Abstract
About 90% of the people infected with Mycobacterium tuberculosis carry latent bacteria that are believed to get activated upon immune suppression. One of the fundamental challenges in the control of tuberculosis is therefore to understand molecular mechanisms involved in the onset of latency and/or reactivation. We have attempted to address this problem at the systems level by a combination of predicted functional protein∶protein interactions, integration of functional interactions with large scale gene expression studies, predicted transcription regulatory network and finally simulations with a Boolean model of the network. Initially a prediction for genome-wide protein functional linkages was obtained based on genome-context methods using a Support Vector Machine. This set of protein functional linkages along with gene expression data of the available models of latency was employed to identify proteins involved in mediating switch signals during dormancy. We show that genes that are up and down regulated during dormancy are not only coordinately regulated under dormancy-like conditions but also under a variety of other experimental conditions. Their synchronized regulation indicates that they form a tightly regulated gene cluster and might form a latency-regulon. Conservation of these genes across bacterial species suggests a unique evolutionary history that might be associated with M. tuberculosis dormancy. Finally, simulations with a Boolean model based on the regulatory network with logical relationships derived from gene expression data reveals a bistable switch suggesting alternating latent and actively growing states. Our analysis based on the interaction network therefore reveals a potential model of M. tuberculosis latency.
Introduction
Increased rate of tuberculosis (TB) infection and the emergence of multi and extensively-drug-resistant tuberculosis [1] have necessitated urgent efforts towards enhanced understanding of its causative agent, Mycobacterium tuberculosis. The complete genome sequence of the common laboratory strain M. tuberculosis H37Rv was unraveled in 1998 [2] and the sequence was later re-annotated [3]. The genome annotation combined with comparative genomic studies has revealed several novel features including gene families that are unique to this bacterium such as PE and PPE genes and the eukaryotic like serine threonine protein kinases (STPKs). The genome also harbors unique genes involved in lipid biosynthesis, drug resistance and pathogenesis [2].
Owing to its distinct characteristics and the disease causing ability, M. tuberculosis has been studied widely across the world. For example, different experiments have accumulated information about the biochemical and structural properties of a number of proteins of this organism [4], [5]. There is also large amount of data on mycobacterial gene expression under a variety of growth conditions [6]. Genome-wide surveys of essential genes of M. tuberculosis, both in vitro and those potentially involved in pathogenesis, have been carried out [7]–[9]. Similarly, large-scale proteome profiling to classify M. tuberculosis proteins into different cellular compartments has been carried out [10]. Such varied studies have helped in understanding not only the unique genetic makeup of the bacillus, but also the possible roles of individual genes during different steps of pathogenesis.
M. tuberculosis has also been the centre of attention for a few studies executed at the systems level. In this regard, there have been attempts to model gene regulation, protein interactions and metabolic pathways of the organism. Functional linkage maps of M. tuberculosis have been defined by using genome context methods [11]. In another study, Balaszi et al have assembled gene regulation information and studied transcriptional changes that might mediate switch to dormancy [12]. Flux balance analysis (FBA) on the model of mycolic acid biosynthesis pathway has revealed potential drug targets pertaining to this pathway [13]. Nonetheless, the systems level understanding of M. tuberculosis remains inadequate. One of the major obstacles being that a large fraction of genes are either putative or are unannotated, and thereby coherence among different pathways that contribute to virulence remains to be defined systematically. There is therefore a pressing need to integrate different approaches to understand tuberculosis and perceive mechanisms by which M. tuberculosis enters a dormant phase, or emerges out of it. A new promise in this aspect is the availability of a number of genome sequences of clinical strains and the data gathered by individual and high-throughput experiments which permit studies at the systems level.
One of the promising approaches to understand complex functional associations of the molecules and their organization is by analyzing genome-wide protein∶protein interactions by means of graph theoretical representations. In this regard, there have been a few attempts to generate protein interaction maps of M. tuberculosis, all of them being in silico predictions. Strong et al used genome context methods to derive a functional linkage map of 4,886 interactions among 1,958 proteins followed by clustering of the network in order to reconstruct some of the biochemical pathways in M. tuberculosis [11]. Another study involved translating high confidence interactions of E. coli to M. tuberculosis [14]. A set of 6,091 interactions among 793 proteins was obtained in this study to identify proteins involved in signaling pathways. The interaction database STRING [15] houses protein interactions of M. tuberculosis derived by literature curation and other methods. The highest confidence interactions in STRING which are supported by multiple methods or curation include around 6,403 interactions between 1,653 proteins. However, none of the above sources represents a comprehensive collection of M. tuberculosis protein interactions.
We have attempted to combine genome context methods, namely phylogenetic profile [16], [17], gene distance [18] operonic co-occurrence [19], [20] with the available high throughput gene expression studies for the prediction of genome-wide functional linkages. The predictive features of these were combined using a Support Vector Machine (SVM). These functional linkages were analyzed to understand the phenomenon of latency, thus providing an integrated perspective of latency. In addition, a Boolean model is employed to investigate the patterns of expression of genes identified as being up and down-regulated during latency. Boolean or logical models [21] are known to be an effective and straightforward means of reproducing the behaviour of gene regulatory networks over time [22]–[24], especially for systems where only qualitative data is available. The model simulations strengthen our understanding of the regulatory control exerted by a small number of genes in order to sustain the latency stage.
Results
Generation of the Protein Functional Linkages
We have reported earlier that the genome context methods, namely phylogenetic profile, operonic frequency and gene distance can be effectively combined using a SVM to predict protein functional linkages in E. coli [25]. Apart from the genome context methods it is interesting to note that the proteins which have functional relations are also known to show good correlation in their gene expression [26]. We therefore attempted to include correlation in gene expression among the genes as a feature for SVM training.
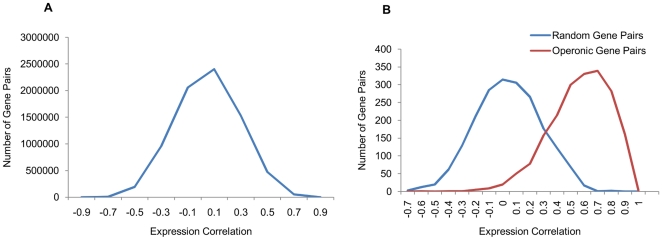
It is well known that large-scale gene expression datasets are expected to be noisy [27]. However, the expression levels between gene pairs, if functionally related, are likely to be correlated across different experimental conditions. In general, no gene pair is anticipated to exhibit high expression correlation except when the two genes are coregulated thereby suggesting a functional relationship between the two. This conjecture was sought to be tested using known operonic gene pairs. Significantly, the operonic gene pairs show high expression correlation compared to the same number of randomly generated non-operonic pairs (Figure 1). This observation, therefore, strengthened the confidence in using the microarray data for interaction predictions. Thus, we have supplemented genome context methods with correlations in gene expression to generate genome-wide protein functional linkages map of M. tuberculosis.
Figure 1. Pearson correlation coefficient between expression values of pairs of genes.
A) The genome-wide gene expression correlations of all the gene pairs and B) expression correlation between operonic gene pairs and randomly paired non-operonic gene pairs. As anticipated, it is evident that the genes on operons exhibit higher correlation in their expression compared to the non-operonic gene pairs.
