (Ser98, His123, and Asp172) are aligned in all 14 subunits to form two hydrogen bridges, which, taken together, classify the structure as the active conformation of SaClpP (see Fig. 1, A and D).
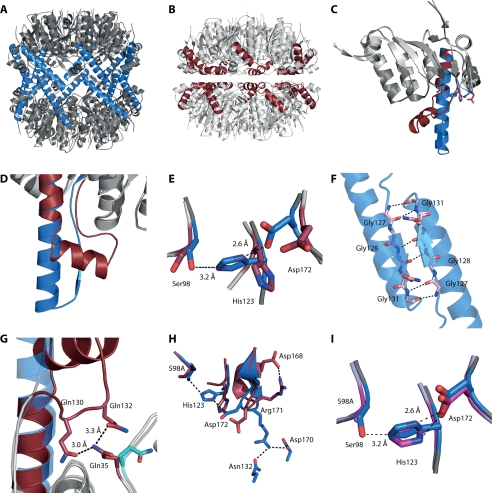
FIGURE 1.
A, tetradecameric SaClpP in the active, extended conformation with the E helix residues colored in blue. B, previously reported (21) structure of SaClpP in the inactive, compressed state with the E helix residues colored in red. C and D, superimposition of a monomer of the two states of SaClpP. E, comparison of the active side residues. F, two handle domains of subunits of different rings interact via an antiparallel β-sheet. G, kinked state stabilized by contacts of Gln130 and Gln132 of handle domain and Gln35 of head domain. H, impact of different conformations of the Arg171 residue in the two states on the orientation of the catalytic residues. I, superimposition of active site residues of the active, extended structure of wild-type SaClpP and of the S98A mutant structure.
While this paper was in preparation, a similar structure of the extended conformation was reported (22). In this structure, however, the residues of the catalytic triad are not aligned because His123 is tilted and not engaging in any contacts. We suspect this to be due to protonation of His123 caused by a lower pH of 6.5 in the crystallization conditions as opposed to the ideal pH of 7.0.
Our observation allowed comparison of the two states of the ClpP protease within one species (see Fig. 1, A–E). The two head domains (residues 17–122 and 146–192) could perfectly be aligned with an r.m.s.d. of 0.3 Å using PyMOL (31). The main differences between the two states could be found in the handle domain. Structural differences originate at the carbonyl groups of Ile122 which point into opposite directions. This shift in orientation also induces different conformations of the adjacent active site His123. The following residues Gln124 to Gln132 of the handle domain protrude out of the head domain and form an antiparallel β-sheet. In this motif, three highly conserved glycines (Gly127, Gly128, Gly131) engage with the respective residues of a monomer on the other ring (see Fig. 1F). Residues Ala133 to Lys145 form an α-helix that directs the strand back to the head domain. The backbone atoms of Thr146 of both states are in good alignment. In the compressed state, the helix is kinked at this point, and residues His123 and Glu135 are linked via a loop structure. This fold is stabilized by a hydrogen bridge network that involves the conserved residues Gln130 and Gln132 in the loop and the nonconserved Gln35 in the head domain (Fig. 1G).
A comparison of the active site residues of both conformations is shown in Fig. 1E. In the extended state, the distance between the active site Ser98Oγ and the His123Nϵ is 3.2 Å, and the distance between His123Nδ and a carbonyl oxygen of Asp172 is 2.6 Å, which shows that the existence of activating hydrogen bonds is highly probable. In the compressed state, however, Asp172 is located with an outbound orientation due to the drastically changed position of the Arg171 side chain and, hence, cannot engage in hydrogen bonding (Fig. 1H). The imidazole ring of His123 is thereby rotated and shifted by ∼3.5 Å.
To find out whether these conformations can be attributed to the different orientations of Arg171 rather than to the different positions of the active site serine, we crystallized the S98A mutant protein and obtained the structure in the extended conformation to 2.8 Å resolution with Rfree = 24.0%. The wild-type and the S98A mutant structures show no significant differences and can be aligned with an r.m.s.d. of 0.2 Å (Fig. 1I). Importantly, the active site His123 adopts the same conformation in both structures proving that the alignment of the active site residues is independent from the active site serine hydrogen bond donor capacities and, therefore, must be due to the conformation of the Arg171 residue.
