Abstract
JAK-STAT-activated gene expression is both rapid and transient and requires dynamic post-translational modification of the chromatin template. Previously, we showed that following IFN-γ treatment, trimethylation of histone H3 at lysine 79 (H3K79me3) is rapidly and highly induced in the 5′-end of the STAT1-dependent gene interferon regulatory factor 1 (IRF1), but the role of this histone modification was unexplored. Here we report that DOT1L, the non-SET domain containing methyltransferase that modifies Lys-79, is localized across IRF1 in the uninduced state and is not further recruited by IFN-γ induction. RNAi-mediated depletion of DOT1L prevents the induction of H3K79me3 and lowers the transcription of IRF1 2-fold, as expected. Surprisingly, STAT1 binding to its DNA recognition element near the IRF1 promoter is diminished 2-fold in the DOT1L-depleted cell line. In vivo and in vitro protein interaction assays reveal a DOT1L-STAT1 interaction. Domain mapping identifies the middle region of DOT1L (amino acids 580–1183) as the STAT1 interaction domain. Overexpression of the DOT1L STAT1 interaction domain represses IRF1 transcription (2-fold) and interferes with STAT1 DNA binding at IRF1 and endogenous DOT1L histone methyltransferase activity. Collectively, our findings reveal a novel STAT1-DOT1L interaction that is required for the regulation JAK-STAT-inducible gene expression.
Keywords: Chromatin Histone Modification, Gene Transcription, Histone Methylation, Protein-Protein Interactions, STAT Transcription Factor
Introduction
Signal transducers and activators of transcription (STATs) regulate growth, differentiation, and the immune response in a wide variety of cells (1). These transcription factors are recruited to cell surface receptors by extracellular ligand binding. STATs are then activated by the Janus kinases (JAKs), allowing them to translocate to the nucleus and drive the transcription of target genes.
In the nucleus, the eukaryotic genome is organized as chromatin. DNA is wrapped around octamers composed of the core histone proteins (H2A, H2B, H3, and H4) to produce nucleosomes. The core histones are post-translationally modified, via acetylation, methylation, sumoylation, ubiquitination, and phosphorylation. These dynamic nuclear signaling events work in concert to regulate transcription by affecting nucleosomal interactions and the recruitment of an array of regulatory proteins (2, 3).
Histone lysine methylation is particularly complex. It is associated with both activated and repressed chromatin states, depending on the site of modification. The degree of the methylation, i.e. mono-, di-, or trimethylation, correlates with different functional outputs. In addition, in mammalian systems, there are multiple histone methyltransferase-containing complexes. To date, all the known histone methyltransferases (HMTases)2 identified contain a SET domain, with the exception of disruptor of telomere silencing 1 (Dot1). Instead of a SET domain, Dot1 contains sequence motifs similar to ones found in DNA methyltransferases and protein arginine methyltransferases (4). Although most HMTases catalyze the addition of methyl groups on the protruding histone tails, Dot1 singularly methylates at lysine 79 (H3K79), which is located within the globular domain of histone H3 (5).
Like H3K4 methylation, H3K79 methylation depends on the monoubiquitination of H2B (6–8). A lysine-rich region of Dot1 (amino acids 101–140) directly interacts with ubiquitin and is important for ubH2B-mediated H3K79 methylation (9). Precisely how ubiquitin affects Dot1 in this cross-talk mechanism is not known; allostery, surface recruitment, and nucleosomal sterics are all possible mechanisms (10).
In yeast, H3K79 methylation is depleted at telomeric, mating-type, and ribosomal DNA but is ubiquitous elsewhere in the genome (11–13). Furthermore, methylation of H3K79 by Dot1 restricts the recruitment of silent information regulator (SIR) proteins to heterochromatic regions (11). Thus, Dot1 and H3K79 methylation generally defines euchromatin as permissive for transcription elongation in yeast (14).
The role of mammalian H3K79 methylation is less clear because it has been globally linked to both active and repressed chromatin states (11, 15). Steger et al. (16) attempted to clarify its role by profiling H3K79 methylation in several mammalian cell lineages. They found that it correlated with actively transcribed chromatin and that its function was mainly similar to that described in yeast (11, 17). Nevertheless, H3K79 methylation can be repressive of transcription at some gene loci. For example, when found in the promoters of aldosterone-regulated genes (18), such as epithelial Na+ channel (ENaCα) (19) and connective tissue growth factor (CTGF) (20), H3K79me3 inhibits basal transcription.
Mammalian Dot1 (DOT1L) is involved in several cellular processes including ES cell differentiation and embryonic development (21, 22), cardiac function (23), WNT signaling (24), parental allele discrimination (25), intestinal homeostasis (26), DNA damage response (27, 28), and erythropoiesis (29). In mixed lineage leukemia, mistargeting of DOT1L by several different MLL fusion proteins causes aberrant H3K79me3 and subsequent activation of target genes (reviewed in Ref. 30). The mechanistic basis for DOT1L function, however, remains largely undefined.
We investigated the contribution that DOT1L and histone H3K79 methylation make to the rapid and transient gene expression induced downstream of JAK-STAT signaling. We present evidence for a novel interaction between STAT1 and the middle region of DOT1L (amino acids 586–1138) that is required for proper IRF1 gene expression.
EXPERIMENTAL PROCEDURES
Antibodies
The antibodies used were: H3K79me3 (Abcam ab2621 for Western blots and Invitrogen 491020 for chromatin immunoprecipitation (ChIP)), Pan H3 CT (Millipore 07-690), RNA polymerase II (Santa Cruz Biotechnology sc-899), IgG (Jackson ImmunoResearch Laboratories), STAT1 (Santa Cruz Biotechnology sc-345X for ChIP, sc-346 for co-immunoprecipitation (co-IP) and Western blots), DOT1L (Bethyl Laboratories A300-953A), FLAG (Sigma F1804), GAPDH (Abcam ab9485), dynamin (Santa Cruz Biotechnology sc7988), phospho-STAT1 (Santa Cruz Biotechnology sc-6402), and anti-rabbit or anti-mouse horseradish peroxidase (HRP) (Jackson ImmunoResearch Laboratories).
Plasmids and shRNAmir Vectors
Dr. Yi Zhang (University of North Carolina) generously provided full-length DOT1L (isoform 2) in pcDNA3β. The DOT1L fragment expression plasmids were PCR-cloned into a pcDNA3.0 vector that incorporates an N-terminal FLAG tag using DOT1L primers containing 5′-EcoRI and 3′-XhoI sites. The fragments were generated with the following primers (5′ to 3′): N-terminal fragment, forward, ATAGTAGAATTCACATGGGGGAGAAGCTGGAGC, reverse, ATAGTACTCGAGCTACTAGTCCTGCTCCAGCTGCTCCGACTGC; middle fragment, forward, ATAGTAGAATTCACCAGTCGGAGCAGCTGGAGCAGGAC, reverse, ATAGTACTCGAGCTACTACAGGGGCTGGTTGATGTTACTGACCATC; C-terminal fragment, forward, ATAGTAGAATTCACGTCAGTAACATCAACCAGCCCCTG; reverse, ATAGTACTCGAGCTACTAGTTACCTCCAACTGTGCCGCCTGCCAC; ΔC fragment, forward, ATAGTAGAATTCACATGGGGGAGAAGCTGGAGC, reverse, ATAGTACTCGAGCTACTACAGGGGCTGGTTGATGTTACTGACCATC.
