Abstract
Chemoreceptor arrays are supramolecular transmembrane machines of unknown structure that allow bacteria to sense their surroundings and respond by chemotaxis. We have combined X-ray crystallography of purified proteins with electron cryotomography of native arrays inside cells to reveal the arrangement of the component transmembrane receptors, histidine kinases (CheA) and CheW coupling proteins. Trimers of receptor dimers lie at the vertices of a hexagonal lattice in a “two-facing-two” configuration surrounding a ring of alternating CheA regulatory domains (P5) and CheW couplers. Whereas the CheA kinase domains (P4) project downward below the ring, the CheA dimerization domains (P3) link neighboring rings to form an extended, stable array. This highly interconnected protein architecture underlies the remarkable sensitivity and cooperative nature of transmembrane signaling in bacterial chemotaxis.
Keywords: protein structure, hybrid methods, two-component systems
Chemotactic bacteria sense their surrounding conditions through an array of transmembrane chemoreceptors (methyl-accepting chemotaxis proteins, or MCPs), which are found with histidine kinases (CheA) and couplers (CheW) in polar clusters (1–3) and along the sides of cells (4, 5). Repellents and attractants bind to the periplasmic domains of the MCPs either directly (6, 7) or via periplasmic binding proteins (8). The status of the binding domain is transmitted along the length of the receptors through the transmembrane region, across one or more HAMP (histidine kinases, adenyl cyclases, MCPs, and some phosphatases) domain(s), and down the coiled-coil cytoplasmic signaling domain where they ultimately regulate the activity of the histidine kinase CheA located at the receptors’ cytoplasmic tips (1–3, 9). CheA is a large, five-domain (P1–P5) protein. P1 contains the substrate histidine, P2 is the docking site for the response regulator CheY, P3 is the dimerization domain, P4 binds ATP and is the kinase, and P5 binds CheW. P1, P2, and P3 are connected to each other by flexible linkers (1, 2). Crystal structures of all domains from Thermotoga maritima CheA are already available (10–13).
In the model system Escherichia coli, the addition of attractants or removal of repellents results in kinase inactivation, causing the flagella to rotate counterclockwise. In that case, the multiple flagella form one large bundle that propels the cells smoothly forward and the cells “run.” In contrast, addition of repellents or removal of attractants activates CheA, which autophosphorylates and then transfers the phosphoryl group to the second messenger CheY, which in turn binds to the flagellar motors and changes the direction of flagellar rotation to clockwise (CW). This switch results in disassembly of the flagellar bundle and causes the cells to “tumble” (14). CheA also regulates the activity of the receptor-modifying enzyme CheB (a methylesterase), which together with CheR (a methyltransferase) controls the methylation state of residues in the MCP adaptation region (1). Methylation tunes receptor sensitivity and kinase activity in a feedback cycle that allows cells to adapt to current conditions and follow gradients by modulating their run and tumble frequencies (15–17).
Cooperativity underlies the remarkable high sensitivity, gain, dynamic range, and feedback control of this system (18–24). Knowledge of how the component proteins arrange and interact within the arrays is therefore essential to understanding all aspects of signal transduction. Here we describe the detailed arrangement of receptor dimers within the hexagonal arrays of chemoreceptors and present the crystallographic structure of the ternary complex formed among a receptor signaling domain, the CheA kinase and regulatory domains, and the coupling protein CheW. From these combined data, we derive a model for the extended architecture of chemoreceptor arrays.
Results and Discussion
Electron cryotomography has previously shown that MCPs form extended hexagonal lattices at the poles of cells linked at their cytoplasmic tips by a CheA/W “baseplate” (25, 26). Moreover, a recent cryotomographic study showed that the basic architecture of the lattice is universally conserved throughout chemotactic bacteria (9). By correcting tilt series for the contrast transfer function of the microscope before 3D reconstruction, here we have substantially increased the resolution of the tomograms so that individual MCP dimers are now clearly visible in subtomogram averages (Fig. 1). In all cases imaged so far, including both Gram-negative (E. coli, Helicobacter hepaticus, and Salmonella enterica) and Gram-positive (Bacillus subtilis) cells, trimers of receptor dimers are located at the vertices of the hexagonal lattice facing their three neighboring trimers in a “two-on-two” orientation. As seen previously in Caulobacter crescentus (25), the higher-resolution tomograms confirm that the arrays are well-ordered near the CheA/W baseplate, but become less so in the HAMP, transmembrane, and periplasmic domains.
Fig. 1.
