Abstract
The ubiquitous and abundant cytoplasmic poly(A) binding protein (PABP) is a highly conserved multifunctional protein, many copies of which bind to the poly(A) tail of eukaryotic mRNAs to promote translation initiation. The N-terminus of PABP is responsible for the high binding specificity and affinity to poly(A), whereas the C-terminus is known to stimulate PABP multimerization on poly(A). Here, we use single-molecule nanopore force spectroscopy to directly measure interactions between poly(A) and PABPs. Both electrical and biochemical results show that the C-C domain interaction between two consecutive PABPs promotes cooperative binding. Up to now, investigators have not been able to probe the detailed polarity configuration (i.e., the internal arrangement of two PABPs on a poly(A) streak in which the C-termini face toward or away from each other). Our nanopore force spectroscopy system is able to distinguish the cooperative binding conformation from the noncooperative one. The ∼50% cooperative binding conformation of wild-type PABPs indicates that the C-C domain interaction doubles the cooperative binding probability. Moreover, the longer dissociation time of a cooperatively bound poly(A)/PABP complex as compared with a noncooperatively bound one indicates that the cooperative mode is the most stable conformation for PABPs binding onto the poly(A). However, ∼50% of the poly(A)/PABP complexes exhibit a noncooperative binding conformation, which is in line with previous studies showing that the PABP C-terminal domain also interacts with additional protein cofactors.
Introduction
Translation initiation is a rate-limiting multistage cascade of events that regulate gene expression. With the exception of histone-encoding mRNAs, the translation of all eukaryotic mRNAs is stimulated by two cis-acting elements: the 5′-cap and the 3′-poly(A) tail. The 5′ m7GpppN cap structure recruits the eukaryotic initiation factor complex 4F (eIF4F), which is composed of the cap-binding eukaryotic initiation factor eIF4E, the ATP-dependent RNA helicase eIF4A, and eIF4G, a scaffolding protein that binds eIF4E and eIF4A. eIF4G also interacts with the poly-adenine (poly(A)) binding protein (PABP) that is bound to the mRNA 3′ terminal poly(A) tail (1–3). PABP is conserved in species ranging from yeast to mammals, and many copies of PABP cover the length of the mRNA poly(A) tail. The eIF4E-eIF4G-PABP complex circularizes the mRNA (4), which stimulates translation initiation (5) by promoting recruitment of the 40S ribosomal subunit (3). This closed-loop model, created by both the cap structure and the poly(A) tail, exerts a synergistic enhancement of the translation initiation process (6).
PABP consists of four consecutive and highly conserved RNA recognition motifs (RRMs) in its N-terminal domain connected to a C-terminal helical domain (PABC) by an unstructured proline- and glutamine-rich linker (1). The N-terminal RRMs, specifically RRMs 1 and 2, bind with high specificity and affinity to the poly(A) (7), and interact with eIF4G to promote the closed-loop mRNA structure (8,9). In contrast, the less-conserved C-terminal domain, which consists of approximately a third of PABP, contains an unstructured region called the linker and PABC, an α-helical peptide-binding domain (10). This C-terminal domain facilitates PABP multimerization on the poly(A) tail (7,11), and is also responsible for binding to several PABP-interacting proteins (10,12). Recently, the cocrystal structure of the first two RRMs of human PABP bound to an 11-mer poly(A) oligomer at 2.6 Å resolution was determined (13). The two RRMs appear in a linear arrangement in which the two β-sheets and linker localize between the two RRM domains, forming a continuous 2.5 nm (width) × 3.6 nm (height) × 4.7 nm (length) RNA-binding trough. The poly(A) oligomer adopts an extended conformation throughout the molecular trough, in contrast to the more stable helical structure in the free solution. A full-length PABP-poly(A) binding model is proposed based on the latter cocrystal structure and the conservation of RNA-binding residues within the four RRMs: a single PABP binds to poly(A) with its four RRMs arranged in a linear manner along the poly(A) molecule, whereas the PABP C-terminal domain is free to interact with either a second PABP or other ancillary proteins regulating posttranscription and translation.
In contrast to the RRMs, the role of the PABP C-terminal domain in the stabilization of the PABP-poly(A) interaction remains unclear. On its own, the C-terminal domain does not display any detectable binding affinity to poly(A); however, it mediates homophilic protein-protein interactions that promote PABP dimerization on the poly(A) oligomer (7,11). Because both nuclear and cytoplasmic PABPs are known to be abundant, enabling sufficient coverage of all poly(A)s in their respective cellular compartments (14,15), it has been suggested that PABP-PABP interactions may play an important role in their function by enhancing the binding activity to coat the entire poly(A) tail of an mRNA transcript. Therefore, it is essential to determine whether the PABP C-terminal domain affects the binding activity of PABP, and to what extent the C-C domain interactions promote PABP dimerization on poly(A). Moreover, the preferred binding polarity of multiple copies of PABP along the poly(A) tail remains unknown. We refer to the C-terminus as the head and the N-terminus as the tail. Specifically, two scenarios are possible: 1), a cooperative binding of two PABPs, whereby the C-termini of adjacent PABPs interact directly with each other in a head-to-head mode, thereby stabilizing the overall complex; and 2), a noncooperative binding mechanism in a head-to-tail or tail-to-tail mode.
