Background: PP6c3 and SAPSR1 bind to DNA-PK and are required for DNA-PK activation following ionizing radiation.
Results: R1 associates with DNA-PK independently of PP6c through two distinct regions.
Conclusion: Contact between R1 and DNA-PK involves two binding sites: one dominant, the other supporting.
Significance: DNA-PK-R1 interface may be exploited as a drug target to inhibit DNA-PK activation and radiosensitize cells.
Keywords: DNA Damage, DNA Repair, Phosphoprotein Phosphatase, Protein Phosphatase, Protein-Protein Interactions, DNA-PK, NHEJ, PP6c, SAPS, Coimmunoprecipitation
Abstract
DNA-dependent protein kinase (DNA-PK) becomes activated in response to DNA double strand breaks, initiating repair by the non-homologous end joining pathway. DNA·PK complexes with the regulatory subunit SAPSR1 (R1) of protein phosphatase-6 (PP6). Knockdown of either R1 or PP6c prevents DNA-PK activation in response to ionizing radiation-induced DNA damage and radiosensitizes glioblastoma cells. Here, we demonstrate that R1 is necessary for and bridges the interaction between DNA-PK and PP6c. Using R1 deletion mutants, DNA-PK binding was mapped to two distinct regions of R1 spanning residues 1–326 and 522–700. Either region expressed alone was sufficient to bind DNA-PK, but only deletion of residues 1–326, not 522–700, eliminated interaction of R1 with DNA-PK. We assign 1–326 as the dominant domain and 522–700 as the supporting region. These results demonstrate that R1 acts as a bidentate anchor to DNA-PK and recruits PP6c. Targeting the dominant interface with small molecule or peptidomimetic inhibitors could specifically prevent activation of DNA-PK and thereby sensitize cells to ionizing radiation and other genotoxic agents.
Introduction
DNA double strand breaks (DSBs)2 are one of the most lethal forms of DNA damage. DNA DSBs arise from normal cellular processes such as V-D-J recombination and the production of free radicals as well as from exogenous stimuli such as exposure to DNA-damaging agents like IR. In mammalian cells, DNA DSBs are repaired by two major pathways: homologous recombination (HR) and non-homologous end joining. A critical component of the non-homologous end joining-mediated DNA DSB repair pathway is DNA-dependent protein kinase (DNA-PK). DNA-PK is a nuclear Ser/Thr protein kinase and a member of the PI3K-related kinase superfamily, which includes the human ataxia telangiectasia mutated (ATM) and ATM-Rad3 related proteins as well as other PI3-like kinases (1, 2). DNA-PK is composed of a catalytic subunit (DNA-PKcs) and two Ku heterodimers (Ku70 and Ku80) that act as regulatory subunits (2). Multiple lines of evidence demonstrate the central role of DNA-PK in and non-homologous end joining-mediated DNA DSB repair and cell survival. DNA-PK deficient glioblastoma cells (M059J) are highly sensitive to DNA-damaging agents (3). siRNA-mediated silencing of DNA-PK or chemical inhibition of DNA-PK inhibits DNA repair and sensitizes multiple tumor cell types to DNA-damaging agents (4–9). DNA-PKcs−/− mice have high levels of unrepaired DSBs after treatment with genotoxic agents and are hypersensitive to irradiation (10). DNA-PK activity is regulated by phosphorylation in two major clusters denoted as ABCDE and PQR (11–17). Some of these residues are DNA-PK autophosphorylation sites and some are targets of ATM (11–16, 18). In addition to regulation by kinases, an accumulating body of evidence suggests that DNA-PK activity is also regulated by phosphatases. Phosphatase inhibitors okadaic acid and microcystin, at concentrations that inhibit PP2A and PP2A-like phosphatases, have been shown to activate DNA-PK and promote DNA repair activity (19, 20). Wechsler et al. (21) have demonstrated that DNA-PK interacts with and is dephosphorylated by the phosphatase PP5. Our previous work (22) has demonstrated that DNA-PK interacts with the protein phosphatase PP6.
PP6 is a Ser/Thr protein phosphatase classified as a type 2A phosphatase family member on the basis of its sequence homology to the catalytic subunit of protein phosphatase 2A (PP2A) and its sensitivity to active site inhibitors such as okadaic acid, microcystin, and calyculin A (23–25). PP6 is conserved among all eukaryotic species from yeast to humans, attesting to its fundamental importance. The Saccharomyces cerevisiae homologue, Sit4, is required for G1 to S progression, expression of G1 cyclins, and several other cellular processes (23, 26). The human PP6 also promotes G1to S progression, predominantly by regulating the levels of cyclin D1 (27). Recently, PP6 has been shown to play a role in regulation of spindle checkpoint by inhibiting Aurora A activation (28).
PP6 belongs to a class of phosphatases known as phosphoprotein phosphatases (phosphoprotein phosphatases), which function as multimeric phosphatases (29, 30). phosphoprotein phosphatase holoenzymes typically consist of a catalytic subunit and one or more regulatory subunits (29, 30). The regulatory subunits impart substrate specificity, determine subcellular localization, and sometimes activate or inhibit the phosphatase holoenzyme (29, 30). The PP6 holoenzyme consists of a trimer including the PP6 catalytic subunit (PP6c), one of the three regulatory subunits (R1, R2, or R3), and one of the three ankyrin repeat subunits (ARS-A, ARS-B, or ARS-C) (31, 32). The regulatory subunits of PP6 (R1, R2, and R3) contain an evolutionarily conserved, highly homologous N-terminal domain designated as the sit4-associated protein sequence (SAPS) domain (31). In fact, R1, R2 and R3 were originally identified by the sequence homology of their SAPS domains to their yeast counterparts (31). Although the different PP6c regulatory subunits may have some redundant functions, as in the case of Aurora A dephosphorylation (28), there is evidence that each subunit may also have specific non-redundant functions (26, 31). Knockdown of R1, but not R3, enhances degradation of endogenous IκBϵ in response to TNF-α (31). Similarly, knockdown of R1, but not R3, abrogates DNA-PK activation in response to IR (22). Identifying the binding partners of different PP6 regulatory subunits will provide insight into how PP6 regulates various cellular processes.
We have demonstrated previously that DNA-PK binds to both the PP6 catalytic subunit and its regulatory subunit, R1. Knockdown of PP6c resulted in reduced DNA-PK activity, impaired DNA repair, and sensitization of glioblastoma cells to irradiation, suggesting that PP6c is required for DNA-PK function. Knockdown of R1 mimicked the PP6c knockdown phenotype, suggesting that R1 is necessary for activation of DNA-PK by PP6c in response to IR. Although DNA-PK, PP6c, and R1 are known to exist in a complex, the interface between DNA-PK and PP6 has not been defined. We present data demonstrating that R1 functions as a bridge between DNA-PK and PP6c. In addition, we have mapped two distinct regions of R1, one necessary but both sufficient for binding to DNA-PK.