Architecture of native chemoreceptor arrays as seen by electron cryotomography. (Upper) Tomographic slice through the top of a S. enterica minicell. OM, outer membrane; IM, inner membrane. (Scale bar: 100 nm.) (Lower) Subtomogram averages of E. coli, H. hepaticus, S. enterica, and B. subtilis (from left to right) chemoreceptor arrays after application of sixfold symmetry. In all cases, the individual receptor dimers (asterisks) are clearly resolved, revealing a two-facing-two packing arrangement: A pair of dimers faces another pair of dimers at each interface around the ring, or to describe it in another way, trimers are oriented such that one receptor dimer points toward the center of each hexagon. The conserved architecture also shows that the cell lysis used to thin the E. coli and B. subtilis samples for high-resolution ECT did not perturb the arrays. (Scale bars: 12 nm.)
The shape of the MCP complexes in the EM maps resembles the “trimer-of-dimers” crystal structure of the truncated cytoplasmic region of the E. coli serine receptor Tsr (27). The cryotomograms show, however, that the receptor dimers retain their four-helix-bundle quaternary structure all the way from the CheA/W baseplate to the HAMP domains, and therefore allow a more complete modeling of the cytoplasmic domains (Fig. 2). In addition, the stalks of the receptor dimers appear straighter adjacent to the baseplate and diverge to a lesser extent than those of the crystal structure. A bend is seen, however, near a conserved glycine hinge that is known to be important for proper receptor function (28). Baseplate densities are also clear, but none of the existing crystal structures, including the dimer of three subdomains of CheA (P3, P4, and P5) (10) or the complex of two CheA subdomains (P4 and P5) and CheW (29) could be unambiguously fit into the EM maps.
Fig. 2.
Model of a receptor trimer within the EM map. Two isosurfaces of the receptor region of the EM map are shown as blue and magenta grids (low and higher density, respectively) with an all-atom model of a receptor trimer fit to the map, seen from the side (Left, with back dimer removed for clarity) and in cross-section at three different positions (Right). The atomic model is based on a crystal structure of a truncated E. coli Tsr MCP which crystallized in a similar configuration (27). To fit that structure into the EM map, the four-helix coiled-coil was extended (based on the crystal structure of receptor Tm1143; ref. 29) to the junction of the HAMP domain (residues 264–514), separated slightly at the tips to better fit the electron density, and then refined against the EM data in reciprocal space (see Materials and Methods). The density clearly confirms the trimers-of-dimers architecture in vivo, but compared to the crystal structure, the receptors bend in the glycine hinge region and the four-helix coiled-coil extends to the level of the HAMP domain. The hexagonal order decreases toward the membrane. The additional density seen around the receptor tips (asterisks) is where the receptor bundle connects with the CheA/W baseplate.
To define the interactions among the receptors, CheA and CheW at higher resolution, crystals were therefore grown of a ternary complex of Thermotoga maritima proteins. The ternary complex crystals contain the CheA kinase (P4) and regulatory (P5) domains, CheW, and the highly conserved signaling domain of a Thermotoga MCP (30). Although the crystals diffract to only 4.5-Å resolution and have a large unit cell (Table 1), their high solvent content and relatively simple asymmetric unit allowed for an unambiguous placement of the secondary structure elements in each component, whose high-resolution structures have all been previously determined (29, 30).
Table 1.
Data collection and refinement statistics for ternary complex
| Wavelength, Å | 0.97700 |
| Space group | R32 |
| Cell parameters | a = 213.99, b = 213.99, c = 208.19 |
| Resolution, Å | 30–4.5 (4.58–4.50) |
| No. of observations | 59,703 |
| No. of unique reflections | 10,933 |
| Completeness, % | 98.3 (96.6) |
| Rsym* | 0.108 (0.65) |
| I/σ(I) | 15.7 (1.9) |
| Refinement statistics | |
| Resolution range | 50–4.5 (4.66–4.50 Å) |
| R factor, % | 24.5 (32.5) |
| Rfree, % | 29.6 (35.7) |
| Molecules/asym unit | 1 P4-P5, 1 CheW, 2 Tm14s |
| Residues/asym unit | 572 |
| Atoms | |
| Protein | 4,497 |
| Solvent content, % | 84 |
| Mean B values, Å2 | |
| CheA P5 | 185 |
| CheW | 197 |
| Tm14 | 206 |
| CheA P4 | 345 |
| Rmsd from ideal geometry | |
| Bonds | 0.002 Å |
| Angles | 0.8° |
| Ramachandran plot, % | |
| Most favored | 67.6 |
| Additionally allowed | 29.3 |
| Generously allowed | 3.1 |
| Disallowed | 0.0 |
| Missing residues | P4 residues 451–507 (ATP-lid) |
Data for outermost resolution shell are given in parenthesis. Asym unit, asymmetric unit.
*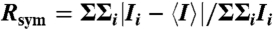 .
.