Here, we use nanopore force spectroscopy (NFS) (16) to quantify the strength of the interactions between either single or multiple PABPs with a poly(A) oligomer, and to decipher the effect of binding polarity and preference on the complex stability. In a nanopore experiment, a controlled electrical force is applied directly onto a charged biopolymer threaded through a molecular-sized constriction made in a thin, insulated lipid membrane separating two reservoirs of buffered salt solution. The electric field applied across two electrodes placed on either side of the bilayer membrane generates a steady ionic countercurrent consisting of negative and positive ions passing through the nanopore. The latter's passage can be measured with a sensitive electrometer. When charged biopolymers randomly approach the nanopore vicinity, they are pulled into the pore region by a residual component of the electrical field (17,18). Once an end of the biopolymer is threaded into the nanopore, the biopolymers are subjected to a stronger force, which causes them to slide from one side of the membrane to the other. This process is usually referred to as polynucleotide translocation (19). If the biopolymer's cross section is not uniform (due to, e.g., a hairpin structure or additional bound proteins), such that a local cross section is larger than the pore diameter, the translocation process will be interrupted. Formally, removal of these obstacles (i.e., unzipping of the hairpin or stripping-off of ancillary proteins) is described by crossing a large energy barrier, much higher than that associated with moving an unstructured single biopolymer through the pore (16,20). Because the waiting time associated with the crossing of an energy barrier increases exponentially with the energy barrier height, the characteristic translocation times of unstructured biopolymers (i.e., single-stranded DNA (ssDNA), ssRNA, or polypeptides) are expected to be orders of magnitude smaller than the typical time required, for example, to unzip a hairpin loop of similar length. These features make NFS particularly useful for characterizing both DNA duplexes (20–25) and interactions between enzymes and ssDNA (26–29).
Materials and Methods
RNA and protein synthesis
We transcribed C50A25 and C50A50 RNA samples in vitro using a procedure similar to that described by Akeson et al. (30). Briefly, for C50A25 RNA, the double-stranded DNA (dsDNA) template was synthesized from a purified single-stranded 107-mer template (5′-TAATACGACTCACTATAGGGC50A25GCTACCACACAC-3′) and 12-mer primer (5′-GTGTGTGGTAGC-3′) purchased from Integrated DNA Technologies (Coralville, IA) using Sequenase (USB, Cleveland, OH). This dsDNA template was used to synthesize RNA using the T7 RNA polymerase in vitro transcription kit Megashortscript (Ambion, Austin, TX). After RNA synthesis was completed, the product was incubated in the presence of Turbo DNase I (Ambion, Austin, TX) to remove DNA, and with RNase T1 (Fermentas, Glen Burnie, MD) to cleave the phosphodiester bond between the 3′-G residues and the 5′-OH residues of the adjacent nucleotide, thus rendering the final RNA product with an additional G-residue at its 3′-end (C50A25Gp). The RNA was loaded onto an 8% polyacrylamide gel and run in 1 × Tris-borate-EDTA (TBE). The desired single band was cut out of the polyacrylamide gel and eluted via a crush-and-soak method, followed by ethanol precipitation. The purified RNA was finally resuspended in Tris-EDTA (TE) buffer (pH 8.0). The same protocol was applied to prepare C50A50 RNA.
The two plasmids encoding the full-length human PABP (designated PABP, 71 kDa) and the C-terminus truncated PABP (designated PABPc−, 42 kDa) were transformed into BL21(DE3) (Stratagene, Santa Clara, CA) Escherichia coli expression strains. The His-tagged PABP was purified using Ni-NTA and heparin-Sepharose chromatography in tandem. The glutathione S-transferase (GST)-tagged PABPc− was purified with the use of a glutathione column. The GST tag was then cleaved off with the use of PreScission protease (GE Healthcare, Waukesha, WI) according to the manufacturer's instructions. The eluted untagged fractions were further loaded onto a heparin-Sepharose chromatography column to remove any nucleic acid contaminants. Both PABP and PABPc− recombinant proteins were loaded onto a Millipore filter to further concentrate and remove lower molecular mass contaminants. The final protein concentrations were determined by means of a Bradford assay. The purity of each protein was ascertained by means of SDS-PAGE or the Experion (BioRad, Hercules, CA) microfluidic gel electrophoresis system. The typical concentration was ∼50 μM with >90% purity.
