Abstract
The transcription factor WUSCHEL (WUS) acts from a well-defined domain within the Arabidopsis thaliana shoot apical meristem (SAM) to maintain a stem cell niche. A negative-feedback loop involving the CLAVATA (CLV) signaling pathway regulates the number of WUS-expressing cells and provides the current paradigm for the homeostatic maintenance of stem cell numbers. Despite the continual turnover of cells in the SAM during development, the WUS domain remains patterned at a fixed distance below the shoot apex. Recent work has uncovered a positive-feedback loop between WUS function and the plant hormone cytokinin. Furthermore, loss of function of the cytokinin biosynthetic gene, LONELY GUY (LOG), results in a wus-like phenotype in rice. Herein, we find the Arabidopsis LOG4 gene is expressed in the SAM epidermis. We use this to develop a computational model representing a growing SAM to suggest the plausibility that apically derived cytokinin and CLV signaling, together, act as positional cues for patterning the WUS domain within the stem cell niche. Furthermore, model simulations backed by experimental data suggest a previously unknown negative feedback between WUS function and cytokinin biosynthesis in the Arabidopsis SAM epidermis. These results suggest a plausible dynamic feedback principle by which the SAM stem cell niche is patterned.
Keywords: signal transduction, cell division
Within a properly patterned stem cell niche, domains of accessory cells produce maintenance signals that support pluripotent stem cells. In plants, the shoot apical meristem (SAM) is the aboveground stem cell niche. It is patterned into functionally distinct domains that interact through cell–cell communication mediated by diffusive signals (1, 2). Unlike animal cells, which can migrate, plants cells are fixed, and their movement is driven by cell proliferation. Therefore, as cell divisions push daughter cells of pluripotent stem cells away from the central zone (CZ) of the SAM into other functional domains, cells must sense their relative position within the niche and adjust their gene expression profile according to the differentiation program of that domain.
A central player in the maintenance of CZ stem cells is the transcription factor WUSCHEL (WUS). WUS-expressing cells reside in the rib meristem (RM) domain of the SAM, just below the CZ, and originate from a central group of multipotent stem cells in the corpus (L3 and lower layers below the anticlinally dividing L1 and L2 layers). WUS is required for the production of a non-cell autonomous proliferative signal to determine the number of overlying pluripotent stem cells in the CZ (3, 4). The CZ cells express the CLAVATA3 gene product, which is processed into a signaling peptide that activates a set of receptor kinases, which, in turn, repress WUS expression in the RM (5–8). Thus, the CLV3-expressing CZ stem cells regulate the strength of the non-cell autonomous proliferative signal produced by WUS in the RM of the SAM (1, 9). Through this feedback loop, the size of the apical pluripotent stem cell population and WUS-expressing cell population are mutually regulated. Although this paradigm adequately explains how the numbers of stem cells are maintained in the SAM, it fails to explain how the relative position of the WUS domain is maintained.
Crosstalk exists between WUS function and the action of the plant hormone cytokinin in the SAM. WUS has been found to repress members of the type A ARABIDOPSIS RESPONSE REGULATOR (ARR) family, which negatively regulate cytokinin signaling (10). In addition, cytokinin was shown to induce expression of WUS (11). The expression domains for WUS and the cytokinin receptor ARABIDOPSIS HISTIDINE KINASE 4 (AHK4) overlap in the SAM (11). These data support a model whereby AHK4 and WUS function within the RM to establish a group of cytokinin sensitized cells (11). Therefore, cytokinin may play a role in patterning the RM within the SAM.
The fixed spatial relationship of the WUS expression domain proximal to the shoot epidermis throughout growth suggests the presence of an inductive signal to position it within the SAM. Given the positive role cytokinin has on WUS expression, it is a likely candidate. Analysis of cytokinin distribution within the Sinapis alba L. SAM by immunohistochemistry suggests a potential gradient of the hormone from the epidermis into the basal cells (12). Several studies support a role for local cytokinin synthesis in the maintenance of a functional SAM during growth (13, 14). In rice, loss of LONELY GUY (LOG)-mediated cytokinin biosynthesis within the upper layers of the SAM results in progressive termination of the stem cell niche reminiscent of the wus mutant phenotype (13). In Arabidopsis, there are nine LOG family members. Analysis of higher order mutant log alleles indicates that AtLOG gene function is redundantly distributed between the activity of multiple family members (15).
In this study, we demonstrate that the cytokinin biosynthetic enzyme LOG4 is expressed in the epidermal layer (L1) of the SAM and floral meristem. Based on this, we formulate a cell-based computational model involving growth and division in the apical–basal axis. We demonstrate the plausibility of our model that epidermally derived cytokinin, together with the CLV-WUS genetic network, regulates cell division and positions the WUS expression domain within the SAM during growth. Lastly, using the model in conjunction with experiments, we reveal a feedback principle whereby WUS negatively regulates epidermally produced cytokinin biosynthesis in the SAM. This leads to an updated picture of how mechanisms of feedback control, which occur over space and time, pattern and maintain the SAM stem cell niche.