The ΔN fragment was generated with an HindIII and XhoI restriction enzyme digestion of the DOT1L pcDNA3β vector, which was then ligated into FLAG-pcDNA3.0 at these same sites. All constructs were confirmed by DNA sequencing. The pTRIPZ vector expressing an shRNAmir directed against DOT1L (RHS4696-100903376), as well as pTRIPZ non-silencing vector (RHS4743), were acquired from Open Biosystems. Another pTRIPZ vector expressing an shRNAmir directed against DOT1L (RHS4696-1009032641) showed the same transcriptional results described here.
Cell Culture and Transfection
2fTGH and U3A reconstituted cell lines (31) were cultured in HyClone Dulbecco's modified/high glucose medium supplemented with 10% cosmic calf serum and 10% antibiotic/antimycotic (Fisher Scientific). Interferon-γ (IFN-γ) treatment involved adding IFN-γ (R&D Systems, 5 ng/ml) to the medium for 30 min, replacing with fresh medium, and harvesting the cells at the indicated times. Transfection was carried out using the Arrest-In reagent according to the manufacturer's protocol (Open Biosystems). Stable cell lines were selected and maintained with puromycin dihydrochloride (MP Biologicals, 3 μg/ml), and RNAi knockdown was induced with doxycycline hydrochloride (MP Biologicals, 1 μg/ml) for 24 h. Individual clones were characterized for DOT1L expression by quantitative reverse transcription-PCR (qRT-PCR) and Western blotting. Transient transfection of knockdown cell lines was carried out 24 h after doxycycline treatment, and doxycycline was maintained in the medium throughout.
qRT-PCR
Total RNA was collected using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. RNA was DNase I (Sigma)-treated, and 2 μg of RNA was converted to cDNA using the high capacity RNA-to-cDNA kit (Applied Biosystems). The cDNA was used as template for SYBR Green quantitative PCR (qPCR) with a 7500 fast real time PCR system (Applied Biosystems). Genomic DNA contamination was assessed using the reverse transcriptase negative control and was undetectable in all experiments. Biological replicates were plotted, and S.E. was calculated (32). Student's t test determined significance. DOT1L, IRF1, GAPDH, POLR2F, and GBP2 qPCR primers used were as follows (5′ to 3′): DOT1L, forward, TCGTCCACACTTGAAAAGCAGAT, reverse, CACCGAGCCAGCGTAGGA; IRF1 (3500), forward, AAAGGAGCCAGATCCCAAGAC, reverse, GGTGGAAGCATCCGGTACAC; IRF1 (1700), forward, GCGAGACCCTTACAAACATACACA, reverse, CCAGCAGCATCCACTTCTCA; GAPDH, forward, GGCATCCTGGGCTACACTGA, reverse, GCCCCAGCGTCAAAGGT; POLR2F, forward, GGCCAACCAGAAGCGAATC, reverse, GCGGGCTCGCTCGTACT; GBP2, forward, GCCTTGGCCCAGATAGAGAACT, reverse, TCTGCTGTTCATAGTGGGCAAT.
Western Blotting, Co-IP, and GST Pulldown Assays
These assays were performed with at least two biological replicates. For Western blotting, whole cell extract (30–50 μg), prepared as described (33), was subjected to SDS-PAGE and transferred to a nitrocellulose membrane. Immunodetection was performed using the antibodies indicated at the dilution recommended by the supplier. A horseradish peroxidase anti-species secondary antibody (1:10,000, Jackson ImmunoResearch Laboratories) was applied, and immunoreactive proteins were visualized using chemiluminescence reagent (Thermo Scientific). Acid extraction of histone proteins was done as described (34). Bands were quantified with ImageJ. Ponceau S staining verified that equal amounts of protein were loaded in each lane.
For co-IP assays, 0.5–1 mg of whole cell extract, prepared from 2fTGH cells or the shRNAmir cells, was immunoprecipitated with 2 μg of α-STAT1, α-DOT1L, or IgG on ice for 90 min. Immunocomplexes were bound to protein A-salmon sperm DNA beads (Millipore) and washed. Proteins were eluted by boiling in SDS loading buffer, separated by 5% SDS-PAGE, transferred to a nitrocellulose membrane, and immunodetected. Input samples were 5% of total sample. The same protocol was performed using the STAT1 isoform reconstituted U3A cells that were transfected with pcDNA3.0 FLAG-DOTL1 or empty vector, except that α-FLAG was used instead of α-DOT1L.
For glutathione S-transferase (GST) pulldown assays, GST-STAT1 and GST were expressed in BL21 bacteria and purified using glutathione-Sepharose 4B beads (GE Healthcare). Equal amounts (determined by Coomassie Brilliant Blue staining of an SDS-PAGE gel) of each were rocked overnight with whole cell extract (750 μg) prepared from 2fTGH cells transiently expressing FLAG-tagged DOT1L protein fragments, in GST binding buffer (10 mm Hepes, pH 7.4, 200 mm NaCl, 5 mm EDTA, pH 8.0, 0.1% Nonidet P-40, 1 mm DTT, and 5 mg/ml BSA). Beads were washed three times with GST binding buffer. Proteins were eluted from the beads by boiling in SDS sample buffer and analyzed by SDS-PAGE and Western blotting with α-FLAG. Input samples were 5% of total sample.
ChIP
ChIP analyses were carried out as described previously (35). shRNAmir-expressing cells were grown to 80% confluency and were treated with 5 ng/ml IFN-γ for the indicated times or left untreated. 1 μl of purified DNA was analyzed using SYBR Green qPCR. PCR efficiencies were determined for all primer pairs. All experiments were performed in duplicate, if not triplicate, but one replicate is shown in the figure. Pan H3 (positive control) and IgG (negative control) were used in every experiment. To ensure the statistical significance of differences reported in the ChIP assays, standard errors were calculated for the multiples and, if necessary, a Student's t test confirmed significance, p ≤ 0.05. The ChIP qPCR primers are included in supplemental Table 1.
RESULTS
DOT1L Is Constitutively Associated with the IRF1 Gene
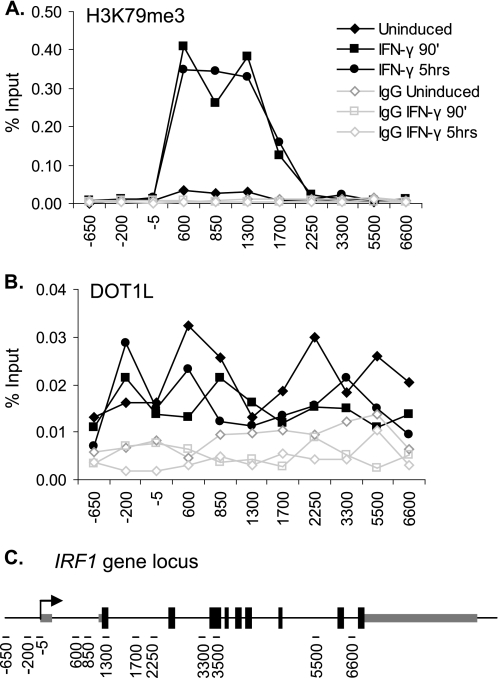
To study H3K79me3 and DOT1L in JAK-STAT-induced gene expression, we profiled H3K79me3 levels at the STAT1-actived gene interferon regulatory factor 1 (IRF1). ChIP assays were performed before, during, and after the induction of IRF1 by IFN-γ treatment. H3K79me3 increased ∼20-fold in response to IFN-γ, remained this high even at the time point (5 h) when IRF1 gene expression returned to basal levels, and was limited to the 5′-end of the gene (Fig. 1A). A graphic depiction of the IRF1 gene and the locations of the qPCR primer pairs used in this study is shown in Fig. 1C. These data recapitulated our previous results using a different H3K79me3 antibody (36).