Gel-shift analysis
An electrophoretic mobility shift assay (EMSA) was used to test the PABP or PABPc− binding activity in a correlative binding buffer (600 mM KCl, 50 mM HEPES, 1 mM EDTA, pH 8.0). Protein and RNA molecules were incubated in a binding buffer at 23°C for 20 min. The reaction mixture was then loaded onto an 8% native polyacrylamide gel. The gel was run at 200 mV for 1 h, stained with SYBR Green II (Invitrogen, Grand Island, NY), and then scanned with a Pharos gel scanner (BioRad, Hercules, CA). Because SYBR Green II stains only the RNA, the protein-bound RNA has a reduced staining efficiency. Therefore, quantitative information on the binding ratio is available only for the unbound RNA and not for the single- and double-bound C50A50 RNA.
Nanopore assays
The apparatus and experimental method used to reconstitute the α-hemolysin (α-HL) pore in a horizontally supported planar lipid bilayer were described previously (31). The two chambers of the cell were filled with binding buffer, which was the same as in the EMSA. After formation of the pore was completed, RNA molecules and PABP were added into the cis chamber, and incubated at 23°C for 20 min before the data were sampled. All experiments were carried out at 23°C, which typically produced an open pore current of 120 pA at 180 mV. The current signal was filtered at 40 KHz using a low-pass filter, recorded at 125 kHz/16 bits.
The dimensionless residual current level is defined as IB = iB/iO, where iB is the residual (or blocked) ion current level when a polymer is threaded through the pore, and iO is the open pore current levels (Fig. 1 C). This definition facilitates a comparison of experiments performed under different voltages, which shift iO and iB but not their ratio. Our experiments exploit the finding that the residual current of poly-cytosine polymers (poly(C)) sliding through the α-HL nanopore is ∼50% of the poly(A)'s current (30,32). This provides a convenient way to monitor the progression of polymers consisting of poly(C)-poly(A) segments during their translocation through the pore. A corollary feature is that the effective charge, q∗, of the polynucleotides residing in the pore can vary slightly due to the different structural forms of the poly(C) or poly(A) segments.
Figure 1.
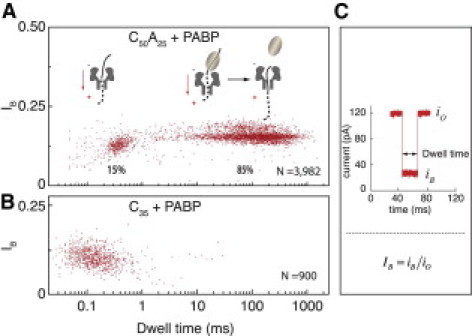
(A) Scatter plot of the dwell time versus IB, when C50A25 is incubated with equal amounts of PABP at 23°C. The left cluster represents free RNA molecule translocation, and the widely distributed population on the right is due to the detachment of a single PABP from the RNA. (B) Scatter plot of the dwell time versus IB, when C35 is incubated with equal amounts of PABP at 23°C. The single cluster overlaps with the characteristic IB and dwell time of the C35 alone. (C) Variables describing nanopore traces: iO is the open pore current, iB is the residual current in the presence of a molecule inside the nanopore, and IB is the normalized blocked current level. The dwell time is the duration in which a molecule is sustained inside the nanopore.
Results and Discussion
A C50A25 ssRNA oligomer is synthesized to enable the binding of a single PABP molecule onto its poly(A) tail, restricted by the poly(C) sequence (7). When C50A25 molecules (1 μM) are incubated in the presence of PABP (1 μM) before the nanopore measurements are obtained, two distinct populations appear event-wise (Fig. 1 A): 1), a ∼15% minority population, presenting a residual current and translocation time similar to the translocation characteristics of the ssRNA oligos alone; and 2), an ∼85% bulk population, showing a marked increase in the characteristic dwell times by nearly 3 orders of magnitude as compared with the dwell time of solely ssRNA translocations. A typical example of an experiment consisting of ∼4000 events is shown as a scatter plot in Fig. 1 A, where each spot represents the IB versus dwell-time values. In contrast, nanopore experiments performed using poly(C) molecules and PABP with the same concentration as in Fig. 1 A result in a single event population, which overlaps with the characteristic residual current and translocation time of the poly(C) RNAs alone (Fig. 1 B).
Considering that a single PABP covers ∼20–25 adenine residues of the poly(A) oligomer and has no binding affinity toward the poly(C) sequence (as indicated by Fig. 1 B and in the literature (7)), it is most likely that the short events in Fig. 1 A correspond to the minority population of unbound ssRNA translocation events, whereas the long and broadly distributed bulk population events are due to a single PABP being bound to the ssRNA oligomer. The cocrystal structure of the N-terminal RRM1 and RRM2 of PABP bound to a poly(A) supports this interpretation, because it suggests that the cross-section size of the complex is on average > 3 nm (13), which is larger than the ∼2.6 nm opening of the α-HL's vestibule (33) through which the ssRNA is inserted, whereas the PABP cannot translocate through (see Fig. S1 of the Supporting Material). By measuring the dependence of the characteristic dwell times on the applied voltage, we were able to rule out the possibility that the RNA/PABP complex is retracted from the nanopore against the voltage gradient, after the poly(C) is threaded through the nanopore, because this would lead to an increase in the dwell time with increasing voltage rather than a decrease in dwell times (Fig. S2). Thus, we can unambiguously conclude that the bulk population (85%) of events in Fig. 1 A corresponds to the detachment of a single PABP molecule per event from the RNA molecules, followed by the subsequent rapid sliding of the C50A25 ssRNA through the nanopore into the trans chamber. Moreover, attempts to conduct translocation measurements using PABP alone do not yield any noticeable blockade episodes in the ion current.