EXPERIMENTAL PROCEDURES
Cell Lines and Transfections
HEK293 and HEK293T cells were maintained in DMEM (Invitrogen) containing 10% (v/v) fetal bovine serum (Invitrogen) at 37 °C in a humidified 5% (v/v) CO2 atmosphere. Plasmids were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. siRNA smart pools were purchased from Dharmacon. siRNAs were transfected in 293 cells at a final concentration of 100 nm using Lipofectamine RNAiMax according to the manufacturer's instructions.
Plasmids, PCR, and Cloning
FLAG-R1, FLAG-1–521, and FLAG-522–881 have been described previously (31). FLAG-522–700 was PCR amplified from FLAG-R1 using primers 5′-GAATTCGTGAACACCCACCACCTACA-3′ and 5′-ATGCCTCGAGCTAGTAGGACAGAGGGGTGGCCC-3′. FLAG-701–881 was PCR-amplified from FLAG-R1 using primers 5′-ATGCGAATTCCCCAGCCCTGGCCCTCAGCC-3′ and 5′-CTCGAGCTATTGGGAGCCTGGGGATG-3′. FLAG-522-700 and FLAG-701–881 PCR amplicons were cloned into pcDNA-FLAG using the EcoRI-XhoI sites. FLAG-Δ522–700, FLAG-Δ1–326, FLAG-1–521-HIS, FLAG-1–326-HIS, FLAG-R1 (4DE-KK), FLAG-1–521-HIS (4DE-KK), and FLAG-1–326-HIS (4DE-KK) were generated using the Phusion site-directed mutagenesis kit (Finnzymes) according to the manufacturer's instructions. FLAG-1–521-HIS was PCR-amplified from FLAG-1–521 using primers 5′-TAGCTCGAGCATGCATCTAGA-3′ and 5′-AAGCTTATGGTGATGGTGATGGTGCAGGTCCACCATGTTCTTCTT-3′. FLAG-1–326-HIS was PCR-amplified from FLAG-1–326 using primers 5′-TAGCTCGAGCATGCATCTAGA-3′ and 5′-AAGCTTATGGTGATGGTGATGGTGGCAGCTGAGCCGCGGGCGTAG-3′. Charge reversal mutants (4DE-KK mutants) of FLAG-R1, FLAG-1–521-HIS, and FLAG-1–236-HIS were generated from the wild-type versions of these constructs by sequential site-directed mutagenesis using primers 5′-AGCAGATCCACCCGTCGAAGAAGAAGAATCAACATTCCAACGCATC-3′ and 5′-CAATCAGCCGCTGGACGATCT-3′ followed by 5′-AGCTCTTAAGCAACATGTTCAAGGGGAAGCAGAGCCAGTCTGTCATCGT-3′ and 5′-GCTCAATCGTCTCCTGCTTCT-3′. FLAG-Δ522–700 was PCR-amplified from FLAG-R1 using primers 5′-CCCAGCCCTGGCCCTCAGCC-3′ and 5′-CAGGTCCACCATGTTCTTCTT-3′. FLAG-Δ1–326 was PCR-amplified from FLAG-R1 using primers 5′-GAATTCATGTTCCACCAGCTCCTGCTGGAG-3′ and 5′-CTCGAGCTAGTTGACGACCTTGCACTCCTG-3′ followed by digestion with EcoRI and circularization of the plasmid. HA-tagged wild-type PP6c has been described previously (33). HA-PP6c-H114A was generated from HA-WT-PP6c using the Stratagene site-directed mutagenesis kit with primers 5′-CGTATTACACTTTTGCGAGGAAATGCAGAGAGTAGACAGATTACAC-3′ and 5′-GTGTAATCTGTCTACTCTCTGCATTTCCTCGCAAAAGTGTAATACG-3′. HA-PP6c-HR-QA was generated from HA-PP6c-WT by sequential site-directed mutagenesis using primers 5′-GAGATATCCAGGGACAGTTTT-3′ and 5′-CACACACTGTTACTGGTGTTG-3′ followed by 5′-GATTTTGTAGACGCAGGTTACTATAGT-3′ and 5′-ACCCATAAATATGTAGTTTGT-3′.
Immunoprecipitations and Western Blotting
Whole cell lysates were prepared in EBC lysis buffer as described previously (34). Lysates were cleared by centrifugation at 20,000 × g for 15 min at 4 °C. Extracts were precleared by incubation with protein-G-Sepharose antibodies for 30 min at 4 °C. One milligram of precleared extracts was incubated with the indicated antibodies overnight followed by incubation with protein-G-Sepharose beads for 2 h at 4 °C. Immune complexes immobilized on protein-G-Sepharose beads were washed three times with cold NET-N as described previously (34). Immunoprecipitates were resolved on 4–20% precast gradient gels (Bio-Rad) and transferred to a 0.22 μm nitrocellulose membrane using a wet transfer assembly. Membranes were incubated with the indicated primary antibodies overnight at 4 °C, followed by incubation with secondary antibodies at room temperature for 1 h. Antibody signal was captured on x-ray films with multiple exposures using enhanced chemiluminescence as described by the manufacturer (Pierce). Film exposures in the linear range were scanned using a densitometric scanner (Bio-Rad GS800) and analyzed using ImageJ software. Statistical significance was determined using a Student's t test.
Antibodies
For Western blotting, FLAG antibody (Sigma) and HA antibody (Abcam) were used at a dilution of 1:1000. DNA-PK antibody (Labvision) and R1 antibody (Bethyl) were used at a dilution of 1:2000. PP6c antibody has been described previously (31) and was used at a dilution of 1:5000. For immunoprecipitations, FLAG antibody was used at 5 μg/μg of lysate. HA antibody was used at 2 μg/mg of lysate. PP6 antibody (Bethyl) was used at 3 μg/mg of lysate.
RESULTS
Orientation of Components in the R1·PP6c·DNA·PK Complex
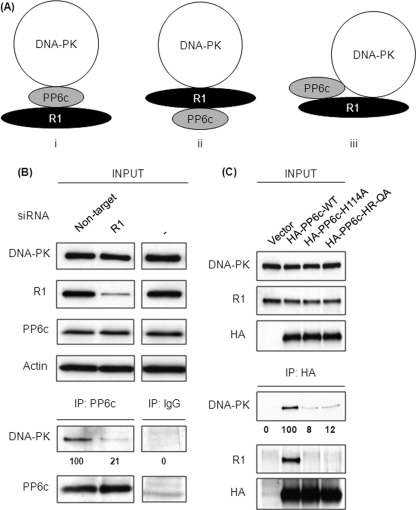
In living cells DNA-PK, R1, and PP6c are present in a complex that can be recovered by immunoprecipitation of any one of the three proteins. To better understand the organization of this complex, we considered three possible models (Fig. 1A). PP6c acts as a bridge to span R1 and DNA-PK (i); R1 bridges between PP6c and DNA-PK (ii); or the three proteins independently interact with one another (iii). To test these models, we knocked down R1 in 293 cells using siRNA (Fig. 1B) and transfected cells with non-targeting siRNA as a control. The knockdown of R1 was effective in reducing R1 protein levels > 80%. Control, knockdown, and untreated cells had the same unperturbed levels of PP6c and DNA-PK on the basis of immunoblotting of cell extracts (Fig. 1B, top panels). Endogenous PP6c was immunoprecipitated in parallel from control and R1 knockdown cells, and the immunoprecipitates were analyzed for the recovery of PP6c and DNA-PK by Western blotting (Fig. 1B, lower panels). Non-immune IgG was used to demonstrate the specificity of the PP6c precipitation. DNA-PK coprecipitated efficiently with PP6c from control cells. However, the amount of DNA-PK coprecipitating with PP6c from R1 knockdown cells was significantly reduced by > 80%, indicating that R1 was required for DNA-PK to associate with PP6c. These results support models ii and iii above (Fig. 1A).
