Background: CRMPs play roles in axon specification and semaphorin 3A-induced growth cone collapse, but their biochemical function is unclear.
Results: CRMPs are found to bind directly to microtubules through a conserved C-terminal region.
Conclusion: CRMPs can stabilize microtubules but are negatively regulated by phosphorylation.
Significance: This work can explain phenotypes associated with loss of CRMPs on axon specification and dendritic arborization.
Keywords: Cell Biology, Cell Signaling, Cytokinesis, Cytoskeleton, Microtubules, Mitotic Spindle, Molecular Cell Biology, Neurobiology, Neurodegeneration
Abstract
Collapsin response mediator proteins are ubiquitously expressed from multiple genes (CRMPs 1–5) and play important roles in dividing cells and during semaphorin 3A (Sema3A) signaling. Nonetheless, their mode of action remains opaque. Here we carried out in vivo and in vitro assays that demonstrate that CRMPs are a new class of microtubule-associated protein (MAP). In experiments with CRMP1 or CRMP2 and their derivatives, only the C-terminal region (residues 490–572) mediated microtubule binding. The in vivo microtubule association of CRMPs was abolished by taxol or epothilone B, which is highly unusual. CRMP2-depleted cells exhibited destabilized anaphase astral microtubules and altered spindle position. In a cell-based assay, all CRMPs stabilized interphase microtubules against nocodazole-mediated depolymerization, with CRMP1 being the most potent. Remarkably, a 82-residue C-terminal region of CRMP1 or CRMP2, unrelated to other microtubule binding motifs, is sufficient to stabilize microtubules. In cells, we demonstrate that glycogen synthase kinase-3β (GSK3β) inhibition potentiates this activity. Thus, CRMPs are a new class of MAP that binds through a unique motif, but in common with others such as Tau, is antagonized by GSK3β. This regulation is consistent with such kinases being critical for the Sema3A (collapsin) pathway. These findings have implications for cancer and neurodegeneration.
Introduction
Collapsin response mediator proteins (CRMPs)2 are cytosolic proteins containing a dihydropyrimidinase-like domain but lacking enzymatic activity (1). CRMP2 was initially identified and cloned based on its up-regulation during neurogenesis (2, 3) and discovered concurrently as an essential component of Sema3A signaling (2, 3). Subsequent studies have confirmed this role (2, 4–6). Sema3A is a chemo-repellant guidance protein, required in neural, cardiac, and peripheral vascular patterning. Class 3 semaphorins, which include Sema3A, are structurally conserved secreted proteins. Neuropilin-1/PlexinA holoreceptor transmits signals from Sema3A to affect multiple processes including axon and dendrite specification (7, 8). This involves the activation of various kinases including Rho-kinase (ROCK) and CDK5. It has wider effects in neurons; for example, Sema3A is one of the key molecules that prevent CNS remyelination following damage (9, 10). In an adult murine demyelination model, Sema3A has been shown to impair oligodendrocyte precursor cell recruitment to the demyelinated area. Ischemic neurons can prevent vascular regeneration of neural tissue by secreting Sema3A (11). Systemic and targeted delivery of Sema3A has been shown to inhibit tumor angiogenesis (12). The way in which CRMPs might function in this pathway is thus of particular interest.
Human CRMPs are encoded by five genes (CRMPs 1–5), which are alternatively spliced to yield shorter (∼65 kDa) and longer forms (∼80 kDa) containing N-terminal extensions (13, 14). CRMPs likely exist as hetero-tetrameric complexes in vivo (1) and are ubiquitously expressed with the highest levels being found in the brain (where microtubules are most abundant as well), particularly during development (14). Many studies have identified CRMP function in neuronal maturation; in Caenorhabditis elegans, mutations of unc-33 (one of three CRMP genes) result in abnormal axonal guidance (15), as with CRMP loss in mammals. CRMP1 knock-out mice also have defects in dendritic spine development, particularly in cortical layer V of the cerebral cortex (16). CRMP3−/− mice have defective dendritic arborization and spine morphology (17), which have been ascribed to altered microtubule dynamics. In dividing cells such as neuroblastoma and lung cancer cells, antibodies directed to CRMP1 and CRMP2 stain the mitotic spindle and midzone structures (18, 19), but whether this represents direct or indirect binding is unclear.
Microtubules (MTs) are particularly important to drive and sustain neuronal morphology. Microtubules are well established as a target in cancer therapy, and drugs perturbing their physiological rate of turnover such as taxol are widely used in chemotherapy. Distinct classes of interacting proteins coordinate filamentous dynamics, including MT-associated proteins (MAPs, such as Tau), MT destabilizers such as stathmin, and the MT plus-end tracking proteins as reviewed (20). Although CRMP2 has been reported to promote MT assembly in vitro (21), this activity appeared to derive from the N-terminal dihydropyrimidinase-like domain. It was suggested that in neurons, CRMP2 binds soluble tubulin as part of a kinesin-1 dependent transport complex in the axon (22).
Sema3A-induced axonal growth cone collapse requires Cdk5 phosphorylation of CRMP2 Ser-522, which primes for GSK3β phosphorylation at Ser-518, Thr-514, and Thr-509 (4, 6, 23). Studies have also identified other phosphorylation events impinging on CRMP. The ROCK phosphorylation of CRMP2 at Thr-555, downstream of EphrinA5 or lysophosphatidic acid, participates in growth cone collapse in dorsal root ganglion neurons (24, 25). Although Sema3A activates ROCK/myosin II, this is required for (secondary) axon retraction but not growth cone collapse (26).
Here we show that CRMP proteins bind to and stabilize MTs in vivo. CRMP1 and CRMP2 localize to mitotic MTs, and siRNA-mediated knockdown of CRMP2 depletes anaphase astral MTs and alters the position of the mitotic spindle relative to the cell periphery. The minimal MT binding region of CRMPs, determined by their in vivo association midzone MTs, is the C-terminal “tail” region of CRMP1 (conserved between the CRMPs). Expression of CRMP1/2 or the GST-C termini of CRMP1 or CRMP2 (82 amino acid residues) effectively stabilizes MTs against nocodazole-induced disassembly. Using this in vivo assay, we show that GSK3 negatively regulates this activity. In cells, CRMP binding to MTs is also blocked by MT-stabilizing drugs such as taxol and epothilone B. Thus, CRMP proteins bind to MTs in a way quite distinct from typical MAPs.
EXPERIMENTAL PROCEDURES
Materials
Anti-HA (Y-11) and anti-β-actin (C4) antibodies were from Santa Cruz Biotechnology. Anti-CRMP2 (C terminus)γ was from ECM biosciences. Anti-α-tubulin, anti-γ-tubulin, anti-polyhistidine, anti-FLAG, anti-FLAG M2 beads, and epothilone B were from Sigma. Anti-Glu tubulin was from Millipore. Anti-β-tubulin was from Cell Signaling. The MuLV RT enzyme was from New England Biolabs, and RNase inhibitor was from Roche Applied Science. A Cdk1 inhibitor RO3306 was from Tocris, and taxol (Paclitaxel, semisynthetic) was from Calbiochem. Purified bovine brain tubulin (TL238) was from Cytoskeleton Inc. HRP-coupled secondary antibodies blot were from Dako. Secondary antibodies used for indirect immuno-fluorescence were from Molecular Probes.
Cell Culture and Synchronization
COS7, OLDN-93, NIH3T3, and N1E-115 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) in a 37 °C incubator with 5% CO2. NIH3T3 or OLDN-93 cells were synchronized overnight with 9 μm RO-3306. The synchronized cells at G2/M phase were released with three washes of media. To determine the effect of taxol/epothilone B on mitotic spindle-bound CRMP2, 25 min after release, 100 nm epothilone B or 200 nm taxol was added to the media, and cells were fixed 15 min later. To determine the effect of the drugs on midzone MT-bound CRMP2, 50 nm epothilone B or 100 nm taxol was added 80 min after release and then fixed 15 min later.
Generation of CRMP, MAP6, Stathmin, and Tau Constructs
Constructs encoding the ORF of rat CRMPs 1–4 and MAP6 were cloned into pXJ-HA via HindIII/XhoI restriction sites. The respective primers are documented in supplemental Table S1. The C-terminal CRMP1(480–572), CRMP1(491–572), and CRMP2(480–572) were cloned into pXJ-GST with 5′-HindIII and 3′-XhoI cloning sites. Full-length CRMP1 was cloned into a modified pET21D His6 vector for expression in bacteria. CRMP1(491–572) was cloned into pGEX4T1 using introduced 5′-BamHI and 3′-XhoI restriction sites. Mouse stathmin was amplified from total cDNA and cloned via 5′-BamHI and 3′-XhoI restriction sites into pGEX4T1. The C-terminal fragment of Tau was amplified from total cDNA and cloned via 5′-BamHI and 3′-XhoI restriction sites into pQE30. The primers employed are listed in supplemental Table S1.