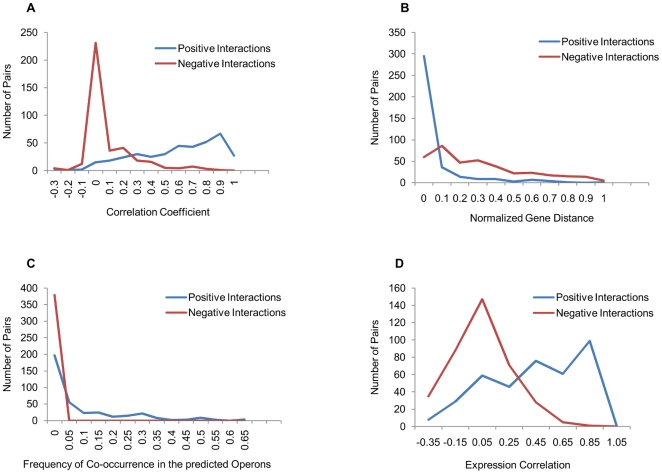
In order to test whether the positive and negative pairs used in training the SVM show characteristic distribution for the data features chosen, students' t-test was performed on the available positive pairs and equal number of randomly chosen negative pairs. Figure 2 depicts the distribution of vectors of the gene pairs used in training, which show a distinctly different distribution with a p value of 2.2e−16. All the data features chosen are therefore capable of distinguishing between positive and negative pairs, suggesting their potential application in the prediction of functional linkages. Thus, using these features, optimization of training for the SVM was performed with 5-fold cross-validation. Furthermore, performance of SVM classification was evaluated by plotting ROC (Figure S1), and the best predictive model showed AUC of 0.834 with 0.85 Mathews Correlation Coefficient (MCC). This optimized model with prediction accuracy and sensitivity of 88% and 76% respectively, was chosen for prediction of functional linkages on the genome-wide scale.
Figure 2. Plots indicating the distinctive behavior of the positive and negative protein interaction pairs with respect to genome context methods and gene expression correlations.
All the four features show statistically significant and distinct distribution for the positive and negative pairs. A) Phylogenetic Profile, B) Gene Distance, C) Operonic Method and D) Expression Correlation.
The Functional Linkages of M. tuberculosis Proteins
The predicted protein functional linkages network has 32,546 interactions among 3,571 proteins (Table S1). The largest connected component of the network comprises of 95% of the nodes and has a diameter of 12. The network shows scale free property with the degree exponent of 1.67. The overall topological parameters of the network are summarized in Table 1. Comparison of the network with previously derived interaction maps shows around 30% overlap (Table S2). Though the overlap appears less, it is observed that protein interaction maps obtained from different sources generally have fewer interactions in common due to inherent bias in the method used or the noise [25].
Table 1. Topological Properties of the predicted protein functional linkages.
| Number of Interactions | 32546 |
| Number of Nodes | 3571 |
| Percentile Core Nodes | 95% |
| Average Degree | 19.2 |
| Degree Exponent | 1.67 |
| Diameter | 12 |
| Average Clustering Coefficient | 0.22 |
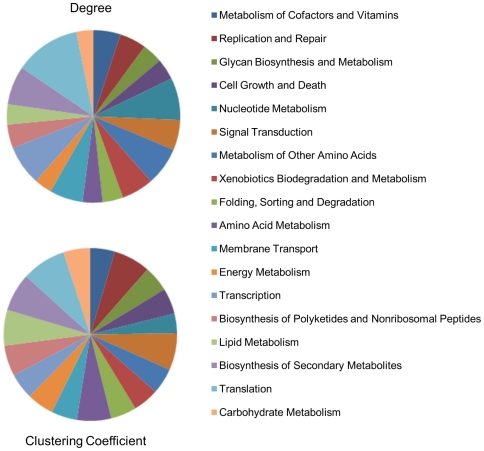
The average degree and clustering coefficient of the proteins classified into different metabolic pathways is shown in Figure 3. Proteins belonging to translation pathway, such as the ribosomal proteins and other translation related proteins, have high degree as well as high clustering coefficient. On the other hand, proteins of the “cell growth and death” pathway have high degree but are less clustered. The membrane transport proteins on the other hand show less degree but are highly clustered. Thus, we observe varied relationships between clustering and degree in different metabolic pathways.
Figure 3. Average clustering coefficient and degree of the proteins across metabolic pathways.
Proteins show high degree or high clustering coefficient depending on their function. Translation pathway proteins have high degree as well as high clustering coefficient whereas membrane transport proteins are highly clustered with less connectivity.
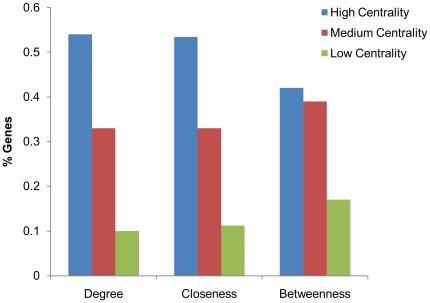
Proteins coded by essential genes in biological networks are known to exhibit high network centrality measures compared to their counterparts [28]. This observation was tested in M. tuberculosis network for the experimentally proposed essential genes [7]. From our functional interaction network, we calculated three centrality values, namely degree, closeness and betweenness, for each of the genes, and divided them into three categories: high centrality (top 30% nodes), medium centrality (between 30–70% nodes) and low centrality (others). The proportion of essential genes for these three centrality parameters was plotted as shown in Figure 4. It is evident from the figure that there is a strong correlation between network centrality and gene essentiality in our proposed network. The proteins of information pathway such as DnaG, DnaB, Rho and ribosomal proteins; proteins belonging to intermediary metabolism and respiration such as ATP synthase subunits, purine and pyrimidine biosynthesis proteins; proteins in the amino acid biosynthesis pathways and cell division proteins such as FtsX, FtsZ and FtsH; and proteins of cell wall formation such as MurA, MurB, MurC and MurD show very high network centrality values. Interestingly, PPE4 and PPE46 of the PPE family proteins have high centrality values, suggesting that these proteins might also be constituents of the essential gene set of M. tuberculosis. The complete list of proteins with high centrality values is given in Table S3.
Figure 4. Proportion of Essential Genes in the bins of decreasing centrality values.
For all the three centrality values calculated, namely degree, closeness and betweenness, it is clearly seen that there exists a good correlation between centrality and lethality.
Proteins involved in closely related functions, or those involved in the same biochemical pathway, are known to cluster together to form functional units in an interaction map [29]. Clustering of our network indicated that it is divided into 184 clusters which we have annotated using the pathway information taken from Sanger and TubercuList databases (Table S4). The largest cluster is enriched with cell wall and cell processes, virulence related proteins and a large number of conserved hypothetical proteins. The ribosomal proteins and other translation related proteins cluster along with DNA replication and repair proteins. A large number of PE-PPE proteins were found to be associated with virulence proteins. It is likely therefore that these proteins play an important role in virulence determination, as has been suggested earlier [2], [30]. Thus, the sub-division of the interaction map into clusters based on the nature of their interaction might lead to a better understanding of cross-talks between different functional categories.
Persistence in M. tuberculosis
One of the least understood phenomena associated with M. tuberculosis is the switch between latent and actively replicating phases. It is interesting to address this phenomenon of latency in M. tuberculosis through the analysis of functional interaction network. The predicted interaction map was therefore used to demarcate the proteins that might play important role in the dormant phase of M. tuberculosis. Towards addressing these questions, we considered several experimental studies that have attempted to simulate the dormancy phase using the in-vitro models to generate gene expression profiles. Such conditions include hypoxia [31]–[35], NO treatment [36], stationary phase [35] and nutrient deprivation [37]. Along with the in-vitro data, murine models of M. tuberculosis dormancy have also been studied [38]–[39]. Since such individual experiments might not capture all the dormancy features of M. tuberculosis, and moreover considering that the microarray experiments might be affected by intrinsic noise, we considered the commonality among these to identify key genes regulating latency.
The gene expression datasets of different persistence models of M. tuberculosis were used to identify up and down-regulated genes during early persistence. The list of different models and the number of up and down-regulated genes are detailed in Table S5. Intriguingly, the overlap among the differentially regulated genes between different models of M. tuberculosis dormancy is not very high. For example, murine model of M. tuberculosis latency has 7%, 19%, 6% and 3% overlap with the other available models of NO treatment, hypoxia, stationary phase growth and starvation model respectively (Table S6). We therefore considered only the genes that are common to at least 5 of the 12 expression conditions related to latency. Using this criterion, we were able to identify 50 genes that show increase in expression levels and 34 genes that are down-regulated during latency (Table S7).