We then move on to examine an RNA oligomer carrying an A50 streak. Unlike the C50A25 RNA, which restricts a single PABP binding onto the poly(A) region, the C50A50 can be bound by either one or two PABPs in tandem. A gel shift analysis confirms this observation, showing two shifted bands of C50A50/PABP, appearing in two locations, versus a single band shift corresponding with the C50A25 oligomer (Fig. 2 A). When a C50A50 RNA is used, the gel shift results depict the possibility of binding either one or two PABPs, where the fraction of two PABP binding increases with the PABP concentration used. A quantitative analysis of A25 ssRNA/PABP binding (first two lanes in Fig. 2 A) reveals a lower ratio of RNA/PABP complex (∼50%) as compared with the 85% in the nanopore measurement. Thus, the nanopore scheme could potentially be less disruptive to the binding equilibrium compared with conventional gel electrophoresis.
Figure 2.
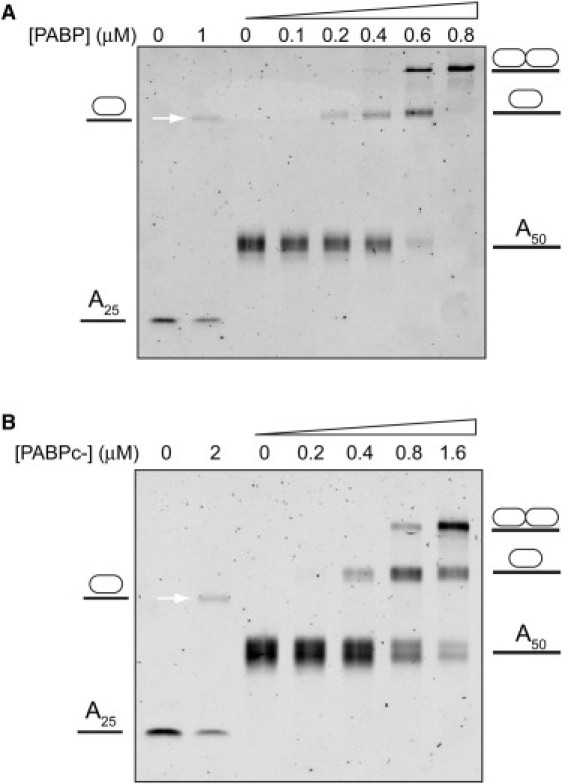
EMSA showing the retardation of poly(A) (A25 and A50) when bound to protein. (A) PABP: The first two lanes depict the band shift of A25 (1 μM) in the presence of PABP, and the rest depict the A50 (0.4 μM) band shift in the presence of increasing PABP concentrations. (B) PABPc−: The first two lanes depict the band shift of A25 (2 μM) in the presence of PABP, and the rest depict the result of an A50 (0.4 μM) band shift in the presence of increasing PABP concentration.
These results naturally lead us to proceed and probe two PABPs binding onto the C50A50 RNA using NFS. Whereas the C50A25/PABP complexes produce but a single type of event (Fig. 3 A), consisting of a single blocked current level (IB = 0.19), the longer RNA molecules (C50A50) preincubated in the presence of PABPs produce two distinct event types (Fig. 3 B): 1) events displaying a single blocked current level (IB = 0.19; 45% frequency of events); and 2), events presenting clear two-state levels (IB = 0.19 followed by IB = 0.24; 55% frequency of events).
Figure 3.

(A) Single PABP detachment of a C50A25/PABP complex, as illustrated on the left-hand side, where the poly(C) segment (dotted line) is inside the nanopore. The middle section shows typical traces highlighting an average IB level of 0.19, and the corresponding dwell-time histogram (right-hand side) with a single exponential fit to the tail, yielding the characteristic time τ. (B) Two detachment modes of the C50A50/PABP complex, illustrated on the left-hand side. The top panel depicts the simultaneous detachment of two PABPs; corresponding single current level traces and the dwell-time histogram are displayed on the top-right. The bottom panel depicts the sequential detachment mode of two PABPs, producing two current level traces (IB = 0.19 when poly(C) is in the nanopore, and IB = 0.24 when poly(A) is in the nanopore).
A statistical analysis of >1500 events, such as those shown in Fig. 3 B, is used to construct the dwell-time distributions of the C50A50/PABP complexes (Fig. 3 B, right-hand side: V = 180 mV). The distributions are tail-fitted to exponential functions to measure the characteristic time scales τ, as indicated. The single-level events of the long C50A50 molecules (RNA/PABP = 1:2) yield τ = 181.1 ± 7.0 ms (Fig. 3 B, top-right histogram), which is ∼2.6 times higher than the τ-value produced by C50A25 binding events of single PABP (Fig. 3 A). We plotted separately the histograms of the first and second current levels (t1 and t2, respectively) for the C50A50 events displaying two current levels (Fig. 3 B, bottom-right histograms). We obtained 86.7 ± 3.7 ms and 141.1 ± 4.8 ms for t1 and t2, respectively.