CheW and the CheA regulatory domain (P5) are paralogs, each composed of two intertwined β-barrels known as subdomains 1 and 2 (31). Up until now, the significance of this relationship has not been fully appreciated. Within the asymmetric unit, P5 subdomain 1 binds CheW subdomain 2 in a pseudosymmetric interaction previously characterized (Fig. 3A, Left) (29, 32). The receptor tip binds alongside CheW at the junction between the two β-barrels with a configuration consistent with previous structural (33) and other studies (34, 35) (Fig. 3A, Right). CheW primarily contacts the receptor on the helix N-terminal to the hairpin tip. Due to the dimeric nature of the receptor, the symmetry-related helix on the adjacent subunit faces the receptor trimer interface. Competition for binding the same N-terminal helix may explain why overexpression of CheW interferes with receptor trimer formation (36).
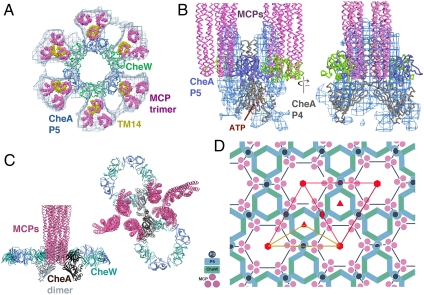
Fig. 3.
Ternary complex crystal structure of T. maritima chemotaxis proteins. (A) Close-ups of the pseudosymmetric interactions made by the opposite ends of CheW (green ribbons) and P5 (blue ribbons), and the interaction between the receptor tip (magenta ribbons) and CheW. Inset shows a schematic of dimeric CheA:CheW, with the crystallized unit boxed. (B) Ring structure formed by the ternary complex crystals. Three CheW domains and three P5 domains generate a ring, and each CheW binds one receptor tip (pink) between subdomains 1 and 2 (Left). Similar interactions between P5 with the distal end of receptors (purple) link rings “head-to-head” in the unit cell (Right). The P4 domains (gray), of which only the core elements are visible, project above and below the double-ring structure at the junction to P5.
Not anticipated, the crystallographic symmetry reveals a remarkable extended assembly state for the ternary complex (Fig. 3B). The crystallographic threefold axis generates a ring structure of the CheW and CheA regulatory domains wherein subdomain 2 of the regulatory domain binds to subdomain 1 of CheW in a contact that mimics the associations made by the analogous surfaces of the opposing β-barrels (Fig. 3A, Center). Furthermore, the distal ends of the receptor helix bundles interact with the regulatory domains in a manner that mimics that of the receptor tip with CheW (Fig. 3B and Fig. S1). Together, these associations generate a large double-ring structure of pseudo-sixfold symmetry with receptors binding alternatively to the CheW and P5 units around the ring (Fig. 3B).
Although alternating receptor bundles around the ring are antiparallel, each of the receptor dimers docks a helix into a groove that is conserved between the two β-barrels of either CheW or the CheA regulatory domain P5 (Fig. S1). The interaction between the regulatory domain and the receptor as found in the crystal is likely nonnative because it would require adjacent receptors around the ring to be oriented in opposite directions, which is implausible because they all traverse the membrane. However, given the residue conservation of P5 and CheW in the binding groove and the similarity in helix side-chain interactions indicated by the two different receptor associations, it is likely that P5 can also bind a receptor tip in the same orientation as CheW does. In support of this important inference, CheA is known to bind receptors without CheW (33, 37), isolated P5 domains are recruited to receptor clusters independent of CheW (38), and CheA and CheW compete for the same or overlapping binding determinants on receptors (37, 39). The P4 kinase domains are not well defined in the crystal structure, but density for the central β-sheet and some peripheral helices is observed projecting above and below the rings at the junction to P5 (Fig. 3B and Fig. S2).
One ring of the crystal structure with its six associated receptors holds a striking relationship in symmetry, dimension, and shape to the CheA/W baseplate density in the cellular tomograms. Superimposing the three receptor bundles associated with one ring of the crystal structure with those fit to the EM maps (Fig. 2) accommodates the CheA P5-CheW ring well within the honeycomb lattice (Fig. 4A). A corresponding ring of density can be seen in the EM maps, although at lower contour levels than the receptor density (Fig. 4A). Thus, the P5-CheW ring is present in cells, but with either lower occupancy or higher disorder than the receptor trimers. Superposition of the P5-CheW unit with one subunit of the dimeric-CheA-bound-to-CheW model from crystallographic and spin-labeling studies (33) places the second P5 subunit within the neighboring hexagon, and rotating about the P3–P4 junction to bring the CheA P5 subunits into planarity aligns CheW and P5 with their expected receptor contacts in the neighboring hexagon (Fig. S3). Without additional manipulation, the kinase domains now project below the rings and between the hexamers in a region of the EM maps that also shows substantial density (Fig. 4B). If the CheA dimerization domain (P3) remains connected to one of either CheA subunit, these manipulations place the dimerization domain in the space between the two-facing-two receptor dimers (Fig. 4C). Rotation about the center of P3 provides reasonable connections to the kinase domains of both subunits and aligns the dimeric axes of P3 with those of the receptors (Fig. 4C and Fig. S3), as indicated by prior studies (33, 40). However, there is little density in this location in the EM maps, which suggests that the dimerization domain does not assume a fixed position against the receptors.