FIGURE 1.
R1 is required for DNA·PK·PP6c interaction. A, schematic representation of the three possible modes of DNA-PK-R1-PP6c interaction. i, DNA-PK binds to PP6c directly, and R1 in the complex is bound to only to PP6c and not to DNA-PK. ii, R1 binds to both DNA-PK and PP6c, thus bringing DNA-PK and PP6c in close proximity. iii, DNA-PK binds to both PP6c and R1 simultaneously. B, 293T cells were transfected with control (non-target) or R1 siRNA (R1). Seventy-two hours post-transfection, endogenous PP6c was immunoprecipitated (IP) from the lysates of these cells using anti-PP6c antibody. The levels of input proteins (top four panels) and immunoprecipitated proteins (bottom two panels) were determined by Western blotting with the indicated antibodies. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with non-targeting siRNA (first lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments. p < 0.001. C, 293T cells were transfected with empty vector (vector) or the indicated HA-tagged PP6c constructs. Forty-eight hours post-transfection, HA-tagged PP6c was immunoprecipitated from the lysates of these cells using anti-HA antibody. The levels of input proteins (top three panels) and immunoprecipitated proteins (bottom three panels) were determined by Western blotting with the indicated antibodies. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with HA-PP6c-WT (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments. p < 0.001 for both HA-PP6c-H114A and HA-PP6c-HR-QA.
Effects of Active Site Mutations in PP6c
We made substitutions of key active site residues in PP6c to test whether phosphatase activity influenced the stability of the heterotrimer complex. Catalysis by phosphoprotein phosphatase phosphatases requires a conserved histidine residue to act as a general acid for protonation of the leaving group (H125 in PP1, H118 in PP2A, and H114 in PP6). Other active site residues are the metal ligand H55 and phosphoryl group binding Arg-85, which we substituted to Glu and Ala, respectively, to produce an HR-QA mutant. The H114A and HR-QA versions of PP6c were expressed in 293T cells at levels identical to the wild-type protein (Fig. 1C, upper panels). Immunoprecipitates were prepared and analyzed by immunoblotting for coprecipitation of endogenous R1 and endogenous DNA-PK (Fig. 1C, lower panels). Wild-type HA-tagged PP6c coprecipitated both R1 and DNA-PK. In contrast, neither of the PP6c mutants, H114A or HR-QA, coprecipitated either R1 or DNA-PK. It was unexpected that single residue substitutions in the active site would completely abrogate association with R1. We suspect that subtle alterations in the catalytic subunit conformation are detected by the SAPS domain of R1. Regardless, these results were consistent with the idea that association of PP6c with DNA-PK is dependent on the ability of PP6c to bind R1.
PP6c Is Not Required for R1 Association with DNA-PK
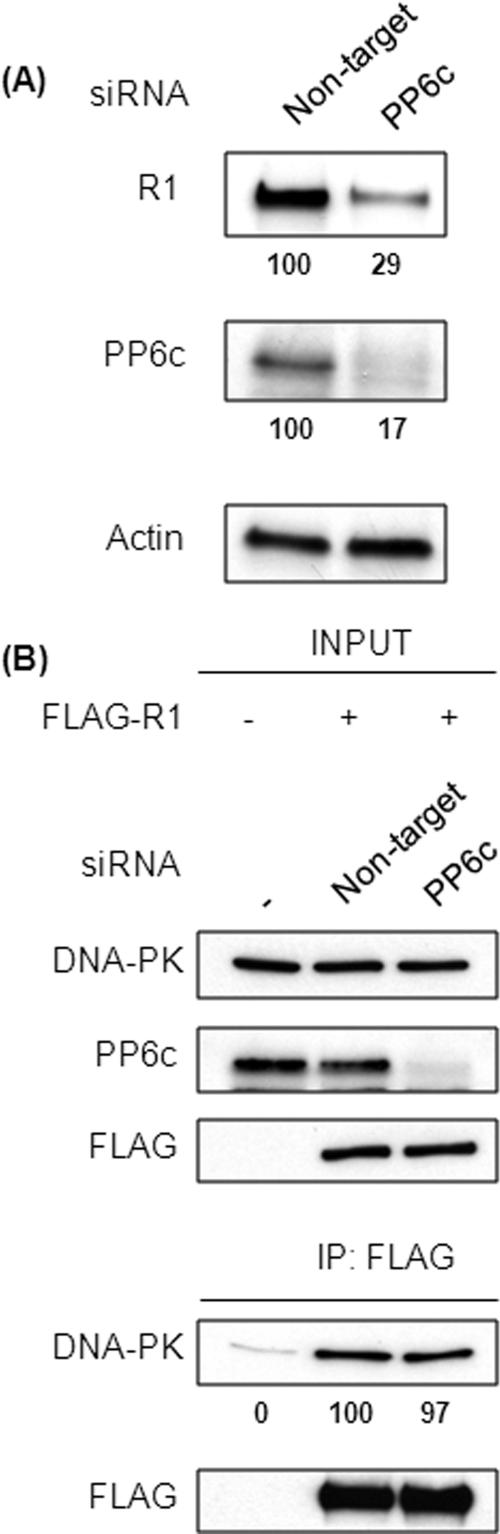
If R1 binding simultaneously to DNA-PK and PP6c is responsible for heterotrimer formation, we would predict that the complex does not depend on interactions between DNA-PK and PP6c. If so, then the ability of R1 to coprecipitate DNA-PK would not be affected in the absence of PP6c. To test this hypothesis, PP6c was knocked down in 293 cells using siRNA, but this also resulted in a reduction in the levels of R1 (Fig. 2A), as we reported previously (31). The expression levels of R subunits are dependent on the expression of PP6c, suggesting that the R subunits are more stable when present in a complex with PP6c as compared with individual free proteins, as discussed by Stefansson et al. (31). As an alternative approach, we expressed FLAG-tagged R1 in control and PP6c knockdown cells. This allowed us to achieve equivalent levels of FLAG-R1 in cells, with or without PP6c (Fig. 2B, upper panels). The FLAG-R1 was immunoprecipitated, and the amount of endogenous DNA-PK recovered was determined by Western blot analysis (Fig. 2B, lower panels). The results showed that FLAG-R1 coprecipitated the same amount of DNA-PK either in the presence or absence of PP6c, demonstrating that PP6c is not required for stable association of DNA-PK with R1. Taken together, these results show that R1 is essential for DNA-PK-PP6c interaction. R1 can mediate this interaction in two possible ways, as shown in models ii and model iii (Fig. 1A).