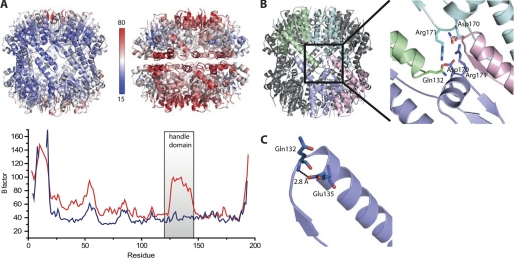
A comparison of the crystallographic B factors, indicative for structural flexibility, shows that the N-terminal loops (residues 1–20) as well as adjacent loops at the top of the cylinder (residues 55–60) in all structures are highly flexible (see Fig. 2A). Interestingly, the handle domain of the compressed state is also flexible at the ring-ring interface (with His142 showing an alternative conformation) whereas the corresponding residues in the extended state are rather rigid. The highly different B factors are an inherent feature of the two conformations and cannot be attributed to a different crystal packing. The effect can also be seen for residues at the tip of the helix within the chamber of the compressed state where there is no influence of the crystal packing.
FIGURE 2.
A, upper, main chain B factor values mapped on schematic representations of the two states. Lower, main chain B factor values of the two states plotted over the residue number. B, four subunits on both rings are linked via a hydrogen bond network that involves residues Gln132, Asp170, and Arg171. C, tip of the extended helix E stabilized by an interaction of Glu135 and Gln132.
Salt Bridge Network of Asp170, Arg171, and Gln132 at Inter-ring interface Is Essential for Both Tetradecamer Formation and Activity
In the extended state the two heptameric rings engage in two different types of interactions. For the first interaction, the handle domains of two monomers on different rings form an antiparallel β-sheet. For the second interaction, a salt bridge network links Arg171 of one monomer with Asp170 of the opposite ring (see Fig. 2B). The second nitrogen in the guanidinium group of Arg171 forms a contact with Gln132 of a monomer adjacent to it. Arg171 thereby links three different subunits and both rings. In the compressed state, however, Arg171 adopts a different conformation in which it forms a hydrogen bond with the conserved Glu168 of the same subunit (Fig. 1H). Asp170 is flipped inward and forms an interaction with Arg147. In essence, multiple cross-subunit interactions that are present in the extended state and thereby are closing the side walls of the cylinder are not existent in the compressed state.
To evaluate the contributions of these residues to the structural organization of ClpP, we individually mutated Gln132, Asp170, and Arg171 to alanine. We furthermore mutated Arg171 to lysine to see whether one of the two acceptor functions is sufficient. We first checked the overall fold of the mutant proteins by circular dichroism spectroscopy which showed only minor changes (data not shown). We next characterized the oligomeric organization of the mutant proteins by size exclusion chromatography as well as static light scattering. Whereas D170A, R171A, and R171K eluted as single heptameric species (Table 2), Q132A eluted predominantly as heptamers with a small percentage of a tetradecameric population. We further investigated whether the transition from a tetradecamer to heptamers could be induced thermally. Protein melting curves were measured using the hydrophobic reporter dye SYPRO Orange. The wild-type protein showed a single unfolding event with a melting temperature of 58.3 ± 0.3 °C which is similar to the values of the mutant proteins (D170A, 60.8 ± 0.5 °C; R171A, 58.5 ± 0.4 °C; R171K, 58.9 ± 0.3 °C). Due to the absence of a second unfolding event at lower temperature only for wild-type but not for heptameric mutant protein, we concluded that the global unfolding of the wild-type protein does not proceed through a heptameric intermediate.
TABLE 2.
Analysis of the oligomeric state of ClpP mutants
The molecular mass was determined by calculation from the retention time of a calibrated size exclusion chromatography column (SEC) as well as by static light scattering (SLS). The peak area was determined by integration of the respective UV absorption signal at 280 nm. The expected mass of a tetradecameric complex is 316 kDa.
| Protein | Molelcular mass |
Peak area | Oligomeric state | |
|---|---|---|---|---|
| SEC | SLS | |||
| kDa | kDa | % | ||
| WT | 290 | 304 | 100 | Tetradecamer |
| Q35A | 303 | 304 | 88 | Tetradecamer |
| 69 | 50 | 12 | Dimer? | |
| S98A | 302 | 325 | 100 | Tetradecamer |
| S98C | 308 | 319 | 100 | Tetradecamer |
| S98T | 317 | NDa | 35 | Tetradecamer |
| 173 | NDa | 65 | Heptamer | |
| G127A/G128A/G131A | 295 | 287 | 20 | Tetradecamer |
| 170 | 167 | 80 | Heptamer | |
| Q130A | 273 | 260 | 5 | Tetradecamer |
| 176 | 185 | 95 | Heptamer | |
| Q132A | 283 | 316 | 7 | Tetradecamer |
| 82 | 63 | 93 | Trimer? | |
| E135A | 148 | 176 | 100 | Heptamer |
| E135R | 281 | 317 | 100 | Tetradecamer |
| E137A | 289 | 292 | 100 | Tetradecamer |
| L144E | 168 | 142 | 47 | Heptamer |
| 68 | 22 | 53 | Monomer | |
| L144G | 164 | 131 | 75 | Heptamer |
| 109 | 84 | 25 | Tetramer? | |
| L144M | 300 | 316 | 100 | Tetradecamer |
| L144R | 260 | 319 | 100 | Tetradecamer |
| D170A | 170 | 163 | 100 | Heptamer |
| R171A | 168 | 151 | 100 | Heptamer |
| R171K | 162 | 146 | 100 | Heptamer |
a ND, not determined.