Fig. 4.
Structure of native chemoreceptor arrays. (A) Superposition of one ring of the crystal structure (P5 blue, CheW green) with its six receptor dimers (yellow helices) on the EM map (blue mesh) with its previously fit 18 receptor dimers (pink). The EM density in the CheA/W ring is only about half as intense as within the receptors, suggesting either lower occupancy or higher disorder. (B) Side view of the EM density (blue mesh) in the region of the CheA-P4 kinase domain (gray). (C) The CheA dimer links CheW/P5 rings. The two subunits of the CheA dimer (black and gray) provide one P5 to each of two neighboring rings. The P3 dimerization domain resides between the receptor bundles at the center of one hexagonal edge and the P4 domains reside beneath the interlocked rings. Views shown are in the plane of the rings (Left) and looking down from the membrane (Right). (D) The arrangement of components within the receptor array produces P6 point symmetry (P6 unit cell boxed in red, with the asymmetric unit in yellow; six-, three-, and twofold symmetry axes are designated in red). The lattice gives a CheA:CheW:MCP subunit stoichiometry of 1∶1∶6. If the “empty” hexagons instead contain six CheW proteins, the ratio would become 1∶2∶6. Movie S1 summarizes in a step-by-step animation how the new electron cryotomographical and X-ray crystallographic data were used to elucidate the array structure shown in D.
The position of the CheA/W complex bridging two trimers of receptor dimers is consistent with the finding that two receptor trimers, one dimeric CheA, and two CheWs are the minimal unit needed for kinase activation (41). The structure of the array precludes CheA, however, from being present in three copies in every ring of the lattice (Fig. 4D). Thus the lattice has P6 point symmetry (Fig. 4D) and in terms of CheA/CheW content comprises one empty hexagon surrounded by six occupied hexagons, each containing three CheA and three CheW subunits. This arrangement produces a CheA:CheW:MCP subunit stoichiometry of 1∶1∶6. Not all CheA or CheW nodes in the lattice need be filled to produce an extended, stable structure, however, which may explain the lower density of the rings in the EM maps. Measured CheA:CheW:MCPs subunit ratios depend on sample and preparation, with the reported stoichiometries varying among 1∶4∶6 (37); 1∶0.8∶6.8 (42); and 1∶3∶6–9 (43). Greater than 1∶1 CheW to CheA ratios may be due to CheW substituting for CheA at certain positions within the lattice or even composing complete rings. If six CheW proteins were to fill the empty hexagon of the lattice, the subunit stoichiometry becomes 1∶2∶6. The completeness of the native arrays may also vary under different conditions, thereby leading to a range of measured ratios.
Previous work supports the notion that different CheW/P5-type domains can compete for similar positions within the arrays. CheA and CheW recognize overlapping sites on receptors with comparable dissociation constants (within a factor of approximately 10), but they also bind synergistically and in a manner that depends on the receptor stoichiometry (37, 39, 43). This competitive, yet cooperative behavior is consistent with a lattice structure where interactions among CheA and CheW subunits organize receptor binding surfaces that are similar on the two proteins. Furthermore, CheW and the P5 regulatory domain may substitute for each other within the rings, with different compositions producing different aggregate levels of kinase activity. Structural data have demonstrated that the P5 domains can self-associate through a symmetric contact that mimics the interaction observed with CheW in the ternary crystal structure (10, 29). Many bacteria also contain CheV, which is a fusion between a CheW and a CheY domain, the latter of which can be phosphorylated by CheA (44). The function of CheV varies among organisms, but generally overlaps with that of CheW (44). It follows that CheV proteins may also replace CheA P5 and/or CheW within the hexagonal lattice and thereby influence coupling between receptor and kinase.
Thus, the precise composition of the rings in terms of CheW, CheA, and CheV may vary in different signaling states, while still maintaining the interlocking nature of the baseplate, which would explain the ultrastability of the arrays (43) and provide the structural connections needed for highly cooperative responses. This model is also consistent with the idea that signal amplification derives from kinase coupling within the extended lattice (45); however, interactions among receptors, CheW, and CheA all may contribute to cooperativity. Finally, because the cells imaged here had adapted to their surrounding conditions, they are expected to contain both active and inactive CheA; hence, the modeled network likely reflects a mixture of these two states (46).
Materials and Methods
Cell Growth and Sample Preparation for Electron Cryotomography (ECT).