FIGURE 1.
Profile of H3K79me3 and DOT1L at the IRF1 gene. A and B, ChIP using antibodies to H3K79me3 (A) and DOT1L (B) in 2fTGH cells treated with IFN-γ for the indicated times or left untreated. qPCR, using primers spanning the IRF1 gene locus, quantified the precipitate yield reported as the percentage of input. IgG served as the negative control. C, graphic depiction of the IRF1 gene (∼9 kb), showing the location of the qPCR primers (black dashes) used in this study.
In human cell lines, DOT1L occupancy correlates with constitutive gene activity (16). In addition, it is well established that monoubiquitination of H2B (ubH2B) is required for H3K79 methylation and H3K4 methylation during transcriptional initiation (37). At IRF1, ubH2B is highly but transiently induced in response to IFN-γ (36, 55). It is unknown, however, whether DOT1L is recruited to the gene locus during inducible gene expression. To address this question, we examined the occupancy of endogenously expressed DOT1L along the IRF1 gene before, during, and after its induction with IFN-γ using ChIP. In these assays, DOT1L levels were the same regardless of gene activity (Fig. 1B), demonstrating that DOT1L is not recruited. Thus, the increase in H3K79me3 in the 5′-end of the gene depends upon an event(s) triggered by IFN-γ, most likely the monoubiquitination of H2B.
RNAi-mediated Depletion of DOT1L Decreases IRF1 Transcription
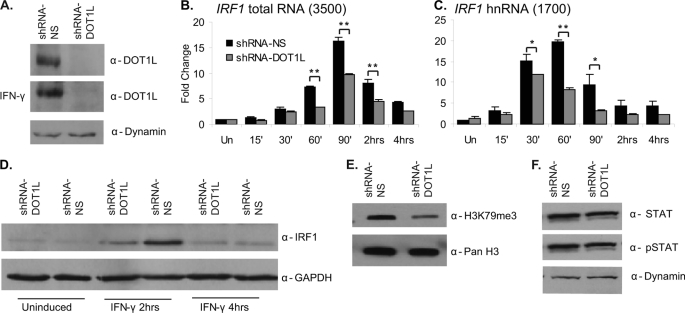
To evaluate the function of DOT1L and H3K79me3 in STAT1-activated transcription, we generated a cell line depleted of DOT1L. A tetracycline-inducible shRNAmir vector targeting DOT1L (shRNAmir-DOT1L, supplemental Fig. 1A) was transfected into 2fTGH cells, and stably selected clones were characterized as to their DOT1L expression levels. A control cell line expressing a non-silencing shRNAmir (shRNAmir-NS) vector was also selected. Western blotting indicated a complete loss of DOT1L expression (Fig. 2A), although qRT-PCR analysis revealed that 10–15% of DOT1L mRNA remains expressed in this cell line (supplemental Fig. 1B). DOT1L expression was not affected by IFN-γ induction (Fig. 2A). More importantly, STAT1 expression and activation levels, as measured by total and phospho-STAT Western blotting, were not affected by DOT1L depletion (Fig. 2F).
FIGURE 2.
RNAi-mediated depletion of DOT1L decreases IRF1 gene expression. A, DOT1L Western blot of whole cell extracts collected from 2fTGH cells stably expressing the pTRIPZ shRNAmir non-silencing (shRNA-NS) or shRNAmir-DOT1L (shRNA-DOT1L) vectors with or without IFN-γ. Dynamin served as a loading control. B and C, qRT-PCR to quantitate IRF1 mRNA and heteronuclear RNA (hnRNA) levels in the shRNAmir non-silencing and shRNAmir-DOT1L cell lines treated with IFN-γ for the times indicated or left untreated (Un). IRF1 was normalized to GAPDH, and expression is presented as -fold change relative to the uninduced, shRNAmir-NS condition. Error bars indicate S.E. (n = 3). Student's t test determined significance; **, p ≤ 0.01, *, p ≤ 0.05. D, IRF1 Western blot of whole cell extracts of shRNAmir non-silencing or shRNAmir-DOT1L cells treated with IFN-γ for the indicated times or left untreated. GAPDH served as a loading control. E, H3K79me3 Western blot of acid-extracted histones from the shRNAmir non-silencing and shRNAmir-DOT1L cell lines. Pan H3 served as a loading control. F, STAT1 and phospho-STAT1 Western blots of whole cell extracts collected from 2fTGH cells stably expressing the shRNAmir non-silencing or shRNAmir-DOT1L vectors. The ratio of phospho-STAT1 (pSTAT) to total STAT1 is the same in both cell lines. Western blot bands were quantified with ImageJ.
Using qRT-PCR, we determined how DOT1L depletion affected STAT1-activated transcription of IRF1 in a time course of IFN-γ treatment. In the shRNAmir-DOT1L cell line, both IRF1 total RNA and pre-mRNA levels were ∼2-fold lower in response to IFN-γ as compared with the non-silencing control cell line (Fig. 2, B and C), indicating that the transcription rate is slower when DOT1L is depleted (38). IFN-γ-induced transcription of two other STAT1 target genes, POLR2F and GBP2, was also ∼2-fold lower (supplemental Fig. 2, A and B). Because protein levels typically correlate with mRNA levels, we performed Western blots on cellular extracts collected from the non-silencing and DOT1L-depleted cell lines. IRF1 expression is ∼70% lower (Fig. 2D) in the shRNAmir-DOT1L cell line. Next, we asked whether overexpressing DOT1L could reverse the reduction in IRF1 transcript and protein expression observed in the shRNAmir-DOT1L cell line. The cells were transiently transfected with a vector expressing FLAG-tagged DOT1L or an empty control vector (supplemental Fig. 2E). Both IRF1 transcript and protein were restored to levels observed in the non-silencing cell line, demonstrating the specificity of the shRNAmir targeting DOT1L (supplemental Fig. 2, C, D, and F). A second shRNAmir vector depleted DOT1L to similar levels and also lowered IRF1 transcription ∼2-fold (data not shown). Taken together, these data indicate that DOT1L is necessary for proper STAT1-activated transcription of IRF1.
Lastly, we asked whether DOT1L depletion affected global levels of H3K79me3. Acid-extracted histones collected from the shRNAmir-DOT1L and control cell lines were subjected to Western blotting with the H3K79me3 antibody (Fig. 2E). H3K79me3 was reduced ∼70% in the shRNAmir-DOT1L cell line.