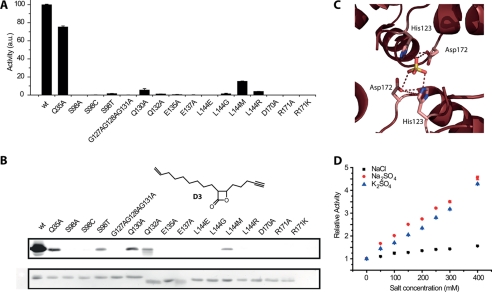
In contrast to ClpP from E. coli (32), SaClpP does not degrade full-length proteins like casein as determined by a FITC-casein assay (data not shown). We therefore determined the peptidase activity of all mutants relative to the wild-type protein with the fluorescent substrate assay. D170A, R171A, and R171K proteins were inactive, whereas Q132A showed a significantly reduced activity (see Fig. 3A).
FIGURE 3.
A, peptidase activity of all mutant proteins relative to the wild-type protein. B, SaClpP-specific activity-based probe lactone D3. Upper, fluorescence gel of all mutant proteins that were incubated with lactone D3 and subsequently labeled with rhodamine azide via click chemistry. Lower, loading control via Coomassie staining. C, close-up view on the inter-ring interface in a previously reported (22) data set of the compressed state of SaClpP showing a sulfate ion bridging active site residues. D, kinetic analysis with a fluorescent substrate assay shows a linear increase in activity when sodium sulfate and potassium sulfate are added to the assay buffer.
To probe the structural organization of the active site, we made use of the ClpP-specific probe D3 (9). This electrophilic β-lactone is subjected to nucleophilic attack by the active site serine. This leads to ring opening of the β-lactone and a covalent modification of the active site serine. We subjected the recombinant proteins to probe D3 (see Fig. 3B), incubated them at room temperature for 30 min, and then attached a fluorescent dye to the probe via an alkyne handle and click chemistry. We separated the free probe from the labeled protein via SDS-PAGE and fluorescently scanned the gel (see Fig. 3B). D170A, R171A, and R171K showed no signal, which is consistent with the previous results from the fluorescent substrate assay. The activity defect of these mutants can therefore be explained by a malformation of the active site which impairs the nucleophilic attack of the active site serine. In comparison, Q132A shows a week band, consistent with its reduced peptidase activity.
We further investigated whether the tetradecameric organization also allowed the nucleophilic activation of a threonine or a cysteine instead of a serine. The S98C mutant protein was completely inactive, possibly due to the larger size of the sulfur atom. The S98T mutant was significantly acylated with D3; however, it was not active in the fluorescent substrate assay. We suspect that a shielding of the enzyme-acyl intermediate by the additional methyl group is responsible for this defect of product release.
The inter-ring interface in the compressed state, in contrast to the extended state, lacks any specific hydrogen bridge, salt bridge, or hydrophobic loop-groove interactions. This led us to the question of how the rings are held together. We analyzed the interfaces of both states with the PDBePISA webserver operated by EMBL-EBI (34). For the extended state, the free energy of dissociation (ΔGdiss) of the tetradecameric assembly is computed to be 81.8 kcal/mol. For a heptameric assembly with the same extended conformation, ΔGdiss is estimated to be around 44 kcal/mol. As expected, the free energy of dissociation for the tetradecamer in the kinked state is much lower (54.3 kcal/mol) whereas the heptameric assembly becomes more stable (106 kcal/mol).
Upon closer inspection of the inter-ring interface of a recently published second data set of the compressed SaClpP structure (22), we noticed two sulfate ions bridging the two rings by means of hydrogen bonds with active site residues His123 and Asp172 and thereby stabilizing the tetradecameric assembly (Fig. 3C). With the high content of ammonium sulfate (1.8 m) present in the crystallization condition, it was not surprising to find a sulfate ion in the structure; however, an increase of peptidase activity triggered by sodium sulfate was also reported (22). Intrigued by this observation, we systematically investigated the effect on peptidase activity of different cations and anions including sodium, potassium, calcium, magnesium, nitrate, chloride, bromide, hydrogen carbonate, hydrogen phosphate, and sulfate at 200 mm concentration. Most of the salts do not alter the peptidase activity, whereas sodium sulfate accelerates the reaction velocity. As shown in Fig. 3D, we observed a concentration-dependent, linear increase of ClpP peptidase activity up to 480 mm salt concentration which led to an almost 4-fold increase in activity. As a control, we showed that potassium sulfate causes the same effect whereas sodium chloride only shows a weak effect, thus indicating that the sulfate ion is responsible for this boost of activity. Furthermore, we could show that both negative charges on the sulfate ion are necessary as neutral or singly charged analogs (methanesulfonamide or sodium methanesulfonate, respectively) showed no effect on enzyme activity.