TH17261 is a mini-cell-producing strain of Salmonella enterica Serovar Typhimurium that overexpresses flagellar structures. TH17261 carries a second copy of the ftsZ+ gene expressed from an arabinose promoter (DaraBAD1091∷ftsz+). This strain was constructed by first replacing the araBAD structural genes with the tetracycline-resistance cassette or tetRA element from transposon Tn10 and then replacing the tetRA element with the ftsZ+ gene as described (47). Induction of excess FtsZ by arabinose results in minicell formation. The lhrA, ydiV, and ecnR genes encode negative regulators of the flagellar master operon, flhDC (48, 49). The lhrA and ecnR gene deletions were constructed by insertion of tetRA and oligonucleotide-directed replacement resulting in gene deletion leaving the first and last 15 base pairs of the coding regions as described (47). The construction of the ecnR deletion (DecnR∷FKF, where FKF represents the Flp recombinase target cassette) was previously described (49). The strain also carries promoter-up mutations in the flagellar flhDC master operon as described (48). The various alleles were moved into a single strain by bacteriophage P22-mediated transduction (49).
S. enterica strain 17261 minicells were grown overnight shaking in LB medium at 37 °C. The culture was then diluted 1/100 into fresh LB containing 0.1% l-Arabinose and grown for an additional 3 h. One milliliter aliquots were centrifuged at 3,000 × g for 5 min to remove large cells and then the supernatant was centrifuged at 18,000 × g to collect minicells. The pellets were then resuspended in 50 μL LB.
B. subtilis subsp. subtilis strain 168 was grown overnight shaking in LB at 37 °C. The culture was diluted in fresh LB medium and grown to log phase. One milliliter culture was spun down for 5 min at 4,000 × g and resuspended in protoplast preparation medium (250 mL containing 6.25 g LB, 20 mM MgCl2, and 20 mM sucrose). Lysozyme was added to a final concentration of 100 μg/mL, and 5 mL were incubated without shaking in a 125-mL Erlenmeyer flask until protoplasts were formed.
H. hepaticus American Type Culture Collection strain 51449 was grown and E. coli strain MG1655 was grown and lysed as described previously (9).
Electron Cryotomography.
Right before plunge freezing, the different cell preparations (S. enterica minicells, lysed E. coli and B. subtilis cells, and intact H. hepaticus cells) were each mixed with colloidal gold pretreated with BSA to avoid particle aggregation (50). Four microliters of cell-and-gold solution were applied to R2/2 copper/Rhodium Quantifoil grids™ (Quantifoil Micro Tools), blotted, and plunged in liquid ethane or ethane/propane mixture (50, 51). Images were collected using an FEI Polara™ (FEI), 300 kV field emission gun transmission electron microscope equipped with a Gatan energy filter and a lens-coupled 4,000 × 4,000 Ultracam (Gatan). Tilt series from up to -70° to 70° with an increment of 1°, an underfocus of -8 to -10 μm, and a pixel size on the specimen level of 6.3 Å were recorded using Leginon (52). A cumulative dose of 200 electrons/Å2 or less was used for each tilt series.
Tilt series were aligned and contrast transfer function corrected using the IMOD software package (53). Three-dimensional reconstructions were calculated using IMOD or TOMO3D (53, 54). Subvolume averaging and symmetrizing was done using PEET (55).
Protein Preparation for Crystallography.
Residues 107–191 of T. maritima receptor Tm14s (30) was PCR cloned into vector pET28a (Novagen) and expressed with an N-terminal Histidine6 tag in E. coli strain BL21 (RIL DE3) (Novagen) after induction with IPTG at 18 °C and overnight growth for 21 h. Tm14s was purified first with Ni-nitrilotriacetate chromatography, followed by overnight thrombin digestion, and then size-exclusion chromatography (Superdex 75 Hi-load FPLC column in 50 mM NaCl, 100 mM Tris 7.5, 10% glycerol). T. maritima CheW and CheA Δ354 (P4P5 domain, residues 355–671) were expressed and purified as described previously (29).
Crystallization and Data Collection.
Cubic shaped crystals (50 × 50 × 50 μm3) were grown from a mixture of 520 μM Tm14s (107–191), 457 μM CheA Δ354, and 121 μM CheW after 1 mo by vapor diffusion from a 2-μL drop [1∶1 mixture of protein and reservoir: 500-μL reservoir of 0.2 M sodium acetate trihydrate, 0.1 M Tris (pH 8.5), 15% wt/vol polyethylene glycol 4,000]. SDS-PAGE analysis with mass-spectrometry identification confirmed all components in the crystals. Most crystals diffracted to < 8-Å resolution; however, after extensive screening, several crystals diffracting to higher resolution were found. Crystals were soaked briefly in cryoprotectant consisting of 85/15 (vol/vol) reservoir solution with glycerol prior to data collection in an N2 cold stream. Diffraction data (Table 1) were collected at 100 K with synchrotron radiation at beamline A1 at the Cornell High Energy Synchrotron Source.