Reverse Transcriptase-PCR
Total RNA was obtained from cells according to standard protocol (RNeasy mini kit, Qiagen). The RNA was quantified, and 2 μg was subjected to each reverse transcription reaction. Synthesis of the cDNAs was performed with MuLV RT under standard conditions. The primers employed are given in supplemental Table S1.
DNA and dsRNA Transfection
Cells were transfected with Lipofectamine 2000 (3 μl) and 1 μof g DNA according to the manufacturer's instructions. Cells were fixed 24 h after transfection. For RNAi, the ratio of 20 pmol to 3 μl of the Lipofectamine 2000 transfection reagent was used. The siRNAs were synthesized by Invitrogen. Double-stranded CRMP2 siRNA sequences contained 3′-dTdT overhangs: CRMP2 SiA (5′ACUCCUUCCUCGUGUACAUdTdT-3′) and CRMP2 SiB (5′CCCACUCCAGAAUGGUGAUdTdT-3′). Cells were typically used 48 h after transfection.
Recombinant Protein Purification
pGEX4T1-CRMP1(491-572) or pGEX4T1-stathmin plasmid was transformed to Escherichia coli BL-21 and induced with 10 mm isopropyl-1-thio-β-d-galactopyranoside for 4 h at 25 °C. Bacteria were harvested at 6,000 rpm (10 min at 4 °C). The pellet was resuspended in 1% Triton X-100, 50 mm Tris, pH 8.0, 1 mm EDTA, 0.1 mm DTT, and 10% glycerol with protease inhibitor mixture (Roche Applied Science) and lysed by adding 1 mg/ml lysozyme (4 °C, 30 min). The supernatant (13,500 rpm, 30 min at 4 °C) was added to glutathione-Sepharose (GenScript, 20 ml of lysate to 1 ml of beads) and washed with lysis buffer. The GST fusion proteins were eluted with 20 mm reduced l-glutathione (Sigma) and dialyzed against PEM buffer (80 mm PIPES, 1 mm MgCl2, 1 mm EGTA, pH 6.8). Protein concentration was measured using a modified Bradford assay (Bio-Rad). His6-CRMP1 and His6-Tau CT constructs were transformed into E. coli BL21. The protein was purified on nickel-nitrilotriacetic acid-Sepharose (Qiagen). Briefly, the bacteria pellet was resuspended in 50 mm NaH2PO4 (pH 8.0), 500 mm NaCl, EDTA-free protease inhibitor mixture, 10 mm imidazole, and 0.1% Triton X-100. Lysis was performed as above for pGEX constructs. The supernatant fractions containing His6-tagged proteins were purified according to the manufacturer's conditions. Recombinant His6-tagged proteins were eluted from the beads with 300 mm imidazole and dialyzed against PEM buffer.
Western Analysis
dsRNA transfected cells were washed with cold PBS and lysed in 0.1 m Tris, pH 7.4, 0.15 m NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 5% glycerol, 1 mm PMSF, 20 mm β-glycerophosphate and protease inhibitor mixture. The lysates were spun in a microcentrifuge at 14,000 rpm for 10 min. Protein sample buffer was added into the supernatants and boiled before SDS-polyacrylamide gel electrophoresis. All the protein samples were subjected to SDS-polyacrylamide gel electrophoresis and transferred onto PVDF membrane. The membranes were blocked with 5% skimmed milk in 0.05% Tween 20, PBS buffer for 30 min and incubated with primary and secondary HRP-conjugated antibodies diluted in 1% milk, 0.05% Tween 20, PBS buffer for 2 and 1 h, respectively, at room temperature.
In Vitro Microtubule Co-sedimentation
Purified tubulin was made up to 5 mg/ml in 10% glycerol-PEM buffer. The tubulin was preclarified by high speed centrifugation for 10 min at 4 °C (100,000 × g, TL100 centrifuge, Beckman). The tubulin solution was polymerized at 37 °C for 30 min with 1 mm DTT, 1× protease inhibitor, 1 mm GTP, and the recombinant proteins (GST, His6-CRMP1, and His6-Tau at 0.2 mg/ml), with or without 10 μm taxol. The final tubulin concentration was 2 mg/ml. Microtubules were spun down at 65,000 rpm for 15 min at 25 °C. The pellets were washed with warm PEM buffer. The pellets and supernatants were diluted with SDS sample buffers and subjected to SDS-PAGE and Western blot analysis.
Surface Plasmon Resonance Measurements (BIAcore 2000)
CM5 chips and HEPES-buffered saline were used for analyte (GE Healthcare) with a flow rate of 15 μl/min. A reference flow cell contained immobilized GST protein. Dialyzed recombinant GST CRMP1(491–572), His6-CRMP1, and GST-stathmin were precleared at 90,000 rpm, 10 min (TLA100, Beckman) before coupling to the CM5 chip, and unreacted sites were blocked with 0.1 m ethanolamine, pH 8.5. Pure bovine tubulin (Cytoskeleton Inc.) dissolved in PEM+T (80 mm PIPES, 1 mm MgCl2, 1 mm EGTA, 0.005% Tween 20, pH 6.8) was preclarified at 90,000 rpm for 10 min at 4 °C. The sensor chip was regenerated with 30 μl of 50 mm HEPES (pH 7.4), 300 mm KCl, 3 mm EDTA, and 0.005% Tween 20. Traces were corrected with respect to the GST control sensorgram.
Nocodazole Stability Assay and Immuno-fluorescence Analysis
COS7 cells were treated with 2 μm nocodazole for 45 min, and fixed in a stabilizing buffer (0.15% Triton X-100, 1% paraformaldehyde-PEM buffer) for 5 min, and transferred to 3% paraformaldehyde/PEM (10 min). To inhibit GSK3, cells were serum-starved for 3 h and treated with 20 mm LiCl or 20 mm NaCl for 2 h before adding nocodazole. After fixation, cells were permeabilized with 0.2% Triton X-100/PBS for 10 min and blocked with 1% FBS in 0.1% Triton X-100/PBS for 10 min and then blocked with 10% FBS/PBS. Primary antibodies were diluted in 0.5% Triton X-100/PBS and incubated with the cells at 32 °C and then washed with 0.1% Triton X-100/PBS. Secondary antibodies (anti-mouse/rabbit Alexa Fluor-488 or Alexa Fluor-546; 1:500) were incubated and washed under similar conditions. Far-red phalloidin and Hoechst dye were added in during the secondary antibody incubation step. Cells were mounted and viewed by Zeiss Axioplan2, and images were collected on a CoolSNAP HQ (Roper Scientific) camera using a 63× 1.4 NA oil lens. Confocal imaging was performed using Olympus FluoView 1000 using a 60×/100× oil lens. Images were processed using Adobe Photoshop and Image J. Deconvolution was performed using the Huygens professional software (Scientific Volume Imaging).
RESULTS
Both CRMP1 and CRMP2 Localize to Mitotic Microtubules
CRMPs 1–5 are the products of five genes, some of which are highly expressed during neuronal development, but some of which are also ubiquitously expressed (27, 28). CRMPs are abundant proteins that exist as cytosolic tetramers (14, 29, 30), of which CRMP2 is the best studied as an essential mediator of growth cone collapse induced by Sema3A signaling (2, 3). CRMP2 itself accumulates in growth cones and also promotes multiple axons when ectopically overexpressed (21, 31). CRMP2 is suggested to play this role through tubulin (dimer) transport along axons by binding to kinesin-1 (22). This might allow proper MT assembly at the growth cone (22). CRMP1 and CRMP2 have been at the mitotic spindle and midzone MTs, but the biochemical basis for this localization has not been described (18, 19).