FIGURE 2.
PP6c is not required for DNA-PK-R1 interaction. A, 293T cells were transfected with control (non-target) or PP6c siRNA (PP6c). Seventy-two hours post-transfection, lysates were subjected to Western blotting with the indicated antibodies. The levels of PP6c and R1 were quantified by densitometric analysis and normalized to actin. Values below the Western blot analysis are mean ± S.D. from three independent experiments. p < 0.005 for PP6c and p < 0.05 for R1. B, 293T cells were transfected with control or PP6c siRNA. Twenty-four hours post-transfection, both control and PP6c knockdown cells were transfected with the FLAG-R1 plasmid. Forty-eight hours post-transfection, FLAG-R1 was immunoprecipitated (IP) from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top three panels) and immunoprecipitated proteins (bottom two panels) were determined by Western blotting with the indicated antibodies. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with non-targeting siRNA (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments, difference not significant.
Mapping R1 Regions Required for Binding DNA-PK
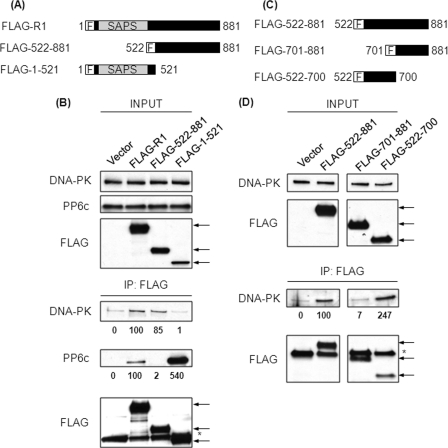
To define which regions of R1 constitute the binding site for DNA-PK, FLAG-tagged constructs of full-length R1 (FLAG-R1), the N terminus (FLAG-1–521), and the C terminus (FLAG-522–881) (Fig. 3A) were transfected into 293T cells. Both FLAG-R1 and FLAG-522–881 were expressed robustly, whereas FLAG-1–521 consistently appeared at lower levels on the basis of anti-FLAG immunoblotting when equivalent amounts of plasmid DNA were transfected (Fig. 3B, upper panels). Transfection of increasing amounts of the plasmid encoding FLAG-1–521 failed to increase the expression of this protein (data not shown). These FLAG-tagged proteins were immunoprecipitated, and the endogenous PP6c and endogenous DNA-PK that coprecipitated were analyzed by immunoblotting with specific antibodies (Fig. 3B, lower panels). FLAG-R1 coprecipitated both PP6c and DNA-PK. FLAG-522–881 coprecipitated DNA-PK but not PP6c. On the other hand, FLAG-1–521, which contains the SAPS domain, did not coprecipitate DNA-PK but coprecipitated significantly higher amounts of PP6c as compared with FLAG-R1. It appears that the SAPS domain is more readily accessible to PP6c in the absence of the C terminus, suggesting that the C terminus in the full-length R1 protein folds back onto the SAPS domain and limits access of PP6c. These results indicated that R1 binds PP6c using its N-terminal SAPS domain (residues 1–521) and depends on residues 522–881 for association with DNA-PK. From this truncation analysis it would appear that the two ends of the R1 protein independently interact with PP6c and DNA-PK.
FIGURE 3.
Residues 522–700 of R1 are sufficient for binding DNA-PK. A, schematic representation of the FLAG-tagged constructs of R1 used. F, N-terminal FLAG tag. B, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in A. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated (IP) from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top three panels) and immunoprecipitated proteins (bottom three panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The asterisk indicates the migration of the antibody heavy chain. The relative amount of coprecipitated DNA-PK and PP6c was quantified by densitometric analysis and compared with the amount of these proteins coprecipitated from cells transfected with FLAG-R1 (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments. For DNA-PK binding to FLAG-522–881, difference not significant and for FLAG-1–521, p < 0.05; for PP6c binding to FLAG-522–881, p < 0.001 and for FLAG-1–521, p < 0.05. C, schematic representation of the FLAG-tagged constructs of R1 used. F, N-terminal FLAG tag. D, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in C. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top two panels) and immunoprecipitated proteins (bottom two panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The asterisk indicates the migration of antibody heavy chain. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-522–881 (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments. For FLAG-701–881, p < 0.05 and for FLAG-522–881, p < 0.05.
Interaction of DNA-PK with residues 522–881 of R1 was delineated further by deletion using FLAG-tagged constructs expressing residues 522–700 or residues 701–881 (Fig. 3C). These proteins were expressed in 293T cells and immunoprecipitated with anti-FLAG antibody (Fig. 3D). FLAG-522–881 was used as a positive control that coprecipitated DNA-PK and FLAG-1–521 as a negative control. We observed equal or slightly better coprecipitation of DNA-PK with FLAG-522–700 compared with FLAG-522–881 but no coprecipitation with FLAG-701–881 (Fig. 3D). Our conclusion was that the DNA-PK binding site in R1 was between residues 522 and 700 because this region alone was sufficient for coprecipitation.
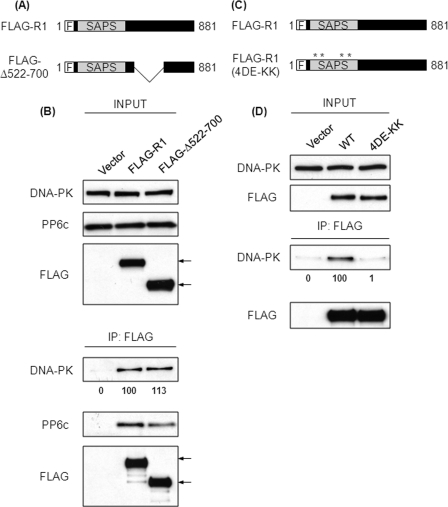
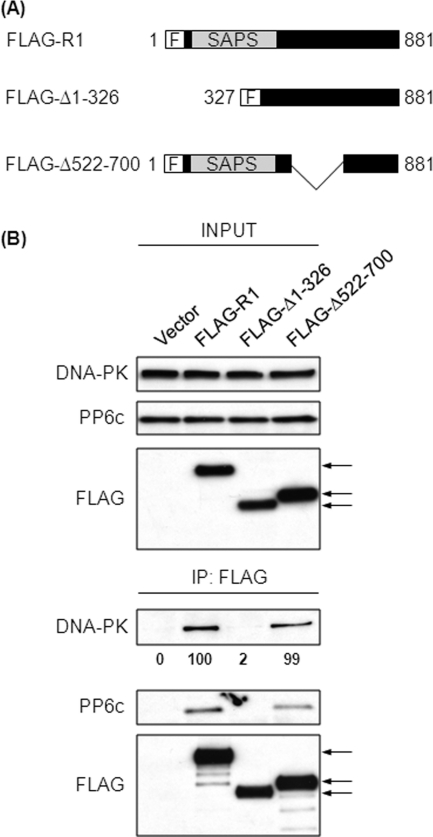
These results led us to predict that residues 522–700 of R1 would be necessary for the association with DNA-PK, so we deleted this region from R1 (Fig. 4A). FLAG-tagged constructs expressing either full-length R1 (FLAG-R1) or the deleted R1 (FLAG-Δ522–700) were expressed in 293T cells (Fig. 4B, upper panels). As expected, wild-type R1 (FLAG-R1) coprecipitated DNA-PK (Fig. 4B). Surprisingly, the R1 lacking residues 522–700 (FLAG-Δ522–700) coprecipitated a nearly equivalent amount of DNA-PK (Fig. 4B, lower panels). This showed, contrary to our expectations, that residues 522–700 were not necessary for association of R1 with DNA-PK. Therefore, we suspected that there was an additional site in R1 that was necessary for association with DNA-PK. The second DNA-PK binding site in R1 was suggested by a different loss of function R1 mutant. We introduced charge reversal substitutions of Lys for four acidic residues (Asp-204, Glu-205, Glu-257, and Glu-59) in the SAPS domain of R1 (Fig. 4C). Substitutions of the corresponding acidic residues in R3 resulted in loss of PP6c binding (35), and indeed, this mutated form of R1 showed loss of PP6c binding (supplemental Fig. S1). These mutations in R3 do not cause perturbations in the overall structure of R3, and because the SAPS domain is highly conserved across R subunits, there is no reason to believe that mutations in R1 would affect R1 conformation. Here we found that the charge reversal substitutions in the SAPS domain eliminated association with DNA-PK (Fig. 4D, lower panels). Taken together, deletion of 522–700 and charge reversal substitutions in the SAPS domain suggested that there was a second DNA-PK binding site in the N terminus of R1 (residues 1–521).
