Abstract
Different lines of evidence point to the resettlement of much of western and central Europe by populations from the Franco-Cantabrian region during the Late Glacial and Postglacial periods. In this context, the study of the genetic diversity of contemporary Basques, a population located at the epicenter of the Franco-Cantabrian region, is particularly useful because they speak a non-Indo-European language that is considered to be a linguistic isolate. In contrast with genome-wide analysis and Y chromosome data, where the problem of poor time estimates remains, a new timescale has been established for the human mtDNA and makes this genome the most informative marker for studying European prehistory. Here, we aim to increase knowledge of the origins of the Basque people and, more generally, of the role of the Franco-Cantabrian refuge in the postglacial repopulation of Europe. We thus characterize the maternal ancestry of 908 Basque and non-Basque individuals from the Basque Country and immediate adjacent regions and, by sequencing 420 complete mtDNA genomes, we focused on haplogroup H. We identified six mtDNA haplogroups, H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1, which are autochthonous to the Franco-Cantabrian region and, more specifically, to Basque-speaking populations. We detected signals of the expansion of these haplogroups at ∼4,000 years before present (YBP) and estimated their separation from the pan-European gene pool at ∼8,000 YBP, antedating the Indo-European arrival to the region. Our results clearly support the hypothesis of a partial genetic continuity of contemporary Basques with the preceding Paleolithic/Mesolithic settlers of their homeland.
Main Text
The settlement history of Europe has been punctuated by several major episodes over the past 50,000 years. These include the first arrival of modern humans from Africa during the Upper Paleolithic, the Late Glacial repeopling of Europe from southern refugia, the Postglacial recolonization of deserted areas after the end of the Younger Dryas, the farming-related population expansion of Near Easterners into Europe during the Neolithic, and the small-scale migrations along continent-wide economic exchange networks beginning from the Copper Age onward.1–3 Furthermore, a number of Urheimat hypotheses suggest the proto-Indo-European culture completed its expansion to Europe between the fifth and first millennia BCE, leaving the Basque language the only remnant of the antedating culture in Western Europe.4 The way in which these events have left their genetic footprints on the current European gene pool has been the focus of intense genetic research over the last decades. At the genome-wide scale, European populations appear rather homogeneous, and there are small existing differences that broadly correlate with geography.5,6 The most noticeable pattern uncovered was a clear distinction between northern and southern Europeans.5,7–9 Despite the contribution of these genome-wide data sets in evaluating the degree of population structure and admixture, they are of more limited use than uniparentally inherited genomes for the detection of subtle population migrations, the timing of such episodes, and gender-biased events. Indeed, the study of Y chromosome and mitochondrial DNA (mtDNA) variation has extensively improved our understanding of the different spatial and temporal sources that contributed to the genetic structure of modern Europeans.3 For example, recent Y chromosome data suggest that most present-day European Y chromosomes were contributed by the Near Eastern Neolithic package,10 although alternative, more complex scenarios have been proposed.11,12 Conversely, founder analyses of mtDNAs suggest that less than 15% of European haplogroups can be traced back to the Neolithic expansion.3
The Late Glacial and Postglacial reoccupation of Europe from refugial areas has played a major role in shaping the gene pool of modern European populations and indicates the critical role of climate change in European demographic history. Several, independent lines of evidence point to the resettlement of much of western and central Europe after the Ice Age from the Franco-Cantabrian region, an area that includes the southern half of France and the northern strip of Spain looking at the Bay of Biscay. On the basis of archaeological data, the Franco-Cantabrian refuge seems to have been the most densely populated region of Europe throughout all the Upper Paleolithic;1 it has a strong signal of both range and size expansions at ∼15,000 years before present (YBP) as the Magdalenian industry spread from the southwest into the rest of Europe.2 Genome-wide studies have revealed that the clinal distributions of some parameters are compatible with a prehistoric population expansion from southern to northern Europe,5,9 but no indication of the time-depth of the underlying events has been provided. Here again, phylogeographic studies based on mtDNA variation have, by and large, supported the Franco-Cantabrian refuge scenario that all mtDNA haplogroups expanded from this region and present Late Glacial or Postglacial expansion times, that is H5 at ∼13,000 YBP and V, H1, and H3 at ∼11,000 YBP.3,13–16 In this context, the study of the Basques, unquestionably the most emblematic population inhabiting the Franco-Cantabrian region, can be particularly informative.
Above all, Basques are characterized by their unique language, Euskara, a non-Indo-European isolate that is not considered a member of any extant language family. The present-day Basque population is located on both sides of the western Pyrenees, and Basque is currently spoken by around 25% of the population. The corresponding area is divided into seven provinces, four in Spain and three in France (Figure 1). Toponymic data suggest a larger territory was previously occupied by ancient Basques, stretching from the Garonne River in the north to the Ebro River in the south. The linguistic isolation of the Basques, together with their outlier position with respect to a large set of classical genetic markers,17–19 has strengthened the view of Basques as being a genetic isolate with the greatest degree of genetic continuity with the early European, Paleolithic hunter-gatherers. However, this popular vision has been challenged by a number of recent, differing studies.
Figure 1.