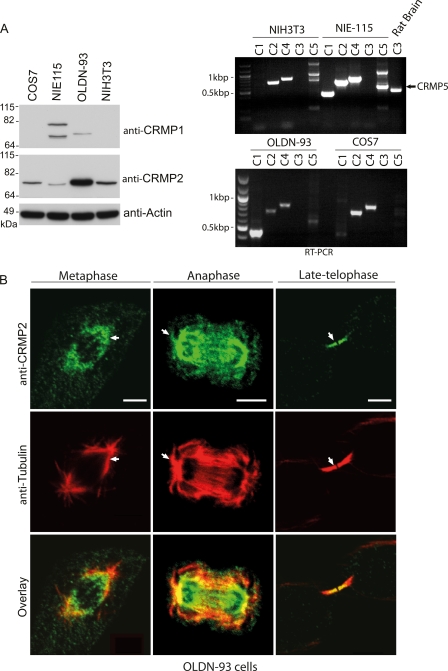
To investigate further the relationship between endogenous CRMP family proteins and MTs in mitotic cells, we first examined CRMP isoforms 1–5 in several cell types using RT-PCR and then CRMP1- and CRMP2-specific antibodies. COS7 (monkey large-T transformed fibroblast line), mouse N1E-115 neuroblastoma cells, oligodendrocytic OLDN-93, and normal mouse NIH3T3 fibroblasts all expressed CRMP2 (Fig. 1A). CRMP1 protein was not detected in the fibroblastic COS7 and NIH3T3 lines. All these cell lines express CRMP2 and CRMP4 mRNA at comparable levels; CRMP4 has recently been investigated in the context of cell mitosis (32).
FIGURE 1.
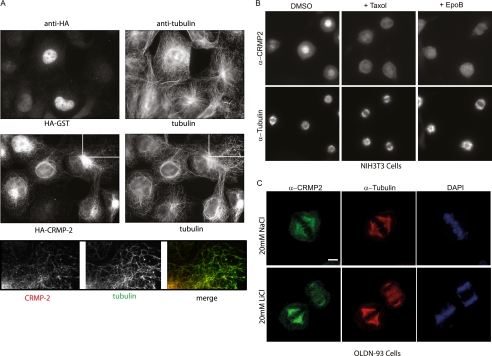
CRMP1 and CRMP2 localize to mitotic microtubules. A, left, representative immunoblots for CRMP1 and CRMP2 expression in various cell lines. Right, RT-PCR CRMP products from mRNA purified from cell lines or rat brain as indicated. The black arrow points to the expected band size of about 500 bp representing CRMP5. B, representative images of endogenous CRMP2 in OLDN-93 cells. RO-3306-synchronized OLDN-93 cells were fixed 45 or 95 min after release to enrich cells with mitotic spindles or midzone MT structures. The white arrows point to regions of CRMP2 colocalization with astral MTs. Scale bar = 5 μm.
OLDN-93 cells had the highest relative amounts of CRMP2 protein (Fig. 1A), facilitating staining of the endogenous CRMP2 by indirect immuno-fluorescence. Here CRMP2 was present in mitotic structures and in puncta along the mitotic spindle at metaphase and on the midzone MTs in late telophase (Fig. 1B). Exogenous HA-tagged CRMP1 was similarly localized on the mitotic spindle and midzone MTs in these cells (data not shown). CRMP2 puncta were also detected on astral MT (Fig. 1B, white arrows; anaphase). In summary, we find that both CRMP1 and CRMP2 proteins closely associate with mitotic structures that are enriched for MTs, suggesting that this is a general feature of CRMPs.
Microtubule Binding Determinants Lie outside the Dihydropyrimidinase-like Domain
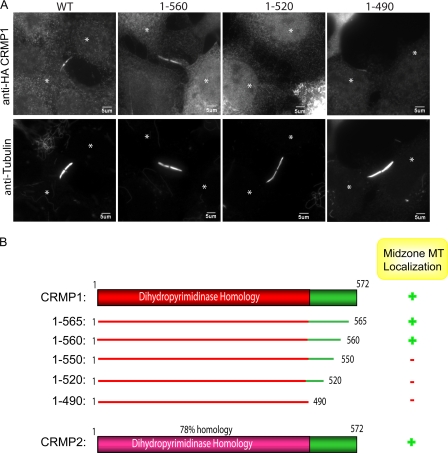
The shorter ∼65-kDa isoforms of CRMP1 and CRMP2 are polypeptides of 572 residues (Fig. 2B). Structural analyses of CRMP2 (residues 13–490) reveal the secondary structure of the tetrameric dihydropyrimidinase-like domain, but not of the flexible C-terminal 82 residues (5, 33). It has been reported that central residues 323–381 within the dihydropyrimidinase domain function to promote MT assembly by binding to tubulin dimers (21). However, HA-CRMP1(1–490) failed to bind to midzone MTs in vivo (Fig. 2, A and B), instead implicating the C-terminal region (residues 490–572) in this interaction. We generated a series of C-terminal truncated proteins to delineate the requirements in this region (Fig. 2B). Microtubule association tolerated the deletion of 12 C-terminal residues (i.e. CRMP1 1–560), but not the further deletion of 10 residues (CRMP1 1–550) (Fig. 2, A and B).
FIGURE 2.
Microtubule binding determinants lie outside the dihydropyrimidinase-like domain. A, representative images of full-length CRMP1 (WT) or C-terminal deleted constructs as shown expressed in COS7 cells. Cells expressing the various constructs were partially pre-permeabilized prior to fixation and stained with anti-HA and anti-α-tubulin. The white asterisk marks centers of the two dividing cells. B, a schematic diagram of CRMP1 and CRMP2 constructs used. CRMP colocalization with midzone MTs is scored as + for high and − for none.
CRMP2 Depletion Affects Astral Microtubules in Mitosis
The mitotic spindle allows proper segregation of chromosomes during cell division, a function targeted by a number of drugs that affect MT dynamics. It is assembled as an anti-parallel array of MTs with their “minus” ends fixed onto the centrosomes and their dynamic “plus” ends projecting toward the chromosomes. Because CRMPs were clearly localized to mitotic MT arrays, we reasoned that these proteins could contribute to their dynamics. In the course of this work, a new study showed that loss of CRMP4 disrupts chromosomal alignment and mitotic progression (32). We investigated siRNA-mediated knockdown of CRMP2 in OLDN-93 cells, in which it is abundantly expressed (Fig. 1A). After siRNA treatment (supplemental Fig. S1A), ∼60% of the cells showed no detectable CRMP2 by immuno-staining (data not shown), allowing us to pinpoint affected cells. Consistent with a cell cycle effect, we noted that cells without CRMP2 took more time to go through mitosis (from cell rounding to re-attachment, data not shown), although we did not observe an effect on chromosome attachment as reported for CRMP4 in HeLa cells (32).
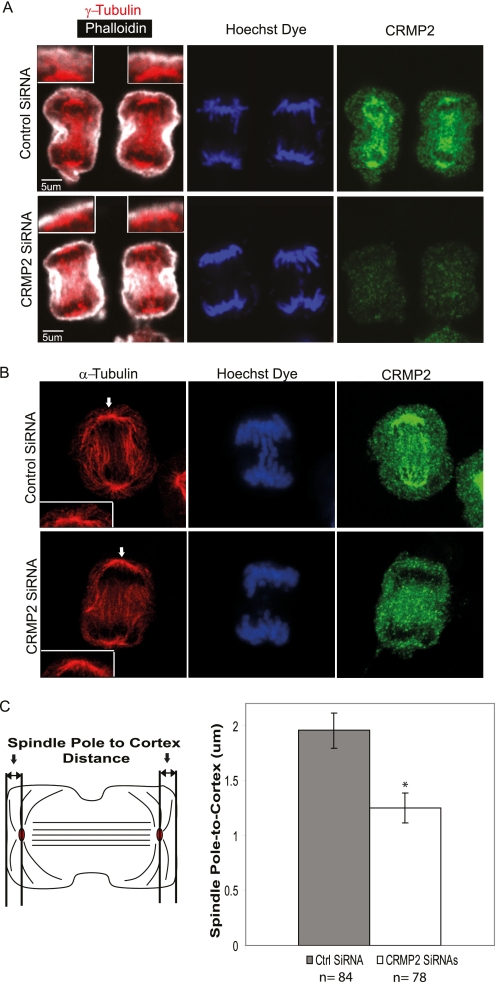
To examine the effects of CRMP2 knockdown, mitotic CRMP2-depleted OLDN-93 cells were stained for tubulin (α and γ), endogenous CRMP2, and F-actin (with phalloidin) (Fig. 3) In control OLDN-93 cells during anaphase, CRMP2 exhibited punctate staining around the area of the centrosome (stained for γ-tubulin, Fig. 3A) and along the elongating central MT spindle (stained for α-tubulin, Fig. 3B). In mitotic cells stained for α-tubulin, MTs that emanate from the spindle away from the plane of division, termed astral MTs, are detected (using deconvolution image analysis; Huygens Professional software). In CRMP2 knockdown cells, there was significant reduction in the density of astral MTs (Fig. 3B), and the spindle lies closer to the cortical cell membrane at anaphase (Fig. 3A and schematic in Fig. 3C). Quantification of this spindle pole-to-cortex distance, stained respectively with γ-tubulin antibody and phalloidin, revealed an ∼37% reduction in this distance in CRMP2 knockdown cells (mean = 1.25 μm) versus control cells (mean = 1.95 μm). There was also a diminished density of astral MTs (compare Fig. 3B, inset), which probably reflects a general effect on integrity of MT structures. A similar effect on the spindle pole-to-cortex distance was also observed in NIH3T3 cells with CRMP2 knockdown (data not shown) suggesting that this is a general role of CRMP2. Thus, CRMP2, perhaps in the context of hetero-dimers with CRMP1 or CRMP4 (1), promotes MT stabilization during mitosis, and this is manifested in knockdown cells as a reduction in astral MTs. This is also consistent with the need for CRMP4 in proper chromosomal alignment (32).