FIGURE 4.
Residues 522–700 of R1 are not required for binding to DNA-PK. A, schematic representation of the FLAG-tagged constructs of R1 used. F, N-terminal FLAG tag. The mutant lacking residues 522–700 is denoted as FLAG-Δ522–700. B, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in A. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated (IP) from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top three panels) and immunoprecipitated proteins (bottom three panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-R1 (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments, difference not significant. C, schematic representation of the FLAG-tagged constructs of R1 used. The asterisks indicate the position of the charge reversal mutations. The charge reversal mutant is denoted as FLAG-R1 (4DE-KK). D, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in C. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top two panels) and immunoprecipitated proteins (bottom two panels) were determined by Western blotting with the indicated antibodies. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-R1 (second lane). Values below the Western blot analysis are mean ± S.D. from three independent experiments. p < 0.001.
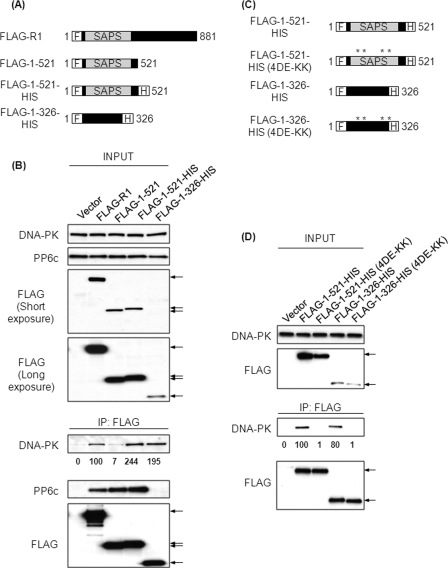
This second site in R1 for binding to DNA-PK was mapped to the N-terminal 326 residues. In an attempt to enhance the expression levels of FLAG-1–521, we added a hexahistidine tag (HIS) to the C terminus (Fig. 5A). This tag did not increase levels of the protein seen in transfected cells when compared with full-length R1 (FLAG-R1) (Fig. 5B). However, although FLAG-1–521 and FLAG-1–521-HIS were expressed at similar levels (Fig. 5B), we discovered that HIS-tagged FLAG-1–521 effectively coprecipitated DNA-PK (B) in contrast to the same protein without the HIS tag. Both the FLAG-1–521 and FLAG-1–521-HIS proteins coprecipitated comparable amounts of PP6c, suggesting that the structure of the proteins, predicted to be predominantly α helical, was not grossly altered by the HIS tag (Fig. 5B). We expressed a truncated protein comprising R1 residues 1–326 that was dual-tagged with an N-terminal FLAG tag and a C-terminal HIS tag (Fig. 5A). This FLAG-1–326-HIS construct coprecipitated DNA-PK but not PP6c (Fig. 5B). These results show that a region for association with DNA-PK lies within the N terminus of R1 and that this region does not support binding to, or require involvement of, PP6c.
FIGURE 5.
Residues 1–326 of R1 are sufficient for binding DNA-PK. A, schematic representation of the FLAG-tagged constructs of R1 used. F, N-terminal FLAG tag; H, C-terminal 6X-histidine tag. B, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in A. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated (IP) from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top four panels) and immunoprecipitated proteins (bottom three panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-R1 (second lane). Values below the Western blot are mean ± S.D. from three independent experiments. For FLAG-1–521, FLAG-1–521-HIS, and FLAG-1–326-HIS, p < 0.05. C, schematic representation of the FLAG-tagged constructs of R1 used. The asterisks indicate the position of the charge reversal mutations. The charge reversal mutants are denoted as 4DE-KK. D, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in C. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top two panels) and immunoprecipitated proteins (bottom two panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-1–521-HIS (lane 2). Values below the Western blot are mean ± S.D. from three independent experiments. For FLAG-1–521-HIS (4DE-KK) and FLAG-1–326-HIS (4DE-KK), p < 0.005; difference not significant for FLAG-1–326-HIS.
To rule out the possibility that the binding of DNA-PK to the FLAG-1–521-HIS and FLAG-1–326-HIS was due to the HIS tag itself, we introduced the same charge reversal substitutions in these two proteins by mutating Asp-204, Glu-205, Glu-257, and Glu-259 to lysine (Fig. 5C). The dual-tagged wild-type and charge reversal mutant versions of 1–521 and 1–326 were expressed in 293T cells and immunoprecipitated with anti-FLAG antibody (Fig. 5D). Western blotting revealed that either of the wild-type proteins coprecipitated DNA-PK but not the charge reversal mutants (FLAG-1–521-HIS (4DE-KK) and FLAG-1–326-HIS (4DE-KK)) (Fig. 5D). Thus, the HIS tag itself was not sufficient for interaction with DNA-PK.
Furthermore, we entirely removed the HIS tag from the C terminus of FLAG-1–521 and replaced it with sequences from two different regions of R1 (supplemental Fig. S2). Most of these proteins were expressed to only low levels, as compared with wild-type FLAG-R1, although some demonstrated higher-level expression (supplemental Fig. S3). Irrespective of the expression level, FLAG-1–521 with residues fused to the C terminus formed a complex with DNA-PK, whereas the FLAG-1–521 protein itself did not (supplemental Fig. S3). All versions of FLAG-1–521, with or without extensions at the C terminus, bound PP6c in the same way (supplemental Fig. S3).