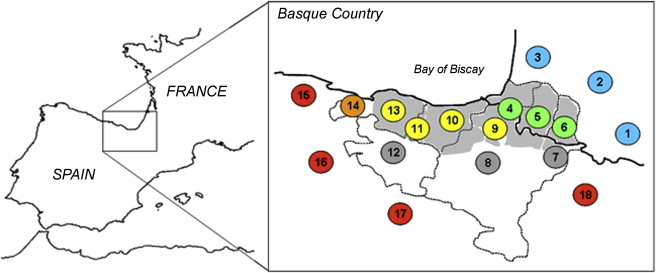
Geographic Location of the Studied Populations
The sampling strategy adopted was centered on the Basque-speaking provinces and included the immediately adjacent non-Basque-speaking regions. Grey shaded areas correspond to regions where the Basque language is currently spoken. Individuals were sampled in 18 different regions that, on the basis of geographic location, linguistic affiliation and surnames, were grouped into six major zone categories. These include Zone A (in blue), French-speaking regions that historically spoke Gascon (Bigorre, Bearn, Chalosse); Zone B (in green), Basque-speaking regions located in France (Lapurdi/Baztan, Lapurdi Nafarroa, Zuberoa); Zone C (in gray), regions where Basque was spoken up to the last century (Roncal and Salazar valleys, central western Nafarroa, Araba); Zone D (in yellow), Basque-speaking regions located in Spain (northwestern Nafarroa, Gipuzkoa, southwestern Gipuzkoa, Bizkaia); Zone E (in orange), a Spanish-speaking region in the Basque country (western Bizkaia); and Zone F (in red), four Spanish-speaking regions (Cantabria, northern Burgos, La Rioja, northern Aragon). Details on sample size per region are provided in Table S1.
At the genome-wide scale, Basques are close to other Europeans,20–22 and the genetic component dominating European populations is observed at the highest proportions among Basques.20,23 This quantitative distinctiveness of Basques has been nevertheless contested by another study.24 From a Y chromosome perspective, Basques are differentiated from other populations, with the exception of the neighboring Gascons,25,26 but the Mesolithic status of Basques has been challenged by a more recent study.10 From an mtDNA perspective, conflicting patterns have also emerged. All major mtDNA haplogroups observed among contemporary Basques are shared with the general European maternal pool.27–33 Although haplogroup H has been shown to be the most dominant among Basques,13,14 very different frequencies of other minor haplogroups have been reported. Indeed, several rare mtDNA variants within H2a5, U8a, J1c1, and J2a have been suggested to be autochthonous to some Basque groups but are absent in others.31,34,35 Likewise, distances between Basque populations have been found to be larger than between Basques and other Iberian groups, pointing to strong local isolation and limited gene flow between Basques and surrounding populations.32 This plethora of conflicting results, probably attesting to the complex demographic matrilineal history of Basques, have led to differing opinions regarding the putative genetic continuity between present-day Basques and Paleolithic Europeans27–29,31,34,35 and, more generally, with respect to the role played by the Franco-Cantabrian refuge in the postglacial recolonization of Europe.13,32
We hypothesized that part of the uncertainty regarding the origin of the Basques might be attributed to the unsatisfying level of mtDNA resolution achieved so far. Indeed, most studies of the Basque population have been based solely on the sequencing of the hypervariable segment (HVS)-I or on complete mtDNA sequences of targeted, low-frequency haplogroups in few individuals.34,35 Furthermore, because the time-depth of the major European haplogroups is in the range of 10,000–50,000 YBP,3 the study of their variation is less relevant to the case of the Basques and the spread of Near Eastern farmers and Indo-European languages, which is considered to have occurred 3,000–7,000 YBP.4 It is possible, though, that a distinguishing feature will exist at the level of complete mtDNA sequences of specific haplogroups. Here, we aimed to increase knowledge of the origins of the Basque people, the putative genetic continuity between present-day Basques, and pre-Indo-European Neolithic peoples and, more generally, the role of the Franco-Cantabrian refuge in the Postglacial repopulation of Europe. To this end, we characterized the maternal ancestry of Basque- and non-Basque-speaking populations from the Franco-Cantabrian region and, by sequencing complete mtDNA genomes, we focused on haplogroup H—the most dominant haplogroup in Europeans and in Basques in particular (∼45%).13,14,34,36 In contrast to genome-wide analysis and Y chromosome data where the problem of poor time estimates remains, a new timescale has been recently established for the mtDNA,37 making this genome at present the most informative marker for studying European prehistory.
We sampled a large collection of 908 individuals from 18 different geographic locations (Figure 1 and Table S1, available online), from whom we collected detailed ethnological and geolinguistic data. Our sampling strategy was centered on the seven Basque-speaking provinces in France and Spain, here referred to as the “Basque Country,” and included the immediately adjacent non-Basque-speaking regions. Inclusion criteria comprised an autochthonous origin confirmed by surnames and birthplaces of each individual, their parents and grandparents. Furthermore, we favored the inclusion of participants of more than 60 years old to increase the probability of a deep local ancestry. On the basis of geographic location, linguistic affiliation, and surnames, individuals were attributed to six major zone categories: French-speaking regions that historically spoke Gascon (Zone A), Basque-speaking regions located in France (Zone B), regions where Basque was spoken up to the last century (Zone C), Basque-speaking regions located in Spain (Zone D), a Spanish-speaking region in the Basque Country (Zone E), and Spanish-speaking regions (Zone F) (Figure 1 and Table S1). It is worthy of mention that in the Spanish-speaking regions E and F, Basque languages have most likely been spoken up to medieval times (Zuazo38 and references therein). For all individuals, written informed consent was obtained, and Ethics Committees at Institut Pasteur (n° RBM 2005.012) and Université Michel de Montaigne Bordeaux 3 approved all procedures.