FIGURE 3.
Knockdown of CRMP2 diminishes spindle pole-to-cortex distance and astral MTs. A, representative confocal images of RO-3306-synchronized cells released for 40 min before fixation. Panels show control siRNA- or CRMP2 siRNA-treated OLDN-93 cells stained with anti-γ tubulin, phalloidin, Hoechst dye, and anti-CRMP2 antibodies. The inset represents an enlarged image of the γ-tubulin-rich spot and the cortical actin. Scale bar = 5 μm. Cells were imaged using a 60× oil lens of a confocal microscope. B, representative deconvoluted confocal images of RO-3306-synchronized, control siRNA-, or CRMP2 siRNA-treated OLDN-93 cells fixed at 40 min after release. The cells were stained with anti-α-tubulin, Hoechst dye, and anti-CRMP2 antibodies. The inset represents an enlarged image of the region pointed out by the white arrow. Cells were imaged using a 100× oil lens of a confocal microscope. Deconvolution was performed, and Z projections are shown. C, left, a schematic diagram illustrating the spindle pole-to-cortex distance measurement between the γ-tubulin spot and the cortical actin. Right, a graph representing the average spindle pole-to-cortex distance for control and CRMP2 siRNA-treated cells. Cells with membrane protrusions at the cortex were excluded during the quantitation. Error bars represent S.D. between three independent experiments with the number of cells (n) counted for each condition labeled below the graph. (*, p < 0.05, Student's t tests when compared with control). Ctrl SiRNA, control siRNA.
CRMP Binds Exclusively Assembled Microtubules in Vitro
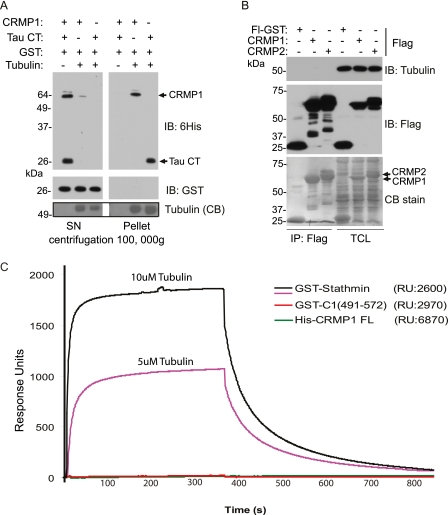
Current models of CRMP function include an interaction with tubulin dimers to regulate their intracellular (axonal) transport (22) or promote their assembly (21). To test whether CRMP could bind directly to polymeric MTs assembled in vitro, we generated full-length purified CRMP1 in bacteria, to avoid co-purifying MAPs. Purified recombinant His6-tagged protein was added to pure tubulin dimers assembled in GTP (without MAPs) and tested for binding by co-sedimentation. Tau C-terminal fragment (Tau CT) containing MT binding repeats (a positive control) and CRMP1 were each efficiently co-sedimented with MTs into the pellet fraction (Fig. 4A), but not recombinant GST protein.
FIGURE 4.
CRMP binds directly to microtubules. A, co-sedimentation of CRMP1 with in vitro polymerized MTs assembled from pure tubulin. Bovine tubulin (2 mg/ml) was polymerized in the presence of GTP and purified His6-CRMP1 or His6-Tau C-terminal (CT) fragment and GST protein. The high speed pellet and supernatant (SN) fractions were analyzed by Western blot analysis using anti-His6 and anti-GST. A buffer control without tubulin is in lane 1 of each fraction. IB, immunoblot; CB, Coomassie Blue. B, soluble tubulin does not co-immunoprecipitate with CRMP1 or CRMP2. COS7 cells were transfected with vector encoding FLAG-GST, FLAG-CRMP1, or CRMP2, and cells were lysed in a tubulin-stabilizing buffer: 25 mm Tris, pH 7.5, 100 mm KCl, 0.5% Triton X-100, 1 mm MgCl2, 5% glycerol, and 5 mm DTT. The lysate was subjected to clarification (100,000 × g, 20 min) at 4 °C and then incubated with anti-FLAG-Sepharose for 1 h on ice. The immuno-precipitated (IP) fractions and lysates (TCL) were subjected to SDS-PAGE (in duplicate), transferred to PVDF and stained with Coomassie Blue, and immuno-blotted (IB) with anti-β-tubulin or anti-FLAG antibodies. FL-GST, Flag GST. C, BIAcore sensorgram of tubulin binding to immobilized His6-CRMP1 (green line), GST-CRMP1(491–572) (red line), or GST-stathmin (pink and black lines). The amount (response units (RU)) immobilized to the surface is indicated. Purified bovine tubulin was injected at 5 or 10 μm for 360 s (association phase) and then was monitored for 400 s (dissociation phase).
To re-assess the binding of CRMP to soluble tubulin (21), we carried out immuno-precipitation of overexpressed FLAG-CRMP1 or FLAG-CRMP2 from cellular lysates, under conditions to stabilize the tubulin (in buffer containing 5% glycerol, 1 mm MgCl2, and 5 mm DTT). After high speed centrifugation to remove all assembled MTs, FLAG-CRMP1 and FLAG-CRMP2, which were easily visible even in total cell lysates (Fig. 4B, arrows), were recovered on M2-Sepharose (Fig. 4B, left side). No tubulin was detected in CRMP1 or CRMP2 pulldown, although >5 μg each of CRMP was loaded (compare Coomassie Blue staining).
We next immobilized recombinant full-length His6-CRMP1 and GST-CRMP1(491–572) C terminus on a CM5 chip (BIAcore) and tested pure tubulin binding by surface plasmon resonance (Fig. 4C). Parallel assays were performed with stathmin as a positive control (34). In contrast to a previous study (25), injections of freshly clarified assembly-competent tubulin dimers (with E-site charged with either GDP or GTP) failed to generate any signal with either full-length CRMP1 or its C terminus when compared with stathmin. These results based on purified recombinant proteins indicate that CRMPs do not bind unassembled tubulin dimer. The experimental differences with earlier literature are unclear, although in our study, high speed ultracentrifugation was used to remove all the assembled tubulin from lysates, and buffers were selected to stabilize the tubulin during immuno-precipitation (Fig. 4B). Sepharose is reported to non-specifically retain multiple cytoskeletal proteins including tubulin (35).
CRMP1 and CRMP2 Can Associate with Interphase Microtubules
To assess whether CRMPs are MT-associated in interphase cells, cytosolic proteins were first removed through a brief pre-permeabilization step before cell fixation (see “Experimental Procedures”). In OLDN-93 cells, endogenous CRMP2 was observed concentrated in the perinuclear region and in thin processes, which are MT-rich (supplemental Fig. S1B, red arrows); this anti-CRMP2 staining was reduced by 48 h of siRNA treatment. In the same way, interphase COS7 cells showed HA-tagged CRMP2 concentrated around the MT-organizing center (Fig. 5A); in the enlarged area, one can see that there is excellent colocalization between HA-CRMP (red) and anti-tubulin staining (green). The control HA-GST protein was only retained in the nucleus. We conclude that CRMP1 and CRMP2 show significant interactions with both mitotic and interphase MTs in vivo.
FIGURE 5.