Structure-based Mutagenic Studies Identify Key Residues Responsible for Conformational Switch
Next, we addressed the question of whether both conformations are relevant to the catalytic cycle. Based on the structural differences, we designed novel mutations and reinvestigated previously reported mutants that would allow destabilizing one, the other, or both conformations. We noticed that the side chain carbonyl oxygen of Glu135, located close to the tip of the extended E helix, forms a hydrogen bridge with the backbone nitrogen of Gln132, which stabilizes the loop at the beginning of the E helix (see Fig. 2C). Consequently, mutation of Glu135 to alanine abolished activity and yielded purely heptameric assemblies as shown by size exclusion chromatography and static light scattering (see Table 2). Similarly, mutation of Leu144 to aspartate caused repulsion with the side chain of Glu137 and led to the dissociation into inactive heptamers and smaller assemblies. The latter probably occurs as Leu144 also forms contacts in the compressed and possibly heptameric conformation. Mutation of Leu144 to an isosterically hydrophobic methionine retained partial activity (21) whereas, surprisingly, mutation to an arginine led to a stable tetradecameric complex with only basal activity. We investigated individual mutants of Leu144 to glycine and Glu137 as well as Gln130 to alanine because these residues are not involved in any obviously important contacts in the extended state but in the compressed state. Glu137 stabilizes the compressed state by contacts with the nitrogen backbone of Thr143 and Ser70. Unexpectedly, these mutants showed different properties. L144G is not active and exhibits a partly heptameric state in combination with a lower mass molecular assembly. E137A forms a stable tetradecamer but is not active at all. Q130A consists to 95% of heptamers with a small percentage of tetradecamers present. It was found to have a basal activity. Mutation of all three glycines 127, 128, and 131 in the antiparallel β-sheet linking the two rings in the extended conformation led to dissociation into heptamers. Although a tetradecameric species is present to approximately 20%, no activity was detected, presumably due to a steric clash impairing proper formation of the antiparallel β-sheet motif. As described above, we noticed that Gln130 and Gln132 of the handle domain are stabilized in the kinked state by an interaction with Gln35 of the head domain. However, mutation of Gln35 to alanine only slightly reduced the activity of the protease.
To gain further insight into these unexpected results, we measured the protein melting points of all mutants. Although most proteins had a similar or only slightly elevated melting temperature compared with the wild-type protein, the Q132A and the E137A mutants had melting temperatures of approximately 13 °C below the wild type (44.6 ± 1.1 °C and 45.1 ± 0.8 °C). The destabilizing effect of Q132A is possibly due to the role of the residue in intra-ring subunit contacts. The effect of the E137A mutant that forms destabilized, inactive tetradecamers remains enigmatic.
DISCUSSION
ClpP is a promising target for novel ways of treating bacterial infections and malaria (35). An in-depth understanding of its molecular mechanism might therefore pave the way for the development of novel and selective either inhibitory or activating agents (9, 17, 36). A key component of ClpP function seems to be a high degree of conformational flexibility in the handle domain which is found between the two heptameric rings. We recently solved the structure of ClpP from the important nosocomial pathogen S. aureus in its compressed, inactive conformation (21). In this report, we present the wild-type and S98A mutant protein structures of SaClpP in the active, extended conformation. Based on an in-depth comparison of these structures, we designed mutations to pinpoint key residues responsible for the conformational switch and the ring-ring interaction. A detailed characterization of these mutated proteins enabled us to provide the structural basis for a model of ClpP function that links oligomeric organization, structural integrity of the active site, and enzymatic activity.
In line with previous observations (19, 22), we identified a hydrogen bridge network involving the residues Arg171, Asp170, and Gln132 that connects the two heptameric rings. We have, for the first time, carried out a comprehensive mutational analysis of all residues involved in the inter-ring interactions. Mutation to alanine of one of these residues causes the dissociation of the tetradecameric form into either heptamers or, in the case of Gln132 due to its additional role in mediating intra-ring interactions, smaller oligomers with a small population of tetradecamers present. We assessed the activity of the mutated proteins with a fluorescent substrate assay and found them to be inactive except for Q132A, which showed a basal activity that is likely derived from the tetradecameric population. We furthermore probed the structural integrity of the active site residues with a ClpP-specific β-lactone-based inhibitor that showed no labeling in the case of the R171A, R171K, and D170A mutants and only weak labeling for the Q132A mutant. The data analyzed in this paper strongly suggest that a tetradecameric assembly is essential for a proper alignment of the active site residues in ClpP and hence activity (see Table 2 and Fig. 1H).