To characterize the maternal ancestry of all individuals, we first typed a panel of 22 coding region SNPs that are diagnostic of the major haplogroups of the mtDNA phylogeny and sequenced the entire control region, including both HVS-I and HVS-II.39 Variable positions throughout the control region were determined from positions 16,024–16,569 and 1–573. The 22 coding region SNPs were analyzed on all samples by SNaPshot typing with the multiplex GenoCoRe22.40 We used this data set to assign each of the mtDNA genomes to its respective nodal haplogroup. The majority of samples, 420 of the 908 (46.3%), belonged to haplogroup H (Table 1 and Table S2), as expected in populations of western European descent.13,14,34,36 All but three of the remaining samples belonged to haplogroups that characterize the western Eurasian mtDNA landscape, including U5, J1, J2, V, and T (Table S2). One individual from the Basque-speaking region of Guipuzcoa and one from the Spanish-speaking region of Burgos belonged to the sub-Saharan African haplogroups L2 and L3h, supporting previous observations of low frequencies of sub-Saharan African haplogroups in northern Iberia.29,30,32 In addition, one individual originating from the French region of Bigorre belonged to haplogroup C, which mostly characterizes East Asian populations.41,42
Table 1.
Haplogroup H Variation in the Basque Country and Adjacent Regions
|
Geographic Zones |
Total | ||||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| Sample size | 164 | 193 | 175 | 231 | 21 | 124 | 908 |
| H samples | 61 | 82 | 78 | 125 | 14 | 60 | 420 |
| H frequency | 37.2 | 42.5 | 44.6 | 54.1 | 66.7 | 48.4 | 46.3 |
| Autochthonous H Haplogroupsa | |||||||
| H1av1 | 0.0 | 1.0 | 0.6 | 2.6 | 0.0 | 0.8 | 1.1 |
| H1av1a | 0.0 | 1.0 | 0.0 | 2.2 | 0.0 | 0.0 | 0.8 |
| H1e1a1 | 3.1 | 3.1 | 0.6 | 0.0 | 0.0 | 0.0 | 1.3 |
| H1j1 | 0.0 | 0.5 | 4.0 | 2.6 | 9.5 | 0.8 | 1.9 |
| H1j1a | 0.6 | 0.5 | 0.6 | 0.9 | 0.0 | 1.6 | 0.8 |
| H1j1a1 | 0.0 | 0.0 | 0.0 | 1.3 | 0.0 | 0.0 | 0.3 |
| H1j1a2 | 1.2 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.6 |
| H1j1b | 0.0 | 2.6 | 0.6 | 3.9 | 0.0 | 0.0 | 1.7 |
| H1j1c | 0.0 | 0.0 | 0.0 | 2.2 | 0.0 | 0.0 | 0.6 |
| H1t1 | 0.0 | 0.0 | 1.1 | 0.4 | 0.0 | 0.0 | 0.3 |
| H1t1a | 1.8 | 3.6 | 2.9 | 3.0 | 0.0 | 1.6 | 2.6 |
| H1t1a1 | 0.0 | 0.0 | 0.0 | 3.0 | 0.0 | 0.0 | 0.8 |
| H2a5a1 | 2.4 | 0.5 | 0.0 | 2.2 | 0.0 | 0.0 | 1.1 |
| H2a5a1a | 0.6 | 1.0 | 0.6 | 3.5 | 0.0 | 0.0 | 1.3 |
| H3c2a | 0.6 | 1.6 | 2.3 | 1.3 | 0.0 | 0.0 | 1.2 |
| H3c2a1 | 0.0 | 1.6 | 0.0 | 0.0 | 0.0 | 0.0 | 0.3 |
| Total frequency of autochthonous haplogroups | 10.4 | 18.7 | 13.1 | 29.0 | 9.5 | 4.8 | 16.6 |
| Proportion of autochthonous H variation | 27.9 | 43.9 | 29.5 | 53.6 | 14.3 | 10.0 | 36.0 |
Frequency of autochthonous haplogroups in the different geographic zones of the Basque Country and adjacent regions.
To gain insight into the haplogroup that accounts for most of the maternal variation of populations from the Basque Country and immediately adjacent regions, we sequenced the complete mtDNA genomes of all individuals (i.e., 420 samples) belonging to haplogroup H. We then integrated these data into the global haplogroup H phylogeny, inferred from a large data set of complete mtDNA genomes from individuals of pan-European origin, and focused on the phylogenies of the specific haplogroups characterizing our sample from the Franco-Cantabrian region (Figures S1–S6). These phylogenies were reconstructed by evaluating all previously published and novel haplogroup H complete mtDNA sequences in order to identify the most parsimonious solution and aided by the mtPhyl software. The 420 samples could be assigned to 129 different haplogroup H sub-haplogroups, with 59 appearing as singletons (Tables S3 and S4). Remarkably, although three different haplotypes were required to support the labeling of a new haplogroup, only nine (2.1%) samples could not be related to either a previously or a herein described haplogroup and were therefore labeled as H. Numerous haplotypes matched previously reported complete mtDNA sequences sampled throughout the European continent. However, our analyses revealed six dominant haplogroups that display an internal structure within the Basque Country and bordering regions and explain 17% of all mtDNA variation and 36% of haplogroup H variation of our data set (Table 1). These haplogroups, in descending order of frequency, included H1j1, H1t1, H2a5a1, H1av1, H3c2a and H1e1a1 (Figure 2). Only H1j1, H1t1 and H2a5a1 have been previously labeled,32,34,43,44 and the remaining three are baptized herein for the first time. Of these haplogroups, only H2a5a1 has been previously suggested to be autochthonous to the Basque Country.32,34,43,44 Most importantly, these six haplogroups were found to be virtually absent in the comparative data set of more than 7,000 complete mtDNA genomes (D.M.B., unpublished data) from populations of predominantly western European origin.