CRMP binds to microtubules in a taxol-sensitive manner. A, tagged CRMP2 partially colocalizes with interphase MTs. Representative images of GST-HA (control) and HA-CRMP2 expressed in COS7 cells briefly washed with PEM +0.1% Triton X-100 buffer at room temperature (see “Experimental Procedures”) and fixed immediately with 3% paraformaldehyde (10 min) are shown. Immunofluorescent staining was performed using anti-HA and anti-α-tubulin antibodies. The lower panel represents the enlarged images of the white box region. B, taxol and epothilone B (EpoB) displace endogenous CRMP2 from mitotic MTs. Representative staining of CRMP2 in RO-3306-synchronized NIH3T3 cells fixed at 40 min after release and following 15 min of treatment with dimethyl sulfoxide (DMSO), taxol, or epothilone B is shown. Cells were partially pre-permeabilized before fixation and stained using anti-CRMP2 and anti-α-tubulin. C, GSK3 inhibition via LiCl treatment does not displace endogenous CRMP from mitotic MTs. Representative confocal images of CRMP2 in RO-3306-synchronized OLDN-93 cells fixed at 45 min after release are shown. Cells were treated with NaCl or LiCl for a total period of 90 min (inclusive of 45 min after release). Cells were partially permeabilized before fixation and stained using anti-CRMP2 and anti-α-tubulin. The cells were imaged using 100× objective (FV1000, Olympus). Scale bar = 5 μm.
To test whether the isolated C-terminal sequences could function as an autonomous MT binding domain, we expressed GST-CRMP1(491–572) and carried out the same protocol. This protein was retained on the interphase MT network (supplemental Fig. S2). We conclude that the C-terminal domain is sufficient to associate with assembled MTs in vivo. We refer to this conserved region as the C-terminal MT binding domain (CMBD) of CRMPs.
Microtubule-stabilizing Agents Displace CRMP from Microtubules
We noted that CRMP2 was absent from mitotic spindles in cells synchronized by taxol treatment (data not shown). Indeed, in cells synchronized with the CDK1 inhibitor RO-3306 and then treated with taxol or epothilone B for 15 min, CRMP2 was consistently lost from all mitotic spindles (Fig. 5B) and midzone MTs (supplemental Fig. S3). Taxol and the epothilones stabilize MTs through binding to an overlapping binding site on tubulin, which is thought to induce a GTP-like state (36). Thus, in vivo CRMP appears to be sensitive to the tubulin conformation induced by these drugs, in contrast to MAPs, which interact with the acidic C termini of tubulins (37–40). An alternate explanation is that taxol-induced MT stabilization signals to pathways that negatively regulate CRMP binding. The weaker in vitro association of CRMP1 with assembled MTs in the presence of taxol, however, does support a direct effect (supplemental Fig. S4). CRMPs may therefore have an opposite MT binding selectivity to plus-end tracking proteins binders such as EB1, which bind GTP-tubulin (41, 42).
A recent study suggests that GSK3 activity is needed for CRMP4 to bind the mitotic spindle (32). To assess whether the well described CRMP2 modification by GSK3β (32) is similarly required, we investigated CRMP2 in synchronized and mitotic OLDN-93 cells treated with LiCl to inhibit GSK3 (43). CRMP2 association with the mitotic spindle was unaffected under these conditions (Fig. 5C). These data support our subsequent findings that GSK3 activity blocks CRMP2 (and CRMP1) binding to MTs (as described below).
CRMP Expression Generates Stable Interphase Microtubules
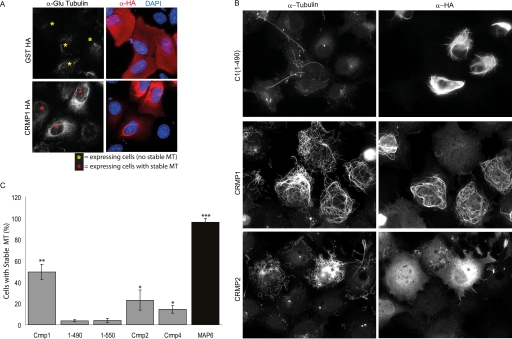
In view of the ability of CRMPs to bind to MTs, we sought a quantitative cell-based assay to measure the outcome of such binding and to investigate cell signaling events related to the phosphorylation of CRMPs. Microtubule co-sedimentation assays were not suitable because these rely on taxol-mediated stabilization of cellular MTs. We also found that CRMP1 expression has no overt effect on overall MT disposition or density in non-mitotic COS7 cells (data not shown). In most cultured cells, only a small subset of MTs (primarily those near the MT-organizing center) are stable with t½ of <15 min (44). However expression of some MAPs can generate extensive arrays of stable MTs, also described as “cold stable.” Such MTs are marked by detyrosinated tubulin (termed Glu-tubulin) and appear in the direction of cell migration (45). In cells expressing HA-CRMP1, we observed increased Glu-tubulin staining on the MT network (Fig. 6A), clearly indicating that CRMP1, like other MAPs, prolongs the lifetime of cellular MTs.
FIGURE 6.
CRMP expression can stabilize microtubules. A, overexpressed CRMP1 promotes Glu tubulin formation in U2OS cells. Cells overexpressing HA-CRMP1 or GST-HA were fixed with methanol and stained with anti-Glu tubulin, anti-HA, and DAPI. Yellow and red asterisks mark transfected cells that exhibited background and higher than background level of stable MTs respectively. B, representative images of the distribution of CRMP1, CRMP1(1–490), and CRMP2 in COS7 cells following treatment with nocodazole (2 μm) for 45 min. The cells were immuno-stained using anti-HA and anti-tubulin antibodies. C, CRMP protects MTs from nocodazole-mediated disassembly. The percentages of COS7 cells with stable MTs after expression of CRMP1, CRMP1(1–490), CRMP1(1–550), CRMP2, CRMP4, or MAP6 (overnight). Positive transfected (medium-high expressing) cells exhibiting substantial levels of stable MTs after nocodazole treatment (45 min) were counted. Three independent experiments were performed for all quantitative analyses involving at least 88 cells for each construct per experiment; error bars represent S.E. between experiments. (*, p < 0.05, **, p < 0.001, ***, p < 0.0001, Student t tests when compared with 1–490).
Stable MTs also show resistance to depolymerization by nocodazole, a simple test of their stability. Both CRMP1, which can form filaments that partially map to the MT network, and CRMP2 (which does not) caused a striking increase in the number of cells exhibiting nocodazole-resistant stable MTs (Fig. 6, B and C, and supplemental Fig. S5). By contrast, CRMP1(1–490) had no effect on MT stability on nocodazole treatment, consistent with its inability to associate with mitotic MTs. This smaller CRMP1 construct also formed filament-like structures independent of MTs. As expected, transfection of the stable tubule-only polypeptide (STOP), or MAP6, which is extremely potent in conferring nocodazole resistance (46), caused a striking increase in nocodazole-resistant MTs. We quantified the fraction of cells expressing moderate levels of CRMPs and containing significant levels of nocodazole-resistant MTs (Fig. 6C). Full-length CRMP1 was consistently a better MT stabilizer than CRMP2 in this assay (∼50% of expressing cells).
The CRMP1-stabilizing activity on MTs was highly robust in this assay on nocodazole-treated cells. The number of cells exhibiting stabilized MTs was >10-fold higher in cells expressing full-length CRMP1 when compared with the inactive truncated mutants (Fig. 6C). This allowed more detailed mutational analysis of the C-terminal CMBD. Finer CRMP1 C-terminal truncation constructs (Fig. 2B) were tested, as illustrated in supplemental Fig. S6. Although CRMP1(1–565) retained low activity, CRMP1(1–550) was completely without activity, as were larger deletions (supplemental Fig. S6). Thus, we find striking correlation between the capacity for mitotic MT localization of CRMP1 deletion mutants (Fig. 2, A and B) and their ability to stabilize interphase MTs (Fig. 6C and supplemental Fig. S6).
We have shown that CRMPs associate directly with MTs in vitro (Fig. 4A) and that the in vivo mitotic spindle association and activity of CRMP on nocodazole-treated interphase MTs correlate well with each other. The nocodazole assay has the key advantage over in vitro binding in that the transfected CRMP proteins can undergo post-translational modifications that occur in mammalian cells. Although the in vivo function of CRMPs is likely to extend beyond MT binding, this MT stabilization in vivo provides a semiquantitative measure of their functional MT association. Similarly, CRMP2 overexpression in primary neurons can generate multiple axons, and this has been used as a functional read-out (21).