Crystal Structure Determination and Refinement.
Diffraction data were processed with HKL2000 (56). Initial phases were obtained by molecular replacement with PHASER (57) using one subunit of the CheA Δ354-CheW complex [Protein Data Bank (PDB) 2CH4 chain A and chain W] as a search model. The truncated receptor dimer (PDB 3G67) was manually built into the resulting electron density maps with XFIT (58). The model was refined to 4.5-Å resolution with the deformable elastic network (DEN) method (59, 60), as implemented in CNS (61). Although not well-resolved, electron density for the core β-sheet of the P4 domain was evident below the connection to P5. Refinement of three different orientations of P4 centered on this density showed little discrimination in Rfree. Electron density at the very tip of the receptors was also weak, and thus the helix register was set by packing constraints at the distal end.
Electron Cryotomography Modeling.
An all-atom model of a Tsr cytoplasmic trimer-of-dimers containing three complete four-helix bundles to the level of the HAMP domain was constructed based on the crystal structures of Tsr (PDB code 1QU7) and Tm1143 (PDB code 2CH7) and then built into ECT density symmetrized about the sixfold axis relating trimers-of-dimers. The model was refined in reciprocal space to 20-Å resolution against vector structure factors from the volume of a single trimer placed in a P1 unit cell, first by rigid body refinement of the three subunits, then by rigid body refinement of nine individual helical sections (three from each dimer) that comprised the signaling tip, stalk to the glycine hinge, and adaptation regions. The threefold symmetry relating the dimers within trimers was not enforced on the ECT maps nor the all-atom model. Geometry optimization in CNS (61) was performed to correct stereochemistry at junctions of the helical segments. Cross-validation methods were applied to monitor the course of refinement. The helical segments were adjusted to difference Fourier maps amidst cycles of refinement. DEN refinement (59, 60) to 20-Å resolution was applied, but produced little improvement in cross-validated refinement statistics. Refined trimers were then related by sixfold symmetry and rigid body refined into a volume composing an entire honeycomb hexameric assembly of receptor trimers. The P5-CheW ring was placed in the residual density. To extend the lattice beyond one ring structure, each subunit from the model of dimeric CheA:CheW (33) was superimposed on P5 domains of adjacent rings, which were positioned in the tomography maps according to the observable density and the receptor trimer positions. This action superimposed the associated CheW domains perfectly and projected the P4 domains down below the receptors. Relative to the model of dimeric CheA:CheW derived from spin-labeling studies, the P4–P5-CheW units have rotated about the hinges to P3 so that they lie in the same plane (Fig. S3). The superposition also placed an associated P3 in the center of the hexagon edges, between the two-facing-two receptor dimers. P3 was then rotated to complete appropriate linkages with each P4. P4 was adjusted slightly about the P4–P5 linkage to optimize overlap with the tomography density.
Supplementary Material
Acknowledgments.
We thank Drs. Morgan Beeby and Songye Chen for collecting some of the tomographic data, Dr. John Heumann for help using the PEET software, Dr. Stanley Maloy for suggesting FtsZ overexpression for minicell production, and A. Vu and Dr. F.W. Dahlquist for advice on choosing constructs for crystallization. We also thank the Cornell High Energy Synchrotron Source for access to data collection facilities. This work was supported by the Howard Hughes Medical Institute, by gifts to Caltech from The Gordon and Betty Moore Foundation, and by National Institutes of Health Grant GM066775 (to B.R.C.).
Note.
Dahlquist and coworkers have recently characterized the interaction between CheW and the T. maritima receptor by NMR (62); the results are in good agreement with the structure presented here.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3UR1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115719109/-/DCSupplemental.