RNA Polymerase II and STAT1 Localization to the IRF1 Promoter Is Reduced in shRNAmir-DOT1L Cells
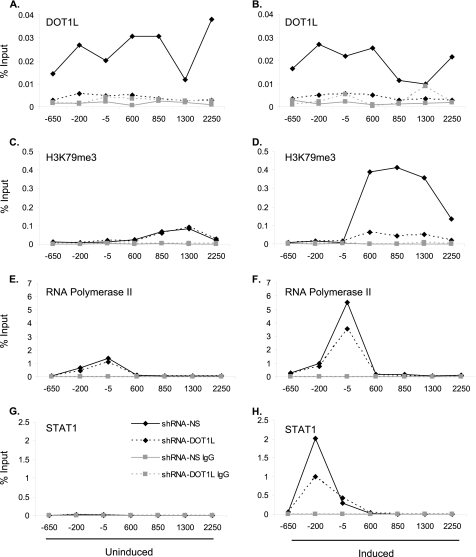
Next, we profiled DOT1L, H3K79me3, and total histone H3 along the IRF1 gene with ChIP assays in the shRNAmir cell lines. As anticipated, DOT1L was not detectable above negative control levels (IgG) in the shRNAmir-DOT1L cell line (Fig. 3, A and B). H3K79me3 was not induced by IFN-γ in the DOT1L-depleted cell line, whereas the non-silencing control cells showed an ∼8-fold increase in H3K79me3 (Fig. 3, C and D). Depletion of DOT1L did not alter the total histone H3 profile across IRF1 (supplemental Fig. 3, A and B).
FIGURE 3.
RNA polymerase II and STAT1 localization to the IRF1 promoter is reduced in shRNAmir-DOT1L cells. A–H, ChIP of shRNAmir non-silencing (shRNA-NS) or shRNAmir-DOT1L (shRNA-DOT1L) cells treated with IFN-γ for 30 min (right panels) or left untreated (left panels). ChIP was performed with the antibodies indicated, and qPCR, using primers spanning the IRF1 gene locus, quantified the precipitate yield reported as the percentage of input. IgG served as the negative control. p ≤ 0.05 for panels A, B, D, F, and H for solid lines with black diamonds versus dotted lines with black diamonds.
We also determined the RNA polymerase II and STAT1 ChIP profiles at the IRF1 promoter. DOT1L knockdown modestly affected the recruitment of RNA polymerase II to IRF1 (Fig. 3, E and F) such that it was 1.5-fold lower at the transcription start site (Fig. 3F). Given the reduction in IRF1 transcription observed via qRT-PCR, this was not an unexpected result. However, it was unexpected that STAT1 recruitment to its DNA recognition element at −200 bp was consistently 2-fold lower (Fig. 3H) in the shRNAmir-DOT1L cell line. (STAT1 homodimers bind to a DNA element known as a γ-activated site (GAS) with the consensus sequence TTCNNNGA when activated by IFN-γ (39).) The same result was observed at the GAS of GBP2 (supplemental Fig. 4C). In other 2fTGH cell lines where we have depleted a histone-modifying complex, we have not observed a change in STAT1 binding at the GAS (36).3 Furthermore, although DOT1L occupies the region just upstream of the transcription start site at IRF1, we did not detect H3K79me3 in this region. Thus, these results suggested that DOT1L might be required to effectively recruit STAT1 to IRF1 in a manner that does not necessarily involve H3K79 methylation.
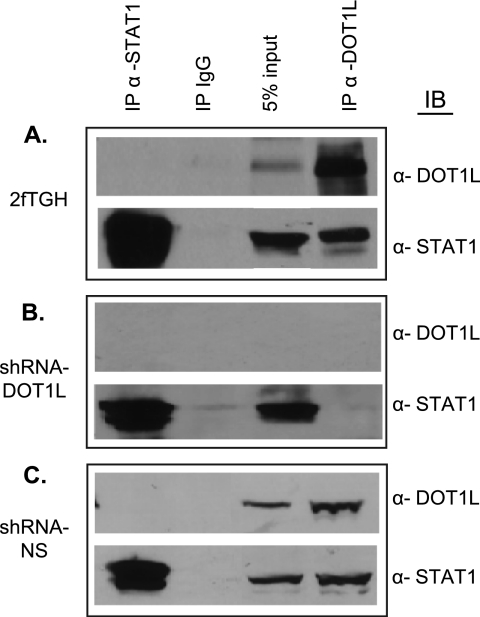
STAT1 Co-immunoprecipitates with DOT1L
To test this hypothesis, we employed co-IP assays to determine whether DOT1L and STAT1 interact endogenously. Whole cell extracts from 2fTGH cells, DOT1L-depleted cells, and control non-silencing cells were immunoprecipitated with antibodies against STAT1, DOT1L, or IgG. Immunoprecipitation of DOT1L followed by Western blotting for STAT1 showed that the two proteins interact (Fig. 4A). As expected, the interaction is lost in the shRNAmir-DOT1L cell line as compared with the non-silencing control cells (Fig. 4, B and C). Performing the co-IP in the reverse direction failed to show an interaction. This might be because the STAT1 antibody interferes with DOT1L binding. IFN-γ treatment did not change the co-immunoprecipitate (data not shown).
FIGURE 4.
STAT1 co-immunoprecipitates with DOT1L. A–C, whole cell extracts prepared from 2fTGH (A), shRNAmir-DOT1L (shRNA-DOT1L) (B), or shRNAmir non-silencing (shRNA-NS) (C) cells were immunoprecipitated (IP) with α-DOT1L, α-STAT1, or IgG (negative control) and then immunoblotted (IB) with the indicated antibodies. A 5% input aliquot of the extracts was included for reference.
In an attempt to define the molecular basis of the STAT1-DOT1L interaction observed, we assayed the ability of two STAT1 isoforms to co-immunoprecipitate with DOT1L. STAT1β is a naturally occurring STAT1 splice variant that is missing the C-terminal transcriptional activation domain (40, 41). ΔNSTAT1 is an artificial STAT1 construct in which the N-terminal 154 amino acids were deleted (42). Both can be phosphorylated and are able to bind the GAS element. Other deletions of the STAT1 core structure will not maintain these properties (43). For the co-IP assays, we used U3A cells reconstituted with vectors stably expressing STAT1, STAT1β, or ΔNSTAT1 (41, 44). U3A is a 2fTGH cell line that was rendered null for STAT1 via genetic mutation (31, 45). These stable cell lines were transiently transfected with a plasmid expressing FLAG-tagged DOT1L or an empty vector, and co-IPs were carried out. All three STAT1 proteins co-immunoprecipitated with DOT1L (supplemental Fig. 5, A–C). Thus, neither the N-terminal domain nor the C-terminal transactivation domain of STAT1 is sufficient to accomplish the interaction with DOT1L.