Figure 2.
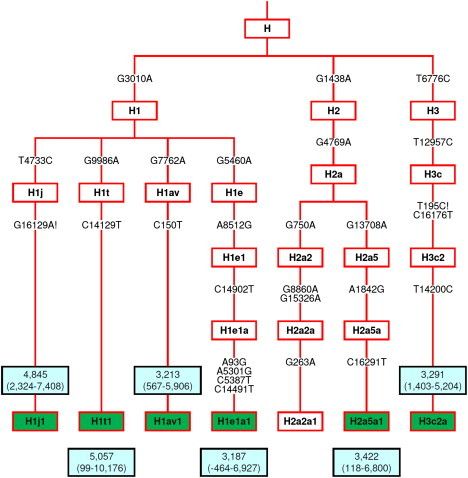
Phylogenetic Tree of the Six Autochthonous Haplogroups
The phylogeny represents the roots of H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1 and their coalescence age estimates based on complete mtDNA sequences. Mutations shown on the branches indicate the actual descendant nucleotide state relative to the root of haplogroup H. Detailed phylogenies of each of the six autochthonous haplogroups are presented in Figures S1–S6.
The most dominant H haplogroup of the Franco-Cantabrian region is H1j1, which accounts for 12.4% of haplogroup H variation of our data set (Figure S1). This haplogroup has only been observed in four previously reported individuals of Basque origin (D.M.B., unpublished data).43 Likewise, haplogroup H1t1, which explains 8.1% of haplogroup H variation, displays its highest frequencies in the Basque-speaking populations from France and Spain, and has been previously observed only among Basques (Figure S2).32 Haplogroup H2a5 accounted for 5.2% of haplogroup H variation. Here, we have further resolved the phylogeny of H2a5, now termed H2a5a1, and all previously reported individuals carrying this haplogroup34,44 belong indeed to the herein refined H2a5a1a (Figure S3). Haplogroup H1av1, which accounts for 4% of H haplogroup variation, has been found so far only in our data set and its distribution is restricted to Basque-speaking populations and immediately adjacent Spanish-speaking populations (Figure S4). Finally, haplogroups H3c2a (Figure S5) and H1e1a1 (Figure S6), which explain 3.3% and 2.9% of haplogroup H variation, respectively, have each been observed only once in a composite sample from Spain.13 Overall, the exclusive geographic distribution of H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1 among Basque-speaking populations and immediately adjacent populations and their absence from a large data set of populations of western European-descent strongly suggest that these haplogroups are indeed autochthonous to the region.
Furthermore, significant matrilineal structure within this geographic region was observed in our database. The frequency of haplogroup H per se was not significantly different between the six geographic zones (Table 1), indicating that the mere comparison of low-resolved haplogroups is not informative enough to detect fine population structure. However, the proportions of the autochthonous haplogroups varied dramatically, and significantly, between Basque- and non-Basque-speaking regions (χ2 test, p < 0.01). These six haplogroups accounted cumulatively for 44%–54% of the total haplogroup H variation among Basque-speaking populations from France and Spain (zones B and D), whereas only for 10%–14% among Spanish-speaking regions (zones E and F) (Table 1). Intermediate frequencies of ∼28% were observed in French-speaking regions and in Spanish regions that historically spoke Basque (zones A and C). The somewhat closer affinity between French speakers (i.e., Gascons) and Basque speakers than between Spanish speakers and Basque speakers is concordant with previous observations based on the Y chromosome.25,26
We next estimated the time-depth of these autochthonous haplogroups within the H phylogeny, by means of the ρ statistic (mean sequence divergence from the inferred ancestral haplotype) by using a mutation rate estimate for the complete mtDNA sequence and a correction for purifying selection.37 All autochthonous H haplogroups coalesce at recent times ranging from 3,187 to 5,057 YBP with an average of 3,836 YBP (Table 2). These time estimates attest to a rather homogeneous signal of expansion for these autochthonous haplogroups, which overlaps with the late Neolithic period in some parts of Europe as well as with the subsequent Copper and Bronze Ages. To further expand our understanding of the temporal origin of these haplogroups, we estimated the ages at which they split from their phylogenetically closest European relatives (Figures S1–S6), not accounting for other possible branches descending from the same root. We found their separation times to range between 5,854 and 14,011 YBP (Table 2). Our findings that the splitting ages of these autochthonous haplogroups precede the putative arrival of the Near Eastern farmers, and possibly the Indo-European languages, to one of the westernmost parts of Europe strongly attest to a genetic continuity of contemporary Basques with the earlier Paleolithic and/or Mesolithic peoples of the region.
Table 2.