In order to understand if the factors leading to dormancy are shared by different prokaryotic species we first probed evolutionary conservation of the 84 genes by constructing a binary phylogenetic profile of these genes across 481 genomes and then counted the numbers of genomes harboring these genes. Interestingly, the 50 genes that are upregulated in M. tuberculosis dormancy are far less conserved across species than the 34 genes that are downregulated. As few as 16 upregulated genes are present in less than 145 of the 481 genomes. On the other hand, as many as 26 down regulated genes are present in at least 336 genomes (Figure S2). The downregulated genes represent those involved in basic cellular processes, such as replication, transcription and translation. The rates of these basic cellular processes being significantly slowed during dormancy offers a possible explanation that the downregulated genes are common to many species. The remarkable observation, however, that the upregulated genes are less conserved than the downregulated genes suggests a possible unique mechanism of dormancy adapted by M. tuberculosis. The 50 upregulated genes therefore appear to constitute a “dormancy signal” that is unique to M. tuberculosis. Such a dormancy signal might then be transmitted to the genes involved in basic cellular processes in order to slow down the overall metabolic rates.
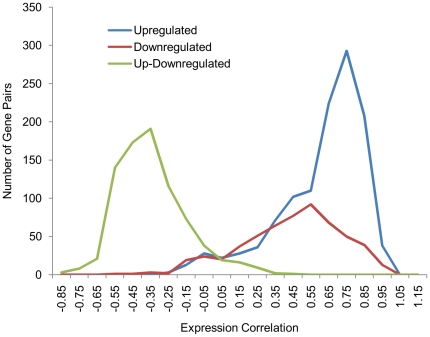
Although the 84 genes showed coordinated regulation during latency-like conditions, it was interesting to probe if similar regulation controlled their expression even under other experimental conditions. The expression correlation among the 50 upregulated genes and the 34 downregulated genes across 154 growth conditions was then computed. Interestingly, the expression values among the 50 upregulated genes correlate highly (Figure 5). Similarly, those among the 34 downregulated genes also show strong correlation. On the other hand, the up and the down-regulated genes exhibit inverse correlation. This observation suggests that the 84 genes are coordinately regulated not only under latency-like conditions, but also are regulated in a controlled manner under most other conditions of growth. The 84 genes thus form a regulon-like structure with an extensive cross talk in their expression. Such a crosstalk can possibly be uncovered using the functional interaction network.
Figure 5. Expression Correlation between up and downregulated genes.
Upregulated genes correlate in their expression and a similar trend is observed for downregulated genes. However, there is inverse correlation in expression between up and downregulated genes.
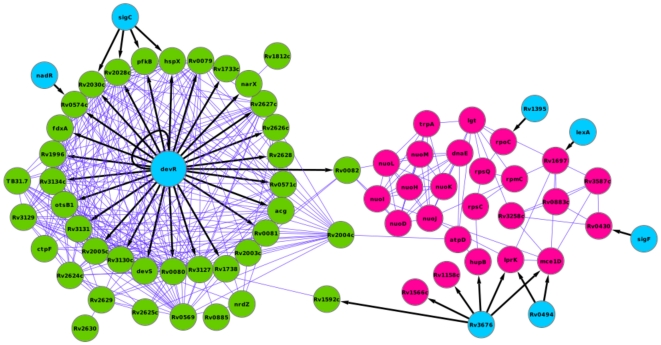
We then integrated our protein functional linkages with the available gene regulatory information in order to understand the crosstalk between the 84 latency-related genes [12]. One of the important observations upon such an integration of the two networks was that DosR, a well studied dormancy associated protein, regulates the expression of about 25 upregulated genes (Figure 6). Further, the genes regulated by DosR also interact extensively among themselves forming a clique-like architecture in the network. Moreover, important latency genes such as hspX, pfkB, Rv2030c and Rv2028c are additionally regulated by sigma factor SigC, implying a multifarious regulatory circuit of latency. On the other hand, Rv3676, a transcriptional regulatory protein of the cAMP receptor protein (CRP) family [40], regulates the expression of downregulated genes such as Rv1566c, Rv1158c, hupB, lprK and mce1D. Thus, a central regulatory circuit controlled by DosR, with degeneracy offered by transcription factors such as SigC and CRP, combined with an extensive network of interactions among the 84 genes appears to control latency in M. tuberculosis.
Figure 6. Graphical representation of functional interactions and regulatory interactions of up and downregulated genes.
DosR acts as a master regulator, regulating the expression of a number of upregulated genes. A probable oxidoreductase Rv0082 under the regulation of DosR associates with the subunits of aerobic respiratory machine NADH oxidoreductase complex. Upregulated proteins are colored in green, downregulated proteins are colored in red and transcription factors are colored in blue. Directed edge denotes gene regulation and undirected edge is a protein functional linkage.
An interesting outcome of the network analysis pertains to the dormany signal detected by the 50 upregulated genes, and its transmission to the 34 downregulated genes. DosR interacts with two-component sensory kinases DosS (Rv3132c), DosT (Rv2027c) and Rv0845. It has been reported previously that DosR acts as a cognate response regulator of both DosS and DosT, which sense hypoxia and NO [41]. Interestingly, a classification based on the region around phosphorylated sensor kinases of M. tuberculosis assigns DosS, DosT and Rv0845 to the same class [42]. Whereas Rv0845 is a possible nitrate/nitrite sensor protein, DosS and DosT function as redox and hypoxia sensors respectively [43]. It is therefore likely that DosS, DosT and Rv0845 sense the dormancy signal and trigger a regulatory cascade controlled by DosR.
The gene regulatory network also suggests that a hypothetical oxidoreductase, Rv0082, is regulated by DosR with increase in its expression level under dormancy conditions. The functional interaction network further suggests that Rv0082 might associate with three of the subunits of NADH-ubiquinone oxidoreductase complex (NDH-1). The subunits of NDH-1 complex are involved in respiration and not surprisingly form a clique in the functional interaction network. Moreover, these are downregulated during latency. The expression correlation between NDH-1 genes spanning from Rv3145–Rv3158 and ATP synthase genes spanning from Rv1304–Rv1311 are correlated in their expression (Figure S3). This suggests a succeeding downregulation of ATP synthesis upon downregulation of NDH-1 genes. Interestingly, NDH-1 proteins interact with ribosomal proteins RpsC and RpsQ, and DNA polymerase DnaE in the subnetwork of functional interactions. The succession of protein connectivity in the subnetwork therefore suggests that DosR possibly communicates latency signals to the respiratory chain through Rv0082, resulting in the shutdown of respiration mediated by NDH-1 followed by growth suppression. Since NAD+ pool is essential for TCA cycle and other biosynthetic pathways, it appears that the downregulation of NDH-1 respiration is critical in the early stages of latency, which might lead to the arrest of cell growth and division subsequently. Interestingly, the mutants of NADH dehydrogenase I of Escherichia coli show competitive disadvantage in the mixed stationary phase cultures [44]. The inability of these mutants to adapt to the stationary phase might arise due to their inefficiency in transmitting signals downstream to slowdown cellular growth processes. Thus, protein interactions and gene regulatory information support the complex regulatory hierarchy of the genes involved in latency.
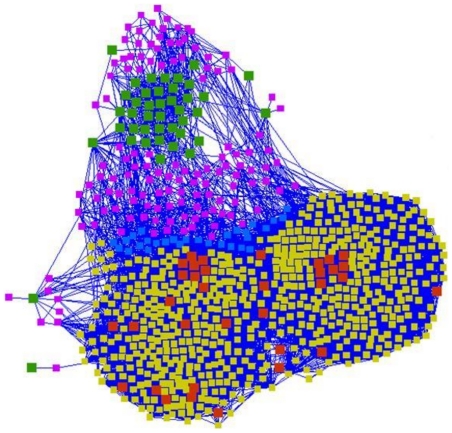
In order to further address the complexity of the cross-talk between the up and the downregulated genes in the network, we expanded the network of the 84 genes to construct a “Dormancy Core” comprised of directly interacting proteins of the up and the downregulated genes (Table S8). The “Dormancy Core” was divided into up or downregulated modules depending on the association of the proteins with the up or the downregulated genes. The number of nodes in the upregulated and downregulated modules was 172 and 632 respectively. There are 29 proteins which are common to both these modules (Figure 7). These 29 proteins might participate in direct signalling between the up and the down regulated modules.
Figure 7. Schematic representation of the up and down-regulated modules.
Node color represents the class as follows: Green – Upregulated proteins; Red – Downregulated proteins; Purple – First neighbors of the upregulated proteins; Yellow – First neighbors of the downregulated proteins; Blue – Proteins interacting with both upregulated and downregulated proteins.