We here propose a role for Asp170 and Arg171 as sensors of the oligomeric state. In our model, binding of Arg171 of one heptamer to Asp170 on the other heptamer is accompanied by a severe conformational change of these residues which presumably resembles a transition from the conformation of the compressed state to the conformation of the extended state. This directly impacts on the conformation of the nearby active site residue Asp172. A rotation of the side chain atoms of Asp172 by 90° and a shift by approximately 3 Å enables the active site His123 to form a bridge between Asp172 and Ser98, thereby establishing a catalytically active triad. The crystal structure of the S98A mutant in the extended conformation shows that the active site His123 adopts the same conformation as in the wild-type structure, which proves the formation of the active site to be independent from the hydrogen bond donor capacities of the serine.
Although mandatory according to our proposal, a tetradecameric assembly is not sufficient for activity as shown with the E137A and the G127A/G128A/G131A mutations. Binding of the two heptamers additionally triggers the formation of the antiparallel β-sheet of two handle domains which, in turn, leads to an unbending of the helix E. It is this movement that enables the active site His123 to adopt the activating conformation. Perturbation of the helix or the β-sheet motif as represented by the mutations mentioned above causes the complete loss of activity due to an improper orientation of the active site residues. The same holds true for disruption of the interaction of the helix residue Gln132 with the oligomeric sensor residue Arg171. Additionally, intrinsic stabilization of the loop at the tip of the handle domain by a hydrogen bridge of Glu135 is critical for activity. Charge-charge interactions between the kinked helix and its cognate head domain as presumably disrupted by the Q35A mutation do not seem critical for activity, but might have facilitated the resolution of the kinked helix in S. aureus compared with ClpP from other organisms. This result is in line with an analysis of the structural B factors that show a high degree of flexibility in the kinked handle domain. In summary, a well defined extended state of the handle domain as well as a molecular hinge motion to a less well defined kinked state of the helix E seem to be essential for ClpP function. However, no conclusion could be drawn from the Q35A mutation regarding the importance of the compressed state for the catalytic cycle.
While we were preparing this manuscript, a report on the extended conformation of SaClpP was published in which the authors state that the heptameric form of SaClpP seems to be the more active conformation (22). Based on the structural and mutational data presented in this work, we suggest tetradecamers as the only active conformation. According to our model, proper orientation of the active site under physiological conditions can only be achieved by interactions between the handle domains of two heptameric rings. This observation is also consistent with studies on ClpP proteins from other organisms. Human ClpP, for instance, forms proteolytically inactive heptamers in solution that dimerize upon binding to a chaperone (26). Furthermore, the hetero-oligomeric ClpP of Listeria monocytogenes displays activity only in tetradecameric assemblies as recently published (37). Hence, mutations of Glu135 and Leu144 abolish activity due to a defect in oligomerization and not, as stated recently, due to a defect of product release (25).
It is, however, intriguing that in nonphysiologically high concentrations of sulfate, ClpP forms heptameric assemblies and, at the same time, shows a higher catalytic turnover. In this context, we find it noteworthy that two sulfate ions bridge the two heptameric rings in the compressed state by interactions with active site residues.
We agree that the compressed conformation might be heptameric in solution and that the tetradecameric assembly thereof might be a crystallographic artifact. A detailed inspection of the inter-ring interface in the heptamer yielded no specific interactions. Our conclusion is supported by computational results from the PDBePISA webserver that show a heptameric assembly of the compressed state to be more stable than one of the extended state.
The physiological relevance of the compressed state still is an unresolved issue. To date, no direct evidence for such a conformation in solution could be obtained. Although the crystals of S. aureus ClpP in the compressed form (21, 22) have been grown at an unphysiologically low pH where SaClpP is not active (as shown in this work), there are a number of indications that this conformation (if not the entire structure then at least the conformation of a single subunit) is of physiological relevance.
First, an elegant quantitative NMR study by Sprangers et al. provided conclusive evidence that ClpP from E. coli exhibits two distinct conformations in solution (23). By mutational analysis, the authors were able to restrict the residues that are subject to these conformational dynamics to lie within the handle domain, i.e. exactly the domain where the differences between the extended and the compressed crystal structures occur (see Fig. 1). Consistent with the two crystal structures, no evidence for major structural changes in the head domain could be found.
Second, it has been shown that ClpP is also active when two chaperones (i.e. one on each side) are bound (33). This excluded the possibility of peptide release via the axial pores. It has moreover been shown that ClpP cleaves substrates processively (32), which excluded the possibility of peptide release via the dissociation of chaperones. This led to the inescapable conclusion that there must be a different way of substrate release out of the closed chamber. In essence, the formation of equatorial pores was proposed because only one conformational state (e.g. the active, extended, and closed form) is not sufficient to describe the catalytic mechanism.