We postulate that the single-level events shown in the NFS assays (Fig. 3 B) correspond to the simultaneous detachment of the two PABP units from the C50A50/RNA in a single step (also termed cooperative detachment), whereas the two-level events correspond to a stepwise removal of the two proteins, as schematically illustrated in Fig. 3 B. The abrupt increase in the residual pore current observed in the two-level events can be explained by the observation that poly(C) fragments induce slightly lower blockade levels than the poly(A) fragments (30,32). In our experiment, the poly(C) tail of the molecule initially resides in the α-HL transmembrane region, but upon detachment of the first PABP, the less-compact poly(A) section of the biopolymer resides inside the pore, resulting in a slight but clearly distinguishable increase in the blocked current level.
Recent studies showed that the effective electrical charge (q∗) of poly(A) coil segments threaded inside the α-HL nanopore is smaller than the corresponding helical poly(A) effective charge, presumably due to the less compact structure of coiled poly(A) inside the nanopore (34). Similarly, the relatively lower IB implies that poly(C) has a more compact structure than coiled poly(A) (30), resulting in a larger effective charge. Because the force acting on the RNA molecule is proportional to q∗ (F = q∗V / l, where V is the voltage applied across the membrane and l is the length of the nanopore), it is expected that poly(A) segments will be subjected to a smaller force as compared with poly(C) segments. Thus, when the C50A50/PABPs complex is threaded through the pore, a larger force is applied on the first PABP than on the second PABP, explaining our clear observation that τ2 > τ1. To check this hypothesis, we performed a titration experiment in which the stoichiometric concentrations of C50A50 to PABP were incrementally increased to the point where most of the events contained either just the C50A50 RNA molecule alone or a single PABP molecule, bound on average in the middle of the A50 region. These conditions roughly mimic the second detachment step shown in Fig. 3 B, because in most cases the poly(A) region will be threaded inside the nanopore before detachment takes place. Fig. 4 shows the dwell-time distribution of C50A50/PABP with a 4:1 ratio, producing τS = 141.9 ± 9.9 ms. This value is identical within error to the distribution of the t2 dwell time (Fig. 3 B, bottom), and approximately twice as high as that of the C50A25-PABP assay (Fig. 3 A). This result supports our assumption that the distributions of t2 dwell times correspond to the detachment of the second PABP molecule, and that the stepwise detachment of the two PABPs in this bulk population of events presents nearly independent occurrences (also termed noncooperative).
Figure 4.
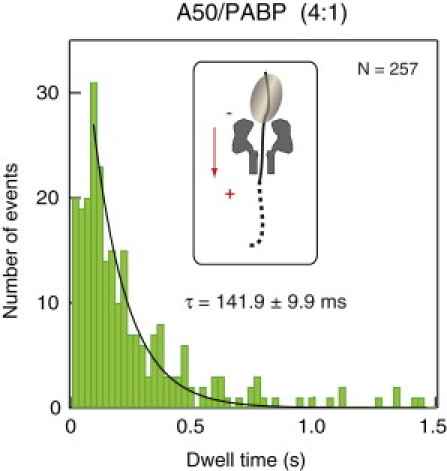
Dwell-time distribution of the C50A50/PABP complex at an RNA/protein ratio of 4:1. A single exponential fit to the tail yields τ = 142 ms. The inset illustrates a C50A50 oligomer being bound by a single PABP, wherein the nanopore is mostly occupied with the poly(A) segment (solid line).
Thus far, our evidence indicates that one can resolve the detachment kinetics of either a single or two PABP units from poly(A) RNA molecules by using NFS and designing di-block poly(C)-poly(A) biopolymers. The choice of the latter streaks, which present a slightly different effective charge, also affects the electrophoretic force, which induces the detachment process. In light of the aforementioned results, we propose the following model based on the possibility that the two PABPs can bind to the poly(A) in four different configurations, which we divide into two modes: cooperative and noncooperative (Fig. 5). The events that show a two-step detachment kinetics appear to be a repetition of the single PABP detachment. The two PABPs are detached one after the other from the C50A50 oligomer (∼70 ms per step, when the poly(C) streaks resides inside the nanopore). It should be noted that the characteristic timescale of t1 (∼87 ms) of the first PABP detachment (Fig. 3 B) is slightly higher than the 70 ms timescale presenting in the single PABP detachment from the shorter C50A25 oligomer (Fig. 3 A), possibly due to the presence of an adjacent PABP, which serves to hinder the detachment process. In contrast, the C50A50 events that show a single current state with a τs of 181 ms (Fig. 3 B) are substantially longer than the postulated ∼140 ms, which is twice the value obtained from a single PABP being detached from the C50A25 oligomer. In the latter case, the detachment kinetics is affected by an internal arrangement of the two PABP molecules, in which the two C-terminal domains are putatively facing each other (cooperative mode). In this mode, the protein-RNA complexes are stabilized via PABP-PABP interactions, further validating the stabilizing prominence of the C-terminal domain of the PABP proteins, the majority of which become further augmented when bound to the poly(A) tails of mRNA transcripts.