Examination of the topological properties of this subnetwork interestingly showed that the nodes in the downregulated module possess higher degree compared to the up-regulated module (Figure S4(A)). As the downregulated module is enriched by proteins of cell growth and division, it is not surprising that these nodes possess higher degree since high degree nodes are more likely to perform essential growth functions [45]. Also, the downregulated module nodes are closer to other proteins in the network as revealed by their closeness centrality compared to the up-regulated module (Figure S4(B)). This suggests their important role in mediating the information flow in the network (Figure S4(C)). However, there is no apparent difference in the clustering coefficients of these two modules (Figure S4(D)). Thus, the known centrality characteristics are able to distinguish between the upregulated and downregulated modules in the network.
The pathway mapping of up and downregulated modules (Figure S5) indicates that the genes from Information pathways are down-regulated during the dormant phase of M. tuberculosis. The examples are replication proteins such as DnaA, DnaB, DnaN and GyrA, translation initiation factors such InfA and InfB, proteins of the ribosomal complex, repair and recombination proteins. Interestingly, the subunits of ATP synthase belong to the downregulated module, suggesting an important role of electron transport during dormancy. The down-regulated module also contains Fad proteins which are involved in the degradation of fatty acids, NADH dehydrogenase subunits, proteins involved in polyketides and non-ribosomal peptide synthesis, and cell envelop proteins from the families Lpr and Mur. The up-regulated module, on the other hand, includes the master regulator of dormancy DosR, DosS which functions coordinately with DosR, nitrate reductases NarG, NarJ and NarX, chaperones such as HspX and HtpG, polyketide synthetases such as MbtB and MbtC. Thus, the analysis of the module composition suggests possible pathways that are activated and repressed during the dormant phase of M. tuberculosis.
Shortest Paths Analysis in the Dormancy Module
An interesting aspect of dormancy is the communication between differentially regulated pathways [46], [47]. We sought to identify the possible communication route between the up and downregulated nodes in the dormancy module. All the possible shortest paths were traced from the 50 upregulated nodes to the 34 downregulated nodes and the most probable shortest paths were derived (Methods). The intermediates in the most probable paths were then ranked based on their frequency of occurrence and the top 25% of the intermediates were selected (Table S9). These proteins, we propose, might be involved in transmitting dormancy signals from the upregulated core proteins involved in dormancy, to the downregulated growth module. Below are the few examples we discuss for their possible role in modulating dormancy.
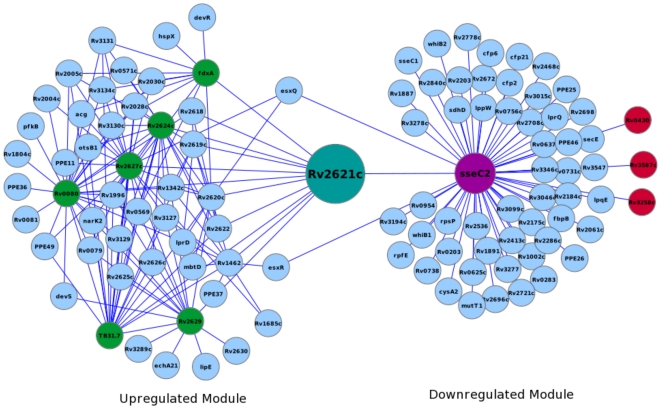
Some of the well known dormancy related proteins such as DosR, DosS and HspX occur frequently in the most probable paths calculated. DosR and DosS are the two-component regulatory proteins that have been shown previously to activate a number of proteins in response to the onset of dormancy [32]. HspX, a chaperone, is one such protein from the dosR regulon that shows significant induction during dormancy [48]. Another protein coded by Rv2621c is a hypothetical transcriptional regulatory protein which interacts with six of the 50 up-regulated nodes. It connects the down-regulated module through SseC2 which is a conserved hypothetical protein thought to be involved in sulphur metabolism. Both these proteins fall frequently in the most probable paths calculated. Figure 8 is a graphical presentation of the interactions of these two proteins. Intriguingly, the interacting partners of SseC2 include WhiB1 and WhiB2, which are transcription factors known to be involved in septation and cell division [49]. They require (4Fe-4S) for the catalytic activity and can function as protein disulfide reductases. The whiB homologue of C. glutamicum is critical for survival after oxidative stress [50]. Another interacting partner of SseC2 is Rv2175c, which is a transcription factor and a substrate of the PknL Ser/Thr kinase. The vicinity of Rv2175c to the dcw (division cell wall) cluster to which pknL belongs [51] suggests its possible role in regulating cell growth and division. SseC2 also associates with antigen 85B (FbpB) which is a mycolyl transferase involved in cell wall biosynthesis [52]. The cutinase precursor Cfp21, which promotes mycobacterial survival and virulence [53], is another interactor of SseC2. Other secreted proteins such as Cfp21, Cfp2, Rv3194c and Rv2672 also show interaction with SseC2. Thus, the cascade laid by Rv2621c and SseC2 in connecting essential proteins of dormancy to the cell division and growth proteins of the downregulated module appears to be important during switch from/to dormancy.
Figure 8. A sub-network depicting the connectivity mediated by proteins Rv2621c and SseC2.
Both these proteins occur most frequently in the paths between up and downregulated genes. The upregulated proteins associating with Rv2621c are colored in green and the downregulated proteins associating with SseC2 are colored in red.
Another interesting protein of a two-component system is Rv1626, the crystal structure of which suggests its possible role in transcriptional antitermination [54]. In our functional linkage network it interacts with PknA, a Ser/Thr protein kinase and NarK2, a nitrate/nitrite reductase. Both these proteins are members of the up-regulated module. Interestingly, Rv1626 interacts with several down-regulated module proteins, some of which are adenylate kinase Adk, tryptophan synthase TrpA, ribosomal proteins RpsA and RplT, and a two-component transcriptional regulator MtrA.
Another example of an important protein in the most probable paths is EsxR, a secreted ESAT-6 like protein. Interestingly, one of the proteins it connects in the down-regulated module is RpfA, which is a resuscitation promoting factor required for resuscitation from dormant state [55]. Notably, Rpfs have been shown to promote growth in Micrococcus luteus and are important for virulence [56]. Thus, the connectivity mediated by EsxR and RfpA between up and downregulated modules appears to be important. The subunits of ATP synthase AtpH, AtpG and AtpC also appear in the most frequent paths. Notably, an inhibitor diarylquinoline targets the proton pump of ATP synthase in M. tuberculosis [57]. We may therefore hypothesize that an alteration in the mode of respiration possibly serves as a signal for growth and it is logical to assume that the proteins of ATP synthase communicate such signals.
Boolean model simulation
In order to determine the main controllers of the dormancy network, a Boolean model simulation was performed using dormancy module. The transcription factors in the dormancy module were sequentially made active at input, individually as well as in various combinations, and the model simulated.
In Boolean terms, a biologically viable steady state of the dormancy model would have the identified upregulated genes in the ON or active state. Likewise, state of the genes identified as down-regulated during dormancy would be OFF or inactive. As mentioned in the methods section, the model included 26 (out of the 50) identified up-regulated genes and 13 (out of 34) down regulated genes. We then set out to determine which of the transcription factors that are known to play important roles in the dormancy network, give rise to a steady state with the required set of genes being active/inactive.
Setting individual transcription factors ON at input resulted in the system having a steady state with all the genes in the inactive or OFF state. However, when three regulatory elements, namely, Rv0081, Rv3132c (DosS) and Rv3133c (DosR), were all set to ON at input, we arrive at a logical steady state, with a number of genes in the active or ON state. Out of these genes, 19 are found to belong to the set of 26 dormancy genes. The simulations therefore confirm that DosR and DosS, along with Rv0081 are important regulators in the model.