Having shown that R1 residues 1–326 were sufficient for association with DNA-PK, we asked whether this region of R1 was necessary for coprecipitation of DNA-PK. We deleted residues 1–326 from R1 (FLAG-Δ1–326) (Fig. 6A). Full-length FLAG-R1 and FLAG-Δ1–326 were expressed in 293T cells and immunoprecipitated with anti-FLAG antibody (Fig. 6B). Western blotting of the immunoprecipitates with anti-DNA-PK antibody revealed that only full-length R1 (FLAG-R1), but not the deletion mutant (FLAG-Δ1–326), coprecipitated DNA-PK, showing that residues 1–326 were necessary for R1 interaction with DNA-PK (Fig. 6B). Thus, there are two regions in R1 involved in association with DNA-PK. One is necessary and sufficient, whereas the other is sufficient but not necessary.
FIGURE 6.
Residues 1–326 of R1 are required for binding DNA-PK. A, schematic representation of the FLAG-tagged constructs of R1 used. F, N-terminal FLAG tag. The mutant lacking residues 1–326 is denoted as FLAG-Δ1–326, and the mutant lacking residues 522–700 is denoted as FLAG-Δ522–700. B, 293T cells were transfected with empty plasmid or plasmids encoding the FLAG-tagged proteins described in A. Forty-eight hours post-transfection, FLAG-tagged proteins were immunoprecipitated (IP) from the lysates of these cells using anti-FLAG antibody. The levels of input proteins (top three panels) and immunoprecipitated proteins (bottom three panels) were determined by Western blotting with the indicated antibodies. The arrows indicate the position of FLAG-tagged proteins. The relative amount of coprecipitated DNA-PK was quantified by densitometric analysis and compared with the amount of DNA-PK coprecipitated from cells transfected with FLAG-R1 (second lane). Values below the Western blot are mean ± S.D. from three independent experiments. For FLAG-Δ1–326, p < 0.001, and difference not significant for FLAG-Δ522–700.
DISCUSSION
Understanding the role of R1 in DNA·PK·PP6c complex formation is a prerequisite for uncovering a druggable target for radiosensitization. Our data demonstrate that R1 is required for DNA·PK·PP6c interaction, as evidenced by the loss of interaction between DNA-PK and PP6c in the absence of R1. Thus, R1 acts a bridge to bring DNA-PK and PP6c in close proximity. The DNA-PK binding site was mapped to two regions of R1, spanning residues 1–326 and residues 522–700. Although each region could associate with DNA-PK independently, only residues 1–326 were critical for the DNA·PK·R1 association. Neither of the DNA-PK binding sites of R1 bound PP6c, further suggesting that the DNA·PK·R1 interaction is independent of the presence of PP6c. We conclude that R1 associates directly with DNA-PK, thus establishing R1 as a targeting subunit that recruits PP6c to DNA-PK.
In agreement with our observations, recent work by Douglas et al. (36) showed the association of DNA-PK with PP6c and its regulatory subunits both in vivo and in vitro. On the basis of the observation that purified recombinant PP6c could interact with purified DNA-PK in an in vitro binding assay, Douglas et al. (36) concluded that PP6c could directly associate with and activate DNA-PK in the absence of PP6 regulatory subunits. There are many explanations for the direct association between DNA-PK and PP6c in vitro but not in vivo, including the absence of posttranslational modifications on the bacterially produced PP6c protein, which may occur normally in human cells and prevent the direct interaction between DNA-PK and PP6c in vivo. It is also possible that in the presence of regulatory subunits in vivo, PP6c has a higher affinity for its regulatory subunits than for DNA-PK.
The results published by Douglas et al. (36) and our unpublished data show that all three PP6 regulatory subunits (R1, R2 and R3) can associate with DNA-PK in vivo, suggesting that these subunits may function redundantly to mediate activation of DNA-PK by PP6c. However, using siRNA mediated knockdown we have previously demonstrated that loss of R1, but not R3, results in impaired DNA-PK activation (22). It is possible that although each R subunit forms a trimeric complex with DNA-PK and PP6c, only the R1-containing complexes are able to promote DNA-PK activation. One possible explanation for this observation is that each R subunit binds to a different region of DNA-PK and regulates different functions of DNA-PK. It is also possible that PP6c is active only when R1, but not R2 or R3, is present in the DNA·PK·PP6c complex. The data presented here and published previously strongly suggest that R1 is critical for DNA·PK·PP6c complex formation and activation of DNA-PK.
Additional evidence that PP6c was not required for DNA·PK·R1 complex formation comes from our mapping experiments. The DNA-PK binding site was mapped to two regions of R1, one comprising residues 1–326 and the other residues 522–700. Both these regions of R1 could interact with DNA-PK in the absence of any detectable PP6c in the immunoprecipitation assay, providing further evidence that R1 directly interacts with DNA-PK. Although both of these regions were sufficient for DNA-PK binding, only 1–326 was necessary for DNA-PK-R1 interaction. We propose that residues 1–326 constitute the primary DNA-PK binding site on R1, whereas residues 522–700 form secondary contacts to strengthen the interaction. We hypothesize that residues 1–326 impart binding specificity, whereas residues 522–700 bind DNA-PK nonspecifically. The nonspecific binding of region 522–700 to DNA-PK is supported by the observation that C-terminal residues of R2 and R3 corresponding to the residues 522–881 (C terminus) of R1 also bind to DNA-PK, as determined by a coimmunoprecipitation assay (supplemental Fig. S4). The absence of significant sequence similarity between the C termini of the three R subunits suggests that the binding is not sequence-dependent. It is possible that the C termini of the three R subunits, as well as residues 522–700 of R1, adopt a conformation that facilitates interaction with DNA-PK nonspecifically. However, this nonspecific interaction may strengthen the DNA-PK-R1 interaction. Another possibility is that the region 522–700 is not a bona fide DNA-PK binding site and that the interaction was an artifact of deletion mutation analysis. For example, the full-length R1 protein may have a conformation, where the region 522–700 is buried deep inside the protein and, hence, not accessible for complex formation with DNA-PK. Then, in deletion mutants containing residues 522–700 or 522–881, residues 522–700 would be accessible to mediate interaction with DNA-PK.