Third, the compressed state is definitely neither a species-specific artifact of S. aureus nor an artifact of a low pH. Structures of ClpP in the compressed state comprise that of M. tuberculosis (pH 8.0), P. falciparum (pH 7.0), and B. subtilis (pH 5.6) (see supplemental Table 2). All of these structures show the characteristic compressed form of the cylinder (18, 20, 25).
Fourth, in the case of B. subtilis, crystals of the compressed and of the extended form have been found next to each other in the same crystallization well (25). This unambiguously points to the fact that both conformations exist in solution.
Fifth, attempts to freeze the extended state via an engineered disulfide bridge (24) for ClpP from E. coli resulted in catalytically inactive tetradecamers that exhibit the compressed conformation.
Sixth, a normal mode analysis (24) and a molecular dynamics simulation (22) suggest a conformational flexibility of the handle domain consistent with the switch from the extended to the compressed state.
In conclusion, a number of indications strongly suggest that the compressed state exhibits a physiological relevance.
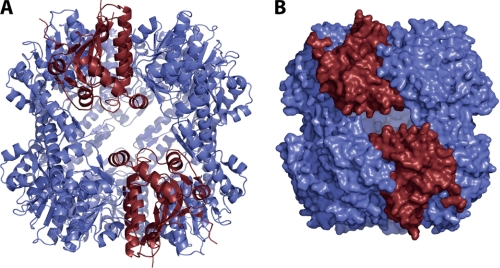
It has been repeatedly argued that a concerted switching of all 14 subunits from the extended conformation to the compressed form during substrate processing would either trigger the formation of equatorial pores or the transient dissociation of the two heptameric rings, both of which enable the release of peptide products formed (21–23, 25). Pores have even been proposed based on a structure in which in this region several residues are not ordered (25). This model, however, would imply bursts in peptide releases and a discontinuous mode of processing which, to our knowledge, have not been observed in single molecule experiments yet. Despite lacking data that would allow for the detection of conformational heterogeneity within one tetradecamer, we find it tempting to speculate about a different model for product release in which only two subunits on opposing rings undergo the conformational rearrangement. We have built a model based on the extended SaClpP structure in which we replaced two interacting subunits on different rings by subunits with a kinked helix based on the position of the respective head domains (see Fig. 4). We find only a minor clash of the kinked helix tip with Phe174 of the subunit adjacent to it which could possibly be accommodated by minor structural changes. It has been shown that ClpP digests proteins to small peptides of approximately 6–8 amino acids (32). Our model of independent transient fluctuations in the handle domain would yield large equatorial pores approximately 12 Å in diameter that, in contrast to the pores observed in the compressed state that are approximately 3 Å in diameter, would allow the release of also larger peptidic fragments. Accumulation of peptides within the chamber would trigger the conformational switch through an unknown mechanism dependent or independent of the catalytic cycle and thereby bring about their release from the protease.
FIGURE 4.
A, tetradecameric model of SaClpP was built on the basis of the extended active conformation (colored in blue) in which two monomers were replaced by monomers in the inactive, compressed state (colored in red). B, surface representation of this model shows large, equatorial pores that could account for the release of product peptides via transient fluctuations of the handle domain.
In essence, we have proposed a model in which the activity of ClpP is tightly controlled in a structural manner by a 2-fold system. First, contact of two heptamers initiates the establishment of an inter-ring bridging Asp170-Arg171 hydrogen bridge network which, secondly, allows interactions that lead to a conformational switch of the handle domain and, ultimately, to the proper formation of the active site. In view of the high degree of unspecificity of the protease, we find this tightly controlled activation mechanism to be biologically sensible. In our model, activity is only present within a properly structured cylinder which, in turn, prevents uncontrolled access to the unspecific active sites of the protease. When ClpP heptamers are present as described for human ClpP, for instance, and free access to the active sites is given, the protease is inactive. Further studies will have to address the question how binding of a chaperone to the N-terminal loops connects to the molecular relay system as described in this work and possibly triggers the conformational switching and assembly of the functional protease.
Supplementary Material
Acknowledgments
We thank Mona Wolf, Arie Geerlof, Sabrina Senz, Sandra Hocke, Martina Müller, Elisabeth Schäfer, and Astrid König for excellent technical assistance and Alma Brodersen for critical evaluation of the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (Emmy Noether program), SFB749, Center for Integrated Protein Science Munich, and the European Research Council (ERC starting grant). Data collection was conducted at the Swiss Light Source at the Paul Scherrer Institute, Villigen, Switzerland.

This article contains supplemental Fig. 1, Tables 1–3, and additional references.