The cAMP receptor protein (CRP) Rv3676 is known to regulate the expression of a few down-regulated genes. In order to determine its effect on the dormancy system, we added CRP along with the earlier input set (Rv3132c, Rv3133c and Rv0081). The simulation gave rise to a logical attractor cycle, with two states. An attractor cycle with two states implies that the system switches between the two states in an alternate manner. In the first state, while 24 out of the 26 up-regulated genes are active, all 13 down-regulated genes are inactive. However, the second attractor state had only 11 of the down-regulated genes inactive, with Rv2986c (hupB) and Rv1158c being active. In this state only 19 of the up-regulated genes were active. Hence when the CRP transcription regulator (Rv3676) is active at input, two of the downregulated genes, namely, Rv1158c and Rv2986c, toggle between active and inactive states (Table S10).
In the dormancy module, sigma factor C (Rv2069) regulates the genes hspX, pfkB, Rv2030 and Rv2028c, which are upregulated during dormancy. In order to test the regulatory effect of SigC in achieving steady state during latency, we included SigC in the set of active input genes along with Rv3132c, Rv3133c, Rv0081 and Rv3676. The simulation with such an input state results in a two-state attractor cycle similar what is observed by activating Rv3132c, Rv3133c, Rv0081 and Rv367. Therefore, it appears that SigC may not be a key regulator of M. tuberculosis latency. Our Boolean modeling with dormancy module hence reveals key regulators namely Rv3132c, Rv3133c, Rv0081 and Rv3676 for activating and maintaining M. tuberculosis latency.
Discussion
One of the enigmatic features of tuberculosis is that only about 5–10% of the infected individuals develop active tuberculosis [58]. In rest of the cases, M. tuberculosis persists in a dormant or a non-replicative state in human tissues for a prolonged time with a potential to resume growth when conditions favor [59]. Understanding the persistent stage of M. tuberculosis has proved challenging and has profound implications in containing the disease as the current anti-tuberculosis drugs target only the cells that are actively growing [59]. Moreover, the micro-environment of the granulomas, where latent M. tuberculosis resides, is impermeable to the drugs. Inevitably, the systems level understanding of persistence to describe key players in this process, and their association with other proteins is not understood in great detail.
There are 84 differentially regulated genes which are common among many gene expression studies of various dormancy models of M. tuberculosis. Our analysis interestingly reveals that these 84 genes are coordinately regulated not only under dormancy-like conditions, but rather form a regulon-like structure. Among these, the 34 downregulated genes show high evolutionary conservation. We argue that the evolutionary conservation of these genes is due to their participation in basic cellular processes. In contrast, the dormancy signal in M. tuberculosis appears to be unique, as evidenced by far less conservation of the upregulated genes. It might therefore appear that different bacteria have adopted different mechanisms of entering dormancy, leading eventually to shutting down of the highly conserved basic metabolic processes.
Having identified two distinct clusters of genes, 50 upregulated and 34 downregulated, it is important to understand how these might be coordinately regulated. We used Boolean modeling to examine such transition to dormant phase in M. tuberculosis through the 84 differentially regulated genes. In Boolean modeling, an attractor state is the terminal vertex of the state transition graph, i.e. the state to which the system will converge given a certain input state. The attractor state can either be a single steady state, or a set of states through which the system cycles [60]. The latency model developed in this study attempts to understand the key regulators, which when turned on, results in an attractor state that mimics the latent phase of the pathogen. The latent state which is derived upon the activation of certain transcription factors is determined by comparing the states of the genes in the model with prior knowledge of their upregulation or repression during latency. Boolean modeling is therefore an attractive approach to address coordinated regulation among the 84 genes.
We observe that the model converges to an attractor cycle in which about 92% of the upregulated genes remain active when four of the transcription factors in the model namely, DosS (Rv3132c), DosR (Rv3133c), Rv0081 and CRP (Rv3676) are activated at the input. This suggests that these transcription factors are required to be expressed in order to maintain other members of the dormancy network in an active state. Several experimental studies have indicated involvement of a number of transcription factors in the regulation of initiation of the dormancy state [59], [61]. Therefore, the above mentioned transcription factors are not necessarily the only regulators of these processes. However, what the model indicates is that they are the minimum set of transcription factors required to obtain a system that exists in a dormancy-like state. While other factors are most likely involved in establishing the latent condition and adapting to external disturbances from the host macrophage, these four regulatory proteins appear to be the core set of regulators for initiating and maintaining signals for latency.
Two of the regulators predicted to be important for M. tuberculosis latency by Boolean modeling are DosR and DosS. Together they form a two-component regulatory system in which DosS is the sensory kinase and DosR is the corresponding response regulator [41]. These regulators have earlier been implicated as key mediators of latency in several experimental studies [59]. For example, rapid induction of dosR and dosS is observed upon reduced oxygen tension [31]. Similarly, targeted disruption of dosR revealed that most of the genes that are induced by hypoxia are regulated by DosR [32]. In addition, DosR and DosS are the members of the ‘dormancy regulon’ identified upon NO treatment [36]. Furthermore, significant load of bacilli and hypervirulence was observed in a SCID mouse model which was infected with M. tuberculosis with dosR deletion [62]. Activation of DosR is mediated by DosS, which has been established as a redox sensor with O2, NO and CO as modulatory ligands [43]. Hence, the two-component system DosR-DosS appears to play a major role in initiating and maintaining latency. Our results using Boolean modeling and network analysis reinforce the importance of these two proteins in establishing latency in M. tuberculosis.
Another regulatory protein predicted by Boolean modeling to be important for latency is Rv3676, a cAMP receptor family protein (CRP), which is a global regulator of number of pathways. Computational identification of possible regulatory sites for CRP has earlier revealed a number of genes, some of which are implicated in starvation and hypoxic conditions [63]. Therefore, the association of Rv3676 in latency appears significant. Although there are no reports on the direct involvement of this protein in latency, it will be interesting to study the regulatory role of Rv3676 in this context. Thus, we hypothesize that Rv3676, by virtue of being a global regulator, might influence cross talk among the genes involved in latency.
The fourth important transcriptional regulator suggested by Boolean modeling in M. tuberculosis latency is Rv0081, which is a regulatory protein from the ArsR/SmtB family [64]. Rv0081 is the first gene in the operonic locus Rv0081–Rv0088, which codes for the components of formate dehydrogenase complex. Rv0081 has been observed to be upregulated in multiple latency models, and is also shown to be regulated by DosR in the transcription regulatory network of M. tuberculosis [12]. Interestingly, the functional interaction network predicted by us in this study places Rv0082 at the intersection of upregulated and downregulated gene clusters. By being on an operon it might be assumed that Rv0081 controls the transcription of Rv0082. Thus, the predicted functional interaction network and Boolean modeling together suggest important roles of Rv0081 and Rv0082 in communicating latency signals between modules of upregulated and downregulated genes in M. tuberculosis latency.
The network based approach supplemented with Boolean modeling in elucidating crosstalk between the upregulated and downregulated genes leads us to propose a fascinating hypothesis of the latency process. We observe that DosR plays an important regulatory role in the dormancy switch. The role of DosR in dormancy is well documented in literature [31], [42]. The dormancy signals sensed by two-component sensor kinases, DosS, DosT and possibly by Rv0845 are transmitted to DosR which is a cognate response regulator. In the downstream, the signal is relayed through Rv0081 to the respiratory chain mediated by the Rv0082. The information flow from Rv0082 then triggers slowing down of ATP synthesis, leading eventually to significant slowing down of replication, transcription and translation processes. Thus, through an intricate communication signal, the basic cellular processes such as cell division and growth are shut down. Some of the hypotheses proposed in our work will obviously need to be tested experimentally.
Methods
Positive and Negative Interaction Pairs
The known interacting protein pairs were obtained by a combination of text mining methods and bi-directional blast against high confident protein interactions for E. coli listed in EcoCyc database [65]. Those obtained by text mining were retrieved from literature by the use of natural language processing methods (Goyal and Mande, unpublished results). Bi-directional BLAST was carried out for each pair listed in the EcoCyc database against the M. tuberculosis H37Rv genome sequence. Only those pairs were further considered which showed a score better than e−10 for both the proteins.
The hypothesized non-interacting data set was obtained by the methods described in [25]. Briefly, the protein pairs which are not colocalised in the same subcellular compartment were considered to be non-interacting. The protein localization was predicted using the SIGCLEAVE tool available at http://mobyle.pasteur.fr/cgi-bin/Mobyle Portal. The top scoring 809 proteins with predicted secretary signal sequence within the first 50 residues of the N-terminus were considered to be extracellular. There are 161 proteins which do not possess any known signal sequence along their entire length and were considered to be cytoplasmic. Such negative interacting protein pairs were generated by randomly pairing the predicted cytoplasmic and extracellular proteins.