Results
Evidence for Apically Produced Cytokinin in the SAM Epidermis.
Previous biochemical work has established that LOG functions as a phosphoribohydrolase to produce active cytokinins (13, 15). Using quantitative PCR (SI Appendix, Fig. S1) and published LOGpro::GUS results (15) as guides, we constructed LOGpro::2XYpet-N7 transcriptional reporters (LOG1, LOG3, LOG4, LOG5, LOG7) and investigated their expression profiles by confocal microscopy in living inflorescence SAM tissue. Our analysis found that expression of the LOG4pro::2XYpet-N7 reporter was restricted to the epidermal layer (L1) of the SAM and floral meristem (Fig. 1A). We also observed weak expression of the LOG7pro::2XYpet-N7 reporter in developing primordia. The expression of LOG4 in the L1 layer suggests this enzyme may function to generate a source of apically derived cytokinin in the SAM proper, whereas LOG7 may function in the developing primordia.
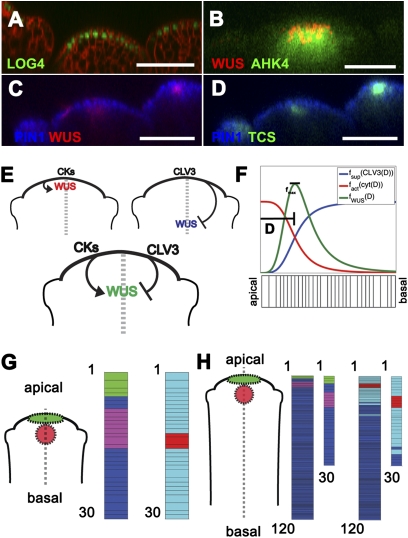
Fig. 1.
Distribution of cytokinin synthesis, perception, and signaling relative to WUS within the SAM and a dynamic model for pattering the apical–basal axis of the SAM during growth. (A) Longitudinal view of LOG4pro::2xYpet-N7 reporter expression (green) and cell membranes (red; FM4-64) in the SAM and floral meristem. (B) Longitudinal distribution of WUSpro::DsRed-N7 (red) within the apical half of the AHK4pro::GFP (green) domain. (C and D) Longitudinal view of WUSpro::WUS-2xVenus (red) (C) and TCSpro::GFP (green) (D) in the SAM and floral meristem. PIN1pro::PIN1-CFP (blue) marks cell membranes. [Scale bars: 50 μm (A–D)]. (E) Schematic of the regulatory interactions between cytokinin/WUS and WUS/CLV within the SAM. (F) WUS expression determined by CLV3 suppression [fsup(CLV3(D))] and activation by cytokinin [fact(cyt(D))] results in a maximum (fmax) for the WUS distribution [fWUS(D)]. (G) Initial pattern of expression of key components in single column of 30 cells spanning the apical–basal axis of the SAM. First column, CLV3 (green) and WUS (magenta); second column, type A ARR (teal) and phosphorylated type B ARR (red). Interspaced regions are in dark blue. (H) Patterns of components after a growth simulation labeled as in G. In H, the larger plot shows all 120 cells after growth, whereas the smaller plots show only the apical-most 30 cells for comparison with patterns in G.
The WUS Expression Domain Corresponds to High Cytokinin Signaling Activity.
We hypothesize that cytokinin produced in the L1 cell layer, through LOG4 function, forms a diffusion gradient within the SAM, whereby it is perceived by cells expressing cytokinin receptors in the underlying cell layers (Fig. 1B). Live imaging of inflorescence SAMs revealed strong expression of a WUS reporter WUSpro::dsRed-N7 (Fig. 1B); WUSpro::WUS-2XVenus (Fig. 1C) in the apical half of the AHK4 receptor domain, marked by AHK4pro::GFP (Fig. 1B), and the cytokinin signaling domain, marked by TCSpro::GFP-ER (Fig. 1D). These live imaging results reveal WUS is expressed within the upper half of the AHK4 domain, which is just below the presumed hormone source, where type B ARR activity is highest.
Mathematical Model for the Positioning of the WUS Domain Within a Growing SAM.
Using these live imaging results, we developed a model of a growing and dividing stem cell niche. Our primary focus was understanding how the WUS domain is positioned proximal to the shoot epidermis. Therefore, we simplified our cellular template to a 1D column of cells representing a transverse apical–basal slice of a growing SAM (dashed line of the schematics in Fig. 1 E, G, and H). Patterning of the modeled stem cell niche occurs as apical cells, marked by CLV3 expression (SI Appendix, Fig. S2B), communicate with underlying cells, marked by WUS expression (SI Appendix, Fig. S2A), through diffusive signals. Induction of WUS by cytokinin alone results in apical expression pattern (red curve in Fig. 1F). In contrast, WUS expression is restricted far into the basal zone of the SAM when regulated by CLV3 suppression alone (blue curve in Fig. 1F). When regulated by both, the peak of WUS expression occurs at (fmax), where the two curves intersect (green curve in Fig. 1F). Therefore, we hypothesize that WUS expression is patterned at a fixed distance from the shoot apex by this molecular network (Fig. 1E). By embedding in each cell of the growing and dividing template, this signaling-genetic network (SI Appendix) maintains the zones of expression and domain identity through cell–cell communication provided by diffusing signals.