GSK3β Phosphorylation of CRMP Negatively Regulates Microtubule Binding
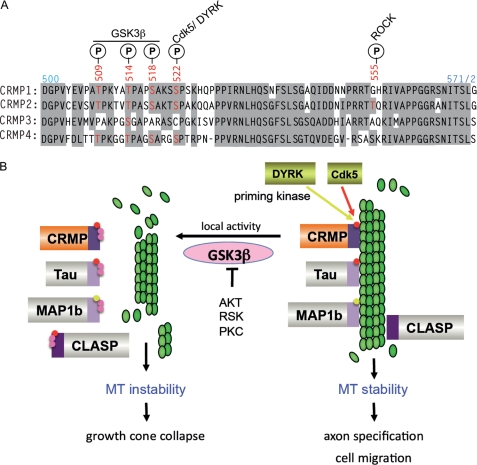
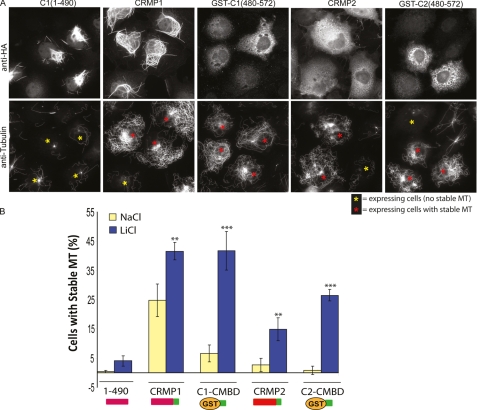
The C-terminal CMBD is highly conserved among the CRMP isoforms (see Fig. 8A) and across species (47). Phosphorylation of Ser-522 (present in CRMP1, CRMP2, and CRMP4), which is a target of CDK5 and other proline-directed kinases, primes the adjacent sites for GSK3β phosphorylation (4, 48). Subsequent GSK3β phosphorylation of CRMP2 at Ser-518, Thr-514, and Thr-509 (all within the CRMP2 CMBD) is necessary for Sema3A-induced growth cone collapse (4). We therefore determined whether the MT-stabilizing activity of CRMP in COS7 cells was affected by GSK3 inhibition. Inhibition of GSK3 by LiCl treatment (43) had a dramatic effect on MT stability in this assay, as has been reported for in vivo activities of Tau (49–51). As LiCl enhanced the region of stable MTs emanating from the MT-organizing center for almost all cells (Fig. 7A) in comparison with control NaCl treatment, we modified the analysis to count as positive only those cells with extensive arrays of stable MTs (arrays emanating from MT-organizing center to the cell edge). Neither GST nor the dihydropyrimidinase-like domain of CRMP1 (residues 1–490) exhibited activity significantly above background in the presence of LiCl (Fig. 7B). Full-length CRMP1 or CRMP2 were moderately enhanced by LiCl treatment. In contrast, the GST-CMBD (compare residues 480–572) showed much greater enhancement (>5-fold) of activity in the presence of LiCl (Fig. 7B) to a level equivalent to the full-length protein. This suggests that GSK3β phosphorylation of the GST-CMBD fusion in vivo is more efficient. Intramolecular interactions between the dihydropyrimidinase-like N-domain and the CMBD might inhibit its phosphorylation. These experiments with the GST-CMBD fusions also suggest that the difference in activities between CRMP1 and CRMP2 (with respect to their efficiency of MT stabilization) relate directly to sequences in the CMBD.
FIGURE 8.
How GSK3β phosphorylation is involved with microtubule destabilization. A, an alignment showing the conserved residues (shaded in gray) of the CMBD region of human CRMPs 1–4. Phosphorylation (P) sites (red) by kinases and GSK3β are indicated. B, multiple MT-binding proteins are co-regulated by phosphorylation. Pathways that lead to activation of priming kinases include Sema3A activation of CDK5 (66) (red) leading to growth cone collapse. The dual-tyrosine-regulated kinase (DYRK) family kinases (yellow) can activate CRMP4 as well as MAP1b (76). Local GSK3β activity leads to secondary phosphorylations (in pink) of MAPs, leading to their dissociation from microtubules and instability of the network. As shown, GSK3β can be antagonized by other protein kinases, which phosphorylate Ser-9 and could serve to terminate this pathway. CLASP, CLIP-associated protein; RSK, ribosomal S6 kinase.
FIGURE 7.
Phosphorylation of CRMP1 by GSK3β diminishes its cellular activity. A, the C-terminal region of CRMP is sufficient to stabilize MTs upon GSK3 inhibition. Representative images of the distribution of CRMP1, CRMP1(1–490), CRMP1(480–572), CRMP2 and CRMP2(480–572) in COS7 cells following treatment with 20 mm LiCl for 2 h and then challenged with nocodazole (2 μm) for 45 min are shown. Red and yellow asterisks mark transfected cells scored as with or without stabilized MTs, respectively. The cells were fixed and stained with anti-HA and anti-tubulin antibodies. B, CRMP1(1–490), GST-CRMP1(480–572), CRMP2, and GST-CRMP2(480–572) were scored according to their ability to generate nocodazole-stable MTs. The yellow bars represent NaCl (control), and blue bars represent LiCl treatment before the nocodazole challenge. Cells with intact MTs that covered more than 50% of the area were scored as positive. (*, p < 0.05, **, p < 0.001, ***, p < 0.0001, one-way analysis of variance with Tukey's honestly significant difference post hoc test when compared with NaCl data).
The effect of LiCl treatment illustrates the importance of GSK3β in antagonizing MT stability in general and points to CRMP as one of several MT-associated proteins that are direct negative targets of GSK3β. To date, these include MAP1b, Tau, ACF7, and CLASP2 (49, 50, 52, 53). It has been suggested that the polarized migration of cells in general requires local GSK3β inhibition at the leading edge (52, 54, 55). Our model (Fig. 8B) illustrates how different classes of MT-associated proteins are coordinately regulated by GSK3β. Further evidence for this is that the priming kinases CDK5 and dual-tyrosine-regulated kinase (DYRK) are involved in regulating CRMPs (56), Tau, CLASP2, and MAP1b. Interestingly, phospho-CRMP2 is normal in the cortex of GSK3α−/− mice, but not detectable in those lacking GSK3β (57).
DISCUSSION
A wide range of physiological activities operates through Sema3A signaling (58–62). It is noteworthy that two recent studies show that Sema3A suppresses axon formation (63) or can convert axons to dendrites (8), activities that are opposite to the effect of ectopically expressed CRMP2 (21, 23, 31). This is consistent with Sema3A acting in part by inactivating CRMP2, leading to a destabilization of MTs. Nishiyama et al. (8) show that secreted Sema3A induces the neurite identity of Xenopus spinal commissural interneurons; the Sema3A-triggered cGMP/PKG signaling here could be involved in CRMP inactivation.
Although existing literature links various CRMPs to the cytoskeleton (21, 22), no studies have advocated that the main function of the protein is to bind assembled MTs, perhaps because the interaction is particularly dynamic. The localization of CRMPs in mitosis was reported (18, 19) but no structure-function analysis was carried out to determine whether this involved direct MT interaction. The primary role of CRMPs as MAPs can explain many of the biological roles ascribed to CRMPs.
Current models suggest a role for CRMP2 in tubulin dimer transport in neurons (21). The in vitro binding experiments described here make this model less attractive (Fig. 4). Our observation that CRMP binding to MTs is taxol- and epothilone B-sensitive (Fig. 5B) is consistent with CRMP also binding directly to assembled MTs in vivo. Because taxol and the epothilones can displace CRMP, it is likely that the CMBD domain recognizes a specific conformation of tubulin when assembled into the 13-protomer lattice.
We provide clear evidence that the CRMP2 stabilizes the mitotic apparatus during cell division. This might explain why CRMP2 knock-out mice have never been described. Using a simple transfection assay of CRMP, we determined that the C-terminal 82-amino-acid domain of CRMPs is critical for MT binding (Figs. 1 and 2) and that CRMP C termini stabilize MTs in vivo (Figs. 3 and 6), an ability that correlates with the in vitro association of recombinant CRMP1 with assembled MTs (Fig. 4). CRMP binding to MTs is sensitive to taxol, making this protein interaction particularly interesting in relation to MT regulation.
What is the basis of this CRMP-tubulin interaction? We anticipate that it is unlikely to rely on interactions with the acidic C termini that protrude from the MT surface and bind MAPs such as Tau. An early study reported that fragments of CRMP including residues 323–381 could promote MT polymerization in vitro (21). Subsequent structural studies of CRMP1 (5) and CRMP2 (33) showed that CRMP organized as a tetramer into a triosephosphate isomerase (TIM)-like barrel. Because residues 323–381 are largely buried in the tetramer, tubulin binding to this region is unlikely without conformational alteration, and we failed to detect any MT binding activity in CRMP1(1–490) (Figs. 2 and 6). This is true although this domain is predisposed to form filaments itself. Given that the CMBD is sufficient for CRMP activities in vivo, we propose that the dihydropyrimidinase-like domain plays a structural and perhaps auto-inhibitory role. It is noted that this structural domain is responsible for the observed ability of CRMP1 to form filaments when overexpressed (Fig. 6A) as can occur with a number of proteins by self-assembly (64).