Selection of the Genomes
The sequences of 763 bacterial genomes were downloaded from NCBI ftp site (ftp://ncbi.nih.gov/genomes/Bacteria). In the initial filter, bacteria with linear or multiple genomes were removed. This resulted in a set of 669 genomes for further analysis. Homologous genes of all the known open reading frames of M. tuberculosis were searched against these 669 genomes using BLASTp with e-value cutoff of e−04. For the species with complete genome sequences of more than one strain, the one which shared maximum number of ORFs with M. tuberculosis was chosen. This resulted in a list of 481 genomes for further consideration (Table S11).
Prediction Features
Phylogenetic Profile
BLAST with e-value cutoff of e−04 was used to obtain bit scores for the ORFs of M. tuberculosis against 481 selected genomes. The resulting profile was doubly normalized as in [25]. Pearson Correlation Coefficient (PCC) was calculated for each gene pair and used as a feature for training the SVM.
Intergenic Distance
For using minimum gene distance as a training feature, top 100 organisms sharing maximum number of ORFs with M. tuberculosis were considered. For each genome, distances (in base pairs) of transcriptional start sites between all the gene pairs were calculated in both clockwise and anticlockwise directions and the minimum of these was normalized by the total genome length. A similar profile was constructed for M. tuberculosis genome as well. For each gene pair of M. tuberculosis, the minimum distance in its genome and its orthologs in other genomes was considered as a feature vector.
Frequency of co-occurrence in the predicted operons
Operon predictions for 267 organisms were obtained from [25]. The frequency of co-occurrence of protein pairs as operonic across all the genomes was calculated.
Expression Correlations
Gene expression data for M. tuberculosis was downloaded from NCBI-Geo [6]. The expression values for the multiple trails were normalized. The conditions which had expression variance of more than 5 were not considered for the analysis. The expression ratio in 154 growth conditions for each gene was compiled and Pearson Correlation Coefficient was derived for each gene pair. The list of selected conditions is detailed in Table S12.
Protein Interactions Prediction
Support Vector Machine (SVM) tool LibSVM [66] was used for the prediction of genome-wide functional linkages. All possible gene pairs in M. tuberculosis carried the feature vector labels, namely correlation coefficient of the phylogenetic profile, minimum intergenic distance, frequency of co-occurrence in predicted operons and expression correlations. The machine was trained on positive and negative data sets using these data features. Since the number of expected interacting pairs was likely to be much lower than the non-interacting pairs, the ratio of negative interacting pairs and the positive interacting pairs was increased for each trial. Each test included five fold cross validation followed by calculations of sensitivity and specificity. The interactions were predicted with each of the model files obtained and the final network was selected based on sensitivity, specificity and the accuracy of prediction. The final predictions are available on our web server (http://www.nccs.res.in/MtbPPI/).
Network Analysis
Different topological parameters of the network that included degree exponent, clustering coefficient and diameter were calculated according to [67]. Centrality measures were calculated as in [28]. Clusters in the network were detected using the Infomap tool [68]. Functional annotations of M. tuberculosis proteins were derived from KEGG [69], TubercuList (http://genolist.pasteur.fr/TubercuList/) and Sanger (http://www.sanger.ac.uk/) databases. Clustering coefficient or degree for a pathway is considered to be high if the average clustering coefficient or the average degree for the proteins in the pathway was more than the average of all the proteins. The shortest paths were calculated using Dijkstra's algorithm [70]. Since there can be more than one shortest path for a pair of nodes, most probable paths were derived by considering most frequently occurring proteins in the shortest paths at each position.
Combined subnetwork was constructed by merging protein functional linkages and gene regulatory interactions [12]. If the interaction for two proteins is represented as both protein functional linkage as well as gene regulatory interaction, the latter was considered for the analysis.
Boolean modeling
Boolean models consist of components or nodes with logical rules or transfer functions governing their interactions. The state of each node in a typical boolean network is usually only a logical value, namely ‘ON’/‘1’ or ‘OFF’/‘0’. In the case of a gene regulatory network, the ‘ON’ state can be considered to represent a gene being activated or expressed and the ‘OFF’ state as a gene being inactivated or unexpressed. The state of a node at time t+1 is derived from states of all the nodes which interact with it at time t, by means of a logical function or rule [71]. The states are updated by means various updating schemes, the simplest one being synchronous updating where all the nodes are updated simultaneously at a given time point. This is in contrast to asynchronous updating, where a random node is selected at each time step and updated. The model dynamics comprise of various states the different components pass through to converge at an attractor state (fixed point or cycle). The attractor states of a model correspond to various biological states of the system under study.
Model of Mycobacterium tuberculosis dormancy network
A large-scale boolean network model was constructed based on a subset of genes identified as being differentially regulated and involved during the early dormancy state in M. tuberculosis. A number of criteria were considered when selecting components for the Boolean model. To begin with, genes annotated as transcription factors or regulatory elements which were found to be expressed in a minimum of 5 experimental latency conditions were identified. The expression correlation values for these transcription factors and their corresponding interactions were also obtained. To this list, the regulatory interactions determined in [12] were also added.
This list of genes was then narrowed down by discarding genes which were activated or inhibited by only a single transcription factor, with the interaction being insignificant. An interaction was considered to be insignificant if the expression correlation between the two members was between −0.3 and +0.3, and significant otherwise. Furthermore, if a given gene was regulated by a single transcription factor and that transcription factor itself had no regulatory elements acting on it, then that gene was removed as well.
The resulting subset consisted of genes or nodes, and 543 interactions or arcs, with 46 found to be annotated as regulatory elements such as transcription factors. Among the 304 genes, 26 belonged to the group of 50 genes identified as being upregulated during dormancy (Table S7). Similarly, 13 out of the 34 genes identified as being downregulated were present in the model.
The network was built by obtaining the interactions identified for each of the 304 genes from the protein functional linkages. As we had selected only transcription factors and the genes the regulated, the interactions were all directed, i.e. those originating from a transcription factor or regulatory element. We then derived the logical rule for each gene using the calculated expression correlation values for these interactions, and employing the following criteria:
For a given gene X,
If a set of transcription factors were positively correlated with X, then the correlation between them was detemernied. If the correlation was significant, then all of them were considered to be required for the activation or expression of X. A logical AND was used in this case.
If a set of transcription factors were positively and signifcantly correlated with X but insignificantly correlated with each other, then any one of them was sufficient for the activation of X. A logical OR was used in this case.
If a set of transcription factors were negatively correlated with X and positively correlated with each other, then all of them were considered to be required for the inhibition or inactivation of X. A logical NOT AND was used in this case.
If a set of transcription factors were negatively correlated with X and insiginificantly correlated with each other, then any one of them was considered to be sufficient for the inactivation of X. A logical NOT OR was used in this case.
The detailed list of logical rules is given in Table S13.
In our model, we do not differentiate between the expression and regulation of a gene but represent them in an equivalent manner. This biological simplification is necessary given that only expression correlation values were available to us. Furthermore, while significant expression correlation between the a gene and transcription factor might arise due to various forms of activation or regulation, we feel safe to represent it as direct in this case, given that the genes were obtained from the set determined to form a regulon-like structure. The model was simulated by setting as ON (i.e ‘1’) the individual transcription factors and updating all the genes in a synchronous manner. The simulations were done with the transcription factors being set as ON simultaneously, as well as one by one.
The boolean model was developed and simulated using the logical modelling, analysis and simulation software GINsim [72].
Supporting Information
Plot of ROC depicting the performance of SVM.
(TIF)
Plot denoting enrichment of Up and Downregulated dormancy genes to different conservation bins. Upregulated genes are less conserved than downregulated genes.
(TIF)
Expression correlation between genes coding for NDH-1 subunits and the genes coding for ATP synthase subunits. The plot indicates that NDH-1 genes and ATP synthase genes are correlated in their expression.