References
- 1.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33:9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 3.Wadhams GH, Armitage JP. Making sense of it all: Bacterial chemotaxis. Nat Rev Mol Cell Biol. 2004;5:1024–1037. doi: 10.1038/nrm1524. [DOI] [PubMed] [Google Scholar]
- 4.Greenfield D, et al. Self-organization of the Escherichia coli chemotaxis network imaged with super resolution light microscopy. PLoS Biol. 2009;7:e1000137. doi: 10.1371/journal.pbio.1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sourjik V, Berg HC. Localization of components of the chemotaxis machinery of Escherichia coli using fluorescent protein fusions. Mol Microbiol. 2000;37:740–751. doi: 10.1046/j.1365-2958.2000.02044.x. [DOI] [PubMed] [Google Scholar]
- 6.Englert DL, Adase CA, Jayaraman A, Manson MD. Repellent taxis in response to nickel ion reqires neither Ni2+ transport nor the periplasmic NikA binding protein. J Bacteriol. 2010;192:2633–2637. doi: 10.1128/JB.00854-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milburn MV, et al. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991;254:1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- 8.Tam R, Saier MHJ. Structural, functional, and evolutionary relationships among extracellular solute-binding receptors of bacteria. Microbiol Rev. 1993;57:320–346. doi: 10.1128/mr.57.2.320-346.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Briegel A, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bilwes AM, Alex LA, Crane BR, Simon MI. Structure of CheA, a signal-transducing histidine kinase. Cell. 1999;96:131–141. doi: 10.1016/s0092-8674(00)80966-6. [DOI] [PubMed] [Google Scholar]
- 11.Bilwes AM, Park SY, Quezada CM, Simon MI, Crane BR. In: Histidine Kinases in Signal Transduction. Inouye M, Dutta R, editors. San Diego: Academic; 2003. pp. 48–74. [Google Scholar]
- 12.Park S-Y, Quezada CM, Bilwes AM, Crane BR. Subunit exchange by CheA histidine kinases from the mesophile Escherichia coli and the thermophile Thermotoga maritima. Biochemistry. 2004;43:2228–2240. doi: 10.1021/bi0352419. [DOI] [PubMed] [Google Scholar]
- 13.Quezada CM, Gradinaru G, Simon MI, Bilwes AM, Crane BR. Helical shifts generate two distinct conformers in the atomic resolution structure of the CheA phosphotransferase domain from Thermotoga maritima. J Mol Biol. 2004;341:1283–1294. doi: 10.1016/j.jmb.2004.06.061. [DOI] [PubMed] [Google Scholar]
- 14.Turner L, Ryu WS, Berg HC. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleene SJ, Hobson AC, Adler J. Attractants and repellents influence methylation and demethylation of methyl-accepting chemotaxis proteins in an extract of Escherichia coli. Proc Natl Acad Sci USA. 1979;76:6309–6313. doi: 10.1073/pnas.76.12.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 17.Toews ML, Goy MF, Springer MS, Adler J. Attractants and repellents control demethylation of methylated chemotaxis proteins in Escherichia coli. Proc Natl Acad Sci USA. 1979;76:5544–5548. doi: 10.1073/pnas.76.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duke TAJ, Bray D. Heightened sensitivity of a lattice of membrane receptors. Proc Natl Acad Sci USA. 1999;96:10104–10108. doi: 10.1073/pnas.96.18.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endres RG, Wingreen NS. Precise adaptation in bacterial chemotaxis through “assistance neighborhoods”. Proc Natl Acad Sci USA. 2006;103:13040–13044. doi: 10.1073/pnas.0603101103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gestwicki JE, Kiessling LL. Inter-receptor communication through arrays of bacterial chemoreceptors. Nature. 2002;415:81–84. doi: 10.1038/415081a. [DOI] [PubMed] [Google Scholar]
- 21.Li G, Weis RM. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell. 2000;100:357–365. doi: 10.1016/s0092-8674(00)80671-6. [DOI] [PubMed] [Google Scholar]
- 22.Li M, Hazelbauer GL. Adaptational assistance in clusters of bacterial chemoreceptors. Mol Microbiol. 2005;56:1617–1626. doi: 10.1111/j.1365-2958.2005.04641.x. [DOI] [PubMed] [Google Scholar]
- 23.Sourjik V, Berg H. Receptor sensitivity in bacterial chemotaxis. Proc Natl Acad Sci USA. 2002;99:123–127. doi: 10.1073/pnas.011589998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sourjik V, Berg HC. Functional interactions between receptors in bacterial chemotaxis. Nature. 2004;428:437–441. doi: 10.1038/nature02406. [DOI] [PubMed] [Google Scholar]
- 25.Briegel A, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim KK, Yokota H, Kim SH. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature. 1999;400:787–792. doi: 10.1038/23512. [DOI] [PubMed] [Google Scholar]
- 28.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44:7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S-Y, et al. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol. 2006;13:400–407. doi: 10.1038/nsmb1085. [DOI] [PubMed] [Google Scholar]