Time Estimates of the Six Autochthonous Haplogroups
| Haplogroup | Na | Percentage | Rho | Standard Error | Age Estimate (in Years) | 95% Confidence Interval (in Years) |
|---|---|---|---|---|---|---|
| Coalescence Age | ||||||
| H1j1 | 52 | 12.4% | 1.86 | 0.49 | 4845 | 2324 − 7408 |
| H1t1 | 34 | 8.1% | 1.94 | 0.97 | 5057 | 99 - 10176 |
| H2a5a1 | 22 | 5.2% | 1.33 | 0.65 | 3422 | 118 - 6800 |
| H1av1 | 17 | 4.0% | 1.24 | 0.52 | 3213 | 567 - 5906 |
| H3c2a | 14 | 3.3% | 1.27 | 0.37 | 3291 | 1403 - 5204 |
| H1e1a1 | 12 | 2.9% | 1.23 | 0.72 | 3187 | −464 - 6927 |
| Splitting Age | ||||||
| H1j1 | 52 | 12.4% | 2.86 | 1.11 | 7514 | 1764 - 13470 |
| H1t1 | 34 | 8.1% | 2.94 | 1.39 | 7730 | 554 - 15227 |
| H2a5a1 | 22 | 5.2% | 2.33 | 1.19 | 6094 | −6 - 12434 |
| H1av1 | 17 | 4.0% | 2.24 | 1.13 | 5854 | 65 - 11860 |
| H3c2a | 14 | 3.3% | 2.27 | 1.07 | 5934 | 443 - 11619 |
| H1e1a1 | 12 | 2.9% | 5.23 | 2.13 | 14011 | 2729 - 26000 |
Number of individuals.
Collectively, our analyses of a large sample of complete mtDNA genomes demonstrate the need for such high-resolution sequence-based studies and large comparative databases to uncloak maternal ancestry patterns and subtle levels of population stratification. These patterns were previously masked in studies based on the typing of preascertained panels of coding region SNPs and/or HVS-I/II sequence variation. Specifically, the study of Basque populations, located at the epicenter of the region that was a major source for the repopulation of Europe after the Ice Age, further improves our understanding of the maternal ancestry of the modern inhabitants of the Franco-Cantabrian region and, more generally, of European prehistory. First, the haplogroup H dissection indicates that populations from the Basque Country and adjacent regions, rather than the Basque population per se, are characterized by numerous low-frequency autochthonous haplogroups, each explaining ∼2%–6% of the region's contemporary maternal ancestry, along with other H haplogroups that present a pan-European distribution and are observed at even lower frequencies. However, the autochthonous haplogroups are clearly more frequent within contemporary Basque speakers. Second, the identification of these autochthonous haplogroups attests to some period of genetic isolation of populations inhabiting the region from the rest of Europe. Interestingly, such a partial genetic isolation of the Basques well parallels their linguistic affiliation, as they speak a language that is considered a linguistic isolate. Third, our findings suggest that the contemporary haplogroup H variation observed among Basque populations can be traced back to at least two different temporal sources. One ancient deme, preceding the arrival of Indo-European peoples and languages to the region, is represented today by a number of surviving autochthonous haplogroups, H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1, which diverged from the pan-European mtDNA pool ∼8,000 years ago and experienced successive expansions at ∼4,000 YBP. A more recent genetic contribution to the region is attested to by the seemingly punctuated introgression of pan-European H haplogroups, which are shared with other European populations, through the historical events affecting this region in the last millennia.
More generally, the end of the glacial period marked the beginning of a flourishing way of life in Europe, where populations from the Franco-Cantabrian region became more sedentary and underwent considerable population growth.2 This left some postglacial imprints in the current genetic landscape of neighboring populations, as suggested by the various mtDNA haplogroups originating in the region (e.g., H1, H3, V, and U5b), many derived subhaplogroups of which are observed today not only among most Europeans3,13–15 but also among North Africans.45–47 However, our data show that the postglacial period also left some genetic footprints in the source populations of such major population expansions. This is clearly demonstrated by the various H haplogroups, including H1j1, H1t1, H2a5a1, H1av1, H3c2a, and H1e1a1, which are today restricted to the Franco-Cantabrian region and, more specifically, to Basque-speaking populations. These autochthonous haplogroups are shared, in some cases, with their immediate neighbors, suggesting gene flow between Basques and non-Basque-speaking peoples of the Franco-Cantabrian region and/or that the geographic extension of the Basque-speaking peoples used to be larger than thought today. We anticipate, however, that the same level of resolution used in the case study of the Basques at the European scale will reveal the presence of low-frequency autochthonous haplogroups in other regions of Europe, such as the case of the U5b3a1a clade that distinctively characterizes the people of Sardinia,48 shedding further light on the demographic history of the European continent. However, our results are particularly meaningful in the case of the Basques because they are concordant with a genetic continuity since postglacial times to the present day, as a possible vehicle for the preservation of the unique Basque language.
In conclusion, our study has identified six autochthonous haplogroups, which explain 36% of the contemporary variation of haplogroup H in the region, restricted to Basque-speaking peoples and their immediate neighbors and virtually absent in the rest of Europe. In light of this, our data provide support for the hypothesis of a partial genetic continuity of contemporary Basques lato sensu—the historical Basque Country—with the earlier settlers of their homeland since pre-Neolithic times.