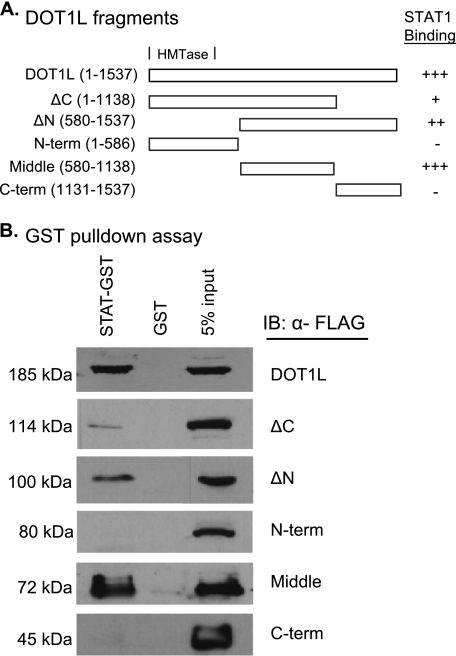
The Middle Region of DOT1L (Amino Acids 580–1138) Specifically Interacts with STAT1 in GST Pulldown Assays
Amino acids 1–332 of DOT1L comprise the H3K79 methyltransferase domain and are homologous with the C-terminal half of the yeast Dot1 protein (5, 46). A lysine-rich region (amino acids 101–140) in yeast Dot1 has been shown to be important for both nucleosomal and ubiquitin binding and for di- and trimethylation of H3K79 (9). In DOT1L, a similar region is found between amino acids 391 and 416. In addition, although the regions between residues 479–659 and 829–972 of DOT1L mediate a repressive interaction with AF9 at the ENaCα gene promoter (18), the function of the remaining C-terminal sequence, which is unique to mammalian DOT1L proteins and absent in yeast Dot1, is unknown.
Using a domain mapping strategy, we sought to determine the region of DOT1L that is responsible for its interaction with STAT1. DOT1L was divided into three regions and cloned into an expression vector so that each would be N-terminally FLAG-tagged. The three regions were (i) the N-terminal 586 amino acids; (ii) a middle fragment, which includes amino acids 580–1138; and (iii) a C-terminal region composed of amino acids 1131–1537. We also created ΔN and ΔC truncation mutations (Fig. 5A). 2fTGH cells were transiently transfected with each of these vectors, as well as a vector expressing FLAG-tagged full-length DOT1L. The protein products of each of these constructs were examined by SDS-PAGE to ensure that they were the correct molecular weight and were expressed at similar levels (data not shown).
FIGURE 5.
Mapping of the DOT1L region that interacts with STAT1 in GST pulldown assays. A, graphic depiction of the DOT1L fragments and their relative STAT1 binding affinities. The presence (+) or absence (−) of an interaction is shown, with +, ++, and +++ indicating weak, modest, and strong interactions, respectively. N-term, N-terminal; C-term, C-terminal. B, GST pulldown assays using whole extracts prepared from 2fTGH cells transiently transfected with pcDNA3β FLAG-tagged DOT1L or FLAG-tagged DOT1L fragment vectors and then immunoblotted (IB) with α-FLAG. A 5% input aliquot of the extracts was included for reference.
To test whether the DOT1L fragments retained the ability to interact with STAT1, we performed GST pulldown assays. STAT1 fused to GST or GST alone was coupled to glutathione beads. The beads were then incubated with extracts prepared from 2fTGH cells expressing the FLAG-tagged DOT1L proteins. The STAT1 interaction was lost when the N-terminal region containing the HMTase domain or the C-terminal domain of DOT1L was expressed (Fig. 5B). In contrast, GST-STAT1 was able to pull down the middle region of DOT1L. This region is present in both the ΔN and the ΔC truncation variants as well, and these proteins retain the STAT1 interaction, albeit less robustly. Thus, we defined the middle region of DOT1L (amino acids 580–1138) as the STAT1 interaction domain (SID). Interestingly, the SID overlaps with the regions of DOT1L (479–659 and 829–972) that were previously shown to interact with AF9 for the repression of the ENaCα gene (18).
Overexpression of the SID of DOT1L Represses Transcription, STAT1 Binding, and H3K79me3 at IRF1
We hypothesized that the SID region of DOT1L most likely would act in a repressive manner on IRF1 gene expression. It could interfere with STAT1 binding at the GAS at the IRF1 promoter, or it could have no impact on STAT1, or even facilitate its binding but interfere with endogenous DOT1L by failing to promote effective H3K79me3 in response to IFN-γ because it lacks the HMTase domain. To distinguish among these possible scenarios, we performed IRF1 transcription and ChIP assays and measured global H3K79me3 levels in the shRNAmir-DOT1L and shRNAmir-NS cell lines overexpressing the DOT1L fragments.
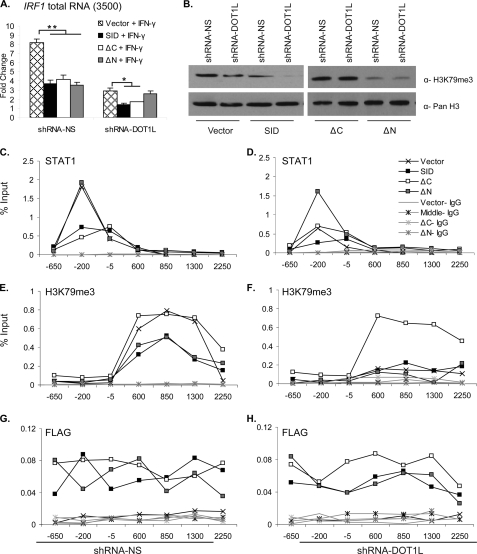
The shRNAmir-DOT1L and shRNAmir-NS cells were transiently transfected with pcDNA3.0 constructs expressing the SID and the ΔN and ΔC truncation variants of DOT1L or the empty vector and then induced with IFN-γ. Overexpression of these three DOT1L fragments led to decreased IRF1 gene expression (∼2-fold) in the shRNAmir non-silencing cell line (Fig. 6A). Overexpression of the SID and the ΔC truncation variant also decreased the remaining IRF1 present in the DOT1L knockdown cell line. This transcriptional activity might reflect the transcription that is possible independent of DOT1L or the fact that a small amount of DOT1L remains in the knockdown cell line (supplemental Fig. 1B). However, overexpression of the ΔN truncation variant did not further decrease IRF1 levels. The N-terminal and C-terminal fragments, which both lack the SID, did not have any effect (supplemental Fig. 6A), but full-length DOT1L rescued the transcriptional defect observed in the shRNAmir-DOT1L cell line (supplemental Fig. 2C).
FIGURE 6.
Overexpression of the SID of DOT1L represses IRF1 gene expression and alters STAT1 binding. A, qRT-PCR measured IRF1 mRNA expression in 2fTGH, shRNAmir non-silencing (shRNA-NS), and shRNAmir-DOT1L (shRNA-DOT1L) cell lines that were transiently transfected with pcDNA3.0 DOT1L SID, ΔC, ΔN, or empty vector. IFN-γ induction was for 30 min. IRF1 was normalized to GAPDH, and expression is presented as -fold change relative to the uninduced, shRNAmir-NS cell line transfected with empty vector. Error bars indicate S.E. (n = 2). Student's t test determined significance, **, p ≤ 0.01, *, p ≤ 0.05. B, Western blots of acid-extracted histones from the shRNAmir non-silencing and shRNAmir-DOT1L cell lines that were transiently transfected as in panel A. PanH3 served as a loading control. C–H, ChIP, using the indicated antibodies in shRNAmir-DOT1L (right panels) or shRNAmir non-silencing (left panels) cells transiently transfected with pcDNA3.0 DOTL1 SID, ΔC, ΔN, or empty vector and treated with IFN-γ for 30 min. p ≤ 0.05 for black lines with an X or gray squares versus black lines with black squares or white squares in panel C; black lines with gray squares versus others in panel D; black lines with an X or white squares versus black lines with black squares or gray squares in panel E; black lines with white squares versus others in panel F.