- 30.Pollard AM, Bilwes AM, Crane BR. The structure of a soluble chemoreceptor suggests a mechanism for propagating conformational signals. Biochemistry. 2009;48:1936–1944. doi: 10.1021/bi801727m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griswold IJ, et al. The solution structure and interactions of CheW from Thermotoga maritima. Nat Struct Mol Biol. 2002;9:121–125. doi: 10.1038/nsb753. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Parkinson JS. Mutational analysis of the chemoreceptor-coupling domain of the Escherichia coli chemotaxis signaling kinase CheA. J Bacteriol. 2006;188:3299–3307. doi: 10.1128/JB.188.9.3299-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhatnagar J, et al. Structure of the ternary complex formed by a chemotaxis receptor signalling domain, the CheA histidine kinase, and the coupling protein CheW as determined by pulsed dipolar ESR spectroscopy. Biochemistry. 2010;49:3824–3841. doi: 10.1021/bi100055m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boukhvalova M, VanBruggen R, Stewart RC. CheA kinase and chemoreceptor interaction surfaces on CheW. J Biol Chem. 2002;277:23596–23603. doi: 10.1074/jbc.M202288200. [DOI] [PubMed] [Google Scholar]
- 35.Underbakke ES, Zhu YM, Kiessling LL. Protein footprinting in a complex milieu: Identifying the interaction surfaces of the chemotaxis adaptor protein CheW. J Mol Biol. 2011;409:483–495. doi: 10.1016/j.jmb.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Studdert CA, Parkinson JS. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci USA. 2005;102:15623–15628. doi: 10.1073/pnas.0506040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levit MN, Grebe TW, Stock JB. Organization of the receptor-kinas signaling array that regulates the Escherichia coli chemotaxis. J Biol Chem. 2002;277:36748–36754. doi: 10.1074/jbc.M204317200. [DOI] [PubMed] [Google Scholar]
- 38.Kentner D, Thiem SMH, Sourjik V. Determinants of chemoreceptor cluster formation in Escherichia coli. Mol Microbiol. 2006;61:407–417. doi: 10.1111/j.1365-2958.2006.05250.x. [DOI] [PubMed] [Google Scholar]
- 39.Asinas AE, Weis RM. Competitive and cooperative interactions in receptor signaling complexes. J Biol Chem. 2006;281:30512–30523. doi: 10.1074/jbc.M606267200. [DOI] [PubMed] [Google Scholar]
- 40.Miller AS, Kohout SC, Gilman KA, Falke JJ. CheA kinase of bacterial chemotaxis: Chemical mapping of four essential docking sites. Biochemistry. 2006;45:8699–8711. doi: 10.1021/bi060580y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li M, Hazelbauer GL. Core unit of chemotaxis signaling complexes. Proc Natl Acad Sci USA. 2011;108:9390–9395. doi: 10.1073/pnas.1104824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Hazelbauer GL. Cellular stoichiometry of the components of the chemotaxis signalling complex. J Bacteriol. 2004;186:3687–3694. doi: 10.1128/JB.186.12.3687-3694.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erbse AH, Falke JJ. The core signaling proteins of bacterial chemotaxis assemble to form an ultrastable complex. Biochemistry. 2009;48:6975–6987. doi: 10.1021/bi900641c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alexander RP, Lowenthal AC, Harshey RM, Ottemann KM. CheV: CheW-like coupling proteins at the core of the chemotaxis signaling network. Trends Microbiol. 2010;18:494–503. doi: 10.1016/j.tim.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldman JP, Levin MD, Bray D. Signal amplification in a lattice of coupled protein kinases. Mol Biosyst. 2009;5:1853–1859. doi: 10.1039/B903397a. [DOI] [PubMed] [Google Scholar]
- 46.Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karlinsey JE. λ-red genetic engineering in Salmonella enterica serovar Typhimurium. Methods Enzymol. 2007;421:199–209. doi: 10.1016/S0076-6879(06)21016-4. [DOI] [PubMed] [Google Scholar]
- 48.Erhardt M, Hughes KT. C-ring requirement in flagellar type III secretion is bypassed by FlhDC upregulation. Mol Microbiol. 2010;75:376–393. doi: 10.1111/j.1365-2958.2009.06973.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wozniak CE, Lee C, Hughes KT. T-POP array identifies EcnR and PefI-SrgD as novel regulators of flagellar gene expression. J Bacteriol. 2009;191:1498–1508. doi: 10.1128/JB.01177-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iancu CV, et al. Electron cryotomography sample preparation using the Vitrobot. Nat Protoc. 2007;1:2813–2819. doi: 10.1038/nprot.2006.432. [DOI] [PubMed] [Google Scholar]
- 51.Tivol W, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167:11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 54.Agulleiro JI, Fernandez JJ. Fast tomographic reconstruction on multicore computers. Bioinformatics. 2011;27:582–583. doi: 10.1093/bioinformatics/btq692. [DOI] [PubMed] [Google Scholar]
- 55.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 56.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 57.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McRee D, Israel M. XtalView, protein structure solution and protein graphics, a short history. J Struct Biol. 2008;163:208–213. doi: 10.1016/j.jsb.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 59.Schröder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schröder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1222. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brunger AT. Version 1.2 of the Crystallography and NMR System. Nat Protoc. 2007;2:2728–2788. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 62.Vu A, Wang X, Zhou H, Dahlquist FW. The receptor CheW binding interface in bacterial chemotaxis. J Mol Biol. 2012;415:759–767. doi: 10.1016/j.jmb.2011.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.