The SAM is a dynamic tissue where the continual turnover of cells from each domain of the stem cell niche drives growth. This makes cell division a fundamental aspect of growth in our model. Therefore, our model simulations were performed on a growing and dividing template of cells (SI Appendix and Movies S1 and S2). Fig. 1G shows the initial patterns of the model components of the proposed regulatory network before growth and cell division (initial template of 30 cells). Based on live imaging results, WUS is expressed three to four cells below the tip and extends five to six cells, whereas CLV3 is primarily expressed in the top two to three cells. A peak of phosphorylated type B ARR activity overlaps with the WUS domain, whereas the type A ARR expression domain flanks the WUS expression domain (SI Appendix). Using these starting parameters, model simulations were performed for a defined period corresponding to >100 cell divisions. Fig. 1H shows the final patterns at the end of the simulation. To verify the crucial point that the essential expression patterns of the network components is maintained throughout growth relative to the initial starting pattern, we plotted the detailed distributions of network components over the same number of cells after model simulations (SI Appendix). This reveals that the pattern of most components is robust even after extensive growth and cell division (compare plot 1–30 in Fig. 1 G and H).
CLV3 Gain of Function Terminates the Theoretical Stem Cell Niche As Observed in Vivo.
To test the accuracy of our computational model, we considered the case of CLV3 gain of function, which leads to the loss of meristem function (1, 16). To compare our model with published observations, we performed a simulation where model parameters were altered such that CLV3 represses WUS more strongly, mimicking gain of function. Comparing the two plots in SI Appendix, Fig. S3 A and B, which show cytokinin signaling (Bp), WUS, CLV3, and cytokinin for the wild type and CLV3 gain of function (SI Appendix), the model accurately simulates the low amounts of WUS that cause eventual meristem termination. Hence, our model recapitulates the experimental observations of meristem loss for the CLV3 gain of function.
Cytokinin Can Partially Rescue Floral Meristem Function in a Hypomorphic wus Mutant.
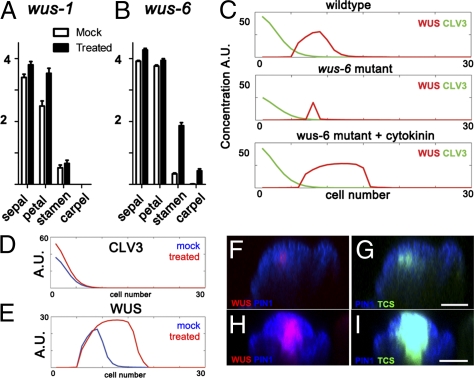
To validate the functional relevance of the inductive effect of cytokinin on WUS expression, we performed rescue experiments on the wus-1 loss-of-function allele and wus-6 hypomorphic allele (containing a T-DNA insertion in the proximal promoter of the gene) (17, 18). The wus-1 and wus-6 alleles result in a similar floral meristem termination phenotype. Exogenous cytokinin treatments failed to rescue the inner whorl organ phenotype in wus-1 mutant plants (Fig. 2A) but did occasionally induce supernumerary outer whorl organs (SI Appendix, Fig. S4 A and B). In contrast, cytokinin application partially restored inner whorl organ development in the wus-6 mutant (Fig. 2B and SI Appendix, Fig. S4 C and D). This is consistent with the observed expression of the TCSpro::GFP-ER reporter within the center of the floral meristem (Fig. 1D and SI Appendix, Fig. S2 C and D). Next, we used our model to determine whether the partial rescue of meristem activity could be attributable to increased expression of WUS in the wus-6 allele. Fig. 2C shows predicted distributions of WUS and CLV3 along the apical–basal axis for wild type and the wus-6 mutant before and after cytokinin treatment. These model simulations demonstrate that exogenous cytokinin treatments could restore sufficient levels of WUS transcription to maintain meristematic function, even in the hypomorphic wus-6 allele (Fig. 2C).
Fig. 2.
Model tests for two cases: first, cytokinin-induced rescue of inner whorl floral organ development requires WUS function; and second, cytokinin affects WUS apical–basal positioning. (A and B) Sepal, petal, stamen, and carpel number for mock-treated and cytokinin-treated wus-1 (n = 55, 81) (A) and wus-6 (n = 353, 182) (B) flowers. (C) Simulations of CLV3 (green) and WUS (red) abundance within a column of cells along the apical–basal axis of the SAM (apical cell, 0; basal-most cell, 30) in wild type (upper plot), wus-6 mutant (center plot), and the wus-6 mutant treated with cytokinin (lower plot). (D and E) Simulations of CLV3 concentration along a column of cells (D) and WUS concentration along the same column of cells (E) (cytokinin perturbation simulations; blue, mock; red, cytokinin-treated). (F and G) Longitudinal view of WUSpro::WUS-2xYFP (red) and TCSpro::GFP (green) in mock-treated SAM. (H and I) Longitudinal view of WUSpro::WUS-2xYFP (red) and TCSpro::GFP (green) in cytokinin-treated SAM. (Scale bars: 50 μm.)