Acknowledgments
We warmly thank all study participants. This work was supported by the Institut Pasteur, National Geographic, and the Histoire des populations et variation linguistique dans les Pyrénées de l'Ouest project, which received funding from the Conseil Régional d'Aquitaine, the Conseil Général des Pyrénées-Atlantiques, the Conseil des Elus du Pays-Basque, and the Centre National de la Recherche Scientifique interdisciplinary program Origine de l'Homme, des Langues et du Langage. This study also benefited from the support of Department of Hematology, Centre Hospitalier de la Côte Basque, in Bayonne, and Association Sang 64. We are grateful to the many contributors for their valuable help with the recruitment of samples, especially to Estibaliz Montoya, and to David Basterot and Tristan Carrère for technical assistance.
Supplemental Data
Web Resources
The URL for data presented herein is as follows:
mtPhyl, http://eltsov.org/mtphyl.aspx
Accession Numbers
The GenBank accession numbers for the 420 complete mtDNA sequences reported in this paper are JQ324516–JQ324935.
References
- 1.Gamble C., Davies W., Pettitt P., Hazelwood L., Richards M. The archaeological and genetic foundations of the European population during the Late Glacial: Implications for ‘agricultural thinking’. Cambridge. Archaeol. J. 2005;15:193–223. [Google Scholar]
- 2.Gamble C., Davies W., Pettitt P., Richards M. Climate change and evolving human diversity in Europe during the last glacial. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004;359:243–253. doi: 10.1098/rstb.2003.1396. discussion 253–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soares P., Achilli A., Semino O., Davies W., Macaulay V., Bandelt H.J., Torroni A., Richards M.B. The archaeogenetics of Europe. Curr. Biol. 2010;20:R174–R183. doi: 10.1016/j.cub.2009.11.054. [DOI] [PubMed] [Google Scholar]
- 4.Diamond J., Bellwood P. Farmers and their languages: The first expansions. Science. 2003;300:597–603. doi: 10.1126/science.1078208. [DOI] [PubMed] [Google Scholar]
- 5.Lao O., Lu T.T., Nothnagel M., Junge O., Freitag-Wolf S., Caliebe A., Balascakova M., Bertranpetit J., Bindoff L.A., Comas D. Correlation between genetic and geographic structure in Europe. Curr. Biol. 2008;18:1241–1248. doi: 10.1016/j.cub.2008.07.049. [DOI] [PubMed] [Google Scholar]
- 6.Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A.R., Auton A., Indap A., King K.S., Bergmann S., Nelson M.R. Genes mirror geography within Europe. Nature. 2008;456:98–101. doi: 10.1038/nature07331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seldin M.F., Shigeta R., Villoslada P., Selmi C., Tuomilehto J., Silva G., Belmont J.W., Klareskog L., Gregersen P.K. European population substructure: Clustering of northern and southern populations. PLoS Genet. 2006;2:e143. doi: 10.1371/journal.pgen.0020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauchet M., McEvoy B., Pearson L.N., Quillen E.E., Sarkisian T., Hovhannesyan K., Deka R., Bradley D.G., Shriver M.D. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 2007;80:948–956. doi: 10.1086/513477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auton A., Bryc K., Boyko A.R., Lohmueller K.E., Novembre J., Reynolds A., Indap A., Wright M.H., Degenhardt J.D., Gutenkunst R.N. Global distribution of genomic diversity underscores rich complex history of continental human populations. Genome Res. 2009;19:795–803. doi: 10.1101/gr.088898.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balaresque P., Bowden G.R., Adams S.M., Leung H.Y., King T.E., Rosser Z.H., Goodwin J., Moisan J.P., Richard C., Millward A. A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cruciani F., Trombetta B., Antonelli C., Pascone R., Valesini G., Scalzi V., Vona G., Melegh B., Zagradisnik B., Assum G. Strong intra- and inter-continental differentiation revealed by Y chromosome SNPs M269, U106 and U152. Forensic Sci. Int. Genet. 2011;5:e49–e52. doi: 10.1016/j.fsigen.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 12.Myres N.M., Rootsi S., Lin A.A., Järve M., King R.J., Kutuev I., Cabrera V.M., Khusnutdinova E.K., Pshenichnov A., Yunusbayev B. A major Y-chromosome haplogroup R1b Holocene era founder effect in Central and Western Europe. Eur. J. Hum. Genet. 2011;19:95–101. doi: 10.1038/ejhg.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Achilli A., Rengo C., Magri C., Battaglia V., Olivieri A., Scozzari R., Cruciani F., Zeviani M., Briem E., Carelli V. The molecular dissection of mtDNA haplogroup H confirms that the Franco-Cantabrian glacial refuge was a major source for the European gene pool. Am. J. Hum. Genet. 