To assess how the DOT1L fragments impacted global H3K79me3, histones were acid-extracted from the transfected shRNAmir-DOT1L and the shRNAmir-NS cell lines and Western blotted with α-H3K79me3 antibody. Overexpression of the SID decreased the level of H3K79me3 globally: 50% in the shRNAmir-NS cell line and nearly completely in the shRNAmir-DOT1L cell line (Fig. 6B). These data indicated that overexpression of the middle region of DOT1L antagonizes native DOT1L HMTase function. Overexpression of the ΔN DOT1L truncation variant also lowered H3K79me3. Meanwhile, the ΔC truncation variant increased global H3K79me3 such that its levels were restored in the shRNAmir-DOT1L cell line and were slightly enhanced in the non-silencing cells (120%). A similar result was observed when the N-terminal fragment was overexpressed, whereas the C-terminal fragment had no effect on H3K79me3 levels (supplemental Fig. 6B).
Finally, we performed ChIP assays in shRNAmir-DOT1L and shRNAmir-NS cells transiently transfected with these same three DOT1L fragments to determine whether and how their overexpression altered the STAT1 and H3K79me3 profiles at IRF1 during IFN-γ induction. STAT1 was displaced ∼2-fold in the non-silencing cells overexpressing the middle and ΔC fragments of DOT1L, whereas the ΔN fragment had no effect (Fig. 6C). In the shRNAmir-DOT1L cell line, where STAT1 localization to the GAS is already compromised, the ΔN fragment restored STAT1 binding to near normal levels (Fig. 6D). The SID and ΔC fragments had no effect on STAT1 binding to the GAS.
The H3K79me3 ChIP profile was 2-to-3-fold lower when non-silencing cells overexpressed the SID and the ΔN fragment, but the ΔC fragment had no effect (Fig. 6E). The loss of H3K79me3 seen in the DOT1L-depleted cell line was not significantly changed by overexpression of the SID or the ΔN fragment (Fig. 6F). However, the ΔC fragment restored H3K79me3 to normal levels.
ChIP with a FLAG antibody consistently showed that the DOT1L fragments were localized to the IRF1 gene (Fig. 6, G and H). This was somewhat unexpected for the SID and ΔN fragments because these proteins lack the lysine-rich domain (391–416) that is the only known Dot1 nucleosome interaction domain (9). One possible explanation for this observation is that there is a second nucleosomal binding domain contained between residues 580 and 1138 of DOT1L. ChIP assays using the DOT1L antibody also confirmed that the middle fragment of DOT1L is localized to the IRF1 gene. The epitope this antibody recognizes maps to a region between residues 1000 and 1050 of DOT1L. Thus, the ChIP signals in the non-silencing cells reflected both endogenous DOT1L and the middle fragment (supplemental Fig. 6E, black squares) and were higher than the levels in the shRNAmir-DOT1L cells (supplemental Fig. 6F, black squares). Total histone H3 levels were consistent in all the experimental conditions (supplemental Fig. 6, C and D).
The results of the transcription and ChIP assays demonstrated that overexpression of the SID of DOT1L acts in an antimorphic manner during IFN-γ-induced IRF1 expression, antagonizing native DOT1L function both in STAT1 binding at the GAS and in H3K79 methylation. In contrast, the ΔN and ΔC fragments of DOT1L disrupt only one or the other of these functions. The ΔN fragment lacks the HMTase domain and thus cannot generate full H3K79me3 in the non-silencing cells, nor ameliorate H3K79me3 in the knockdown cell line. However, it does restore STAT1 binding in the knockdown cell line. Meanwhile, the ΔC fragment can generate proper H3K79me3 in the non-silencing cells and can even restore H3K79me3 to normal levels in the knockdown cells. However, STAT1 is displaced by overexpression of this DOT1L fragment. Thus, the C-terminal residues (1139–1537) of DOT1L seem to modulate the SID and counteract its dominant negative effect on STAT1 binding to GAS. Collectively, the data identify a novel interaction between STAT1 and a region within the non-conserved C terminus of DOT1L that is required for proper JAK-STAT-inducible gene expression.
DISCUSSION
DOT1L is the only non-SET domain containing HMTase, and it the only known HMTase that modifies Lys-79 of histone H3. Despite its unique features, DOT1L is enigmatic in its function. In this study, we exploited the highly rapid and transient gene expression that is induced by JAK-STAT signaling to investigate DOT1L and more fully define its role in the regulation of transcription.
Enzymatically Active DOT1L Is Induced, Not Recruited, to IRF1 in Response to IFN-γ
In this study, we have shown H3K79me3 is highly induced in the 5′-end of the IRF1 when the transcription of this gene is triggered by IFN-γ. This induction, however, does not require the recruitment of DOT1L to the IRF1 gene locus. Instead, DOT1L appears constitutively associated with the IRF1 nucleosomal template and its catalytic activity as an HMTase is likely promoted by the well established, trans-histone cross-talk with H2B ubiquitination (36).4
Three mutually non-exclusive mechanisms for how ubH2B controls H3K79 methylation have been proposed (reviewed in Ref. 47). In the first mechanism, a direct interaction between ubiquitin and Dot1 is required for H3K79 methylation. In another mechanism, additional factors, such as those associated with the Set1-containing COMPASS complex, work as a bridge to recruit Dot1. In addition, in a third mechanism, ubiquitination of H2B alters the chromatin structure to make H3K79 more accessible to Dot1. We have previously established that a COMPASS-like complex is, in fact, recruited to IRF1 in response to IFN-γ (36). Thus, of these three possibilities, the mechanism in which DOT1L is recruited to the IRF1 nucleosomal substrate appears the least likely. Our ongoing studies are aimed at confirming the predicted dependence of H3K79me3 on ubH2B at IRF1 and determining whether this mechanistic relationship is direct, as imagined in the first scenario, or indirect, as suggested by the third possibility.
DOT1L-STAT1 Interaction
DOT1L is localized to the chromatin both upstream and downstream of the IRF1 transcription start site, and its RNAi-mediated depletion affects STAT1 binding to the GAS element. Given that DOT1L appears to be integrated into the chromatin structure, possibly in part via the lysine-rich region found between residues 380 and 428 (5, 9), we speculated that DOT1L might have a role in STAT1 binding that was separate from its function as the Lys-79 HMTase.
Using in vivo and in vitro approaches, we demonstrated that STAT1 and DOT1L interact robustly. Furthermore, the middle region of DOT1L was indentified as the SID. We determined the consequences of overexpressing the SID, on its own or as part of the larger ΔN- and ΔC-terminal fragments, for IRF1 gene transcription, STAT1 binding at the GAS, and H3K79me3 both globally and at IRF1. The results of these experiments suggest that DOT1L can be divided into three functional domains when considering its role in STAT1-activated gene expression. First, the data confirm that the N-terminal region (amino acids 1–580) provides the H3K79 methyltransferase activity. A portion of this region (amino acids 1–332) is conserved with the C-terminal half of yeast Dot1 (5), which contains the enzymatic activity of that protein. Next, the middle region (amino acids 580–1138) functions as the SID. Overexpression of this protein interfered with STAT1 chromatin binding. It also lowered H3K79me3, likely because it competes with endogenous DOT1L throughout the chromatin template. Finally, the remaining C-terminal residues (amino acids 1130–1537) work to modulate the interactivity of the SID. Overexpression of the SID, on its own or as part of the ΔC variant, disrupted STAT1 binding. In contrast, when the C-terminal region was included, whether in the context of full-length DOT1L or the ΔN variant, STAT1 binding was normal in the non-silencing cells and even restored in the DOT1L-depleted cell line.