How does serine/threonine phosphorylation affect CRMP function? The Sema3A pathway signaling to CRMP2 (2, 4, 65) includes activation of Cdk5 (66) and GSK3β (67), which phosphorylate CRMP2. We find that LiCl can aid MT stabilization in COS7 cells (Fig. 7), but given that GSK3 functions in numerous pathways (68), the MT dynamics likely result from multiple targets including CRMPs. There is consistent evidence that phosphorylation of the CRMP C-terminal region negatively regulates their biological function in various contexts, as reviewed (47, 69), which fits with the notion that the primary biological role of CRMPs is MT regulation.
CRMP2 is widely expressed, so what is its role in non-neuronal cells? Our data suggest that CRMP2 is involved in stabilizing astral MTs during mitosis. CRMP2 knockdown was also associated with asymmetrical cell division in NIH3T3 cells (data not shown). Astral MT integrity is involved in the positioning of the cleavage plane during cytokinesis (70, 71), which could explain the asymmetry observed. Hyper-phosphorylation of CRMP2 has been reported to correlate with aberrant cell proliferation (72). However, this may represent a bystander effect of kinase activation.
The role of CRMPs suggested in this study is consistent with ability of CRMP2 to promote supernumerary axon formation in primary neurons, which can be phenocopied by low doses of MT-stabilizing drugs (73). Hyper-phosphorylated CRMP2 is found in disease states such as Alzheimer disease, where the C-terminal GSK3β sites show increased phosphorylation (74, 75), and the role of GSK3β in promoting neurofibrillary tangles of Alzheimer disease involving Tau has been suggested (47). Tau and CRMP2 share several similarities; both drive MT stabilization, both are substrates of Cdk5 (for priming) and GSK3β, and both are hyper-phosphorylated in Alzheimer disease (47). The antibody 3F4, which recognizes phosphorylated Tau in Alzheimer disease neurofibrillary tangles, also stains this phospho-CRMP2 (6). It seems more than a coincidence that CDK5 and GSK3β function inhibits multiple MAPs, and this suggests that these protein families are co-regulated. Enhancing CRMP (or Tau) activity by inhibiting relevant protein kinase(s) is an attractive therapeutic target.
In summary, our results indicate that CRMPs represent a new class of MT-binding proteins, which is consistent with the known biological roles of CRMPs. This interaction is regulated by phosphorylation at sites in the C-terminal binding domain of CRMP2 by Cdk5 and GSK3β. In non-neuronal cells, the CRMP proteins likely promote stability of MT elements that include astral MTs and the mitotic spindle.
Acknowledgment
We thank Dr. Graham Wright of Institute of Medical Biology (IMB) imaging unit for performing the deconvolution process of the confocal images.
This work was partially supported by the GSK-IMCB fund.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. S1–S6.
- CRMP
- collapsin response mediator protein
- CMBD
- CRMP C-terminal MT binding domain
- MT
- microtubule
- CDK5
- cyclin-dependent kinase 5
- GSK3
- glycogen synthase kinase-3
- ROCK
- Rho-associated coiled-coil kinase
- Sema3A
- semaphorin 3A.
REFERENCES
- 1. Wang L. H., Strittmatter S. M. (1997) J. Neurochem. 69, 2261–2269 [DOI] [PubMed] [Google Scholar]
- 2. Goshima Y., Nakamura F., Strittmatter P., Strittmatter S. M. (1995) Nature 376, 509–514 [DOI] [PubMed] [Google Scholar]
- 3. Minturn J. E., Fryer H. J., Geschwind D. H., Hockfield S. (1995) J. Neurosci. 15, 6757–6766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown M., Jacobs T., Eickholt B., Ferrari G., Teo M., Monfries C., Qi R. Z., Leung T., Lim L., Hall C. (2004) J. Neurosci. 24, 8994–9004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Deo R. C., Schmidt E. F., Elhabazi A., Togashi H., Burley S. K., Strittmatter S. M. (2004) EMBO J. 23, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Uchida Y., Ohshima T., Sasaki Y., Suzuki H., Yanai S., Yamashita N., Nakamura F., Takei K., Ihara Y., Mikoshiba K., Kolattukudy P., Honnorat J., Goshima Y. (2005) Genes Cells 10, 165–179 [DOI] [PubMed] [Google Scholar]
- 7. Erskine L., Reijntjes S., Pratt T., Denti L., Schwarz Q., Vieira J. M., Alakakone B., Shewan D., Ruhrberg C. (2011) Neuron 70, 951–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Nishiyama M., Togashi K., von Schimmelmann M. J., Lim C. S., Maeda S., Yamashita N., Goshima Y., Ishii S., Hong K. (2011) Nat. Cell Biol. 13, 676–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piaton G., Aigrot M. S., Williams A., Moyon S., Tepavcevic V., Moutkine I., Gras J., Matho K. S., Schmitt A., Soellner H., Huber A. B., Ravassard P., Lubetzki C. (2011) Brain 134, 1156–1167 [DOI] [PubMed] [Google Scholar]
- 10. Syed Y. A., Hand E., Möbius W., Zhao C., Hofer M., Nave K. A., Kotter M. R. (2011) J. Neurosci. 31, 3719–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Joyal J. S., Sitaras N., Binet F., Rivera J. C., Stahl A., Zaniolo K., Shao Z., Polosa A., Zhu T., Hamel D., Djavari M., Kunik D., Honoré J. C., Picard E., Zabeida A., Varma D. R., Hickson G., Mancini J., Klagsbrun M., Costantino S., Beauséjour C., Lachapelle P., Smith L. E., Chemtob S., Sapieha P. (2011) Blood 117, 6024–6035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Casazza A., Fu X., Johansson I., Capparuccia L., Andersson F., Giustacchini A., Squadrito M. L., Venneri M. A., Mazzone M., Larsson E., Carmeliet P., De Palma M., Naldini L., Tamagnone L., Rolny C. (2011) Arterioscler. Thromb. Vasc. Biol. 31, 741–749 [DOI] [PubMed] [Google Scholar]
- 13. Hamajima N., Matsuda K., Sakata S., Tamaki N., Sasaki M., Nonaka M. (1996) Gene 180, 157–163 [DOI] [PubMed] [Google Scholar]
- 14. Wang L. H., Strittmatter S. M. (1996) J. Neurosci. 16, 6197–6207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hedgecock E. M., Culotti J. G., Thomson J. N., Perkins L. A. (1985) Dev. Biol. 111, 158–170 [DOI] [PubMed] [Google Scholar]
- 16. Yamashita N., Morita A., Uchida Y., Nakamura F., Usui H., Ohshima T., Taniguchi M., Honnorat J., Thomasset N., Takei K., Takahashi T., Kolattukudy P., Goshima Y. (2007) J. Neurosci. 27, 12546–12554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Quach T. T., Massicotte G., Belin M. F., Honnorat J., Glasper E. R., Devries A. C., Jakeman L. B., Baudry M., Duchemin A. M., Kolattukudy P. E. (2008) FASEB J. 22, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shih J. Y., Lee Y. C., Yang S. C., Hong T. M., Huang C. Y., Yang P. C. (2003) Clin. Exp. Metastasis 20, 69–76 [DOI] [PubMed] [Google Scholar]
- 19. Gu Y., Ihara Y. (2000) J. Biol. Chem. 275, 17917–17920 [DOI] [PubMed] [Google Scholar]
- 20. Poulain F. E., Sobel A. (2010) Mol. Cell. Neurosci. 43, 15–32 [DOI] [PubMed] [Google Scholar]
- 21. Fukata Y., Itoh T. J., Kimura T., Ménager C., Nishimura T., Shiromizu T., Watanabe H., Inagaki N., Iwamatsu A., Hotani H., Kaibuchi K. (2002) Nat. Cell Biol. 4, 583–591 [DOI] [PubMed] [Google Scholar]
- 22. Kimura T., Watanabe H., Iwamatsu A., Kaibuchi K. (2005) J. Neurochem. 93, 1371–1382 [DOI] [PubMed] [Google Scholar]
- 23. Cole A. R., Knebel A., Morrice N. A., Robertson L. A., Irving A. J., Connolly C. N., Sutherland C. (2004) J. Biol. Chem. 279, 50176–50180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arimura N., Inagaki N., Chihara K., Ménager C., Nakamura N., Amano M., Iwamatsu A., Goshima Y., Kaibuchi K. (2000) J. Biol. Chem. 275, 23973–23980 [DOI] [PubMed] [Google Scholar]