Relative Strengths of Cytokinin and CLV3 Signaling Position the WUS Domain.
To explore the interaction between the antagonistic effects of cytokinin and CLV3 on positioning of the WUS domain, we simulated cytokinin treatments in our model. Fig. 2 D and E shows the predicted distributions of CLV3 (upper plot) and WUS (lower plot) before (blue) and after (red) cytokinin treatment. Whereas CLV3 expression is largely unaffected, WUS expression extends basally upon cytokinin treatment. To validate these results, we observed the expression patterns for WUSpro::WUS-2xVenus and TCSpro::GFP-ER by live imaging floral meristems before and after cytokinin treatment. Both the WUS expression and cytokinin-signaling domains primarily enlarge in the basal direction into the RM after cytokinin treatment (Fig. 2 F–I). Therefore, in both model and experimental perturbations, when cytokinin signaling is increased, the mean length of the WUS domain from the shoot apex is extended primarily in the basal direction. In the model simulations, expansion of WUS expression in the apical direction is suppressed because of activation of CLV3 by the non-cell autonomous stem cell signal. We used our model to simulate how impaired clv3 function would impact the WUS expression domain. In this case, WUS expression extended apically, in response to clv3 loss of function (SI Appendix, Fig. S5), as reported previously (9). WUS does not appear in the L1 layer, because AHK4 is not significantly expressed in the upper layers of the SAM (Fig. 1 B and C). Therefore, clv3 loss of function decreases the mean length of the WUS zone from the shoot apex. The above results suggest that WUS is positioned proximal to the shoot apex through the antagonistic activities of cytokinin and CLV3 function.
CLV3 Function Buffers the Positive Effects of Cytokinin on Meristem Growth.
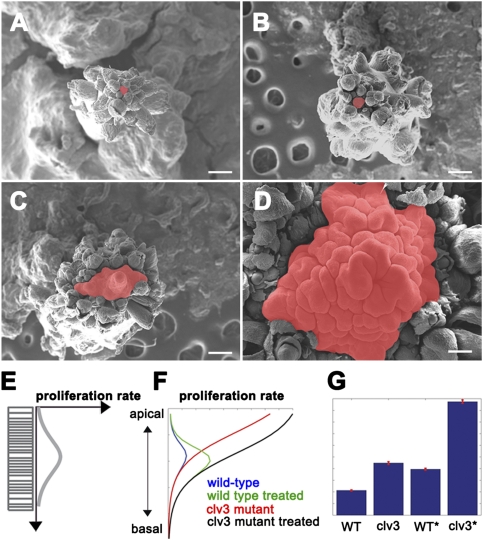
Cytokinin has been proposed to have a positive effect on SAM growth and cell division (19, 20). In contrast, the CLV pathway has a negative non-cell autonomous influence on cell division and growth in the SAM (2). To determine the relationship between these two antagonistic effects on SAM growth, we quantified wild-type and clv3-2 SAM area, as measured by scanning electron microscopy before and after cytokinin treatment. Cytokinin treatment of wild-type plants caused a small expansion in SAM surface area (1.5-fold by scanning electron microscopy) (Fig. 3 A and B) and occasionally induced formation of additional inner whorl floral organs (SI Appendix, Fig. S6 A and B). In comparison, cytokinin treatment of clv3-2 mutant plants resulted in a striking 15-fold increase in SAM surface area (Fig. 3 C and D) without altering cell size (SI Appendix, Fig. S6 E and F). We also observed enlargement of floral meristems and floral organ numbers in clv3-2-treated plants compared with mock-treated clv3 and cytokinin-treated wild type (SI Appendix, Fig. S6 C, D, G, and H). This drastic enhancement of SAM size in the clv3 loss-of-function background indicates that the CLV3 signaling pathway acts to buffer the positive effect of cytokinin on SAM growth and cell division.
Fig. 3.
Cytokinin promotes an increase in meristem size in a CLV3-dependent manner. (A–D) Scanning electron images of wild type mock-treated (A), wild-type cytokinin-treated (B), clv3-2 mutant mock-treated (C), and clv3-2 mutant cytokinin-treated (D) SAMs. (E) Schematic of rates of proliferation along the 1D column of cells for wild type. (F) Simulated proliferation rates along the 1D column of cells for wild type (blue), wild-type cytokinin-treated (green), clv3 mutant (red), and cytokinin-treated clv3 mutant (black). (G) Simulated cell numbers for wild type, clv3 mutant, cytokinin-treated wild type, and cytokinin-treated clv3 mutant. [Scale bars: 200 μm (A–D).]