The atomic coordinates and structure factors (codes 3V5E and 3V5I) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- ClpP
- caseinolytic protease P
- AMC
- 7-amino-4-methylcoumarin
- MPD
- 2-methyl-1,3-propanediol
- r.m.s.d.
- root mean square deviation
- SaClpP
- ClpP from Staphylococcus aureus
- Suc-LeuTyr-AMC
- N-succinyl-leucine-tyrosine-7-amino-4-methylcoumarin
- TBTA
- tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine
- TCEP
- tris(2-carboxyethyl)phosphine hydrochloride.
REFERENCES
- 1. Yu A. Y., Houry W. A. (2007) ClpP: a distinctive family of cylindrical energy-dependent serine proteases. FEBS Lett. 581, 3749–3757 [DOI] [PubMed] [Google Scholar]
- 2. Sauer R. T., Bolon D. N., Burton B. M., Burton R. E., Flynn J. M., Grant R. A., Hersch G. L., Joshi S. A., Kenniston J. A., Levchenko I., Neher S. B., Oakes E. S., Siddiqui S. M., Wah D. A., Baker T. A. (2004) Sculpting the proteome with AAA+ proteases and disassembly machines. Cell 119, 9–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Baker T. A., Sauer R. T. (2012) ClpXP, an ATP-powered unfolding and protein-degradation machine. Biochim. Biophys. Acta 1825, 15–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Katayama-Fujimura Y., Gottesman S., Maurizi M. R. (1987) A multiple-component, ATP-dependent protease from Escherichia coli. J. Biol. Chem. 262, 4477–4485 [PubMed] [Google Scholar]
- 5. Gottesman S., Roche E., Zhou Y., Sauer R. T. (1998) The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system. Genes Dev. 12, 1338–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Michel A., Agerer F., Hauck C. R., Herrmann M., Ullrich J., Hacker J., Ohlsen K. (2006) Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 188, 5783–5796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Flynn J. M., Neher S. B., Kim Y. I., Sauer R. T., Baker T. A. (2003) Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683 [DOI] [PubMed] [Google Scholar]
- 8. Maurizi M. R., Thompson M. W., Singh S. K., Kim S. H. (1994) Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 244, 314–331 [DOI] [PubMed] [Google Scholar]
- 9. Böttcher T., Sieber S. A. (2008) β-Lactones as specific inhibitors of ClpP attenuate the production of extracellular virulence factors of Staphylococcus aureus. J. Am. Chem. Soc. 130, 14400–14401 [DOI] [PubMed] [Google Scholar]
- 10. Böttcher T., Sieber S. A. (2009) Structurally refined β-lactones as potent inhibitors of devastating bacterial virulence factors. ChemBioChem 10, 663–666 [DOI] [PubMed] [Google Scholar]
- 11. Frees D., Qazi S. N., Hill P. J., Ingmer H. (2003) Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol. Microbiol. 48, 1565–1578 [DOI] [PubMed] [Google Scholar]
- 12. Frees D., Sørensen K., Ingmer H. (2005) Global virulence regulation in Staphylococcus aureus: pinpointing the roles of ClpP and ClpX in the sar/agr regulatory network. Infect Immun. 73, 8100–8108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang J., Hartling J. A., Flanagan J. M. (1997) The structure of ClpP at 2.3 A resolution suggests a model for ATP-dependent proteolysis. Cell 91, 447–456 [DOI] [PubMed] [Google Scholar]
- 14. Joshi S. A., Hersch G. L., Baker T. A., Sauer R. T. (2004) Communication between ClpX and ClpP during substrate processing and degradation. Nat. Struct. Mol. Biol. 11, 404–411 [DOI] [PubMed] [Google Scholar]
- 15. Kolygo K., Ranjan N., Kress W., Striebel F., Hollenstein K., Neelsen K., Steiner M., Summer H., Weber-Ban E. (2009) Studying chaperone-proteases using a real-time approach based on FRET. J. Struct. Biol. 168, 267–277 [DOI] [PubMed] [Google Scholar]
- 16. Kim D. Y., Kim K. K. (2008) The structural basis for the activation and peptide recognition of bacterial ClpP. J. Mol. Biol. 379, 760–771 [DOI] [PubMed] [Google Scholar]
- 17. Lee B. G., Park E. Y., Lee K. E., Jeon H., Sung K. H., Paulsen H., Rübsamen-Schaeff H., Brötz-Oesterhelt H., Song H. K. (2010) Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nat. Struct. Mol. Biol. 17, 471–478 [DOI] [PubMed] [Google Scholar]
- 18. Vedadi M., Lew J., Artz J., Amani M., Zhao Y., Dong A., Wasney G. A., Gao M., Hills T., Brokx S. (2007) Genome-scale protein expression and structural biology of Plasmodium falciparum and related apicomplexan organisms. Mol. Biochem. Parasitol. 151, 100–110 [DOI] [PubMed] [Google Scholar]