Figure 5.
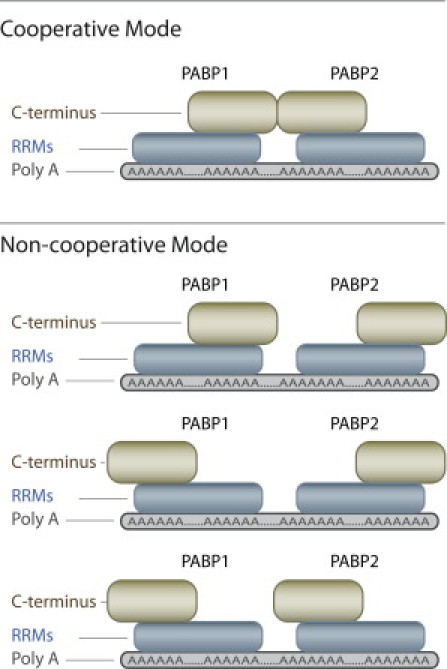
Schematic illustration of the cooperative and noncooperative modes of binding of two PABP molecules to poly(A). Stabilization of the PABP-PABP interactions through C-termini contacts is possible only in the cooperative mode.
The two postulated modes of A50C50/PABP detachment (Fig. 5) are related to the ability of the C-terminus of the protein to interact with its neighboring protein's C-terminus. Therefore, we further examined our hypothesis by expressing a PABP mutant protein deleted of its entire C-terminal domain (PABPc−) (7,11). We performed gel-shift assays to check this mutant's affinity to the RNA molecules used. The results show that the PABPc− binding to the C50A50 molecules is weaker than the binding of the wild-type (WT) PABP (Fig. 2 B). Specifically, even at a PABPc−/C50A50 4:1 ratio, significant amounts of unbound or singly bound RNA molecules are still observed (the A50 band size is still apparent even at the highest PABPc− concentration). In contrast, no free or singly bound RNA is observed in the WT PABP assays, even at a smaller PABP/RNA 2:1 stoichiometric ratio (Fig. 2 A).
As previously mentioned, we postulate that the C-terminal domain of PABP facilitates cooperative detachment by interaction of the first PABP C-terminal domain with its follower. Hence, a cleavage of the C-terminal domain is expected to produce a higher frequency of the noncooperative detachment mode of events at the expense of the single-state mode (the cooperative one). This hypothesis is further corroborated by the data shown in Fig. 6. Of nearly 1000 nanopore detachment events (performed under identical conditions to those employed for the WT PABP) in the presence of C50A50/PABPc− complexes, <20% of the overall events display a single-current level, whereas >80% display clear-cut two-level events. Moreover, because our gel-shift data indicate that even at the ratio of PABP/RNA 4:1, a substantial percentage of the RNA C50A50 is bound to a single PABPc−, we speculate that a large fraction of the 20% cooperative events correspond to the detachment of a single PABPc− of the C50A50 oligomer. A closer inspection of the current states in the single-level-events mode (Fig. 6, top trace) reveals a value of IB = 0.24 corresponding to poly(A) residing inside the nanopore—an impossible postulated configuration, in case the poly(A) segment is entirely apprehended by a couple of PABPc− protein mutants. Thus, we can conclude that the minority one-level events largely correspond to a single PABPc− being bound to the C50A50, whereas the bulk two-level-events population represents sequential noncooperative detachment of the two PABPc− proteins.
Figure 6.
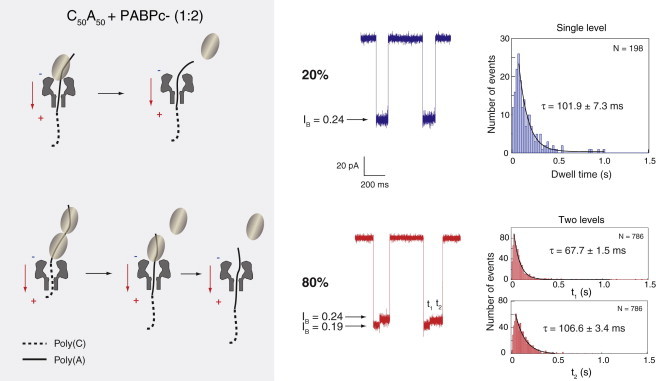
Two types of detachment observed in the presence of the C50A50/PABPc− complex. Top panel: Single-current level (IB = 0.24) events corresponding to a single PABP being bound to the RNA oligomer, as illustrated at the top-left. Botton panel: Two-level (IB = 0.19 and IB = 0.24) events due to a sequential detachment of the two bound PABPs, as illustrated at the bottom-left. The corresponding dwell-time histograms are depicted on the right-hand side with single exponential fits to the tails.