- 25. Arimura N., Ménager C., Kawano Y., Yoshimura T., Kawabata S., Hattori A., Fukata Y., Amano M., Goshima Y., Inagaki M., Morone N., Usukura J., Kaibuchi K. (2005) Mol. Cell. Biol. 25, 9973–9984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gallo G. (2006) J. Cell Sci. 119, 3413–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Byk T., Ozon S., Sobel A. (1998) Eur. J. Biochem. 254, 14–24 [DOI] [PubMed] [Google Scholar]
- 28. Shih J. Y., Yang S. C., Hong T. M., Yuan A., Chen J. J., Yu C. J., Chang Y. L., Lee Y. C., Peck K., Wu C. W., Yang P. C. (2001) J. Natl. Cancer Inst. 93, 1392–1400 [DOI] [PubMed] [Google Scholar]
- 29. Vincent P., Collette Y., Marignier R., Vuaillat C., Rogemond V., Davoust N., Malcus C., Cavagna S., Gessain A., Machuca-Gayet I., Belin M. F., Quach T., Giraudon P. (2005) J. Immunol. 175, 7650–7660 [DOI] [PubMed] [Google Scholar]
- 30. Charrier E., Reibel S., Rogemond V., Aguera M., Thomasset N., Honnorat J. (2003) Mol. Neurobiol. 28, 51–64 [DOI] [PubMed] [Google Scholar]
- 31. Nishimura T., Fukata Y., Kato K., Yamaguchi T., Matsuura Y., Kamiguchi H., Kaibuchi K. (2003) Nat. Cell Biol. 5, 819–826 [DOI] [PubMed] [Google Scholar]
- 32. Ong Tone S., Dayanandan B., Fournier A. E., Mandato C. A. (2010) PLoS One 5, e14345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stenmark P., Ogg D., Flodin S., Flores A., Kotenyova T., Nyman T., Nordlund P., Kursula P. (2007) J. Neurochem. 101, 906–917 [DOI] [PubMed] [Google Scholar]
- 34. Jourdain I., Lachkar S., Charbaut E., Gigant B., Knossow M., Sobel A., Curmi P. A. (2004) Biochem. J. 378, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trinkle-Mulcahy L., Boulon S., Lam Y. W., Urcia R., Boisvert F. M., Vandermoere F., Morrice N. A., Swift S., Rothbauer U., Leonhardt H., Lamond A. (2008) J. Cell Biol. 183, 223–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nettles J. H., Li H., Cornett B., Krahn J. M., Snyder J. P., Downing K. H. (2004) Science 305, 866–869 [DOI] [PubMed] [Google Scholar]
- 37. Vallee R. B. (1982) J. Cell Biol. 92, 435–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Serrano L., de la Torre J., Maccioni R. B., Avila J. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 5989–5993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maccioni R. B., Rivas C. I., Vera J. C. (1988) EMBO J. 7, 1957–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cross D., Dominguez J., Maccioni R. B., Avila J. (1991) Biochemistry 30, 4362–4366 [DOI] [PubMed] [Google Scholar]
- 41. Hayashi I., Ikura M. (2003) J. Biol. Chem. 278, 36430–36434 [DOI] [PubMed] [Google Scholar]
- 42. Galjart N. (2010) Curr. Biol. 20, R528–537 [DOI] [PubMed] [Google Scholar]
- 43. Klein P. S., Melton D. A. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 8455–8459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schulze E., Kirschner M. (1987) J. Cell Biol. 104, 277–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gundersen G. G., Bulinski J. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 5946–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bosc C., Cronk J. D., Pirollet F., Watterson D. M., Haiech J., Job D., Margolis R. L. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2125–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Soutar M. P., Thornhill P., Cole A. R., Sutherland C. (2009) Curr. Alzheimer Res. 6, 269–278 [DOI] [PubMed] [Google Scholar]
- 48. Yoshimura T., Kawano Y., Arimura N., Kawabata S., Kikuchi A., Kaibuchi K. (2005) Cell 120, 137–149 [DOI] [PubMed] [Google Scholar]
- 49. Wagner U., Utton M., Gallo J. M., Miller C. C. (1996) J. Cell Sci. 109, 1537–1543 [DOI] [PubMed] [Google Scholar]
- 50. Hong M., Chen D. C., Klein P. S., Lee V. M. (1997) J. Biol. Chem. 272, 25326–25332 [DOI] [PubMed] [Google Scholar]
- 51. Goold R. G., Owen R., Gordon-Weeks P. R. (1999) J. Cell Sci. 112, 3373–3384 [DOI] [PubMed] [Google Scholar]
- 52. Wu X., Shen Q. T., Oristian D. S., Lu C. P., Zheng Q., Wang H. W., Fuchs E. (2011) Cell 144, 341–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kumar P., Lyle K. S., Gierke S., Matov A., Danuser G., Wittmann T. (2009) J. Cell Biol. 184, 895–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Etienne-Manneville S., Hall A. (2003) Nature 421, 753–756 [DOI] [PubMed] [Google Scholar]
- 55. Sun T., Rodriguez M., Kim L. (2009) Dev. Growth Differ. 51, 735–742 [DOI] [PubMed] [Google Scholar]
- 56. Cole A. R., Causeret F., Yadirgi G., Hastie C. J., McLauchlan H., McManus E. J., Hernández F., Eickholt B. J., Nikolic M., Sutherland C. (2006) J. Biol. Chem. 281, 16591–16598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Soutar M. P., Kim W. Y., Williamson R., Peggie M., Hastie C. J., McLauchlan H., Snider W. D., Gordon-Weeks P. R., Sutherland C. (2010) J. Neurochem. 115, 974–983 [DOI] [PubMed] [Google Scholar]
- 58. Miao H. Q., Soker S., Feiner L., Alonso J. L., Raper J. A., Klagsbrun M. (1999) J. Cell Biol. 146, 233–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Spriggs M. K. (1999) Curr. Opin. Immunol. 11, 387–391 [DOI] [PubMed] [Google Scholar]
- 60. Yamada T., Endo R., Gotoh M., Hirohashi S. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 14713–14718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yazdani U., Terman J. R. (2006) Genome Biol. 7, 211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pasterkamp R. J., Verhaagen J. (2006) Philos. Trans. R. Soc. Lond. B Biol. Sci. 361, 1499–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shelly M., Cancedda L., Lim B. K., Popescu A. T., Cheng P. L., Gao H., Poo M. M. (2011) Neuron 71, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noree C., Sato B. K., Broyer R. M., Wilhelm J. E. (2010) J. Cell Biol. 190, 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Uchida Y., Ohshima T., Yamashita N., Ogawara M., Sasaki Y., Nakamura F., Goshima Y. (2009) J. Biol. Chem. 284, 27393–27401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sasaki Y., Cheng C., Uchida Y., Nakajima O., Ohshima T., Yagi T., Taniguchi M., Nakayama T., Kishida R., Kudo Y., Ohno S., Nakamura F., Goshima Y. (2002) Neuron 35, 907–920 [DOI] [PubMed] [Google Scholar]
- 67. Eickholt B. J., Walsh F. S., Doherty P. (2002) J. Cell Biol. 157, 211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Meijer L., Flajolet M., Greengard P. (2004) Trends Pharmacol. Sci. 25, 471–480 [DOI] [PubMed] [Google Scholar]
- 69. Schmidt E. F., Strittmatter S. M. (2007) Adv. Exp. Med. Biol. 600, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rappaport R. (1961) J. Exp. Zool. 148, 81–89 [DOI] [PubMed] [Google Scholar]
- 71. Eckley D. M., Ainsztein A. M., Mackay A. M., Goldberg I. G., Earnshaw W. C. (1997) J. Cell Biol. 136, 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tahimic C. G., Tomimatsu N., Nishigaki R., Fukuhara A., Toda T., Kaibuchi K., Shiota G., Oshimura M., Kurimasa A. (2006) Biochem. Biophys. Res. Commun. 340, 1244–1250 [DOI] [PubMed] [Google Scholar]
- 73. Witte H., Neukirchen D., Bradke F. (2008) J. Cell Biol. 180, 619–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gu Y., Hamajima N., Ihara Y. (2000) Biochemistry 39, 4267–4275 [DOI] [PubMed] [Google Scholar]
- 75. Cole A. R., Noble W., van Aalten L., Plattner F., Meimaridou R., Hogan D., Taylor M., LaFrancois J., Gunn-Moore F., Verkhratsky A., Oddo S., LaFerla F., Giese K. P., Dineley K. T., Duff K., Richardson J. C., Yan S. D., Hanger D. P., Allan S. M., Sutherland C. (2007) J. Neurochem. 103, 1132–1144 [DOI] [PubMed] [Google Scholar]
- 76. Scales T. M., Lin S., Kraus M., Goold R. G., Gordon-Weeks P. R. (2009) J. Cell Sci. 122, 2424–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]