In our initial deletion mutation analysis we failed to detect an association of DNA-PK with region 1–521 of R1. Our inability to detect the interaction was not due to low levels of the FLAG-1–521 protein because HIS-tagged FLAG-1–521 expressed at similar levels but could bind DNA-PK robustly. We demonstrated that the association of DNA-PK with FLAG-1–521 was not a function of the HIS tag because HIS-tagged charge reversal mutants failed to bind DNA-PK. Furthermore, replacement of the C-terminal HIS tag with two different regions of R1 resulted in detectable binding of DNA-PK to FLAG-1–521. The dramatic difference in the DNA-PK binding efficiency of native FLAG-1–521 and FLAG-1–521 with added residues at its C terminus could be a result of differences in the secondary structure of these peptides. We have reported previously reported that the SAPS domain of R3 forms α-helical repeats (35). Because the sequences of the SAPS domains of the three R subunits are highly conserved, the SAPS domain of R1 can be predicted to consist of similar α-helical repeats. The residues 1–521 of R1 contain the SAPS domain. It is possible that the stability of the α helices in region 1–521 of R1 is different in the presence and absence of additional residues at the C terminus of this region. Indeed, α helices are known to be stabilized through helix capping residues that flank the N and C termini of α helices and stabilize the helices through hydrogen bonding and hydrophobic interactions (37–39). Although certain amino acids are preferred as capping residues, there is some degree of variability in terms of the type and number of amino acids in the cap (37). Thus, histidine residues in the HIS tag or amino acids in the sequences from the two regions of R1 added to the C terminus of FLAG-1–521 could function as C-terminal helix capping residues and stabilize the α helices in this region. Indeed, histidine residues in the cap have been shown to contribute to the stability of α helices (40, 41). In the wild-type, full-length R1 protein, the C-terminal residues immediately following the α helices of the SAPS domain would act as helix capping residues to stabilize the helices in the SAPS domain and enable interaction of residues in region 1–521 with DNA-PK. In the absence of helix capping residues, the α helices in region 1–521 are destabilized so that the resulting peptide can either no longer bind DNA-PK or the interaction is too weak to be detected by a coimmunoprecipitation assay. Because the binding of PP6c to FLAG-1–521 is not affected by the presence or absence of additional C-terminal residues, we believe that the interaction between PP6c and FLAG-1–521 is resistant to changes in helix stability.
In summary, we have identified two regions of R1 that mediate its interaction with DNA-PK. Blocking either one or both of these interfaces could potentially be a means to prevent the association of DNA-PK with R1 and, hence, prevent activation of DNA-PK by PP6c. However, we believe that the N-terminal region of R1 is the primary interface. Solving the crystal structure of DNA-PK in complex with either one or both of the binding sites of R1 could aid in the rational development of small molecule or peptide inhibitors to block the DNA·PK·R1 association. Targeting the DNA·PK·R1 interface would specifically prevent radiation-induced DNA-PK activation without affecting its other cellular functions. This approach can be exploited to specifically impair DNA repair in tumor cells by using targeted radiation delivery to improve tumor cell killing. The advantage of this approach would be that by specifically blocking the radiation-induced DNA-PK activation, the toxic effects associated with general DNA-PK inhibition could be circumvented. Also, because this approach does not involve knockdown or chemical inhibition of either R1 or PP6c, the normal functions of these proteins would also remain unaffected. Although we have defined the DNA-PK-R1 interface, further investigations are necessary to determine whether this interface can be exploited as a druggable target.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health grant 5R01CA142823-02 (to J. M. L.).

This article contains supplemental Figs. S1–S4.
- DSB
- double strand break
- IR
- ionizing radiation
- HR
- homologous recombination
- DNA-PK
- DNA-dependent protein kinase
- ATM
- ataxia telangiectasia mutated protein
- PP
- protein phosphatase
- PP6c
- protein phosphatase 6 catalytic subunit
- SAPSR1
- R1, PP6 regulatory subunit R1
- SAPSR2
- R2, PP6 regulatory subunit R2
- SAPSR3
- R3, PP6 regulatory subunit R3
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- SAPS
- sit4-associated protein subunit.
REFERENCES
- 1. Chan D. W., Lees-Miller S. P. (1996) The DNA-dependent protein kinase is inactivated by autophosphorylation of the catalytic subunit. J. Biol. Chem. 271, 8936–8941 [DOI] [PubMed] [Google Scholar]
- 2. Lees-Miller S. P. (1996) The DNA-dependent protein kinase, DNA-PK. 10 years and no ends in sight. Biochem. Cell Biol. 74, 503–512 [DOI] [PubMed] [Google Scholar]
- 3. Allalunis-Turner M. J., Barron G. M., Day R. S., 3rd, Dobler K. D., Mirzayans R. (1993) Isolation of two cell lines from a human malignant glioma specimen differing in sensitivity to radiation and chemotherapeutic drugs. Radiat. Res. 134, 349–354 [PubMed] [Google Scholar]
- 4. Collis S. J., Swartz M. J., Nelson W. G., DeWeese T. L. (2003) Enhanced radiation and chemotherapy-mediated cell killing of human cancer cells by small inhibitory RNA silencing of DNA repair factors. Cancer Res. 63, 1550–1554 [PubMed] [Google Scholar]
- 5. Peng Y., Zhang Q., Nagasawa H., Okayasu R., Liber H. L., Bedford J. S. (2002) Silencing expression of the catalytic subunit of DNA-dependent protein kinase by small interfering RNA sensitizes human cells for radiation-induced chromosome damage, cell killing, and mutation. Cancer Res. 62, 6400–6404 [PubMed] [Google Scholar]
- 6. Durant S., Karran P. (2003) Vanillins. A novel family of DNA-PK inhibitors. Nucleic Acids Res. 31, 5501–5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ismail I. H., Mårtensson S., Moshinsky D., Rice A., Tang C., Howlett A., McMahon G., Hammarsten O. (2004) SU11752 inhibits the DNA-dependent protein kinase and DNA double-strand break repair resulting in ionizing radiation sensitization. Oncogene 23, 873–882 [DOI] [PubMed] [Google Scholar]
- 8. Shinohara E. T., Geng L., Tan J., Chen H., Shir Y., Edwards E., Halbrook J., Kesicki E. A., Kashishian A., Hallahan D. E. (2005) DNA-dependent protein kinase is a molecular target for the development of noncytotoxic radiation-sensitizing drugs. Cancer Res. 65, 4987–4992 [DOI] [PubMed] [Google Scholar]
- 9. Veuger S. J., Curtin N. J., Richardson C. J., Smith G. C., Durkacz B. W. (2003) Radiosensitization and DNA repair inhibition by the combined use of novel inhibitors of DNA-dependent protein kinase and poly(ADP-ribose) polymerase-1. Cancer Res. 63, 6008–6015 [PubMed] [Google Scholar]
- 10. Taccioli G. E., Amatucci A. G., Beamish H. J., Gell D., Xiang X. H., Torres Arzayus M. I., Priestley A., Jackson S. P., Marshak Rothstein A., Jeggo P. A., Herrera V. L. (1998) Targeted disruption of the catalytic subunit of the DNA-PK gene in mice confers severe combined immunodeficiency and radiosensitivity. Immunity 9, 355–366 [DOI] [PubMed] [Google Scholar]