Additional confirmation of our hypothesis comes from a statistical analysis of the detachment kinetics of the C50A50/PABPc− complexes. The single step detachment of the mutant PABPc− yields a typical timescale of 101.9 ± 7.3 ms, which is identical within error to the typical t2 timescale in the two-level detachment 106.6 ± 3.4 ms of the same proteins (Fig. 6). This further corroborates the model in which most of the single-step events correspond to a single PABPc− detachment. Moreover, the timescale produced (67.7 ms and 106.6 ms for t1 and t2, respectively) is shorter than the corresponding timescale obtained upon detachment of the WT PABP (86.7 ms and 141.1 ms, respectively). This suggests that the C-terminal domain alone stabilizes the PABP-RNA interactions, yielding a slower dissociation kinetics, in contrast to the previous observation that the C-terminus does not contribute to binding affinity. This highlights the distinction between the kinetic and thermodynamic properties of biomolecular interactions.
Conclusion
In this work, we were able to use single-molecule NFS measurements to directly monitor the interactions between RNA and multiple protein copies by designing a di-block copolymer of poly(C) and poly(A) in sequence. Because NFS does not require molecular tagging or chemical conjugations, it is well suited for a broad range of biophysical studies of nucleic acids–protein interactions (26). In particular, we show that the nanopore method can be used to quantified the fraction of bound poly(A)/PABP complexes at equilibrium (Fig. 1), as well as the complexes' dissociation kinetics under force (Figs. 3, 4, and 6).
Two PABP proteins can bind to a sufficiently long poly(A) oligomer presenting four optional polarity combinations, only one of which yields a possible C-C domain interaction that promotes cooperative binding (Fig. 5). We have shown qualitatively that C-C domain interactions between PABPs can promote cooperative binding, whereas neither C-N domain nor N-N domain interactions can (7,11). Moreover, whereas previously the polarity configuration could not be probed in detail, our NFS system is able to directly observe, for the first time to our knowledge, both the cooperative and noncooperative binding conformations of two PABPs on a poly(A) streak. Considering that the probability that two PABPs on a poly(A) streak will be in the cooperative mode (head-to-head) is 25% if no preferred interaction occurs between the C-terminus, the ∼50% cooperative binding conformation of WT PABPs indicates that the C-terminal domain interaction doubles the cooperative binding probability. Moreover, the C-C domain interaction is also responsible for the longer dissociation time of a cooperatively bound poly(A)-PABP complex as compared with a noncooperatively bound one. However, >50% of the poly(A)-PABP complexes still exhibit a noncooperative binding conformation, corresponding to evidence that the C-terminal domain of PABP also functions to interact with other protein cofactors (10).
Acknowledgments
We acknowledge support from Human Frontier Science Program grant RGP0036-2005 (A.M. and N.S.).
Supporting Material
References
- 1.Mangus D.A., Evans M.C., Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn U., Wahle E. Structure and function of poly(A) binding proteins. Biochim. Biophys. Acta. 2004;1678:67–84. doi: 10.1016/j.bbaexp.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wells S.E., Hillner P.E., Sachs A.B. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell. 1998;2:135–140. doi: 10.1016/s1097-2765(00)80122-7. [DOI] [PubMed] [Google Scholar]
- 5.Sachs A.B. Physical and functional interactions between the mRNA cap structure and the poly(A) tail. In: Sonenberg N., Hershey J.W.B., Mathews M.B., editors. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 2000. pp. 447–465. [Google Scholar]
- 6.Michel Y.M., Poncet D., Borman A.M. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 2000;275:32268–32276. doi: 10.1074/jbc.M004304200. [DOI] [PubMed] [Google Scholar]
- 7.Kühn U., Pieler T. Xenopus poly(A) binding protein: functional domains in RNA binding and protein-protein interaction. J. Mol. Biol. 1996;256:20–30. doi: 10.1006/jmbi.1996.0065. [DOI] [PubMed] [Google Scholar]
- 8.Imataka H., Gradi A., Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kessler S.H., Sachs A.B. RNA recognition motif 2 of yeast Pab1p is required for its functional interaction with eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 1998;18:51–57. doi: 10.1128/mcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozlov G., Trempe J.F., Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc. Natl. Acad. Sci. USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melo E.O., Dhalia R., de Melo Neto O.P. Identification of a C-terminal poly(A)-binding protein (PABP)-PABP interaction domain: role in cooperative binding to poly (A) and efficient cap distal translational repression. J. Biol. Chem. 2003;278:46357–46368. doi: 10.1074/jbc.M307624200. [DOI] [PubMed] [Google Scholar]