We updated our model to include the antagonistic regulatory links whereby CLV3 restricts cell division and cytokinin promotes cell division in the SAM (SI Appendix). This model led to a distribution of cell division rates depicted schematically in Fig. 3E. Model predictions of division rates in wild type and clv3 loss of function before and after simulated cytokinin treatment are shown in Fig. 3F. In wild-type simulations, cytokinin treatment extended the zone over which cells divide in the basal direction. In contrast, clv loss-of-function simulations indicated that not only do the rates of cell division increase but also the peak of cell division moves in the apical direction. We compared repeated simulations of SAM growth over a fixed period and counted the number of cells for the different conditions (Fig. 3G). Therefore, a similar trend was observed between the experimental increase in SAM sizes and predicted rounds of cell division shown in Fig. 3 A–D and G. However, model simulations did not predict the large difference in size as seen when comparing wild type (Fig. 3A) with treated clv3 loss of function (Fig. 3D), which in the experimental case, is much larger (see Discussion).
WUS Regulation of Cytokinin Biosynthesis Emerges As a Possible Feedback Principle.
One goal for building our computational model was to make predictions about the function and interaction of network components of the SAM stem cell niche. In the models mentioned above, we assumed no feedback between WUS function and cytokinin biosynthesis. However, WUS does alter the identity and number of cells in the shoot apex (2). An obvious hypothesis for the cellular overproliferation phenotype of the clv3-2 mutant SAM, where WUS levels are presumed higher is elevated levels of cytokinin in the stem cell niche. To explore the consequence of feedback between WUS function and cytokinin biosynthesis, we modeled two alternative regulatory rules where WUS function in the RM activates (activator model) or represses (repressor model) cytokinin biosynthesis in the L1 layer (SI Appendix).
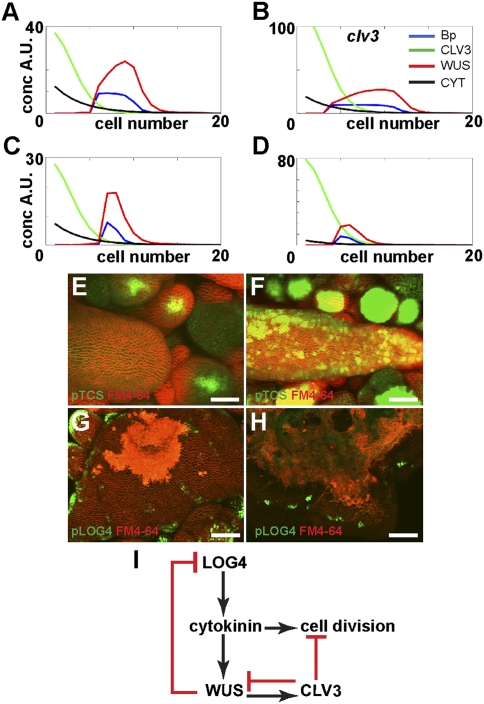
Fig. 4 A and B shows predicted distributions of CLV3, WUS, activated type B ARR (Bp), and cytokinin in wild-type and in clv3 loss-of-function simulations for the activator model. In response to clv3 loss of function, WUS expression extends apically, which increases cytokinin biosynthesis extending the cytokinin gradient further into the shoot, broadening WUS expression in the basal direction as well. Alternatively, Fig. 4 C and D shows distributions of the model components for the wild-type and clv3 loss-of-function simulations for the repressor model. Here, WUS moves apically, but now even more so, because as WUS moves closer to the shoot apex, it causes further suppression of cytokinin biosynthesis and a shallower gradient of cytokinin on which WUS expression depends. In contrast to the activator model, simulations for the repressor model shifts the peak of WUS expression only toward the shoot apex, as seen experimentally (SI Appendix, Fig. S7) (9).
Fig. 4.
Discriminating feedback models between WUS and cytokinin biosynthesis validated by experimental observations. Simulation results for WUS, CLV3, phosphorylated type B ARR (Bp), and cytokinin concentration as a function of cell number, along the apical–basal axis. Wild type (A) and clv3 loss of function (B) corresponding to the model where we include WUS positive feedback on cytokinin synthesis. Wild type (C) and clv3 loss of function (D) corresponding to the model where we include WUS negative feedback on cytokinin synthesis (SI Appendix). (E and F) TCSpro::GFP reporter expression in clv3-2 mutant mock-treated SAM and in clv3-2 mutant cytokinin-treated SAM. (G and H) LOG4pro::2xYpet-N7 reporter expression in clv3-2 mutant mock-treated SAM and in clv3-2 mutant cytokinin-treated SAM. FM4-64 membrane dye marks cell membranes in red (E–H). (I) The schematic shows that cell divisions are controlled antagonistically by CLV3 and cytokinin and that cytokinin biosynthesis is negatively regulated by WUS. This is in addition to the feedback between CLV3 and WUS, as well as WUS induction by cytokinin. (Scale bars: 50 μm.)