- 11. Chan D. W., Chen B. P., Prithivirajsingh S., Kurimasa A., Story M. D., Qin J., Chen D. J. (2002) Autophosphorylation of the DNA-dependent protein kinase catalytic subunit is required for rejoining of DNA double-strand breaks. Genes Dev. 16, 2333–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cui X., Yu Y., Gupta S., Cho Y. M., Lees-Miller S. P., Meek K. (2005) Autophosphorylation of DNA-dependent protein kinase regulates DNA end processing and may also alter double-strand break repair pathway choice. Mol. Cell. Biol. 25, 10842–10852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ding Q., Reddy Y. V., Wang W., Woods T., Douglas P., Ramsden D. A., Lees-Miller S. P., Meek K. (2003) Autophosphorylation of the catalytic subunit of the DNA-dependent protein kinase is required for efficient end processing during DNA double-strand break repair. Mol. Cell. Biol. 23, 5836–5848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Douglas P., Cui X., Block W. D., Yu Y., Gupta S., Ding Q., Ye R., Morrice N., Lees-Miller S. P., Meek K. (2007) The DNA-dependent protein kinase catalytic subunit is phosphorylated in vivo on threonine 3950, a highly conserved amino acid in the protein kinase domain. Mol. Cell. Biol. 27, 1581–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas P., Sapkota G. P., Morrice N., Yu Y., Goodarzi A. A., Merkle D., Meek K., Alessi D. R., Lees-Miller S. P. (2002) Identification of in vitro and in vivo phosphorylation sites in the catalytic subunit of the DNA-dependent protein kinase. Biochem. J. 368, 243–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meek K., Douglas P., Cui X., Ding Q., Lees-Miller S. P. (2007) Trans-autophosphorylation at DNA-dependent protein kinase's two major autophosphorylation site clusters facilitates end processing but not end joining. Mol. Cell. Biol. 27, 3881–3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soubeyrand S., Pope L., Pakuts B., Haché R. J. (2003) Threonines 2638/2647 in DNA-PK are essential for cellular resistance to ionizing radiation. Cancer Res. 63, 1198–1201 [PubMed] [Google Scholar]
- 18. Chen B. P., Uematsu N., Kobayashi J., Lerenthal Y., Krempler A., Yajima H., Löbrich M., Shiloh Y., Chen D. J. (2007) Ataxia telangiectasia mutated (ATM) is essential for DNA-PKcs phosphorylations at the Thr-2609 cluster upon DNA double strand break. J. Biol. Chem. 282, 6582–6587 [DOI] [PubMed] [Google Scholar]
- 19. Douglas P., Moorhead G. B., Ye R., Lees-Miller S. P. (2001) Protein phosphatases regulate DNA-dependent protein kinase activity. J. Biol. Chem. 276, 18992–18998 [DOI] [PubMed] [Google Scholar]
- 20. Merkle D., Douglas P., Moorhead G. B., Leonenko Z., Yu Y., Cramb D., Bazett-Jones D. P., Lees-Miller S. P. (2002) The DNA-dependent protein kinase interacts with DNA to form a protein-DNA complex that is disrupted by phosphorylation. Biochemistry 41, 12706–12714 [DOI] [PubMed] [Google Scholar]
- 21. Wechsler T., Chen B. P., Harper R., Morotomi-Yano K., Huang B. C., Meek K., Cleaver J. E., Chen D. J., Wabl M. (2004) DNA-PKcs function regulated specifically by protein phosphatase 5. Proc. Natl. Acad. Sci. U.S.A. 101, 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mi J., Dziegielewski J., Bolesta E., Brautigan D. L., Larner J. M. (2009) Activation of DNA-PK by ionizing radiation is mediated by protein phosphatase 6. PLoS ONE 4, e4395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bastians H., Ponstingl H. (1996) The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J. Cell Sci. 109, 2865–2874 [DOI] [PubMed] [Google Scholar]
- 24. Mann D. J., Dombrádi V., Cohen P. T. (1993) Drosophila protein phosphatase V functionally complements a SIT4 mutant in Saccharomyces cerevisiae and its amino-terminal region can confer this complementation to a heterologous phosphatase catalytic domain. EMBO J. 12, 4833–4842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Prickett T. D., Brautigan D. L. (2006) The 4 regulatory subunit exerts opposing allosteric effects on protein phosphatases PP6 and PP2A. J. Biol. Chem. 281, 30503–30511 [DOI] [PubMed] [Google Scholar]
- 26. Morales-Johansson H., Puria R., Brautigan D. L., Cardenas M. E. (2009) Human protein phosphatase PP6 regulatory subunits provide Sit4-dependent and rapamycin-sensitive sap function in Saccharomyces cerevisiae. PLoS ONE 4, e6331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stefansson B., Brautigan D. L. (2007) Protein phosphatase PP6 N terminal domain restricts G1 to S phase progression in human cancer cells. Cell Cycle 6, 1386–1392 [DOI] [PubMed] [Google Scholar]
- 28. Zeng K., Bastos R. N., Barr F. A., Gruneberg U. (2010) Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J. Cell Biol. 191, 1315–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shi Y. (2009) Serine/threonine phosphatases. Mechanism through structure. Cell 139, 468–484 [DOI] [PubMed] [Google Scholar]
- 30. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision. Protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 31. Stefansson B., Brautigan D. L. (2006) Protein phosphatase 6 subunit with conserved Sit4-associated protein domain targets IκBϵ. J. Biol. Chem. 281, 22624–22634 [DOI] [PubMed] [Google Scholar]
- 32. Stefansson B., Ohama T., Daugherty A. E., Brautigan D. L. (2008) Protein phosphatase 6 regulatory subunits composed of ankyrin repeat domains. Biochemistry 47, 1442–1451 [DOI] [PubMed] [Google Scholar]
- 33. Prickett T. D., Brautigan D. L. (2004) Overlapping binding sites in protein phosphatase 2A for association with regulatory A and α-4 (mTap42) subunits. J. Biol. Chem. 279, 38912–38920 [DOI] [PubMed] [Google Scholar]
- 34. Dalal S. N., Schweitzer C. M., Gan J., DeCaprio J. A. (1999) Cytoplasmic localization of human cdc25C during interphase requires an intact 14-3-3 binding site. Mol. Cell Biol. 19, 4465–4479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guergnon J., Derewenda U., Edelson J. R., Brautigan D. L. (2009) Mapping of protein phosphatase-6 association with its SAPS domain regulatory subunit using a model of helical repeats. BMC Biochem. 10, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Douglas P., Zhong J., Ye R., Moorhead G. B., Xu X., Lees-Miller S. P. (2010) Protein phosphatase 6 interacts with the DNA-dependent protein kinase catalytic subunit and dephosphorylates γ-H2AX. Mol. Cell Biol. 30, 1368–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Aurora R., Rose G. D. (1998) Protein Sci. 7, 21–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Forood B., Feliciano E. J., Nambiar K. P. (1993) Stabilization of α-helical structures in short peptides via end capping. Proc. Natl. Acad. Sci. U.S.A. 90, 838–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Presta L. G., Rose G. D. (1988) Helix signals in proteins. Science 240, 1632–1641 [DOI] [PubMed] [Google Scholar]
- 40. Anbarasan S., Janis J., Paloheimo M., Laitaoja M., Vuolanto M., Karimaki J., Vainiotalo P., Leisola M., Turunen O. (2010) Effect of glycosylation and additional domains on the thermostability of a family 10 xylanase produced by Thermopolyspora flexuosa. Appl. Environ. Microbiol. 76, 356–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Armstrong K. M., Baldwin R. L. (1993) Charged histidine affects α-helix stability at all positions in the helix by interacting with the backbone charges. Proc. Natl. Acad. Sci. U.S.A. 90, 11337–11340 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.