- 19. Gribun A., Kimber M. S., Ching R., Sprangers R., Fiebig K. M., Houry W. A. (2005) The ClpP double ring tetradecameric protease exhibits plastic ring-ring interactions, and the N termini of its subunits form flexible loops that are essential for ClpXP and ClpAP complex formation. J. Biol. Chem. 280, 16185–16196 [DOI] [PubMed] [Google Scholar]
- 20. Ingvarsson H., Maté M. J., Högbom M., Portnoï D., Benaroudj N., Alzari P. M., Ortiz-Lombardía M., Unge T. (2007) Insights into the inter-ring plasticity of caseinolytic proteases from the x-ray structure of Mycobacterium tuberculosis ClpP1. Acta Crystallogr. D Biol. Crystallogr. 63, 249–259 [DOI] [PubMed] [Google Scholar]
- 21. Geiger S. R., Böttcher T., Sieber S.A., Cramer P. (2011) A conformational switch underlies ClpP protease function. Angew Chem. Int. Ed. Engl. 50, 5749–5752 [DOI] [PubMed] [Google Scholar]
- 22. Zhang J., Ye F., Lan L., Jiang H., Luo C., Yang C. G. (2011) Structural switching of Staphylococcus aureus Clp protease: a key to understanding protease dynamics. J. Biol. Chem. 286, 37590–37601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sprangers R., Gribun A., Hwang P. M., Houry W. A., Kay L. E. (2005) Quantitative NMR spectroscopy of supramolecular complexes: dynamic side pores in ClpP are important for product release. Proc. Natl. Acad. Sci. U.S.A. 102, 16678–16683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kimber M. S., Yu A. Y., Borg M., Leung E., Chan H. S., Houry W. A. (2010) Structural and theoretical studies indicate that the cylindrical protease ClpP samples extended and compact conformations. Structure 18, 798–808 [DOI] [PubMed] [Google Scholar]
- 25. Lee B. G., Kim M. K., Song H. K. (2011) Structural insights into the conformational diversity of ClpP from Bacillus subtilis. Mol. Cells 32, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang S. G., Dimitrova M. N., Ortega J., Ginsburg A., Maurizi M. R. (2005) Human mitochondrial ClpP is a stable heptamer that assembles into a tetradecamer in the presence of ClpX. J. Biol. Chem. 280, 35424–35432 [DOI] [PubMed] [Google Scholar]
- 27. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Cryst. 26, 795–800 [Google Scholar]
- 28. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Turk D. (1992) Improvement of a Program for Molecular Graphics and manipulation of Electron Densities and its application for Protein Structure Determination. Ph.D. thesis, Technische Universität München, Munich Germany [Google Scholar]
- 30. Vagin A. A., Steiner R. A., Lebedev A. A., Potterton L., McNicholas S., Long F., Murshudov G. N. (2004) REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195 [DOI] [PubMed] [Google Scholar]
- 31. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 32. Jennings L. D., Lun D. S., Médard M., Licht S. (2008) ClpP hydrolyzes a protein substrate processively in the absence of the ClpA ATPase: mechanistic studies of ATP-independent proteolysis. Biochemistry 47, 11536–11546 [DOI] [PubMed] [Google Scholar]
- 33. Ortega J., Lee H. S., Maurizi M. R., Steven A. C. (2004) ClpA and ClpX ATPases bind simultaneously to opposite ends of ClpP peptidase to form active hybrid complexes. J. Struct. Biol. 146, 217–226 [DOI] [PubMed] [Google Scholar]
- 34. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 35. Rathore S., Sinha D., Asad M., Böttcher T., Afrin F., Chauhan V. S., Gupta D., Sieber S. A., Mohmmed A. (2010) A cyanobacterial serine protease of Plasmodium falciparum is targeted to the apicoplast and plays an important role in its growth and development. Mol. Microbiol. 77, 873–890 [DOI] [PubMed] [Google Scholar]
- 36. Brötz-Oesterhelt H., Beyer D., Kroll H. P., Endermann R., Ladel C., Schroeder W., Hinzen B., Raddatz S., Paulsen H., Henninger K., Bandow J. E., Sahl H. G., Labischinski H. (2005) Dysregulation of bacterial proteolytic machinery by a new class of antibiotics. Nat. Med. 11, 1082–1087 [DOI] [PubMed] [Google Scholar]
- 37. Zeiler E., Braun N., Böttcher T., Kastenmüller A., Weinkauf S., Sieber S. A. (2011) Vibralactone as a tool to study the activity and structure of the ClpP1P2 complex from Listeria monocytogenes. Angew Chem. Int. Ed. Engl. 50, 11001–11004 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.