In the repressor model, cytokinin concentrations are reduced but not absent, which explains why WUS is expressed in clv3-2 meristems. Furthermore, in Fig. 4D, we see that activated type B ARR signal (cytokinin signaling; Bp) is still present. We further explored how WUS expression could be maintained in the presence of low levels of cytokinin signaling by perturbing the model in two ways: first, we assumed that cytokinin signaling activates WUS even more strongly; and second, we modeled a case with a stronger clv3 loss-of-function mutant (SI Appendix, Fig. S8 A and B and SI Appendix). Comparing WUS in the wild type (SI Appendix, Fig. S8A) with WUS in the clv3 mutant (SI Appendix, Fig. S8B), even where cytokinin signaling is extremely low, we still see WUS expression (SI Appendix, Fig. S8B). Type A ARR and WUS expression both depend on the strength of cytokinin signaling, and, hence, the relative strength of induction by the latter promotes WUS over type A ARR. Furthermore, because WUS represses type A ARR expression, which, in fact, negatively regulates cytokinin signaling, this implements a positive-feedback effect, whereby having stronger WUS activation leads to even more WUS. This aspect of the regulatory network, in combination with the relief from CLV3 suppression, allows WUS to be expressed in the presence of reduced cytokinin biosynthesis (SI Appendix, Fig. S8 C and D).
Live Imaging of a Cytokinin Biosynthesis and Signaling Reporter Supports the Repressor Model.
Of the two models, the counterintuitive repressor model seems the most plausible based on the predicted and observed distribution of WUS expression. To assess the cytokinin signaling status within the clv3-2 SAM, we used the TCSpro::GFP-ER. In a wild-type SAM, TCSpro::GFP-ER signal peaks within the RM (Fig. 1 C and D) (11). However, in the clv3-2 SAM, the TCSpro::GFP-ER signal was extremely weak but still observed within floral meristems (Fig. 4E). Treatment of clv3-2 plants with cytokinin led to reactivation of the TCSpro::GFP-ER within the clv3-2 SAM proper (Fig. 4F). These data indicate that the cells of the clv3-2 mutant SAM are competent to perceive cytokinin and activate the downstream two-component phospho-relay cascade. Taken together, these data suggest that under normal growth conditions cytokinin metabolism may be disrupted in the clv3-2 SAM.
We next investigated the expression status of the LOG4pro::2XYpet-N7 reporter in the clv3-2 SAM. Consistent with the absence of TCS signal, we observed expression of the LOG4pro::2XYpet-N7 reporter is largely absent from the L1 layer of the clv3-2 SAM (Fig. 4G). However, expression of LOG4pro::2XYpet-N7 is still observed in clv3-2 FMs, consistent with the notion that production of the active hormone is present to induce robust TCSpro::GFP-ER signal (Fig. 4E). In addition, we noted expression of the LOG4pro::2XYpet-N7 reporter is not rescued by cytokinin treatment (Fig. 4H). These data indicate that cytokinin synthesis is significantly reduced in the clv3-2 SAM proper and, taken together, are consistent with the model prediction of WUS repression on cytokinin biosynthesis.
Discussion
In this study, we hypothesize that integration of cytokinin signaling and metabolism, which regulates WUS expression (11) with the CLV-WUS feedback loop, acts as a mechanism to position the WUS domain within the stem cell niche. Based on the live imaging data, we propose LOG4 functions in the L1 layer of the SAM and FM to establish a gradient of cytokinin that extends into the RM, providing a molecular cue to those cells. Using this observation, we developed a simplified mathematical model and tested the plausibility that cytokinin could provide an apical cue to position the WUS expression domain within a growing SAM. Using the iteration of model simulations and experimentation, we found that, indeed, cytokinin could perform such a role. Furthermore, the model provided a framework for testing additional hypotheses and for exploring principles of regulation within the SAM stem cell niche. A key experiment could be to test whether apically derived cytokinin is required for the maintenance of WUS activity to promote stem cell survival (SI Appendix, Fig. S9). In Arabidopsis, overexpression of CTYOKININ OXIDASE (CKX) gene family members from the 35S promoter causes a loss of apical dominance (21). More recently, data demonstrate that CKX3 is expressed in a domain similar to WUS and septuple log mutants fail to maintain an inflorescence meristem (22, 23). It may be interesting to test experimentally whether reducing cytokinin synthesis specifically in the epidermis could mimic these phenotypes.
Previous studies have developed informative models to test hypothetical mechanisms involving actions of the CLV-WUS feedback loop in patterning WUS expression within the SAM (24–28). One advance over these previous models is that we have added a regulatory link by explicitly modeling cytokinin biosynthesis and perception as they regulate WUS activity. We believe that our current model represents a plausible set of rules that pattern and maintain gene expression domains in the apical–basal axis. However, to accurately reproduce the large difference in cell numbers observed between cytokinin-treated clv3 and wild-type plants, a more realistic 3D template is needed, perhaps involving a 3D model of the SAM based upon its reconstruction from live imaging data.