(TIF)
Differential Network Properties of the Modules of up and downregulated genes. Proteins in the downregulated module show high degree centrality (Figure S4(A), P<2.2e−16), high closeness centrality (Figure S4(B), P<2.2e−16) and high betweenness centrality (Figure S4(C), P<3.9e−09) compared to the proteins in the upregulated module. However, there is no apparent difference in their clustering coefficients (Figure S4(D), P<0.44).
(TIF)
TubercuList pathway map of the proteins belonging to up and down-regulated modules. Genes belonging to Information pathway are significantly downregulated during dormancy.
(TIF)
Predicted Protein functional linkages of M. tuberculosis.
(TXT)
Comparison of predicted interactions with other available interactions. There is about 30% overlap between predicted interactions and previous reports.
(DOC)
List of high centrality proteins in the network.
(TXT)
List of communities identified in the interaction map using Infomap community detection tool.
(TXT)
List of publications related to dormancy models used in this study and the number of up and down-regulated genes in each model.
(DOC)
Comparison of different dormancy models in terms of number of genes up and downregulated in each.
(DOC)
List of up and downregulated genes during dormancy and their functions.
(TXT)
List of proteins in the Dormancy Core.
(TXT)
List of proteins possibly mediating the dormancy signals between up and down-regulated modules.
(TXT)
Genes which are active at the logical steady state reached by the system when three regulatory elements are ON at input (Rv0081, Rv3132c and Rv3133c) and when four regulatory elements are ON at input (Rv0081, Rv1343c, Rv3132c and Rv3133c).
(DOC)
List of selected genomes for constructing phylogenetic profile.
(TXT)
List of microarray conditions used for calculating expression correlations.
(TXT)
Logical rules derived from expression data for each of the nodes in the simulated model.
(DOC)
Acknowledgments
We thank SUN Centre of Excellence in Medical Bioinformatics, Centre for DNA Fingerprinting and Diagnostics (CDFD), for access to computational facilities. We thank Kshama Goyal for help in the text mined protein interaction pairs of M. tuberculosis.
Footnotes
Competing Interests: The authors have read the journal's policy and have the following conflicts. Although three of the authors work for Tata Consultancy Services, this does not alter the authors' adherence to all the PLoS ONE policies on sharing data and materials.
Funding: The study was funded by the Department of Biotechnology under the Centre for Excellence Grant. SRH is a University Grants Commission Senior Research Fellow. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.WHO Report on Multidrug and extensivelydrug-resistant TB (M/XDR-TB) 2010. 2010: Global Report on Surveillance and Response, World Health Organization.
- 2.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Camus JC, Pryor MJ, Médigue C, Cole ST. Re-annotation of the genome sequence of Mycobacterium tuberculosis H37Rv. Microbiology. 2002;148:2967–2973. doi: 10.1099/00221287-148-10-2967. [DOI] [PubMed] [Google Scholar]
- 4.Smith I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev. 2003;16:463–96. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hett EC, Rubin EJ. Bacterial growth and cell division: a mycobacterial perspective. Microbiol Mol Biol Rev. 2008;72:126–56. doi: 10.1128/MMBR.00028-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–90. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sassetti CM, Boyd DH, Rubin EJ. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol. 2003;48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 8.Sassetti CM, Rubin EJ. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A. 2003;100:12989–94. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rengarajan J, Bloom BR, Rubin EJ. Genome-wide requirements for Mycobacterium tuberculosis adaptation and survival in macrophages. Proc Natl Acad Sci U S A. 2005;102:8327–32. doi: 10.1073/pnas.0503272102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mawuenyega KG, Forst CV, Dobos KM, Belisle JT, Chen J, et al. Mycobacterium tuberculosis functional network analysis by global subcellular protein profiling. Mol Biol Cell. 2005;16:396–404. doi: 10.1091/mbc.E04-04-0329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strong M, Graeber TG, Beeby M, Pellegrini M, Thompson MJ, et al. Visualization and interpretation of protein networks in Mycobacterium tuberculosis based on hierarchical clustering of genome-wide functional linkage maps. Nucleic Acids Res. 2003;31:7099–109. doi: 10.1093/nar/gkg924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balázsi G, Heath AP, Shi L, Gennaro ML. The temporal response of the Mycobacterium tuberculosis gene regulatory network during growth arrest. Mol Syst Biol. 2008;4:225. doi: 10.1038/msb.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raman K, Rajagopalan P, Chandra N. Flux balance analysis of mycolic acid pathway: targets for anti-tubercular drugs. PLoS Comput Biol. 2005;1:e46. doi: 10.1371/journal.pcbi.0010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cui T, Zhang L, Wang X, He ZG. Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Genomics. 2009;10:118. doi: 10.1186/1471-2164-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. STRING 8—a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–6. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc Natl Acad Sci USA. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enault F, Suhre K, Abergel C, Poirot O, Claverie JM. Annotation of bacterial genomes using improved phylogenomic profiles. Bioinformatics. 2003;19:i105–i107. doi: 10.1093/bioinformatics/btg1013. [DOI] [PubMed] [Google Scholar]
- 18.Korbel JO, Jensen LJ, von Mering C, Bork P. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- 19.Dandekar T, Snel B, Huynen M, Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 20.Overbeek R, Fonstein M, D'Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci USA. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kauffman SA. Metabolic stability and epigenesis in randomly connected nets. J Theoret Biol. 1969;22:437–467. doi: 10.1016/0022-5193(69)90015-0. [DOI] [PubMed] [Google Scholar]
- 22.Saez-Rodriguez J, Simeoni L, Lindquist JA, Hemenway R, Bommhardt U, et al. A Logical Model Provides Insights into T Cell Receptor Signaling. PLoS Comput Biol. 2007;3:e163. doi: 10.1371/journal.pcbi.0030163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Davidich MI, Bornholdt S. Boolean Network Model Predicts Cell Cycle Sequence of Fission Yeast. PLoS ONE. 2008;3:e1672. doi: 10.1371/journal.pone.0001672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacLean D, Studholme DJ. A Boolean Model of the Pseudomonas syringae hrp Regulon Predicts a Tightly Regulated System. PLoS ONE. 2010;5:e9101. doi: 10.1371/journal.pone.0009101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yellaboina S, Goyal K, Mande SC. Inferring genome-wide functional linkages in E. coli by combining improved genome context methods: Comparison with high-throughput experimental data. Genome Res. 2007;17:527–535. doi: 10.1101/gr.5900607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 27.Marshall E. Getting the Noise Out of Gene Arrays. Science. 2004;306:630–631. doi: 10.1126/science.306.5696.630. [DOI] [PubMed] [Google Scholar]
- 28.Manimaran P, Hegde SR, Mande SC. Prediction of conditional gene essentiality through graph theoretical analysis of genome-wide functional linkages. Mol Biosyst. 2009;5:1936–1942. doi: 10.1039/B905264j. [DOI] [PubMed] [Google Scholar]
- 29.Rives AW, Galitski T. Modular organization of cellular networks. Proc Natl Acad Sci U S A. 2003;100:1128–1133. doi: 10.1073/pnas.0237338100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Banu S, Honoré N, Saint-Joanis B, Philpott D, Prévost MC, et al. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol. 2002;44:9–19. doi: 10.1046/j.1365-2958.2002.02813.x. [DOI] [PubMed] [Google Scholar]