- 12.Khaleghpour K., Kahvejian A., Sonenberg N. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol. Cell. Biol. 2001;21:5200–5213. doi: 10.1128/MCB.21.15.5200-5213.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deo R.C., Bonanno J.B., Burley S.K. Recognition of polyadenylate RNA by the poly(A)-binding protein. Cell. 1999;98:835–845. doi: 10.1016/s0092-8674(00)81517-2. [DOI] [PubMed] [Google Scholar]
- 14.Görlach M., Burd C.G., Dreyfuss G. The mRNA poly(A)-binding protein: localization, abundance, and RNA-binding specificity. Exp. Cell Res. 1994;211:400–407. doi: 10.1006/excr.1994.1104. [DOI] [PubMed] [Google Scholar]
- 15.Krause S., Fakan S., Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp. Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- 16.Dudko O., Mathé J., Meller A. Nanopore force spectroscopy tools for analyzing single biomolecular complexes. In: Walter N., editor. Methods in Enzymology. Elsevier Academic Press; San Diego: 2010. pp. 565–589. [DOI] [PubMed] [Google Scholar]
- 17.Henrickson S.E., Misakian M., Kasianowicz J.J. Driven DNA transport into an asymmetric nanometer-scale pore. Phys. Rev. Lett. 2000;85:3057–3060. doi: 10.1103/PhysRevLett.85.3057. [DOI] [PubMed] [Google Scholar]
- 18.Meller A., Branton D. Single molecule measurements of DNA transport through a nanopore. Electrophoresis. 2002;23:2583–2591. doi: 10.1002/1522-2683(200208)23:16<2583::AID-ELPS2583>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 19.Meller A. Dynamics of polynucleotide transport through nanometre-scale pores. J. Phys. Condens. Matter. 2003;15:R581–R607. [Google Scholar]
- 20.Dudko O.K., Mathé J., Hummer G. Extracting kinetics from single-molecule force spectroscopy: nanopore unzipping of DNA hairpins. Biophys. J. 2007;92:4188–4195. doi: 10.1529/biophysj.106.102855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mathé J., Visram H., Meller A. Nanopore unzipping of individual DNA hairpin molecules. Biophys. J. 2004;87:3205–3212. doi: 10.1529/biophysj.104.047274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howorka S., Movileanu L., Bayley H. Kinetics of duplex formation for individual DNA strands within a single protein nanopore. Proc. Natl. Acad. Sci. USA. 2001;98:12996–13001. doi: 10.1073/pnas.231434698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vercoutere W., Winters-Hilt S., Akeson M. Rapid discrimination among individual DNA hairpin molecules at single-nucleotide resolution using an ion channel. Nat. Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- 24.Sauer-Budge A.F., Nyamwanda J.A., Branton D. Unzipping kinetics of double-stranded DNA in a nanopore. Phys. Rev. Lett. 2003;90:238101. doi: 10.1103/PhysRevLett.90.238101. [DOI] [PubMed] [Google Scholar]
- 25.Schink S., Renner S., Gerland U. Quantitative analysis of the nanopore translocation dynamics of simple structured polynucleotides. Biophys. J. 2012;102:85–95. doi: 10.1016/j.bpj.2011.11.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hornblower B., Coombs A., Akeson M. Single-molecule kinetic analysis of DNA-protein complexes using nanopores. Nat. Methods. 2007;4:315–317. doi: 10.1038/nmeth1021. [DOI] [PubMed] [Google Scholar]
- 27.Goodrich C.P., Kirmizialtin S., Movileanu L. Single-molecule electrophoresis of β-hairpin peptides by electrical recordings and Langevin dynamics simulations. J. Phys. Chem. B. 2007;111:3332–3335. doi: 10.1021/jp071364h. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Q., Sigalov G., Timp G. Detecting SNPs using a synthetic nanopore. Nano Lett. 2007;7:1680–1685. doi: 10.1021/nl070668c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dorvel B., Sigalov G., Timp G. Analyzing the forces binding a restriction endonuclease to DNA using a synthetic nanopore. Nucleic Acids Res. 2009;37:4170–4179. doi: 10.1093/nar/gkp317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akeson M., Branton D., Deamer D.W. Microsecond time-scale discrimination among polycytidylic acid, polyadenylic acid, and polyuridylic acid as homopolymers or as segments within single RNA molecules. Biophys. J. 1999;77:3227–3233. doi: 10.1016/S0006-3495(99)77153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meller A., Nivon L., Branton D. Voltage-driven DNA translocations through a nanopore. Phys. Rev. Lett. 2001;86:3435–3438. doi: 10.1103/PhysRevLett.86.3435. [DOI] [PubMed] [Google Scholar]
- 32.Butler T.Z., Gundlach J.H., Troll M.A. Determination of RNA orientation during translocation through a biological nanopore. Biophys. J. 2006;90:190–199. doi: 10.1529/biophysj.105.068957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Song L., Hobaugh M.R., Gouaux J.E. Structure of staphylococcal α-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 34.Lin J., Kolomeisky A., Meller A. Helix-coil kinetics of individual polyadenylic acid molecules in a protein channel. Phys. Rev. Lett. 2010;104:158101. doi: 10.1103/PhysRevLett.104.158101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