Recently, Kuroha et al. demonstrated that members of the Arabidopsis LOG family carry out a direct activation step in the cytokinin biosynthetic pathway similar to the rice LOG enzyme (15). Using live imaging and confocal microscopy, we found that transcriptional reporters for LOG4 and LOG7 were the only two LOGpro::2xYpet-N7 reporters analyzed to be identified as being expressed in the SAM proper. These fluorescent reporter expression data agree with the LOG4pro::GUS data in the study by Kuroha et al. showing LOG4 expressed in shoot apical tissue and a published in situ hybridization of LOG7 mRNA in the SAM (29). Within the SAM, LOG4 reporter expression is restricted to the L1 layer, whereas the LOG7 reporter is expressed in developing primordia. However, our data raise the following question: What is the significance of LOG4 function in the maintenance of WUS expression in the SAM? Kuroha et al. found that single or higher-order combinations had minimal phenotypic effects related to SAM function. This is in contrast to mutations in the rice LOG gene, which cause premature termination of the inflorescence meristem. However, a triple Atlog3;log4;log7 mutant has reduced apical dominance with smaller inflorescences and fewer flowers. These results suggest that there is a degree of functional redundancy among the Arabidopsis LOG family members, which differs from rice. Ubiquitous overexpression of LOG4 in Arabidopsis resulted in a semidwarfed phenotype, which suggests that LOG4 expression outside the epidermis of the SAM causes developmental abnormalities. It will be interesting to see how the domains of gene expression for CLV3, WUS, AHK4, and cytokinin signaling are affected by multiple loss-of-function LOG alleles or ubiquitous overexpression of LOG family members (i.e., LOG4 and LOG7).
Our study explored how adding a regulatory link, which states that the CLV-WUS pathway controls cytokinin biosynthesis, affects the dynamics of this feedback network. Our model simulations suggested that in the case where cytokinin biosynthesis is positively regulated by a WUS signal, a CLV3 loss-of-function mutant would have higher levels of cytokinin and that the WUS expression domain should extend further into basal cells compared with wild type. In the second case, with cytokinin biosynthesis negatively regulated by a WUS signal, cytokinin levels are decreased and the WUS zone moves apically, resulting in loss of WUS in basal cells. Given these predictions, we performed a series of live imaging studies to assess cytokinin biosynthesis, via LOG4 expression, cytokinin signaling, as readout by the TCS reporter, and WUS expression in the clv3-2 background. In the clv3 mutant, WUS expression tightly overlaps with an AHK4 expression domain that is compressed closer to the epidermis and not extended further into the RM. Furthermore, the signals from both the LOG4 reporter and cytokinin reporter are drastically reduced in the clv3-2 mutant SAM. Taken together, loss of LOG4 expression is consistent with a lack of sufficient cytokinin biosynthesis to induce observable cytokinin response using the TCS reporter. It is also consistent with reduced expression of the primary cytokinin response gene ARR7 in the clv3 mutant SAM, rather than a direct suppression by increased WUS function (10, 30). These data are, therefore, consistent with the repressor model in which WUS activity produces negative feedback on cytokinin synthesis. Experimentally, WUS expression shifted apically in clv3 loss-of-function SAMs and did not expand basally (9), which is also consistent with the repressor model. It can be argued from a purely theoretical point of view that, in the positive-feedback model, a few cell divisions within the apical zone would push the WUS zone farther away from the cytokinin producing L1 layer, which would lead to reduction in WUS expression and, hence, in turn, to reduced cytokinin biosynthesis. Although the receding L1 layer would lead to reduced levels of CLV3, which would offer a respite from suppression of WUS, WUS levels would still decrease because of lower gradients of cytokinin. This would lead to further reduction in cytokinin levels as well as expression levels of CLV3 and WUS. In the case where cytokinin biosynthesis is negatively regulated by WUS, in a similar scenario, cell divisions in the apical zone move the WUS zone farther away from the apex. However, now, this would lead to increased levels of cytokinin because of decreased negative control of its synthesis, thereby allowing cytokinin to permeate further into the tissue and providing a restoring element to WUS induction. Hence, the negative-feedback rule provides a more robust mechanism for pattern maintenance.
This study, therefore, supports two feedback principles for maintenance of the stem cell niche in the Arabidopsis SAM during growth. The first involves the dynamic positioning of the WUS domain by the combined antagonistic effects of cytokinin and CLV3. The second suggests that cytokinin biosynthesis itself is under negative control by WUS (Fig. 4I). WUS negative feedback on cytokinin biosynthesis in the shoot apex could limit the positive effect of cytokinin on cell division in the enlarged meristems of clv3 mutants. Therefore, this model is also consistent with the synergistic increase in meristem size observed in response to cytokinin in the clv3-2 mutant background. In the absence of the repressive effects of CLV function on cell division, cytokinin provides an unchecked proliferative signal to significantly increase cell division. Experiments in which these regulatory principles are further tested could provide insights into how local cytokinin biosynthesis and signaling affect cell division and growth within the SAM.
Materials and Methods
All details concerning plant growth conditions and hormone treatments can be found in SI Appendix. Standard molecular biology techniques were used for construction of plasmids and reporter transgenes (SI Appendix). Imaging was performed using a Zeiss 510 confocal microscope with a 63× or 40× water-dipping objective (SI Appendix). Computational details are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank Adrienne Roeder, Kaoru Sugimoto, Yun Zhou, and members of the Computable Plant group (http://computableplant.org/) for comments on the manuscript; and A. Garda for technical support. This work was supported by National Science Foundation Grant IOS-0846192 (to E.M.M.) and National Institutes of Health National Research Service Award Postdoctoral Fellowship F32-GM090534 (to P.T.T.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1200636109/-/DCSupplemental.