Together, the data suggest a tripartite description of DOT1L, where the N-terminal region provides the HMTase activity, the middle region is the SID, and the C-terminal region regulates the interactivity of SID. Such a description is supported by the changes in global H3K79me3 (Fig. 6B) observed in the shRNAmir-NS and shRNAmir-DOT1L cell lines overexpressing these DOT1L constructs. The SID and ΔN fragment both lack the HMTase domain and thus decrease global H3K79me3 in both shRNA cell lines. As expected, overexpression of the ΔC fragment increases H3K79me3 globally; it is missing the region of DOT1L that affects the SID, but maintains the HMTase domain.
As mediators of the signal transduction and gene expression triggered by several cytokines and growth factors, STATs interact with several other transcription factors and co-activators (reviewed in Refs. 48 and 49). Of these STAT-interacting proteins, some are known chromatin modifiers, including the Brahma-related gene 1 (BRG1) component of the SWI/SNF complex (50), the histone acetyltransferase CREB-binding protein (CBP)/p300 (51), and the histone deacetylase I (HDAC1) (52). Unphosphorylated STAT interacts with the heterochromatin protein, HP1 in Drosophila (53). The N-terminal domain and transactivation domain (TAD) of STATs participate in many of these interactions, although residues in the other core domains of STATs can be important as well. This seems to be the case for the STAT1-DOT1L interaction, which is the first time an HMTase as been shown to bind a STAT protein because Stat1β and ΔNSTAT1 bind DOT1L as well as full-length STAT1.
Ongoing studies are testing the tripartite hypothesis for DOT1L in STAT1-activated transcription described above. More generally, the results presented here inform the larger issue that remains unresolved for DOT1L. Specifically, what is the purpose of the large C-terminal region (amino acids 330–1537) that is specific to mouse and human DOT1L, but is absent in yeast Dot1?
Non-conserved C Terminus of DOT1L
In regulating normal gene expression, DOT1L contributes to multiple elongation complexes (reviewed in Ref. 30). The elongation complex components AF10, AF9, and ENL all interact with DOT1L. In mixed lineage leukemia, these components are a few of the many proteins (>50) that become fused, via chromosomal translocations, to the H3K4 methyltransferase, MLL. MLL normally controls the transcriptional activity of Hox genes. However, in leukemia, the MLL fusion proteins overtake normal MLL function, resulting in constitutive Hox gene activation and uncontrolled cell proliferation. In many cases, DOT1L also contributes to leukemogenesis as it is mistargeted to gene loci via its interaction with an MLL fusion partner, such as AF4, AF10, AF9, or ENL. In this way, MLL target genes become hypermethylated at H3K79, resulting in their constitutive expression and cellular transformation.
In some instances, a DOT1L interaction domain has been mapped to a region of the partner protein. AF10 and DOT1L interact through the region of AF10 that contains an octapeptide motif and a leucine zipper (OM-LZ) (54). Besides our study, an interaction domain for DOT1L itself has been mapped in just one other study to our knowledge. Zhang et al. (18) have demonstrated that DOT1L interacts with AF9 through the region spanning residues 479 and 972 at aldosterone-regulated genes. Thus, the DOT1L-AF9 interaction region and the SID defined here overlap. The biological outcomes for these interactions, however, do not. In contrast to its role in JAK-STAT-induced transcription, DOT1L interacts with AF9 to promote H3K79 methylation to repress aldosterone-regulated genes. Furthermore, the methylation of H3K79 occurs in the ENaCα promoter, rather than the nucleosomes downstream of the transcription start site. Two important conclusions of that study were that DOT1L can act in a targeted manner and that DOT1L-interacting proteins determine where and when DOT1L HMTase activity is directed across the genome. It is intriguing, therefore, to speculate that the function of the large, non-conserved C-terminal region of DOT1L is to specify DOT1L interactions with various partner proteins so that H3K79 methylation invokes proper and diverse biological outputs. This seems a likely explanation for why the pattern and function of DOT1L and H3K79me3 appear to be more complicated in mammalian systems than in yeast; Dot1 does not possess an interaction domain that can mediate different interactions with proteins for diverse biological effects.
In this study, we have identified a novel DOT1L interaction that is required for the proper transcription of STAT1-activated genes. Identifying other proteins that interact with DOT1L and determining the mechanistic basis of their interactions and how these partnerships play out across the chromatin landscape will greatly improve our understanding of DOT1L in the many biological processes to which it contributes in mammalian systems.
Acknowledgments
We are grateful to Dr. James E. Darnell, Jr. (Rockefeller University) for the 2fTGH and U3A reconstituted cell lines and the GST-STAT1 fusion BL21 bacterial stock. Full-length FLAG-tagged DOT1L was generously provided by Dr. Yi Zhang (University of North Carolina School of Medicine).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–6.
L. J. Buro and E. Chipumuro, personal communication.
E. Chipumuro and M. A. Henriksen, unpublished observations.
- HMTase
- histone methyltransferase
- SID
- STAT1 interaction domain
- ub
- ubiquitin
- DOT1L
- disrupter of telomere silencing 1-like
- MLL
- mixed lineage leukemia
- co-IP
- co-immunoprecipitation
- qRT-PCR
- quantitative reverse transcription-PCR
- qPCR
- quantitative PCR
- GAS
- γ-activated site
- CREB
- cAMP-response element-binding protein.