- 31.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–7539. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park HD, Guinn KM, Harrell MI, Liao R, Voskuil MI, et al. Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol Microbiol. 2003;48:833–843. doi: 10.1046/j.1365-2958.2003.03474.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bacon J, James BW, Wernisch L, Williams A, Morley KA, et al. The influence of reduced oxygen availability on pathogenicity and gene expression in Mycobacterium tuberculosis. Tuberculosis. 2004;84:205–217. doi: 10.1016/j.tube.2003.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Muttucumaru DG, Roberts G, Hinds J, Stabler RA, Parish T. Gene expression profile of Mycobacterium tuberculosis in a non-replicating state. Tuberculosis. 2004;84:239–246. doi: 10.1016/j.tube.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 35.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Voskuil MI, Schnappinger D, Visconti KC, Harrell MI, Dolganov GM, et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 38.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan, et al. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, et al. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med. 2004;200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kumar P, Joshi DC, Akif M, Akhter Y, Hasnain SE, et al. Mapping conformational transitions in cyclic AMP receptor protein: crystal structure and normal-mode analysis of Mycobacterium tuberculosis apo-cAMP receptor protein. Biophys J. 2010;98:305–314. doi: 10.1016/j.bpj.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts DM, Liao RP, Wisedchaisri G, Hol WG, Sherman DR. Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J Biol Chem. 2004;279:23082–23087. doi: 10.1074/jbc.M401230200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tyagi JS, Sharma D. Signal transduction systems of mycobacteria with special reference to M. tuberculosis. Current Science. 2004;86:93–102. [Google Scholar]
- 43.Kumar A, Toledo JC, Patel RP, Lancaster JR, Jr, Steyn AJ. Mycobacterium tuberculosis DosS is a redox sensor and DosT is a hypoxia sensor. Proc Natl Acad Sci U S A. 2007;104:11568–11573. doi: 10.1073/pnas.0705054104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zambrano MM, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 46.Boshoff HI, Barry CE., 3rd Tuberculosis - metabolism and respiration in the absence of growth. Nat Rev Microbiol. 2005;3:70–80. doi: 10.1038/nrmicro1065. [DOI] [PubMed] [Google Scholar]
- 47.Höner zu Bentrup K, Russell DG. Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 2001;9:597–605. doi: 10.1016/s0966-842x(01)02238-7. [DOI] [PubMed] [Google Scholar]
- 48.Hu Y, Movahedzadeh F, Stoker NG, Coates AR. Deletion of the Mycobacterium tuberculosis alpha-crystallin-like hspX gene causes increased bacterial growth in vivo. Infect Immun. 2006;74:861–868. doi: 10.1128/IAI.74.2.861-868.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gomez JE, Bishai WR. whmD is an essential mycobacterial gene required for proper septation and cell division. Proc Natl Acad Sci U S A. 2000;97:8554–8559. doi: 10.1073/pnas.140225297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim TH, Park JS, Kim HJ, Kim Y, Kim P, et al. The whcE gene of Corynebacterium glutamicum is important for survival following heat and oxidative stress. Biochem Biophys Res Commun. 2005;337:757–764. doi: 10.1016/j.bbrc.2005.09.115. [DOI] [PubMed] [Google Scholar]
- 51.Narayan A, Sachdeva P, Sharma K, Saini AK, Tyagi AK, et al. Serine threonine protein kinases of mycobacterial genus: phylogeny to function. Physiol Genomics. 2007;29:66–75. doi: 10.1152/physiolgenomics.00221.2006. [DOI] [PubMed] [Google Scholar]
- 52.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, et al. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997;276:1420–2. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 53.West NP, Wozniak TM, Valenzuela J, Feng CG, Sher A, et al. Immunological diversity within a family of cutinase-like proteins of Mycobacterium tuberculosis. . Vaccine. 2008;26:3853–3859. doi: 10.1016/j.vaccine.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morth JP, Feng V, Perry LJ, Svergun DI, Tucker PA. The crystal and solution structure of a putative transcriptional antiterminator from Mycobacterium tuberculosis. Structure. 2004;12:1595–1605. doi: 10.1016/j.str.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 55.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, et al. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–684. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukamolova GV, Turapov OA, Kazarian K, Telkov M, Kaprelyants AS, et al. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol Microbiol. 2002;46:611–621. doi: 10.1046/j.1365-2958.2002.03183.x. [DOI] [PubMed] [Google Scholar]
- 57.Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 58.Bloom BR, Murray CJ. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 59.Stewart GR, Robertson BD, Young DB. Tuberculosis: a problem with persistence. Nat Rev Microbiol. 2003;1:97–105. doi: 10.1038/nrmicro749. [DOI] [PubMed] [Google Scholar]
- 60.Albert I, Thakar J, Li S, Zhang R, Albert R. Boolean network simulations for life scientists. Source Code Biol Med. 2008;3:16. doi: 10.1186/1751-0473-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chao MC, Rubin EJ. Letting sleeping dos lie: does dormancy play a role in tuberculosis? Annu Rev Microbiol. 2010;64:293–311. doi: 10.1146/annurev.micro.112408.134043. [DOI] [PubMed] [Google Scholar]
- 62.Parish T, Smith DA, Kendall S, Casali N, Bancroft GJ, et al. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect Immun. 2003;3:1134–40. doi: 10.1128/IAI.71.3.1134-1140.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bai G, McCue LA, McDonough KA. Characterization of Mycobacterium tuberculosis Rv3676 (CRPMt), a cyclic AMP receptor protein-like DNA binding protein. J Bacteriol. 2005;187:7795–804. doi: 10.1128/JB.187.22.7795-7804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Campbell DR, Chapman KE, Waldron KJ, Tottey S, Kendall S, et al. Mycobacterial cells have dual nickel-cobalt sensors: sequence relationships and metal sites of metal-responsive repressors are not congruent. J Biol Chem. 2007;282:32298–310. doi: 10.1074/jbc.M703451200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chih-Chung C, Chih-Jen L. 2001. LIBSVM : a library for support vector machines,. Software available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- 67.Dorogovtsev SN, Mendes JF. Evolution of Networks: From Biological Nets to Internet and WWW. Oxford: Oxford University Press; 2003. [Google Scholar]
- 68.Rosvall M, Bergstrom CT. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 2008;105:1118–1123. doi: 10.1073/pnas.0706851105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dijkstra EW. Numerische Math. 1959;1:269–271. [Google Scholar]
- 71.de Jong H. Modeling and Simulation of Genetic Regulatory Systems: A Literature Review. J Comput Biol. 2004;9:67–103. doi: 10.1089/10665270252833208. [DOI] [PubMed] [Google Scholar]
- 72.Gonzalez AG, Naldi A, Sánchez L, Thieffry D, Chaouiya C. GINsim: a software suite for the qualitative modelling, simulation and analysis of regulatory networks. Biosystems. 2006;84:91–100. doi: 10.1016/j.biosystems.2005.10.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Plot of ROC depicting the performance of SVM.
(TIF)
Plot denoting enrichment of Up and Downregulated dormancy genes to different conservation bins. Upregulated genes are less conserved than downregulated genes.
(TIF)
Expression correlation between genes coding for NDH-1 subunits and the genes coding for ATP synthase subunits. The plot indicates that NDH-1 genes and ATP synthase genes are correlated in their expression.
(TIF)
Differential Network Properties of the Modules of up and downregulated genes. Proteins in the downregulated module show high degree centrality (Figure S4(A), P<2.2e−16), high closeness centrality (Figure S4(B), P<2.2e−16) and high betweenness centrality (Figure S4(C), P<3.9e−09) compared to the proteins in the upregulated module. However, there is no apparent difference in their clustering coefficients (Figure S4(D), P<0.44).
(TIF)
TubercuList pathway map of the proteins belonging to up and down-regulated modules. Genes belonging to Information pathway are significantly downregulated during dormancy.
(TIF)
Predicted Protein functional linkages of M. tuberculosis.
(TXT)
Comparison of predicted interactions with other available interactions. There is about 30% overlap between predicted interactions and previous reports.
(DOC)
List of high centrality proteins in the network.
(TXT)
List of communities identified in the interaction map using Infomap community detection tool.
(TXT)
List of publications related to dormancy models used in this study and the number of up and down-regulated genes in each model.
(DOC)
Comparison of different dormancy models in terms of number of genes up and downregulated in each.
(DOC)
List of up and downregulated genes during dormancy and their functions.
(TXT)
List of proteins in the Dormancy Core.
(TXT)
List of proteins possibly mediating the dormancy signals between up and down-regulated modules.
(TXT)
Genes which are active at the logical steady state reached by the system when three regulatory elements are ON at input (Rv0081, Rv3132c and Rv3133c) and when four regulatory elements are ON at input (Rv0081, Rv1343c, Rv3132c and Rv3133c).
(DOC)
List of selected genomes for constructing phylogenetic profile.
(TXT)
List of microarray conditions used for calculating expression correlations.
(TXT)
Logical rules derived from expression data for each of the nodes in the simulated model.
(DOC)