References
- 1.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 2.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 3.Mayer KF, et al. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- 4.Laux T, Mayer KF, Berger J, Jürgens G. The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development. 1996;122:87–96. doi: 10.1242/dev.122.1.87. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 6.Ohyama K, Shinohara H, Ogawa-Ohnishi M, Matsubayashi Y. A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat Chem Biol. 2009;5:578–580. doi: 10.1038/nchembio.182. [DOI] [PubMed] [Google Scholar]
- 7.Müller R, Bleckmann A, Simon R. The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell. 2008;20:934–946. doi: 10.1105/tpc.107.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kinoshita A, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 9.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/s0092-8674(00)80700-x. [DOI] [PubMed] [Google Scholar]
- 10.Leibfried A, et al. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature. 2005;438:1172–1175. doi: 10.1038/nature04270. [DOI] [PubMed] [Google Scholar]
- 11.Gordon SP, Chickarmane VS, Ohno C, Meyerowitz EM. Multiple feedback loops through cytokinin signaling control stem cell number within the Arabidopsis shoot meristem. Proc Natl Acad Sci USA. 2009;106:16529–16534. doi: 10.1073/pnas.0908122106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacqmard A, Detry N, Dewitte W, Van Onckelen H, Bernier G. In situ localisation of cytokinins in the shoot apical meristem of Sinapis alba at floral transition. Planta. 2002;214:970–973. doi: 10.1007/s00425-002-0742-4. [DOI] [PubMed] [Google Scholar]
- 13.Kurakawa T, et al. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature. 2007;445:652–655. doi: 10.1038/nature05504. [DOI] [PubMed] [Google Scholar]
- 14.Yanai O, et al. Arabidopsis KNOXI proteins activate cytokinin biosynthesis. Curr Biol. 2005;15:1566–1571. doi: 10.1016/j.cub.2005.07.060. [DOI] [PubMed] [Google Scholar]
- 15.Kuroha T, et al. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller R, Borghi L, Kwiatkowska D, Laufs P, Simon R. Dynamic and compensatory responses of Arabidopsis shoot and floral meristems to CLV3 signaling. Plant Cell. 2006;18:1188–1198. doi: 10.1105/tpc.105.040444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graf P, et al. MGOUN1 encodes an Arabidopsis type IB DNA topoisomerase required in stem cell regulation and to maintain developmentally regulated gene silencing. Plant Cell. 2010;22:716–728. doi: 10.1105/tpc.109.068296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamada S, et al. Mutations in the WUSCHEL gene of Arabidopsis thaliana result in the development of shoots without juvenile leaves. Plant J. 2000;24:91–101. doi: 10.1046/j.1365-313x.2000.00858.x. [DOI] [PubMed] [Google Scholar]
- 19.Higuchi M, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc Natl Acad Sci USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishimura C, et al. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werner T, Motyka V, Strnad M, Schmülling T. Regulation of plant growth by cytokinin. Proc Natl Acad Sci USA. 2001;98:10487–10492. doi: 10.1073/pnas.171304098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tokunaga H, et al. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. 2012;69:355–365. doi: 10.1111/j.1365-313X.2011.04795.x. [DOI] [PubMed] [Google Scholar]
- 23.Bartrina I, Otto E, Strnad M, Werner T, Schmülling T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell. 2011;23:69–80. doi: 10.1105/tpc.110.079079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jönsson H, et al. Modeling the organization of the WUSCHEL expression domain in the shoot apical meristem. Bioinformatics. 2005;21(Suppl 1):i232–i240. doi: 10.1093/bioinformatics/bti1036. [DOI] [PubMed] [Google Scholar]
- 25.Nikolaev SV, Penenko AV, Lavrekha VV, Melsness ED, Kolchanov NA. [A model study of the role of proteins CLV1, CLV2, CLV3, and WUS in regulation of the structure of the shoot apical meristem] Ontogenez. 2007;38:457–462. [PubMed] [Google Scholar]
- 26.Hohm T, Zitzler E, Simon R. A dynamic model for stem cell homeostasis and patterning in Arabidopsis meristems. PLoS ONE. 2010;5:e9189. doi: 10.1371/journal.pone.0009189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geier F, et al. A quantitative and dynamic model for plant stem cell regulation. PLoS ONE. 2008;3:e3553. doi: 10.1371/journal.pone.0003553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujita H, Toyokura K, Okada K, Kawaguchi M. Reaction-diffusion pattern in shoot apical meristem of plants. PLoS ONE. 2011;6:e18243. doi: 10.1371/journal.pone.0018243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yadav RK, Girke T, Pasala S, Xie M, Reddy GV. Gene expression map of the Arabidopsis shoot apical meristem stem cell niche. Proc Natl Acad Sci USA. 2009;106:4941–4946. doi: 10.1073/pnas.0900843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, et al. Hormonal control of the shoot stem-cell niche. Nature. 2010;465:1089–1092. doi: 10.1038/nature09126. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.