2004;75:910–918. doi: 10.1086/425590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pereira L., Richards M., Goios A., Alonso A., Albarrán C., Garcia O., Behar D.M., Gölge M., Hatina J., Al-Gazali L. High-resolution mtDNA evidence for the late-glacial resettlement of Europe from an Iberian refugium. Genome Res. 2005;15:19–24. doi: 10.1101/gr.3182305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torroni A., Bandelt H.J., D'Urbano L., Lahermo P., Moral P., Sellitto D., Rengo C., Forster P., Savontaus M.L., Bonné-Tamir B., Scozzari R. mtDNA analysis reveals a major late Paleolithic population expansion from southwestern to northeastern Europe. Am. J. Hum. Genet. 1998;62:1137–1152. doi: 10.1086/301822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torroni A., Bandelt H.J., Macaulay V., Richards M., Cruciani F., Rengo C., Martinez-Cabrera V., Villems R., Kivisild T., Metspalu E. A signal, from human mtDNA, of postglacial recolonization in Europe. Am. J. Hum. Genet. 2001;69:844–852. doi: 10.1086/323485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calafell F., Bertranpetit J. Mountains and genes: Population history of the Pyrenees. Hum. Biol. 1994;66:823–842. [PubMed] [Google Scholar]
- 18.Calafell F., Bertranpetit J. Principal component analysis of gene frequencies and the origin of Basques. Am. J. Phys. Anthropol. 1994;93:201–215. doi: 10.1002/ajpa.1330930205. [DOI] [PubMed] [Google Scholar]
- 19.Bertranpetit J., Cavalli-Sforza L.L. A genetic reconstruction of the history of the population of the Iberian Peninsula. Ann. Hum. Genet. 1991;55:51–67. doi: 10.1111/j.1469-1809.1991.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 20.Behar D.M., Yunusbayev B., Metspalu M., Metspalu E., Rosset S., Parik J., Rootsi S., Chaubey G., Kutuev I., Yudkovsky G. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 21.Li J.Z., Absher D.M., Tang H., Southwick A.M., Casto A.M., Ramachandran S., Cann H.M., Barsh G.S., Feldman M., Cavalli-Sforza L.L., Myers R.M. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 22.Zlojutro M., Roy R., Palikij J., Crawford M.H. Autosomal STR variation in a Basque population: Vizcaya Province. Hum. Biol. 2006;78:599–618. doi: 10.1353/hub.2007.0007. [DOI] [PubMed] [Google Scholar]
- 23.Rodríguez-Ezpeleta N., Alvarez-Busto J., Imaz L., Regueiro M., Azcárate M.N., Bilbao R., Iriondo M., Gil A., Estonba A., Aransay A.M. High-density SNP genotyping detects homogeneity of Spanish and French Basques, and confirms their genomic distinctiveness from other European populations. Hum. Genet. 2010;128:113–117. doi: 10.1007/s00439-010-0833-4. [DOI] [PubMed] [Google Scholar]
- 24.Laayouni H., Calafell F., Bertranpetit J. A genome-wide survey does not show the genetic distinctiveness of Basques. Hum. Genet. 2010;127:455–458. doi: 10.1007/s00439-010-0798-3. [DOI] [PubMed] [Google Scholar]
- 25.Adams S.M., Bosch E., Balaresque P.L., Ballereau S.J., Lee A.C., Arroyo E., López-Parra A.M., Aler M., Grifo M.S., Brion M. The genetic legacy of religious diversity and intolerance: Paternal lineages of Christians, Jews, and Muslims in the Iberian Peninsula. Am. J. Hum. Genet. 2008;83:725–736. doi: 10.1016/j.ajhg.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alonso S., Flores C., Cabrera V., Alonso A., Martín P., Albarrán C., Izagirre N., de la Rúa C., García O. The place of the Basques in the European Y-chromosome diversity landscape. Eur. J. Hum. Genet. 2005;13:1293–1302. doi: 10.1038/sj.ejhg.5201482. [DOI] [PubMed] [Google Scholar]
- 27.Bertranpetit J., Sala J., Calafell F., Underhill P.A., Moral P., Comas D. Human mitochondrial DNA variation and the origin of Basques. Ann. Hum. Genet. 1995;59:63–81. doi: 10.1111/j.1469-1809.1995.tb01606.x. [DOI] [PubMed] [Google Scholar]
- 28.Salas A., Comas D., Lareu M.V., Bertranpetit J., Carracedo A. mtDNA analysis of the Galician population: A genetic edge of European variation. Eur. J. Hum. Genet. 1998;6:365–375. doi: 10.1038/sj.ejhg.5200202. [DOI] [PubMed] [Google Scholar]
- 29.Richards M., Côrte-Real H., Forster P., Macaulay V., Wilkinson-Herbots H., Demaine A., Papiha S., Hedges R., Bandelt H.J., Sykes B. Paleolithic and neolithic lineages in the European mitochondrial gene pool. Am. J. Hum. Genet. 1996;59:185–203. [PMC free article] [PubMed] [Google Scholar]
- 30.Richards M., Macaulay V., Hickey E., Vega E., Sykes B., Guida V., Rengo C., Sellitto D., Cruciani F., Kivisild T. Tracing European founder lineages in the Near Eastern mtDNA pool. Am. J. Hum. Genet. 2000;67:1251–1276. [PMC free article] [PubMed] [Google Scholar]
- 31.Alfonso-Sánchez M.A., Cardoso S., Martínez-Bouzas C., Peña J.A., Herrera R.J., Castro A., Fernández-Fernández I., De Pancorbo M.M. Mitochondrial DNA haplogroup diversity in Basques: A reassessment based on HVI and HVII polymorphisms. Am. J. Hum. Biol. 2008;20:154–164. doi: 10.1002/ajhb.20706. [DOI] [PubMed] [Google Scholar]