REFERENCES
- 1. Levy D. E., Darnell J. E., Jr. (2002) Nat. Rev. Mol. Cell Biol. 3, 651–662 [DOI] [PubMed] [Google Scholar]
- 2. Gardner K. E., Allis C. D., Strahl B. D. (2011) J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kouzarides T. (2007) Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 4. Feng Q., Wang H., Ng H. H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. (2002) Curr. Biol. 12, 1052–1058 [DOI] [PubMed] [Google Scholar]
- 5. Sawada K., Yang Z., Horton J. R., Collins R. E., Zhang X., Cheng X. (2004) J. Biol. Chem. 279, 43296–43306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee J. S., Shukla A., Schneider J., Swanson S. K., Washburn M. P., Florens L., Bhaumik S. R., Shilatifard A. (2007) Cell 131, 1084–1096 [DOI] [PubMed] [Google Scholar]
- 7. McGinty R. K., Kim J., Chatterjee C., Roeder R. G., Muir T. W. (2008) Nature 453, 812–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ng H. H., Xu R. M., Zhang Y., Struhl K. (2002) J. Biol. Chem. 277, 34655–34657 [DOI] [PubMed] [Google Scholar]
- 9. Oh S., Jeong K., Kim H., Kwon C. S., Lee D. (2010) Biochem. Biophys. Res. Commun. 399, 512–517 [DOI] [PubMed] [Google Scholar]
- 10. Chandrasekharan M. B., Huang F., Sun Z. W. (2010) Epigenetics 5, 460–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ng H. H., Ciccone D. N., Morshead K. B., Oettinger M. A., Struhl K. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 1820–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ng H. H., Feng Q., Wang H., Erdjument-Bromage H., Tempst P., Zhang Y., Struhl K. (2002) Genes Dev. 16, 1518–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pokholok D. K., Harbison C. T., Levine S., Cole M., Hannett N. M., Lee T. I., Bell G. W., Walker K., Rolfe P. A., Herbolsheimer E., Zeitlinger J., Lewitter F., Gifford D. K., Young R. A. (2005) Cell 122, 517–527 [DOI] [PubMed] [Google Scholar]
- 14. van Leeuwen F., Gafken P. R., Gottschling D. E. (2002) Cell 109, 745–756 [DOI] [PubMed] [Google Scholar]
- 15. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 16. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vakoc C. R., Sachdeva M. M., Wang H., Blobel G. A. (2006) Mol. Cell. Biol. 26, 9185–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang W., Xia X., Reisenauer M. R., Hemenway C. S., Kone B. C. (2006) J. Biol. Chem. 281, 18059–18068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Reisenauer M. R., Anderson M., Huang L., Zhang Z., Zhou Q., Kone B. C., Morris A. P., Lesage G. D., Dryer S. E., Zhang W. (2009) J. Biol. Chem. 284, 35659–35669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu Z., Kong Q., Kone B. C. (2010) Am. J. Physiol. Renal Physiol 298, F617–F624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jones B., Su H., Bhat A., Lei H., Bajko J., Hevi S., Baltus G. A., Kadam S., Zhai H., Valdez R., Gonzalo S., Zhang Y., Li E., Chen T. (2008) PLoS Genet. 4, e1000190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Barry E. R., Krueger W., Jakuba C. M., Veilleux E., Ambrosi D. J., Nelson C. E., Rasmussen T. P. (2009) Stem Cells 27, 1538–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nguyen A. T., Xiao B., Neppl R. L., Kallin E. M., Li J., Chen T., Wang D. Z., Xiao X., Zhang Y. (2011) Genes Dev. 25, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., Zhang Y., Washburn M. P., Florens L., Shilatifard A. (2010) Genes Dev. 24, 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Singh P., Han L., Rivas G. E., Lee D. H., Nicholson T. B., Larson G. P., Chen T., Szabó P. E. (2010) Mol. Cell Biol. 30, 2693–2707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mahmoudi T., Boj S. F., Hatzis P., Li V. S., Taouatas N., Vries R. G., Teunissen H., Begthel H., Korving J., Mohammed S., Heck A. J., Clevers H. (2010) PLoS Biol. 8, e1000539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. FitzGerald J., Moureau S., Drogaris P., O'Connell E., Abshiru N., Verreault A., Thibault P., Grenon M., Lowndes N. F. (2011) PLoS One 6, e14714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huyen Y., Zgheib O., Ditullio R. A., Jr., Gorgoulis V. G., Zacharatos P., Petty T. J., Sheston E. A., Mellert H. S., Stavridi E. S., Halazonetis T. D. (2004) Nature 432, 406–411 [DOI] [PubMed] [Google Scholar]
- 29. Feng Y., Yang Y., Ortega M. M., Copeland J. N., Zhang M., Jacob J. B., Fields T. A., Vivian J. L., Fields P. E. (2010) Blood 116, 4483–4491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Slany R. K. (2009) Haematologica 94, 984–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McKendry R., John J., Flavell D., Müller M., Kerr I. M., Stark G. R. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cumming G., Fidler F., Vaux D. L. (2007) J. Cell Biol. 177, 7–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shuai K., Liao J., Song M. M. (1996) Mol. Cell Biol. 16, 4932–4941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shechter D., Dormann H. L., Allis C. D., Hake S. B. (2007) Nat. Protoc. 2, 1445–1457 [DOI] [PubMed] [Google Scholar]
- 35. Buro L. J., Shah S., Henriksen M. A. (2010) J. Vis. Exp. 41, pii: 2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Buro L. J., Chipumuro E., Henriksen M. A. (2010) Epigenetics Chromatin 3, 16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Weake V. M., Workman J. L. (2008) Mol. Cell 29, 653–663 [DOI] [PubMed] [Google Scholar]
- 38. Elferink C. J., Reiners J. J., Jr. (1996) BioTechniques 20, 470–477 [DOI] [PubMed] [Google Scholar]
- 39. Shuai K., Schindler C., Prezioso V. R., Darnell J. E., Jr. (1992) Science 258, 1808–1812 [DOI] [PubMed] [Google Scholar]
- 40. Schindler C., Fu X. Y., Improta T., Aebersold R., Darnell J. E., Jr. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 7836–7839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Müller M., Laxton C., Briscoe J., Schindler C., Improta T., Darnell J. E., Jr., Stark G. R., Kerr I. M. (1993) EMBO J. 12, 4221–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Haspel R. L., Darnell J. E., Jr. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 10188–10193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen X., Vinkemeier U., Zhao Y., Jeruzalmi D., Darnell J. E., Jr., Kuriyan J. (1998) Cell 93, 827–839 [DOI] [PubMed] [Google Scholar]
- 44. Meraz M. A., White J. M., Sheehan K. C., Bach E. A., Rodig S. J., Dighe A. S., Kaplan D. H., Riley J. K., Greenlund A. C., Campbell D., Carver-Moore K., DuBois R. N., Clark R., Aguet M., Schreiber R. D. (1996) Cell 84, 431–442 [DOI] [PubMed] [Google Scholar]
- 45. Darnell J. E., Jr., Kerr I. M., Stark G. R. (1994) Science 264, 1415–1421 [DOI] [PubMed] [Google Scholar]
- 46. Min J., Feng Q., Li Z., Zhang Y., Xu R. M. (2003) Cell 112, 711–723 [DOI] [PubMed] [Google Scholar]
- 47. Nguyen A. T., Zhang Y. (2011) Genes Dev. 25, 1345–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chatterjee-Kishore M., van den Akker F., Stark G. R. (2000) Trends Cell Biol. 10, 106–111 [DOI] [PubMed] [Google Scholar]
- 49. Brierley M. M., Fish E. N. (2005) J. Interferon Cytokine Res. 25, 733–744 [DOI] [PubMed] [Google Scholar]
- 50. Huang M., Qian F., Hu Y., Ang C., Li Z., Wen Z. (2002) Nat. Cell Biol. 4, 774–781 [DOI] [PubMed] [Google Scholar]
- 51. Zhang J. J., Vinkemeier U., Gu W., Chakravarti D., Horvath C. M., Darnell J. E., Jr. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15092–15096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nusinzon I., Horvath C. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 14742–14747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Shi S., Larson K., Guo D., Lim S. J., Dutta P., Yan S. J., Li W. X. (2008) Nat. Cell Biol. 10, 489–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Okada Y., Feng Q., Lin Y., Jiang Q., Li Y., Coffield V. M., Su L., Xu G., Zhang Y. (2005) Cell 121, 167–178 [DOI] [PubMed] [Google Scholar]
- 55. Chipumuro E., Henriksen M. A. (2011) FASEB J., in press [DOI] [PubMed] [Google Scholar]