- 32.García O., Fregel R., Larruga J.M., Álvarez V., Yurrebaso I., Cabrera V.M., González A.M. Using mitochondrial DNA to test the hypothesis of a European post-glacial human recolonization from the Franco-Cantabrian refuge. Heredity (Edinb) 2011;106:37–45. doi: 10.1038/hdy.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prieto L., Zimmermann B., Goios A., Rodriguez-Monge A., Paneto G.G., Alves C., Alonso A., Fridman C., Cardoso S., Lima G. The GHEP-EMPOP collaboration on mtDNA population data—A new resource for forensic casework. Forensic Sci. Int. Genet. 2011;5:146–151. doi: 10.1016/j.fsigen.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Iglesias V., Mosquera-Miguel A., Cerezo M., Quintáns B., Zarrabeitia M.T., Cuscó I., Lareu M.V., García O., Pérez-Jurado L., Carracedo A., Salas A. New population and phylogenetic features of the internal variation within mitochondrial DNA macro-haplogroup R0. PLoS ONE. 2009;4:e5112. doi: 10.1371/journal.pone.0005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.González A.M., García O., Larruga J.M., Cabrera V.M. The mitochondrial lineage U8a reveals a Paleolithic settlement in the Basque country. BMC Genomics. 2006;7:124. doi: 10.1186/1471-2164-7-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roostalu U., Kutuev I., Loogväli E.L., Metspalu E., Tambets K., Reidla M., Khusnutdinova E.K., Usanga E., Kivisild T., Villems R. Origin and expansion of haplogroup H, the dominant human mitochondrial DNA lineage in West Eurasia: The Near Eastern and Caucasian perspective. Mol. Biol. Evol. 2007;24:436–448. doi: 10.1093/molbev/msl173. [DOI] [PubMed] [Google Scholar]
- 37.Soares P., Ermini L., Thomson N., Mormina M., Rito T., Röhl A., Salas A., Oppenheimer S., Macaulay V., Richards M.B. Correcting for purifying selection: An improved human mitochondrial molecular clock. Am. J. Hum. Genet. 2009;84:740–759. doi: 10.1016/j.ajhg.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zuazo K. Alberdania; Irún, Spain: 2010. El Euskera y sus dialectos. [Google Scholar]
- 39.Behar D.M., Rosset S., Blue-Smith J., Balanovsky O., Tzur S., Comas D., Mitchell R.J., Quintana-Murci L., Tyler-Smith C., Wells R.S., Genographic Consortium The Genographic Project public participation mitochondrial DNA database. PLoS Genet. 2007;3:e104. doi: 10.1371/journal.pgen.0030104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haak W., Balanovsky O., Sanchez J.J., Koshel S., Zaporozhchenko V., Adler C.J., Der Sarkissian C.S., Brandt G., Schwarz C., Nicklisch N., Members of the Genographic Consortium Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 2010;8:e1000536. doi: 10.1371/journal.pbio.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kong Q.P., Yao Y.G., Sun C., Bandelt H.J., Zhu C.L., Zhang Y.P. Phylogeny of east Asian mitochondrial DNA lineages inferred from complete sequences. Am. J. Hum. Genet. 2003;73:671–676. doi: 10.1086/377718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torroni A., Schurr T.G., Cabell M.F., Brown M.D., Neel J.V., Larsen M., Smith D.G., Vullo C.M., Wallace D.C. Asian affinities and continental radiation of the four founding Native American mtDNAs. Am. J. Hum. Genet. 1993;53:563–590. [PMC free article] [PubMed] [Google Scholar]
- 43.Gómez-Carballa A., Cerezo M., Balboa E., Heredia C., Castro-Feijóo L., Rica I., Barreiro J., Eirís J., Cabanas P., Martínez-Soto I. Evolutionary analyses of entire genomes do not support the association of mtDNA mutations with Ras/MAPK pathway syndromes. PLoS ONE. 2011;6:e18348. doi: 10.1371/journal.pone.0018348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Catelli M.L., Alvarez-Iglesias V., Gómez-Carballa A., Mosquera-Miguel A., Romanini C., Borosky A., Amigo J., Carracedo A., Vullo C., Salas A. The impact of modern migrations on present-day multi-ethnic Argentina as recorded on the mitochondrial DNA genome. BMC Genet. 2011;12:77. doi: 10.1186/1471-2156-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Achilli A., Rengo C., Battaglia V., Pala M., Olivieri A., Fornarino S., Magri C., Scozzari R., Babudri N., Santachiara-Benerecetti A.S. Saami and Berbers—an unexpected mitochondrial DNA link. Am. J. Hum. Genet. 2005;76:883–886. doi: 10.1086/430073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cherni L., Fernandes V., Pereira J.B., Costa M.D., Goios A., Frigi S., Yacoubi-Loueslati B., Amor M.B., Slama A., Amorim A. Post-last glacial maximum expansion from Iberia to North Africa revealed by fine characterization of mtDNA H haplogroup in Tunisia. Am. J. Phys. Anthropol. 2009;139:253–260. doi: 10.1002/ajpa.20979. [DOI] [PubMed] [Google Scholar]
- 47.Ottoni C., Primativo G., Hooshiar Kashani B., Achilli A., Martínez-Labarga C., Biondi G., Torroni A., Rickards O. Mitochondrial haplogroup H1 in north Africa: An early holocene arrival from Iberia. PLoS ONE. 2010;5:e13378. doi: 10.1371/journal.pone.0013378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pala M., Achilli A., Olivieri A., Kashani B.H., Perego U.A., Sanna D., Metspalu E., Tambets K., Tamm E., Accetturo M. Mitochondrial haplogroup U5b3: A distant echo of the epipaleolithic in Italy and the legacy of the early Sardinians. Am. J. Hum. Genet. 2009;84:814–821. doi: 10.1016/j.ajhg.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


